Abstract
The highly conserved ADAR enzymes, found in all multicellular metazoans, catalyze the editing of mRNA transcripts by the deamination of adenosines to inosines. This type of editing has two general outcomes: site specific editing, which frequently leads to recoding, and clustered editing, which is usually found in transcribed genomic repeats. Here, for the first time, we looked for both editing of isolated sites and clustered, non-specific sites in a basal metazoan, the coral Acropora millepora during spawning event, in order to reveal its editing pattern. We found that the coral editome resembles the mammalian one: it contains more than 500,000 sites, virtually all of which are clustered in non-coding regions that are enriched for predicted dsRNA structures. RNA editing levels were increased during spawning and increased further still in newly released gametes. This may suggest that editing plays a role in introducing variability in coral gametes.
Keywords: RNA editing, ADAR, evolution, coral
Introduction
RNA editing by adenosine deamination is a simple biochemical process where selected adenosines (A) are converted to inosine (I) within RNA molecules. Inosine is recognized as guanosine (G) during translation (Basilio et al. 1962), so that editing of an adenosine at a nonsynonymous position within a codon results in recoding. Editing can take place in essentially every region of every class of RNA, often with far reaching effects. In addition to recoding, splicing (Rueter et al. 1999), microRNA processing (Kawahara et al. 2007), microRNA targeting (Kawahara et al. 2007), and mRNA stability (Wang et al. 2013) can all be influenced by editing, as are other processes. The A-to-I conversion is catalyzed by the ADAR (Adenosine Deaminase that Acts on RNA) family of enzymes (Bass and Weintraub 1988; Kim et al. 1994; O’Connell et al. 1995; Melcher et al. 1996). Vertebrates have two ADARs; ADAR1 (ADAR) and ADAR2 (ADARB1). To edit a specific A, an ADAR first binds to the surrounding dsRNA structure via N-terminal double-stranded RNA binding motifs (dsRBDs). This positions a C-terminal catalytic domain next to the target A, allowing the deamination reaction to proceed. Thus, from a biochemical perspective, the only factors required for RNA editing to occur are ADAR expression and the presence of a suitable RNA structure.
Early studies on RNA editing focused on recoding sites discovered by chance. Editing was found to affect mRNAs encoding proteins essential for neurotransmission, such as neurotransmitter receptors and ion channels (Burnashev et al. 1992; Burns et al. 1997; Rosenthal and Bezanilla 2002; Bhalla et al. 2004; Rula et al. 2008). Although the specific effects on the nervous system could not be denied, these studies fostered the premature conclusion that the purpose of RNA editing is predominantly to regulate, and generate diversity, within the nervous system. Recent studies using deep sequencing technologies, have cast doubt on these early assumptions. Transcriptome wide screens revealed that recoding editing sites are exceptionally rare, particularly in vertebrates. In mammals, although there are millions of editing sites (Bazaket al. 2014a; Ramaswami and Li 2014), only about 50 conserved recoding sites have been reported (Pinto et al. 2014). The vast majority of editing sites localized in transcribed retrotransposon elements within introns and 3′ untranslated regions. On a functional level, it was suggested that in humans most (non-conserved) recoding by RNA editing is not adaptive (Xu and Zhang 2014). In addition, recent studies have hypothesized that editing plays a key role in regulating innate immunity and mediating the interferon response, and that this function of RNA editing, rather than recoding, may be the primary role (Mannion et al. 2014; Liddicoat et al. 2015; O’Connell et al. 2015). Exploring the editome of basal metazoans may shed light on the question of the ancestral function of editing.
ADARs are ubiquitously expressed among multicellular metazoan (Kohn et al. 2015), however the patterns by which they edit RNAs have only been explored in a handful of organisms outside of the vertebrate lineage. Only very few potential recoding sites have been identified in C. elegans transcripts (Morse et al. 2002; Whipple et al. 2015; Zhao et al. 2015; Goldstein et al. 2016) although editing sites are abundant in untranslated regions (Wheeler et al. 2015). In Drosophila and other insects, recoding sites are more common than in vertebrates, but they still only occur in a small fraction of mRNAs (St Laurent et al. 2013; Li et al. 2014). As with mammals, recoding sites tend to be enriched in mRNAs encoding proteins that play a role in excitability. Recoding in cephalopods, however, appears to be an exception, as it is very common, occurring in the majority of all nervous system transcripts (Albertin et al. 2015; Alon et al. 2015). In both squid and Drosophila, editing appears to be influenced by the environment (Garrett and Rosenthal 2012; Rieder et al. 2015) and in both organisms, there is abundant evidence that editing influences nervous system function.
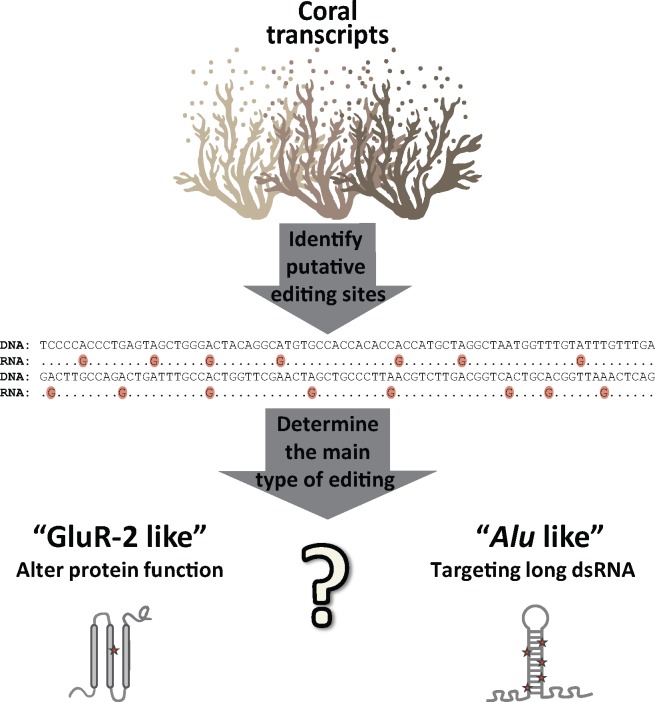
Cnidarians possess the most primitive true nervous system, consisting of a diffuse nerve net. They date back to at least 700 million years, thereby representing one of the most basal groups of metazoans. This makes them ideal organisms to explore the evolutionary relationship between the nervous system and RNA editing, and to investigate whether editing first emerged as a tool to regulate the nervous system through recoding, or to regulate other processes such as innate immunity (fig. 1). To address this question, we chose to explore the landscape of RNA editing in Acropora millepora, a Scleractinian coral (reef building) from the Great Barrier Reef (GBR) of Australia during their reproductive/spawning stages. Coral spawning on the GBR (Kaniewska et al. 2015) is one of nature’s greatest examples of synchronized behavior and the Earth’s largest reproductive event. During this annual occurrence that spans a couple of nights, changes in the intensity of moonlight trigger the spawning of more than 130 species of Scleractinian corals as well as hundreds of other invertebrates. Coral reproductive success is vital to the persistence of coral reef ecosystems. Broadcast spawning requires coral colonies to carefully synchronize the release of egg and sperm into the water column, in order to optimize fertilization success. We used several detection tools to investigate the distribution of editing sites. As with vertebrates, we found that although editing is common, sites in coding regions are extremely rare. Interestingly, our results show that RNA editing levels are temporally regulated, being highest during spawning stages and in newly released gametes. Consistently, ADAR2 expression levels showed a similar trend.
