Abstract
Plant genomes contain numerous transposable elements (TEs), and many hypotheses on the evolutionary drivers that restrict TE activity have been postulated. Few models, however, have focused on the evolutionary epigenomic interaction between the plant host and its TE. The host genome recruits epigenetic factors, such as methylation, to silence TEs but methylation can spread beyond the TE sequence and influence the expression of nearby host genes. In this study, we investigated this epigenetic trade-off between TE and proximal host gene silencing by studying the epigenomic regulation of repressing long terminal repeat (LTR) retrotransposons (RTs) in Oryza sativa. Results showed significant evidence of methylation spreading originating from the LTR-RT sequences, and the extent of spreading was dependent on five factors: 1) LTR-RT family, 2) time since the LTR-RT insertion, 3) recombination rate of the LTR-RT region, 4) level of LTR-RT sequence methylation, and 5) chromosomal location. Methylation spreading had negative effects by reducing host gene expression, but only on host genes with LTR-RT inserted in its introns. Our results also suggested high levels of LTR-RT methylation might have a role in suppressing TE-mediated deleterious ectopic recombination. In the end, despite the methylation spreading, no strong epigenetic trade-off was detected and majority of LTR-RT may have only minor epigenetic effects on nearby host genes.
Keywords: retrotransposon, epigenomics, transposable element, Oryza sativa
Introduction
Almost all eukaryotic organisms harbor transposable elements (TEs) in their genomes (Biémont and Vieira 2006) and the evolution of these elements was initially studied through theoretical and population genetic modeling. A landmark study by Charlesworth and Charlesworth (1983) modeled the dynamics of TEs in a host population by factoring transposition, excision, selection, and drift as main evolutionary parameters determining TE frequencies. With specific evolutionary conditions, a stable TE frequency can be maintained when the loss and generation of TEs cancel each other out. Ultimately, since proliferations of TEs are deleterious to the host, it is in the best interest for both the TE and the host to deter the TE from overproliferating in the host genome.
However, empirical and theoretical studies have showed that selection on the TE itself to self-regulate and reduce its transposition rate is weak when it comes to shaping its genomic copy numbers (Charlesworth and Langley 1986). Rather, the deleterious effects of TEs on the host leads to strong selective pressure on host factors to silence the TE activity and controlling its copy number from increasing in the host population (Charlesworth and Langley 1986, 1989; Lee and Langley 2012). There are three hypothesized drivers of selection on the host that leads to removing TEs from the host population (Charlesworth and Langley 1989; Nuzhdin 1999; Le Rouzic and Deceliere 2005; Blumenstiel 2011; Barrón et al. 2014), and these are selection against 1) the deleterious effect of TE transpositions (Charlesworth 1991; Brookfield 1996), 2) the metabolic cost of expressing TEs (Nuzhdin et al. 1996), and 3) ectopic recombination occurring between TEs positioned in nonhomologous chromosomal regions (Montgomery et al. 1987; Langley et al. 1988). Finally, breeding system is another factor that can determine TE dynamics. In selfing species, depending on the model of selection that removes TEs from the host population, self-fertilization can either increase or decrease TE copy numbers in the host (Charlesworth and Charlesworth 1995; Wright and Schoen 1999; Morgan 2001).
One mechanism that silences TE activity, at least at the posttranscriptional level, is through the generation of small RNAs that targets TE transcripts (Aravin et al. 2007; Malone and Hannon 2009). Since TEs are highly repetitive and accumulating mutations leads to divergent sequences and motifs between TE classes, the generation of small RNAs is crucial for recognizing and regulating TEs (Bousios and Gaut 2016). In plants, these small RNAs (specifically small interfering RNAs [siRNAs]) are further involved in the epigenetic regulation of TEs (Lisch 2009; Matzke et al. 2009). siRNAs are recruited by the RNA-directed DNA methylation (RdDM) pathway to target TE sequences in the host genome and transcriptionally represses their activity (Matzke and Mosher 2014). However, DNA methylation is not limited to the TE sequence and can spread beyond the TE sequence, affecting the methylation status of its surrounding genomic region (Ahmed et al. 2011). Epigenetic modification of euchromatic TEs can lead to methylation of nearby host genes and affect the host gene expression (Lippman et al. 2004; Zhang et al. 2008). Thus, a fourth driver of selection may arise from the spreading of TE-mediated repressive epigenetic modifications (Hollister and Gaut 2009; Lee 2015; Lee and Karpen 2017).