Fig. 1.
Overview and rational of this study. Examining the prevalent editing sites in the coral transcriptome and detecting whether editing is predominantly in intergenic or in coding sites enables us to decipher the basal function of ADAR. The goal is to determine whether ADAR originated as a mechanism of preventing immune responses to dsRNA such as the case of Alu sequences in primates, or as a process of adding diversity through transient point mutations in translated RNA such as in the human GluR-2 gene.
Results
Corals Express ADAR1 and ADAR2 Orthologs
As a first step to characterizing the RNA editing process, we asked whether corals express multiple ADAR isoforms, and if so whether the candidates can be classified as ADAR1 or ADAR2 family members. A previous study, based on transcriptome resources available in 2008, identified an ADAR2 ortholog in Acropora millepora, but both ADAR1 and ADAR2 orthologs are evident in Nematostella vectensis, a sea anemone. A hydrozoan, Hydra magnipapillata, had a single ADAR that could not be classified as either ADAR1 or ADAR2. In the RNA-seq data from adult coral tissue (including both ectoderm and endoderm layers, supplementary fig. S1, Supplementary Material online) we detected two A. millepora ADARs. Based on sequence homology, both of these fulfil the generic ADAR requirement of having at least one N-terminal double-stranded RNA binding motifs (dsRBDs) and a C-terminal catalytic domain.
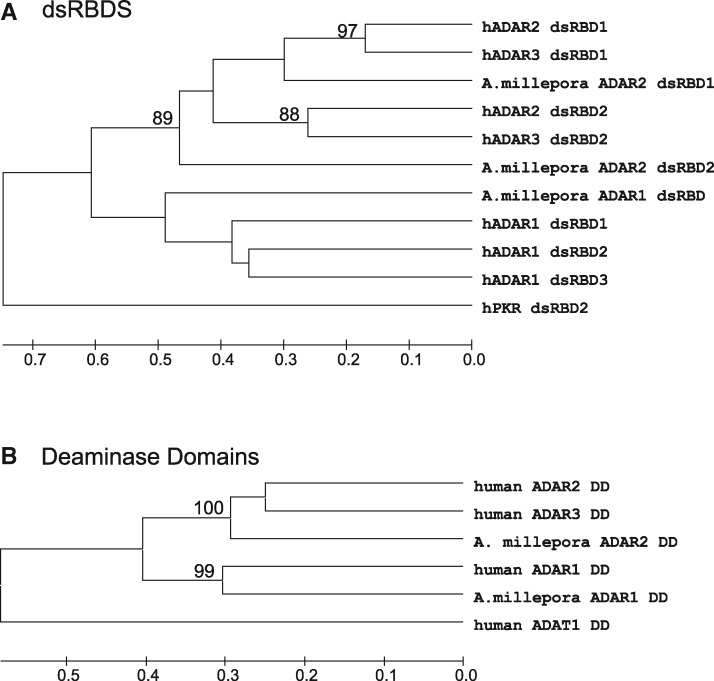
To better identify these sequences, we compared their dsRBDs and deaminase domains with those from human ADAR1, ADAR2, and ADAR3, separately (fig. 2). Based on bootstrap values from neighbor-joining trees, both the dsRBD comparisons, and those for the deaminase domains, suggest that one of the coral sequences is an ADAR1 ortholog and the other is an ADAR2 ortholog. The coral ADAR2 sequence is conventional. As with vertebrate ADAR2s, it contains two dsRBDs and a conserved deaminase domain. Residues known to participate in proton shuttling and zinc coordination within the reaction center are conserved, as are most of those responsible for coordinating the IP6 molecule around which the deaminase domain folds (Macbeth et al. 2005; and supplementary fig. S1, Supplementary Material online). The coral ADAR1 sequence is also conserved although there are some notable differences when compared with the vertebrate ADAR1 family. Unlike vertebrate ADAR1s, which contain three dsRBDs, the coral enzyme contains only one. In addition, vertebrate ADAR1s have both a Zα and Zβ domain at their N-terminus, while the coral ADAR1 has only an identifiable Zα domain. Most of the important catalytic and IP6-binding residues are conserved. Based on this conservation of structure, we hypothesize that both coral clones encode catalytically active ADARs.
Fig. 2.
Phylogenetic analysis of ADAR domains. Coral ADAR1 and ADAR2 are hADAR1 and hADAR2 orthologs. (A) Clustering of the coral and hADAR dsRBD domains. The two coral ADAR2 dsRBDs clustered closest to dsRBD from hADAR2 and hADAR3. The single coral ADAR1 dsRBD clustered closer to the hADAR1 dsRBD domains. (B) Clustering of the deaminase domains. The coral ADAR1 DD clustered with the hADAR1 DD and the coral ADAR2 DD clustered with the hADAR2 and hADAR3 DD.
Detecting A-to-I RNA Hyper-Editing Events in Coral RNA-Seq
To assess the overall level of RNA editing in coral samples we first used a previously published method (Porath et al. 2014) to detect hyper-editing events. This approach allows for direct detection of RNA editing sites without the need for available matched DNA information from the same sample. ADARs can extensively edit RNA transcripts, such that the heavily edited RNA molecules differ widely from their corresponding DNA. Consequently, alignment of the derived RNA reads to the reference genome includes many mismatches. Thus, many extensively edited reads are discarded from consideration, for the lack of a clean alignment. A dedicated tool that circumvents this issue allows us to analyze these hyper-edited reads, resulting in the detection of many more edited sites (Porath et al. 2014).
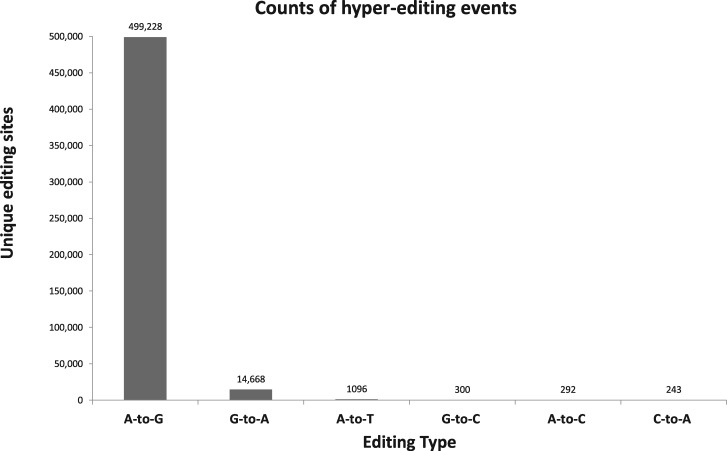
We applied the hyper-editing pipeline using RNA sequencing datasets of A. millepora coral samples collected during different time points: before, during and post spawning event (see (Kaniewska et al. 2015) and materials and methods). Twelve RNA-seq datasets were prepared consisting of ≈403 million reads (of 100 × 2 bp). We used BWA (Burrows–Wheeler Aligner) to align these reads to the A. millepora genome (Moya et al. 2012), resulting in ≈ 131 million reads that were deemed unmapped. The hyper-editing protocol was applied to these unmapped sequences to generate a reference set of potential hyper-edited reads. We detected 1,834,968 editing events (499,228 unique A-to-G sites) (fig. 3, supplementary table S2) within only 195,643 reads. These edited sites clustered to 34,226 distinct regions, consistent with the known ADAR preference for editing in dense clusters.
Fig. 3.
Distribution of hyper-editing events. Unmapped reads were analyzed for hyper-editing events. Results showed that 97% of the unique mismatch sites were A-to-G, indicating a high level of specificity for the hyper-editing screen. The 499,228 detected sites were found within 195,643 reads and clustered to 34,226 regions.
We next examined the 499,228 hyper-edited sites to determine their editing levels, using the reads that did align to the genome (in two representative samples—see Methods). An adenosine site was considered edited only if it was covered by ten or more reads and at least 1% of the covering reads were G. Of the hyper-edited sites, 347,105 were covered by at least one read (from the aligned read set) in at least one of the samples, and only 116,722 sites were covered by ten or more reads in at least one of the samples. Among these, 34,422 sites were edited in one of the samples. The average level of editing was ≈ 20% (median of 9%) in each of the two samples studied.
The specificity of an editing detection routine is usually evaluated by comparing the A-to-G mismatch signal with that from other mismatches. Our screen for hyper-editing in the coral data set achieved high specificity, with 97% of the unique hyper-editing sites being A-to-G (fig. 3). Additional support for the specificity of the A-to-G results comes from the strand information. The RNA-seq data we used was strand-indifferent, and therefore the observed mismatches of edited sites should be either A-to-G or T-to-C at roughly equal frequency. As expected, our detected A-to-G sites have nearly equal distribution between the strands (47%/53% A-to-G/T-to-C). Moreover, in 82% of the A-to-G hyper-edited reads that overlapped with predicted gene regions, the expressed strand shows A-to-G mismatch. The remaining A-to-G reads may be due to lacking a reliable annotation or editing of transcripts expressed from the antisense strand. This high level of strand bias indicates the high level of true A-to-G editing sites.
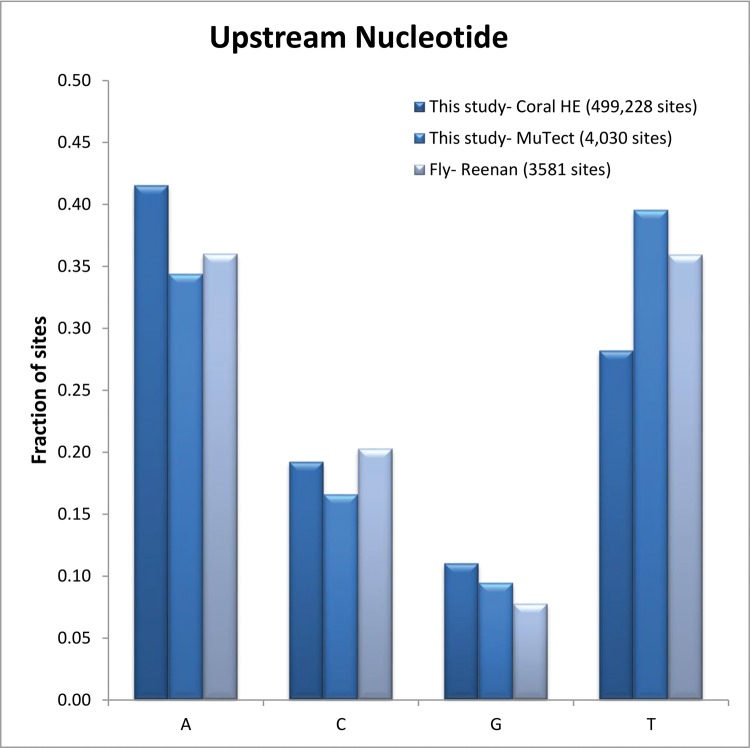
Examining the sequence context of our detected sites showed that even with the ancient divergence of coral ADAR (from the well-characterized vertebrate isoforms), editing sites exhibited the main known ADAR preference for neighboring residues: G’s were depleted on the 5′ side (guanosine was found one base upstream to the edited nucleotide in only ∼10% of the unique A-to-G events, fig. 4) (Eggington et al. 2011; Ramaswami et al. 2013). Next, we examined the region surrounding editing sites for the potential to form dsRNA structures, a structural requirement for ADARs. This was done by using bl2seq (Tatusova and Madden 1999) to align the hyper-edited regions identified in one of the samples (6709 regions) and their flanking (±2 kbp) sequences at reverse orientation (see methods). In 34.8% of the hyper-edited regions, a match was found (defined as ≥ 65% identity along ≥ 80% of the region), compared with only 8.4% of the regions returning a match when aligned to the same flanking sequences but on the same strand (a match that does not make possible the formation of dsRNA). We computed the number of hyper-editing sites per mapped read as an approximation of the true editing rate, and found the coral editing level (1.8 × 10−3) to be even higher than previously shown in human brain (1.3 × 10−3), mouse (1.1 × 10−4), and Drosophila (1.6 × 10−4) (Porath et al. 2014). Overall, our genome-wide screen for hyper-editing sites established that editing by ADAR enzymes is highly abundant in corals.
Fig. 4.
The sequence context of edited A-to-G sites. Guanosines are depleted upstream to the edited As, both in hyper-edited sites and single sites (as calculated by MuTect). This is in agreement with previously shown data concerning edited sites in other organisms such as the fly. No enrichment for G an or C were found at the 3′ base.
Only 79,341 of the 499,228 detected sites overlapped with predicted gene regions. Gene regions were defined by mapping A. millepora transcripts that contained evolutionary conserved regions (by BLAST alignment to Swiss-Prot database) to the A. millepora genome (Moya et al. 2012) (see Methods for more details). Out of 79,341 editing sites in genes, the majority (61,340) were in introns. Of the remaining 18,001 sites that were located in exons, only 3928 resided in coding regions (supplementary fig. S2a, Supplementary Material online). Of these, 337 editing sites were confirmed in at least one of the two samples using the aligned reads (see above). Of these, only 18 sites were identified as true non-synonymous sites (by manual inspection; see Methods), suggesting that recoding by editing is a very rare event. Moreover, only 3.5% of the detected coding sites were edited at levels of 20% or more, in comparison to 10.9% of the sites detected in UTR regions. Overall, these results suggest that while extensive hyper-editing is common and easily detected, only very few sites are located in coding regions, and most of these have low editing levels.