Asian rice Oryza sativa genome is ∼400 Mb with repetitive DNA consisting ∼35% of its genome (Yu et al. 2002; International Rice Genome Sequencing Project 2005). Long terminal repeat (LTR) retrotransposons (RTs) consist of a large part of the Oryza repetitive genome (Copetti et al. 2015). Structurally, LTR-RTs have two LTR sequences flanking the internal sequences and are classified as class I TEs due to its transpositioning via a “copy-and-paste” like mechanism (Wicker et al. 2007). Past O. sativa-TE evolutionary studies have primarily focused on LTR-RTs (Vitte and Panaud 2003; Ma et al. 2004; Vitte et al. 2007; Baucom et al. 2008; Tian et al. 2009), since its canonical structure aids de novo annotation of the element in whole genome sequences (Feschotte et al. 2002), and LTR-RTs are one of the most abundant TEs in plant genomes (Kumar and Bennetzen 1999). These studies have shown for O. sativa, recombination rate and/or gene density (as both are positively correlated with each other) is an important factor determining the LTR-RT density. Further, an epigenetic study by vonHoldt et al. (2012) showed that phylogenetically related LTR-RTs were likely to share methylation status, and younger LTR-RTs had higher methylation levels than older LTR-RTs.
Majority of these previous studies had focused on the japonica subpopulation. Given that genome-wide studies showed independent origin for the two major O. sativa subpopulations (japonica and indica) (Huang et al. 2012; Choi et al. 2017), it is necessary to compare and contrast the LTR-RT evolution in both subpopulations. Further, the effect of LTR-RT mediated spreading of methylation on host gene expression has not been thoroughly examined. In this study, using published genomic, transcriptomic, and methylomic data, we have analyzed the evolutionary genomic consequence of LTR-RT epigenomic regulation in the japonica and indica subpopulation of O. sativa.
Consistent with past study on LTR-RT sequence properties between the two subpopulations (Ma and Bennetzen 2004), our evolutionary epigenomic results between japonica and indica were largely concordant with each other. Epigenetically, there was significant evidence of methylation spreading that originated from the LTR-RT sequence. Methylation spreading was variable and dependent on the LTR-RT family, insertion time, recombination rate, methylation level of the inserted LRT-RT, and the chromosomal position of the LTR-RT. Highly methylated LTR-RTs were more likely to be removed from the host genome, but this was not due to an increase in recombination-mediated excision nor due to the selection against the LTR-RT originating methylation spreading. Despite the evidence of methylation spreading, we did not have evidence that it affected nearby host gene expression, and only LTR-RTs that transposed into the introns of host genes had significantly lower gene expression.
Results
Evolution of O. sativa LTR-RT
Past de novo annotation of LTR-RT elements in the japonica genome predicted a wide range of LTR-RT copy number in its genome, where it ranged as low as 1,000 elements to over 6,000 elements in the genome (McCarthy et al. 2002; Vitte and Panaud 2003; Gao et al. 2004; Ma et al. 2004; Chaparro et al. 2007; Vitte et al. 2007; Baucom et al. 2008; Tian et al. 2009; Xu et al. 2010; vonHoldt et al. 2012; Copetti et al. 2015). This variation could be due to analyzing different genome versions but more likely from using different LTR-RT annotation methods. We used LTRharvest to de novo annotate candidate RTs that have the canonical structures of a LTR-RT and likely to be a full length LTR-RT in structure. LTRharvest has also been shown to have the highest sensitivity for LTR-RT discovery (Lerat 2010; Hoen et al. 2015). In total, we discovered 4,240 LTR-RT elements in the japonica genome and 4,707 LTR-RT elements in the indica genome. Most LTR-RT annotation methods, however, suffer from high false positive rates (Lerat 2010); hence candidate LTR-RTs were further examined for mis-annotated RTs (supplementary fig. 1, Supplementary Material online).
Each LTR-RT was classified into its appropriate family using RepeatClassifier and results are shown in supplementary table 1, Supplementary Material online. For the japonica genome, 3,295 out of 4,240 predicted elements (77.7%) and in indica 2,560 out of 4,707 predicted elements (54.4%) matched known TE elements in the Repbase database. Even though LTRharvest is designed specifically for detecting class I LTR-RTs, there were several class II DNA TEs that were also detected (japonica genome had 59 DNA elements out of 3,295 and indica genome had 273 DNA elements out of 2,560), and these were excluded from downstream analysis.
We then annotated protein-coding domains common to LTR-RTs (Wicker et al. 2007) to the candidate LTR-RTs. More than half of the annotated LTR-RTs had at least one protein-coding domain related to transposing itself (supplementary table 2, Supplementary Material online), suggesting at least half of the LTR-RTs in both japonica and indica genomes were likely to be autonomous elements (Wicker et al. 2007). The LTR-RTs that had no matching identity to Repbase database (hereinafter designated as unknown LTR-RTs) were also searched for known protein coding domains. Results showed that almost all unknown LTR-RTs had no known protein coding sequences (supplementary table 2, Supplementary Material online). This is possible if most unknown LTR-RTs are old and degenerated TEs that are not recognizable when compared with TEs in a reference database or some may even be novel LTR-RTs not listed in Repbase. However, to be conservative, for our downstream analyses, we excluded the unknown class and based our analyses on the 3,213 RT elements in japonica and 2,165 RT elements in indica that had matching identity to known LTR-RTs in Repbase.
Using the rve gene, which was the most common protein domain found across both japonica and indica LTR-RTs, we inferred the evolutionary history of the japonica and indica LTR-RTs by reconstructing their phylogenetic history. The phylogenetic tree indicated that all LTR-RTs could be divided into two major clades, a copia- and gypsy-like clade (fig. 1). When the subpopulation of origin was annotated onto the tree, all major phylogenetic clusters had japonica and indica LTR-RTs with a paraphyletic relationship with each other.
Fig. 1.
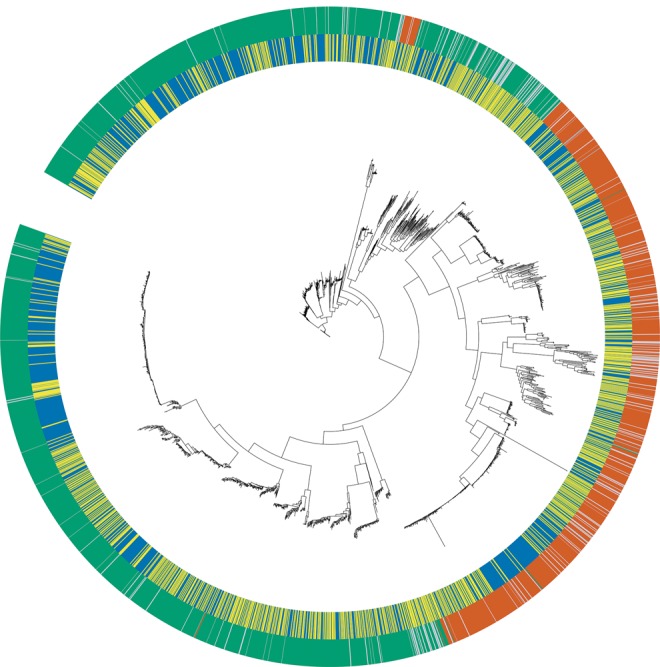
Phylogenetic tree of 3,613 rve genes from japonica and indica LTR-RTs. Outer circle represents RT family, where green represent gypsy-like, orange represents copia-like, and light blue represents LTR-like elements. Inner circle represents subpopulation of origin, where blue represents japonica and yellow represents indica.