Detecting Isolated RNA Editing-Sites
The hyper-editing detection protocol is well-suited for clustered sites, but may miss single recoding sites. Identification of genuine isolated editing events with high sensitivity and specificity is a very challenging task (Bazak et al. 2014a). We have tried to meet this challenge by analyzing RNA and DNA from the same coral sample. RNA-seq and DNA-seq were aligned to the A. millepora genome and analyzed for putative A-to-G editing sites by MuTect (Cibulskis et al. 2013) (see methods for more details). By comparing the transcriptome data to the genome data of the same individual coral specimen, we were able to filter false-positives due to polymorphic variations in the genome. We detected 6,714 sites of mismatches between the DNA and the RNA sequences. Out of the six possible types of mismatches, 4,030 (60%) were A-to-G (Supp. Table 3), indicating a clear A-G bias. Only 15.6% were G-A, the next most common type of mismatch. Here too, A-to-G editing sites exhibited the known ADAR motif, with Gs depleted one base upstream (∼9%; fig. 4) and enriched one base downstream (∼ 34%) of the editing sites (Eggington et al. 2011; Ramaswami et al. 2013).
To search for recoding sites, we looked within the coding regions of genes. Out of the 4,030 A-to-G sites that were detected by MuTect, only 860 (21.3%) were found in predicted gene regions (although those regions are covered by many more reads) (supplementary fig. S2b, Supplementary Material online). Of these, 628 A-to-G sites overlapped with predicted exons. The A-to-G signal within the exons (65%; 628 sites out of 969 mismatches of all types) was comparable to the 60% value found genome-wide. It is well known in other organisms that specific recoding sites are often accompanied by neighboring satellite sites. Thus, to increase the specificity of detection we focused on sites with at least one additional adjacent site within 1000 bp. This increased the specificity of detection of A-to-G mismatches in exons from 65% to 86% (464/539), yielding a total of 464 putative A-to-I editing sites (Supp. Table 4). The residual noise (14%) is likely to be an overestimate, since 61/75 (81%) of the non A-to-G sites are T-to-C sites, which probably represent bona-fide editing sites of transcripts expressed from the antisense strand. The average level of editing in these exonic sites, calculated using REDItools (Picardi and Pesole 2013), was 33% (median 25%). Sites with higher editing levels show a stronger ADAR motif bias than sites with lower editing levels.
The 464 A-to-G sites in exons lie within genes such as WNT5, integrin alpha, cytochrome p450, E3 ubiquitin, prolactin releasing peptide, calmodulin, dopamine receptor, cry 1, and RAS related protein RAB. Thus, some of the edited genes have a role in signal transduction and were also identified in our previous reports as showing high levels of gene expression (Kaniewska et al. 2015; Rosenberg et al. 2017). Specifically, dopamine and its G-protein coupled receptor participate in entraining the endogenous circadian clock and are also involved in the timing of spawning (Isomura et al. 2013). In corals, cry1 is known to act as a photoreceptor integrated within the circadian clock machinery and its expression levels are affected by the phase of the moon (Levy et al. 2007). Six randomly chosen editing regions in exons were successfully validated by Sanger sequencing (see methods and supplementary fig. S3, Supplementary Material online for more details).
However, only four of the 464 exonic sites reside in coding regions and after a manual inspection, they look like false positives. Taken together, the hyper-editing detection scheme including MuTect analysis suggest that the abundance of editing in coding regions is very low compared with the editing in non-coding regions, indicating that the editing process itself may be more important than the resulting protein diversification.
Evidence for Physiological Function of Editing in Coral-Spawning
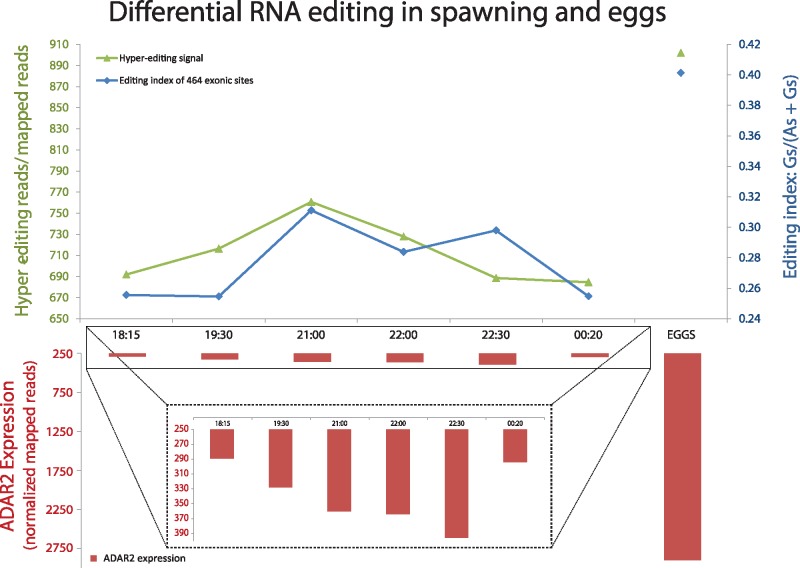
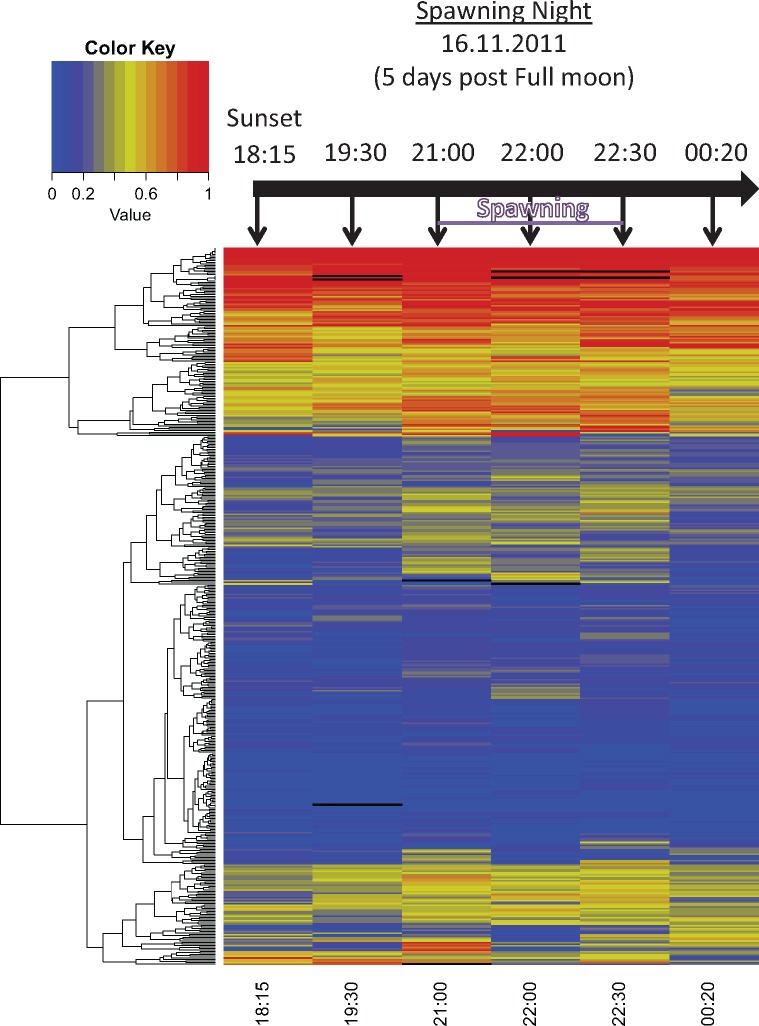
During the spawning night on the 16th of November 2011 at Heron Island Great Barrier Reef, six colonies were transferred to large, outdoor flow-through aquaria and were exposed to ambient conditions: of natural sunlight, moonlight phases, and flow-through seawater from the reef flat spawned in a similar manner to those on the reef. Colonies began to show signs of ‘setting’, while gamete bundles appeared in the polyp mouths at around 19:30 and gametes were released at approximately 21:30–22:30 (Kaniewska et al. 2015). Samples were taken at six time points during this period corresponding to pre-setting (18:15 PM), setting (19:30 PM), spawning (21:00, 22:00, and 22:30 PM), and post spawning (00:20 AM). RNA editing analysis from the different stages of spawning showed a clear increase in RNA editing levels just prior to the “setting” phase and the gametes release phase. For each sample, we calculated the hyper-editing signal (number of detected hyper-edited reads divided by the number of total mapped reads) and the editing index [total number of Gs divided by the total number of Gs + As; see (Bazak et al. 2014b)] for the exonic editing sites. Both measures of editing activity exhibit a peak just prior to the gamete release (figs. 5 and 6). As expected, ADAR2 expression levels (Kaniewska et al. 2015) mirrored the trend in editing levels throughout the sampled time points (fig. 5). Thus, the level of ADAR2 expression may explain the changes observed in editing levels.