Oryza sativa LTR-RT Methylation Analysis
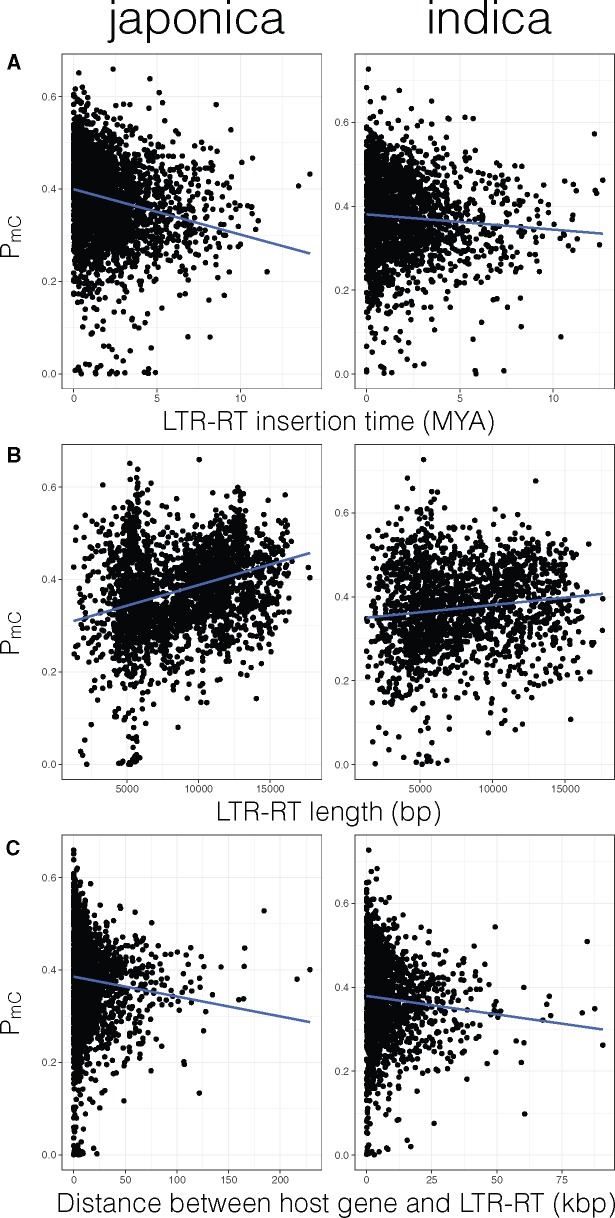
Japonica and indica methylation status of each LTR-RTs were determined using BS-seq reads from stage-matched leaf tissues. We were able to estimate the percentage of methylated cytosines (PmC) in 3,164 LTR-RTs for japonica and 1,991 LTR-RTs for indica. Japonica LTR-RTs had significantly higher PmC values than indica LTR-RTs, but the difference in PmC was marginal (supplementary fig. 2A, Supplementary Material online; median PmC for japonica LTR-RTs = 0.387 [95% BCI: 0.383–0.391] and for indica LTR-RTs = 0.374 [95% BCI: 0.368–0.380]; Mann–Whitney U [MWU] test P = 0.034). Compared with LTR-RTs from indica, LTR-RTs from japonica were significantly younger (supplementary fig. 2B, Supplementary Material online; median insertion time 1.37 Ma [95% BCI: 1.30–1.44] for japonica and 1.55 Ma [95% BCI: 1.47–1.64] for indica; MWU test P = 4.68 × 10−4) and more distantly located from host genes (supplementary fig. 2C, Supplementary Material online; median distance to host gene 5.1 kb [95% BCI: 4.8–5.4] for japonica and 3.8 kb [95% BCI: 3.5–4.1] for indica; MWU test P = 2.43 × 10−10). LTR-RT PmC also varied depending on the tissue type, where the endosperm had the lowest median PmC values and 6-week mature leaf had the highest median PmC values (supplementary fig. 3, Supplementary Material online).
We then looked at factors that determined the methylation level of each LTR-RTs (vonHoldt et al. 2012). For both japonica and indica LTR-RTs, time of LTR-RT insertion was negatively correlated with LTR-RT PmC (fig. 2A; Spearman’s rho [ρ] = −0.277, P = 1.74 × 10−54 for japonica; and ρ = −0.089, P =9.55 × 10−4 for indica). For japonica, this was consistent with previous results (Baucom et al. 2008; vonHoldt et al. 2012) and in indica, albeit weaker correlation than japonica, there was concordant results suggesting recently transposed LTR-RTs were more highly methylated than older elements.
Fig. 2.