Fig. 5.
Editing levels during spawning and in eggs. The hyper-editing signal (green line) increases towards the gamete release stage (21:00) and then decreases until post spawning. The level of editing in exonic sites (blue line) shows a similar pattern, with lower levels of editing pre and post-spawning. The level of ADAR expression (red bars, in inset) increases just prior to and throughout spawning, corresponding to the levels of editing. Editing levels were much higher in eggs, compared to peak spawning times, both in exonic sites and in hyper-edited events. Similarly, the expression level of ADAR was much higher in the gametes than in host tissue analyzed during the “setting” and the spawning stages. The elevated levels of editing may serve to enrich the variability in gametes without necessitating permanent changes in the genome.
Fig. 6.
Editing in exons during spawning. Heat map of editing levels in exonic sites. Each line in the heat map represents an editing site. The color bar represents the range of editing level in all sites that were covered (a black box represents a point which was not covered by reads). More than a quarter of sites showed a mid-to-high level of editing throughout the spawning event. A similar number of editing sites had a low level of editing, which rose during spawning and then decreased post-spawning.
In addition to the spawning samples of the coral host, we also analyzed samples of the egg bundle (comprising sperm and eggs) collected during the gamete release between 21:30 and 22:30. Interestingly, the level of ADAR2 expression in this sample was extremely higher (2900 normalized mapped reads) compared with the samples of spawning adult coral (360–396 normalized mapped reads) and, accordingly, the hyper-editing signal (902) and the index of the exonic editing-sites (0.4) were also much higher (fig. 5). A total of 47,064 hyper-editing sites were identified in the egg samples and were assessed for editing levels. Of these, 3436 sites were covered by at least 10 reads and edited by at least 30%. Moreover, 2363 hyper-editing sites are located in coding regions (1647 recoding sites), about five times more coding sites detected in eggs than in adult sample—308 coding sites in 70 genes were found (covered by at least 5 reads and were edited by at least 1%) (supplementary table S5, Supplementary Material online). Composition of reads (edited and unedited) in two representative coding regions are presented in supplementary figure S4, Supplementary Material online, showing the diversity potential of proteins that are encoded by those regions. Most of the genes detected were previously shown to be highly expressed during spawning, and to be related to the canonical pathways that synchronize the gamete release in those coral species (Kaniewska et al. 2015; Rosenberg et al. 2017). Putative editing in Neuropeptide Y receptor active site is also shown in supplementary figure S5, Supplementary Material online. Neuropeptide Y receptor is part of the rhodopsin-like 7-transmembrane GPCR family. Like many other members of the family, it is highly upregulated during the spawning event and participates in transmitting intracellular signals. Many of the rhodopsin-like family have been shown to react to light-absorbing pigments, becoming activated in response to light to mediate a variety of photoreactive processes in different organisms and participate in entraining the endogenous clock. The function of light sensing receptors is important for regulating physiological actions that depend on light cycles such as the synchronized spawning that occurs in coral reefs. Another gene that had recoding events was Calmodulin. This protein is part of the calcium-signaling pathway. Binding of Ca2+ ions to the intermediate messenger calmodulin-calcium-modulated protein (CAM) transduces the calcium signals and, as the Ca2+/CAM complex accumulates, it can phosphorylate and activate a serine/threonine-specific protein kinase (Ca2+/calmodulin-dependent protein kinase II). The Ca2+/CAM complex also binds nitric oxide synthase and stimulates the production of the free radical NO which binds guanylate cyclase resulting in an increase in cGMP in response to cellular levels of calcium. Further analysis of GO enrichment analysis using DAVID (Huang et al. 2009a,b) detected an enrichment of a few interesting gene groups which have a possible role in the spawning stage and in new gamete release. The spectrin binding genes, such as Ankyrin-2, and actin binding genes, such as Alpha-adducin-2, are involved in cytoskeletal organization and structure—one of the functions upregulated during spawning. In squid, 10% of SPECTRIN’s amino acids (247/2412) are recoded (Alon et al. 2015) showing the ubiquity of editing of this gene in diverse phyla.
To further support our findings of high editing level in Acropora millepora egg bundle, we analyzed another coral species Acropora digitifera (Rosenberg et al. 2017). Here too, we found extremely higher levels of ADAR2 expression in eggs compared to adults. Similarly, enrichment of editing in coding regions was found in eggs, particularly in comparison to the spawning samples where editing in coding regions is quite depleted (supplementary fig. S6, Supplementary Material online). The high number and levels of RNA editing events in newly released gametes of both analyzed corals suggest that the process has an important physiological role at this critical stage of coral life cycle.
Discussion
For many years, it was believed that the main function of RNA editing by ADAR was to recode a few critical sites in the nervous system, such as the Q/R site in glutamate receptors. Recently, it has become clear that ADAR enzymes play a critical role in editing self dsRNA, which when unedited stimulate the innate immune system and trigger the interferon cascade (Liddicoat et al. 2015; Pestal et al. 2015). Evolutionarily, RNA editing by ADAR is ancient. It dates back to around the same time as the emergence of the nervous system (Grice and Degnan 2015). As with all Cnidarians, corals display the simplest nervous system organisation at the tissue level and can offer insights into the origins of the molecular machinery that regulates RNA editing throughout the animal kingdom. In this report, we studied the coral A. millepora to investigate the primordial function of RNA editing.