Japonica and indica subpopulation correlation between LTR-RT methylation with (A) age of LTR-RT insertion, (B) length of LTR-RT, and (C) distance between LTR-RT and nearest host gene.
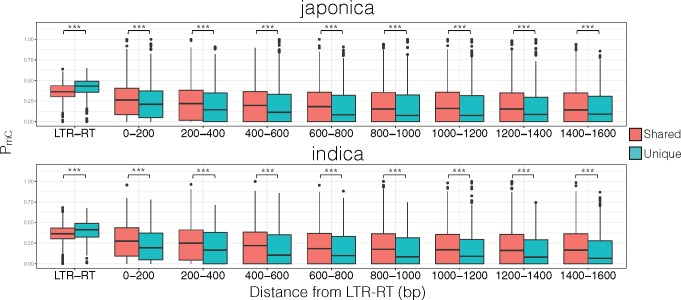
Different TE insertion timing methods can lead to widely different results (Maumus and Quesneville 2014) biasing analyses using LTR-RT sequence features to estimate the insertion time. Hence, we used a method that was unrelated to analyzing the TE structure itself to differentiate recent and ancient insertions. By using genome alignments between japonica and indica, we determined LTR-RTs that were shared or unique to each genome and estimated their methylation levels. In both japonica and indica, unique LTR-RTs had significantly younger time of insertion (supplementary fig. 4, Supplementary Material online; MWU test P value for shared vs. unique LTR-RT insertion time in japonica genome = 2.10 × 10−77; MWU test P value for shared vs. unique LTR-RT insertion time in indica genome = 8.42 × 10−22) suggesting unique LTR-RTs are likely from a recent transposition. Methylation level results showed that LTR-RTs that were unique to the reference genome had significantly higher PmC than LTR-RTs that were shared between the reference and query genome (fig. 4; MWU test P value for shared vs. unique LTR-RT in japonica genome = 5.29 × 10−23; MWU test P value for shared vs. unique LTR-RT in indica genome = 5.14 × 10−5).
Fig. 4.
Japonica and indica subpopulation levels of methylation for shared and unique LTR-RTs, and their surrounding regions. ***P < 0.001.
For japonica, consistent with vonHoldt et al. (2012), a significantly positive correlation was seen between PmC and LTR-RT length (fig. 2B; ρ = 0.335, P = 7.95 × 10−81); and significantly negative correlation was seen between PmC and distance between host gene and LTR-RT (fig. 2C; ρ = −0.139, P = 1.43 × 10−13). Concordant relationships were also seen for the indica genome (fig. 2B and C;PmC vs. LTR-RT length: ρ = 0.140, P = 7.52 × 10−9; PmC vs. distance between host gene and LTR-RT: ρ = −0.092, P = 5.44 × 10−4). These correlations can be explained if LTR-RT mediated ectopic recombination is a strong driver of selection. Ectopic recombination from LTR-RTs in different regions of a homologous chromosome can lead to loss or duplication of chromosomal regions that includes host genes, and longer LTR-RT have more sites to be involved in those deleterious ectopic recombination. Methylation of LTR-RTs silences their activity from proliferating in the genome but we wondered if methylation could also have a role in preventing deleterious ectopic recombination events. We utilized the high-density genetic map of japonica to see if there were any relationship between the host recombination rate and PmC status of the LTR-RT. Interestingly, a significantly positive correlation was observed between PmC and the rate of recombination (fig. 3; ρ = 0.186, P = 6.19 × 10−24).
Fig. 3.
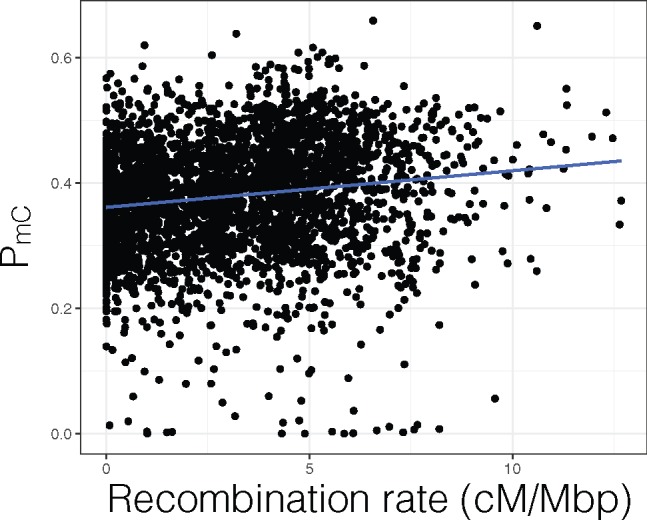
Japonica subpopulation correlation between LTR-RT methylation and recombination rate.
In plants, methylation occurs in three different cytosine contexts: in CG, CHG, and CHH sites (where H is A, T, or C nucleotide) (Law and Jacobsen 2010). Since in maize the proximity of a TE to its host gene determined the cytosine context that got methylated (Gent et al. 2013; Li et al. 2015), we examined whether specific LTR-RT cytosine contexts were enriched for methylation based on its proximity to the host gene. In japonica LTR-RTs, all cytosine contexts had a significantly negative correlation with distance but in indica LTR-RTs, only the CHH sites had a significantly negative correlation while CG and CHG sites had no significant correlations (supplementary table 3, Supplementary Material online).
Methylation Spreading around LTR-RTs
Past O. sativa studies have shown methylation spreading around regions surrounding TE sequences (Li et al. 2008, 2012; Zhang et al. 2015), but the extent of spreading differs between studies and TEs were not the main focus of the study. We examined PmC surrounding the shared or unique LTR-RTs of japonica and indica genomes and results are shown in figure 4. LTR-RTs that were unique within a subpopulation (i.e., recent insertion) had higher PmC than LTR-RTs that were shared between subpopulation (i.e., ancient insertion). Regions surrounding the LTR-RT, however, had opposite results where the spread of methylation dropped significantly more rapidly for unique LTR-RTs. This suggests that recent LTR-RT insertions are strongly silenced and methylation is more regulated to minimize the spreading around the RT sequences, or selection is strongly against highly methylated and highly spreading LTR-RTs from the host population that it is not likely to exists long term.
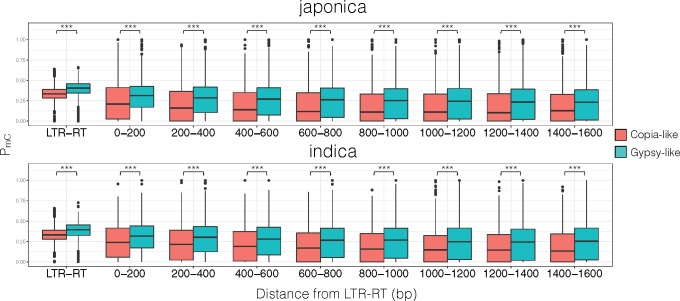
Phylogenetically related LTR-RTs share methylation status (vonHoldt et al. 2012), and we examined PmC of copia- and gypsy-like RT sequences and the PmC in their surrounding genomic regions. Results showed that gypsy-like RTs had significantly higher PmC than copia-like RTs, and the spreading of methylation was also significantly higher across regions surrounding the gypsy-like RTs (fig. 5). Since gypsy-like RTs had higher methylation spreading than copia-like RTs, we compared the distance to nearest host gene for both copia- and gypsy-like RTs. Results showed that in both japonica and indica, host genes were significantly further away for gypsy-like RTs (supplementary fig. 5, Supplementary Material online; MWU test P value for copia- vs. gypsy-like LTR-RT distance to host gene in japonica genome = 2.49 × 10−14; MWU test P value for copia- vs. gypsy-like LTR-RT distance to host gene in indica genome = 6.22 × 10−12).
Fig. 5.
Japonica and indica subpopulation levels of methylation for copia- and gypsy-like LTR-RTs, and their surrounding regions. ***P < 0.001.