While the presence of ADAR has been shown in basal metazoans (Grice and Degnan 2015), its activity has not previously been demonstrated. Here we show RNA editing sites in the cnidarian anthozoan coral A. millepora during different stages of spawning and gamete release. We used computing tools (Cibulskis et al. 2013; Porath et al. 2014) to identify and categorize isolated and hyper-editing sites in RNA-seq transcripts from samples of coral taken before, during, and shortly after a spawning event at the Great Barrier Reef in 2011 (Kaniewska et al. 2015). Evaluating these results clearly demonstrated the presence of numerous edited sites with the A-to-G conversion signal much higher than other mismatches. The computed rate of hyper-editing in coral was found to be even higher than that shown in human, mouse or Drosophila (Porath et al. 2014). Together with the conservation of the ADAR functional domains, this indicates high functionality of ancient ADAR and the presence of RNA editing in a basal organism. As with vertebrates, we found that although editing is very common, sites in coding regions are extremely rare. The abundant editing in non-coding regions compared with the almost non-existing levels in coding regions in the coral point to the original role of ADAR as a non-specific use of opening dsRNA more than as a mechanism of causing transient point mutations. While recoding events do occur, they are limited. A connection between opening dsRNA and inducing the immune response has not yet been shown to occur in corals. Further study is needed to discern whether the non-specific activity of coral ADAR is for inducing a basal form of an immune system or as a primitive defense system against foreign dsRNA.
Interestingly, we observed a clear temporal change in the levels of ADAR2 mRNA expression and the frequency of RNA editing during spawning (figs. 5 and 6), peaking at the time of gamete release. This may imply an important role of RNA-editing during early developmental stages. The coral reef ecosystem depends greatly on the reproductive success of corals. Corals are sessile animals but their early larval life stages are pelagic, and thereby exposed to potentially more dynamic and varied environmental conditions than the adults (Padilla-Gamiño et al. 2012). Our results suggest that during spawning RNA-editing may be used to introduce variability among the numerous gametes being released (fig. 5), generating post-transcriptional diversity without affecting the genome. Although most of the editing is in non-coding regions, such transient mutations could provide a beneficial mutation for selected gametes without relying on somatic changes and indeed, we find much more recoding events in eggs. Another possible mechanism for the high occurrence of editing at eggs is the report that a high level of dsRNA causes RNAi in oocytes but not somatic cells (Nejepinska et al. 2012) suggesting that editing activity might be more important in eggs than in adults because of the need to deal with dsRNA structures. Taken together, these results point to the opening of dsRNA as the primitive function of ADAR. Nonetheless, the recoding events and the higher editing in gametes might suggest that ADAR has already evolved both functions in the last common ancestor of metazoan.
Materials and Methods
Coral Collection and Experimental Design
Ten colonies of Acropora millepora were collected on November 9, 2011 from the Heron Island reef flat (23 33′S, 151 54′E), Great Barrier Reef, Australia. Small branches were cut from the central portion of each colony prior to specimen collection to confirm the presence of pink-colored eggs, which indicate that a colony is reproductively mature. Four of the colonies were transported to the vicinity of the shore but were left in the field on the reef flat. The remaining six colonies were transferred to large, outdoor flow-through aquaria and were exposed to ambient conditions: of natural sunlight, moonlight phases, and flow-through seawater from the reef flat. The experiment was conducted at Heron Island Research Station in an area that was maintained in darkness at night to avoid artificial light contamination from non-experimental sources [for more details see Kaniewska et al. (2015)]. On November 16th (the spawning night), we sampled the corals at noon, 18:15, 19:30, 21:00, 22:00, 22:30, and 00:20. The release of gametes occurred between 21:30 and 22:30, and the colonies in the Ambient treatment (natural sunlight, moonlight phases) began to show signs of ‘setting’ at 19:30. Sampled coral branches were snap-frozen in liquid nitrogen and stored at -80 °C until processed for total RNA extraction.
Total RNA Isolation
Total RNA from the coral branches was isolated by homogenizing 100 mg coral tissue in 1 ml TRIzol (Invitrogen) according to the manufacturer’s instructions. The RNA was then extracted once with 1 volume chloroform and precipitated in ½ volume isopropanol, then washed in 1 volume of 75% ethanol and subsequently dissolved in RNase-free water. These samples were then processed through a 5 M LiCl precipitation overnight at −20 °C, washed three times with 75% ethanol and subsequently dissolved in RNase-free water. The integrity and quality of the total RNA was assessed using a Bioanalyzer (Agilent Technology). Only samples including intact RNA (RNA Integrity number > 8) were used for the RNA-seq analysis.
RNA-Seq Analysis
The Illumina TruSeq protocol was used to prepare libraries from the RNA samples. Overall libraries of samples from the spawning experiment were run on two additional lanes in the Illumina HiSeq2000 machine using the multiplexing strategy of the TruSeq protocol. On average ∼15 million paired-end reads were obtained for each sample in the spawning experiment. The sequencing data of the spawning samples was deposited in the Sequence Read Archive (SRA), under accession number SRP055723. The reads were paired-end 100 bases long. The sequencing data of the one sample coral that was used for the isolated editing sites detection, was deposited in the Sequence Read Archive (SRA), under accession number SRP080785. The reads were paired-end 100 bases long.
DNA-Seq Analysis
Genomic DNA from ambient treated coral sampled at 9 pm on 12/11/2011 was isolated by homogenizing tissue in extraction buffer (100 mM EDTA, 10 mM Tris, 1% SDS, pH 7.5) and incubating the slurry at 65 °C for an hour. Proteinase K was then added to a final concentration of 200 µg/ml and the slurry was incubated at 37 °C overnight. After this time, the samples were extracted once with one volume phenol chloroform isoamyl alcohol (25:24:1 v/v) and once with one volume chloroform. Samples were then precipitated in 0.6 volumes of isopropanol and 1/10 volume of 3 M sodium acetate, washed three times with 70% ethanol before being redissolved in TE (0.01 M Tris-HCl, pH 7.5, 0.001 M EDTA). DNA quality was assessed using gel electrophoresis visualization. The Illumina TruSeq protocol was used to prepare a library from the DNA sample and samples were run on an Illumina HiSeq2000 machine. The sequencing data of the one sample coral was deposited in the Sequence Read Archive (SRA), under accession number SRP080785. The reads were paired-end 100 bases long.
Validation
PCR re-sequencing of results, presented in supplementary figure S3, Supplementary Material online (representative result), was done at the Australian Genome Research Facility using primers for 18 separate reactions (supplementary table S1, Supplementary Material online). Amplicon success was assessed using gel electrophoresis visualization and successful amplicons were sequenced using Sanger Sequencing.
Identification of Hyper-Editing Reads and Sites
Hyper-editing sites were identified with the protocol described previously (Porath et al. 2014). As an input, we used RNA-seq datasets (paired-end 100 bp reads) as described above. We consider the paired-end datasets as two separated single-end data, since the genome of A. millepora is not completely assembled. Running of the hyper-editing protocol was done by using the default parameters.
Prediction of Genes and Coding Regions
To predict gene regions, we used the published transcripts of Acropora millepora from NCBI and considered only transcripts with a conserved region. Conservation was examined by aligning the transcripts to the Swiss-Prot database (downloaded on Sep 2014) using BLASTX (parameters: blastx -evalue 1e−6 -num_descriptions 0 -num_alignments 1). The transcripts that were found to have conserved regions were then mapped to the Acropora millepora genome using BLAT (parameters: blat -mask = lower -noHead -minIdentity = 99 -minScore = 150) (Kent 2002).