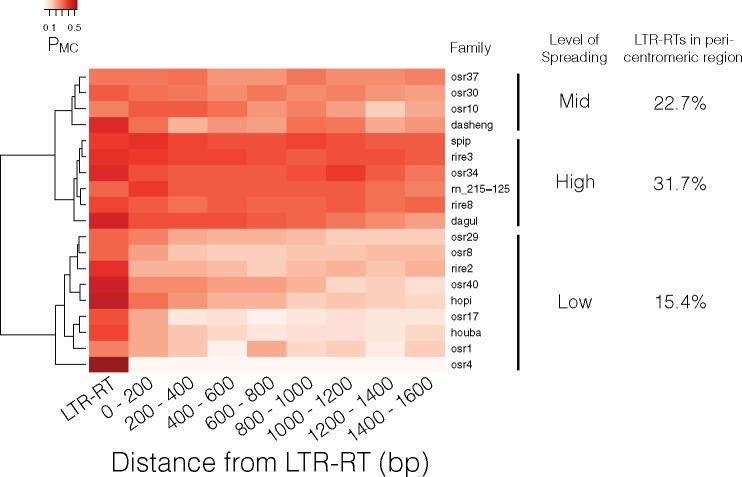
We further classified the LTR-RTs into specific families (Wicker et al. 2007) using the RetrOryza database (Chaparro et al. 2007) and were able to classify 2,292 and 1,040 elements in the japonica and indica genome, respectively (supplementary table 4, Supplementary Material online). Spreading of methylation was plotted for LTR-RT families that had at least 30 copies in the japonica genome and were then grouped using hierarchical clustering (fig. 6). Results showed that methylation spreading could be divided into three major groups of low-, mid-, and high levels of spreading. We then examined whether the activity of each LTR-RT family determined the level of methylation spreading. LTR-RT activity was indirectly measured through the age of each LTR-RT insertion, since active LTR-RTs amplify in burst leading to numerous copies with recent insertion times (Vitte and Panaud 2003; Vitte et al. 2007). However, insertion times for each family showed no clear patterns in relation to the level of methylation spreading (supplementary fig. 6, Supplementary Material online).
Fig. 6.
Heatmap of methylation level and spreading for top 19 most enriched LTR-RT families in the japonica genome. Levels of spreading were grouped into three major groups using Euclidean distance and Ward’s method of aggregation (Murtagh and Legendre 2014).
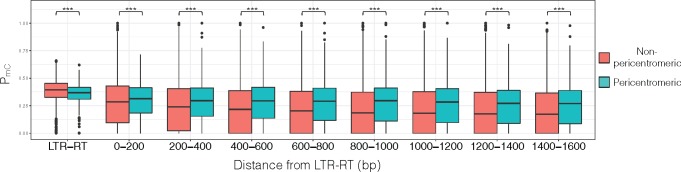
Since previous study had shown contrasting genomic features between LTR-RTs located in the pericentromeric versus nonpericentromeric regions (Tian et al. 2009), we took a closer examination of the genomic environment would have on the LTR-RTs. Consistent with previous observation (Tian et al. 2009), compared with nonpericentromeric regions our estimated pericentromeric region had significantly greater number of LTR-RTs per Mb (supplementary fig. 7, Supplementary Material online; MWU test P value = 7.95 × 10−20). Chromosomal positions of each LTR-RT family showed that families with high levels of methylation spreading had higher proportion of LTR-RTs located within the pericentromeric regions (fig. 6). Further, comparison of all LTR-RTs within the pericentromeric versus nonpericentromeric region indicated that LTR-RTs located within pericentromeric regions had significantly lower PmC, but significantly higher levels of methylation spreading (fig. 7). Because there is no genetic map generated for indica, we could not estimate the pericentromeric regions for the indica genome. However, when the same families of LTR-RT examined in the japonica genome (fig. 6) were examined in indica, the level of methylation and spreading were largely concordant between the japonica and indica LTR-RTs (supplementary fig. 8, Supplementary Material online). This suggests the pericentromeric regions may largely be conserved between japonica and indica.
Fig. 7.
Japonica subpopulation levels of methylation for LTR-RTs and their surrounding regions in pericentromeric and nonpericentromeric environment. ***P < 0.001.