In order to define the coding region, the longest conserved region for each transcript was selected and the ORF was extended in both directions. The 3′ end was extended until the first stop codon or, in the absence of a stop codon, until the end of the transcript. The 5′ end was extended until the MET codon proximate to a stop codon or, in the absence of a MET codon, until a stop codon or until the start of the transcript.
Defining Hyper-Edited Regions and dsRNA Structure
We defined the portion of the edited reads from the first to the last A-to-G mismatch as a cluster of edited mismatches. To identify the hyper-edited regions, we first merged the corresponding genomic coordinates of all overlapping (or with a distance ≤ 20 bp) edited clusters, and then set the boundaries of the region from the first base of the most upstream cluster to the last base of the most downstream cluster. Construction and analysis of the edited regions were carried out using BEDTools (Quinlan and Hall 2010).
To detect potential dsRNA structures formed by hyper-edited RNAs, the DNA sequences of the hyper-edited regions were aligned to the sequences 2 kbp upstream and 2 kbp downstream of the regions. We used bl2seq (Tatusova and Madden 1999) with parameters -F F -W 7-r 2, and a match for alignment required at least 65% identity along 80% of the hyper-edited region length.
Computing Editing Levels
In order to calculate the editing levels of the identified sites, we first aligned the RNA-seq datasets to the A. millepora genome by STAR 2-pass with default parameters (Dobin et al. 2013). Alignments were improved by picard (http://picard.sourceforge.net) and GATK (McCormick et al. 2015) tools and analyzed for editing levels by the REDItools known (Picardi and Pesole 2013) with parameters: -v 1 -n 0.01 -c 1 -T 6-6.
Nonsynonymous Sites
In order to determine the final set of non-synonymous sites (out of the sites that were detected by the hyper-editing procedure), we computed the editing levels in two representative samples using the aligned reads. We consider only recoding sites that were confirmed to be edited in both samples and with at least 10% editing in one of them. Sites were also verified to be “A” in the DNA-sequencing of the one individual coral sample (having at least 50 “A” reads and no “G” reads).
Detecting RNA Editing Using DNA-Seq and RNA-Seq
In order to detect novel editing sites, we first aligned the DNA-seq and RNA-seq datasets to the Acropora millepora genome using default parameters of BWA (Li and Durbin 2009) for the DNA-seq, and STAR 2-pass (Dobin et al. 2013) for the RNA-seq. STAR 2-pass aligner was chosen in order to reduce mapping errors at exon-intron junctions. Alignments were improved by picard (http://picard.sourceforge.net) and GATK (McCormick et al. 2015) tools and analyzed for novel editing sites by MuTect (Cibulskis et al. 2013) version 1.1.4 (default parameters) using the DNA-seq as the normal sample and the RNA-seq as the tumor sample. [MuTect was developed to detect somatic mutations in cancer samples by comparing the normal DNA-seq with the tumor DNA-seq of same individual].
Evolutionary Trees Analysis
Evolutionary trees were inferred using the UPGMA method and amino acid sequences (Sneath and Sokal 1962). The optimal tree with the sum of branch length = 4.94 is shown for figure 2 panel A and branch length = 2.42 for panel B. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (10000 replicates) are shown next to the branches (Felsenstein 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling 1965) and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option). There were a total of 59 positions in the final dataset for figure 2 panel A (dsRBDs) and of 275 positions for panel B (Deaminase Domains). Phylogenetic analyses were conducted using MEGA version 4.0.2 (Tamura et al. 2007).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Heron Island Research Station staff for their help during our spawning experiments. This work was supported by the Australian Research Council (to PK), the European Research Council (grant 311257), the I-CORE Program of the Planning and Budgeting Committee in Israel (grants 41/11 and 1796/12), and the Israel Science Foundation (1380/14). The sequencing data of the one sample coral was deposited in the Sequence Read Archive (SRA), under accession number SRP080785.
References
- Albertin CB, Simakov O, Mitros T, Wang ZY, Pungor JR, Edsinger-Gonzales E, Brenner S, Ragsdale CW, Rokhsar DS.. 2015. The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature 524:220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon S, Garrett SC, Levanon EY, Olson S, Graveley BR, Rosenthal JJC, Eisenberg E, Alon S, Garrett S, Levanon E, et al. 2015. The majority of transcripts in the squid nervous system are extensively recoded by A-to-I RNA editing. Dryad Digit Repos. 4:613–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BASILIO C, WAHBA AJ, LENGYEL P, SPEYER JF, OCHOA S.. 1962. Synthetic polynucleotides and the amino acid code V. Proc Natl Acad Sci U S A. 48:613–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL, Weintraub H.. 1988. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 55: 1089–1098. [DOI] [PubMed] [Google Scholar]
- Bazak L, Haviv A, Barak M, Jacob-Hirsch J, Deng P, Zhang R, Isaacs FJ, Rechavi G, Li JB, Eisenberg E, et al. 2014a. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 24:365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazak L, Levanon EY, Eisenberg E.. 2014b. Genome-wide analysis of Alu editability. Nucleic Acids Res. 42:6876–6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla T, Rosenthal JJC, Holmgren M, Reenan R.. 2004. Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nat Struct Mol Biol. 11:950–956. [accessed 2010 Sep 14]. http://www.ncbi.nlm.nih.gov/pubmed/15361858 [DOI] [PubMed] [Google Scholar]
- Burnashev N, Schoepfer R, Monyer H, Ruppersberg JP, Günther W, Seeburg PH, Sakmann B.. 1992. Control by asparagine residues of calcium permeability and magnesium blockade in the NMDA receptor. Science 257:1415–1419. [DOI] [PubMed] [Google Scholar]
- Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB.. 1997. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387:303–308. [DOI] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G.. 2013. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 31:213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR.. 2013. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggington JM, Greene T, Bass BL.. 2011. Predicting sites of ADAR editing in double-stranded RNA. Nat Commun. 2:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution (N. Y) 39:783. [DOI] [PubMed] [Google Scholar]
- Garrett S, Rosenthal JJC.. 2012. RNA editing underlies temperature adaptation in K+ channels from polar octopuses. Science (80-.) 335:848–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B, Agranat-Tamir L, Light D, Ben-Naim Zgayer O, Fishman A, Lamm AT.. 2016. Dec 28. A-to-I RNA editing promotes developmental-stage-specific gene and lncRNA expression. Genome Res. gr.211169.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice LF, Degnan BM.. 2015. The origin of the ADAR gene family and animal RNA editing. BMC Evol Biol. 15:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA.. 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA.. 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4:44–57. [DOI] [PubMed] [Google Scholar]
- Isomura N, Yamauchi C, Takeuchi Y, Takemura A, Domeier ML, Colin PL, Babcock RC, Harrison PL, Hayashibara T, Babcock RC, et al. 2013. Does dopamine block the spawning of the acroporid coral Acropora tenuis? Sci Rep. 3:698–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniewska P, Alon S, Karako-Lampert S, Hoegh-Guldberg O, Levy O.. 2015. Signaling cascades and the importance of moonlight in coral broadcast mass spawning. Elife 4: 09991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K.. 2007. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep 8:763–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K.. 2007. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science (80-.) 315: 1137–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res 12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K.. 1994. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc Natl Acad Sci USA. 91:11457–11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn AB, Sanford RS, Yoshida M, Moroz LL.. 2015. Parallel evolution and lineage-specific expansion of RNA editing in ctenophores. Integr Comp Biol. 55:1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy O, Appelbaum L, Leggat W, Gothlif Y, Hayward DC, Miller DJ, Hoegh-Guldberg O.. 2007. Light-responsive cryptochromes from a simple multicellular animal, the coral Acropora millepora. Science 318:467–470 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Wang Z, Lian J, Schiøtt M, Jin L, Zhang P, Zhang Y, Nygaard S, Peng Z, Zhou Y, et al. 2014. Caste-specific RNA editomes in the leaf-cutting ant Acromyrmex echinatior. Nat Commun. 5:4943.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, Li JB, Seeburg PH, Walkley CR.. 2015. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science (80-.) 349:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macbeth MR, Schubert HL, Vandemark AP, Lingam AT, Hill CP, Bass BL.. 2005. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 309:1534–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannion NM, Greenwood SM, Young R, Cox S, Brindle J, Read D, Nellåker C, Vesely C, Ponting CP, McLaughlin PJ, et al. 2014. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 9:1482–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick RF, Truong SK, Mullet JE.. 2015. RIG: recalibration and interrelation of genomic sequence data with the GATK. G3 (Bethesda) 5:655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher T, Maas S, Herb a, Sprengel R, Seeburg PH, Higuchi M.. 1996. A mammalian RNA editing enzyme. Nature 379:460–464. [DOI] [PubMed] [Google Scholar]
- Morse DP, Aruscavage PJ, Bass BL.. 2002. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc Natl Acad Sci U S A. 99:7906–7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya A, Huisman L, Ball EE, Hayward DC, Grasso LC, Chua CM, Woo HN, Gattuso J-P, Forêt S, Miller DJ.. 2012. Whole transcriptome analysis of the coral Acropora millepora reveals complex responses to CO2-driven acidification during the initiation of calcification. Mol Ecol. 21:2440–2454. [DOI] [PubMed] [Google Scholar]
- Nejepinska J, Malik R, Filkowski J, Flemr M, Filipowicz W, Svoboda P.. 2012. DsRNA expression in the mouse elicits RNAi in oocytes and low adenosine deamination in somatic cells. Nucleic Acids Res. 40:399–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell MA, Krause S, Higuchi M, Hsuan JJ, Totty NF, Jenny A, Keller W.. 1995. Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase. Mol Cell Biol. 15:1389–1397. [accessed 2016 Jul 13]. http://www.ncbi.nlm.nih.gov/pubmed/7862132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell MA, Mannion NM, Keegan LP.. 2015. The epitranscriptome and innate immunity. PLoS Genet. 11:e1005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla-Gamiño JL, Pochon X, Bird C, Concepcion GT, Gates RD.. 2012. From parent to gamete: vertical transmission of symbiodinium (Dinophyceae) ITS2 sequence assemblages in the reef building coral Montipora capitata.PLoS One 7:e38440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestal K, Funk CC, Snyder JM, Price ND, Treuting PM, Stetson DB.. 2015. Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development. Immunity 43:933–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picardi E, Pesole G.. 2013. REDItools: High-throughput RNA editing detection made easy. Bioinformatics 29:1813–1814. [DOI] [PubMed] [Google Scholar]
- Pinto Y, Cohen HY, Levanon EY.. 2014. Mammalian conserved ADAR targets comprise only a small fragment of the human editosome. Genome Biol. 15:R5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porath HT, Carmi S, Levanon EY.. 2014. A genome-wide map of hyper-edited RNA reveals numerous new sites. Nat Commun. 5:4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G, Li JB.. 2014. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 42:D109–D113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G, Zhang R, Piskol R, Keegan LP, Deng P, O’Connell MA, Li JB.. 2013. Identifying RNA editing sites using RNA sequencing data alone. Nat Methods 10:128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder LE, Savva YA, Reyna MA, Chang Y-J, Dorsky JS, Rezaei A, Reenan RA.. 2015. Dynamic response of RNA editing to temperature in Drosophila. BMC Biol. 13:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg Y, Doniger T, Harii S, Sinniger F, Levy O.. 2017. Canonical and cellular pathways timing gamete release in Acropora digitifera, Okinawa, Japan. Mol Ecol. 26:2698–2710. [DOI] [PubMed] [Google Scholar]
- Rosenthal JJC, Bezanilla F.. 2002. Extensive editing of mRNAs for the squid delayed rectifier K+ channel regulates subunit tetramerization. Neuron 34:743–757. [DOI] [PubMed] [Google Scholar]
- Rueter SM, Dawson TR, Emeson RB.. 1999. Regulation of alternative splicing by RNA editing. Nature 399:75–80. [DOI] [PubMed] [Google Scholar]
- Rula EY, Lagrange AH, Jacobs MM, Hu N, Macdonald RL, Emeson RB.. 2008. Developmental modulation of GABA(A) receptor function by RNA editing. J Neurosci. 28:6196–6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneath PH, Sokal RR.. 1962. Numerical taxonomy. Nature 193:855–860. [DOI] [PubMed] [Google Scholar]
- St Laurent G, Tackett MR, Nechkin S, Shtokalo D, Antonets D, Savva YA, Maloney R, Kapranov P, Lawrence CE, Reenan RA.. 2013. Genome-wide analysis of A-to-I RNA editing by single-molecule sequencing in Drosophila. Nat Struct Mol Biol. 20:1333–1339. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S.. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 24:1596–1599. [DOI] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL.. 1999. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 174:247–250. [DOI] [PubMed] [Google Scholar]
- Wang IX, So E, Devlin JL, Zhao Y, Wu M, Cheung VG.. 2013. ADAR regulates RNA editing, transcript stability, and gene expression. Cell Rep. 5:849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler EC, Washburn MC, Major F, Rusch DB, Hundley HA.. 2015. Noncoding regions of C. elegans mRNA undergo selective adenosine to inosine deamination and contain a small number of editing sites per transcript. RNA Biol. 12:162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple JM, Youssef OA, Aruscavage PJ, Nix DA, Hong C, Johnson WE, Bass BL.. 2015. Genome-wide profiling of the C. elegans dsRNAome. RNA 21:786–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Zhang J.. 2014. Human coding RNA editing is generally nonadaptive. Proc Natl Acad Sci U S A. 111:3769–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H-Q, Zhang P, Gao H, He X, Dou Y, Huang AY, Liu X-M, Ye AY, Dong M-Q, Wei L.. 2015. Profiling the RNA editomes of wild-type C. elegans and ADAR mutants. Genome Res 25:66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L.. 1965. Evolutionary divergence and convergence in proteins In: Bryson V, Vogel HJ, editors. Evolving genes and proteins. New York: Academic Press, p. 97–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.