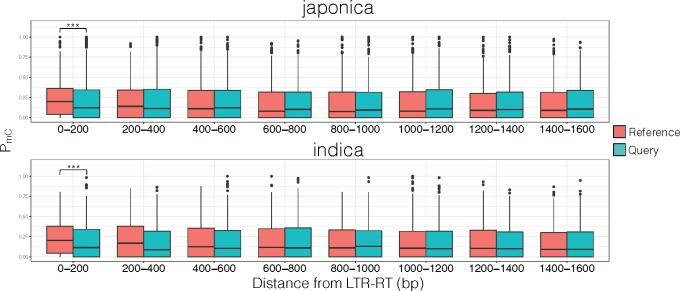
Our results indicated that methylation from LTR-RT sequences was able to spread into the flanking regions, but the extent depended on the age, type of LTR-RT, and chromosomal location. However, our results were also consistent with a different model, where LTR-RTs could have a preference for transposing into genomic regions that were already enriched for methylation. Here, the spreading of methylation would be an artifact of a bias for LTR-RT transposition into a favorable (i.e., highly methylated) epigenomic environment. To differentiate the scenarios, we focused our methylation analysis on flanking regions of unique LTR-RTs and their orthologous positions, which do not have a LTR-RT insertion. Results showed in both japonica and indica genomes, in the absence of an LTR-RT insertion there was a significantly lower PmC levels compared with orthologous regions immediately next to LTR-RT sequences (fig. 8). The spread of methylation from a recent LTR-RT insertion dropped rapidly where after 200 bp from the LTR-RT sequence, methylation levels were not significantly different from an epigenetic environment that did not have any LTR-RT insertion.
Fig. 8.
Japonica and indica subpopulation levels of methylation for regions surrounding LTR-RT (reference) and its genome aligned orthologous positions that do not have a LTR-RT (query). ***P < 0.001.
We then examined the effect of LTR-RT mis-annotation would have on our results. We compared our list of analyzed LTR-RT to that was discovered from Rice TE (RiTE) database (Copetti et al. 2015). The RiTE database had annotated 997 and 984 LTR-RTs in japonica and indica genomes, respectively, which 783 and 585 LTR-RTs were annotated in both this study and the RiTE database (supplementary fig. 9, Supplementary Material online). We compared the level of methylation for the LTR-RT and its surrounding region for those that were only annotated in this study and those that were found in both this study and the RiTE database. Compared with LTR-RTs that were found in both this study and RiTE database, LTR-RTs that were annotated in this study had significantly lower PmC, however PmC in the surrounding regions were not significantly different from each other (supplementary fig. 10, Supplementary Material online). Further, age of LTR-RT and distance to host genes were not significantly different between the LTR-RTs that were found only in this study compared with those that were found in both this study and the RiTE database (supplementary figs. 11 and 12, Supplementary Material online). This suggested possible LTR-RT mis-annotation was unlikely to affect our downstream interpretations of LTR-RT evolution.
Effect of LTR-RT on Host Gene Expression
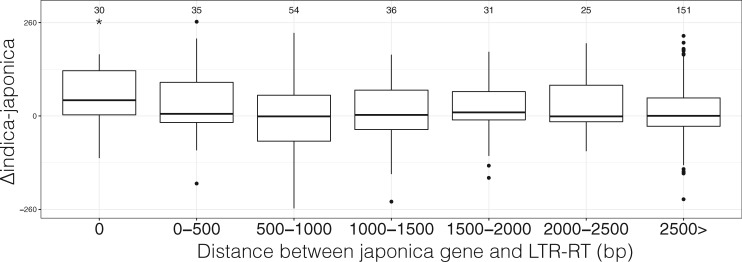
We examined if the methylation spreading from LTR-RT affects host gene expression. We focused on unique LTR-RTs since for these RTs the nearby host genes’ ortholog does not have any LTR-RT insertions nearby; hence it is possible to measure the effect of LTR-RT presence or absence on host gene expression. For japonica, we conducted Wilcoxon Signed-Rank (WSR) test on pairs of orthologous genes where japonica has a LTR-RT insertion nearby whereas indica did not. Results showed that across five different distance bins between the LTR-RT and host gene, only LTR-RTs that were inserted within the japonica gene had significant WSR test result (P = 0.047) and the difference in gene expression indicated that indica had the higher gene expression (fig. 9). For indica, no bin showed any significant WSR test result, however the sample sizes were smaller than japonica and the difference could be due to lower statistical power (supplementary fig. 13, Supplementary Material online). We then examined gene expression differences of host genes near LTR-RTs that were shared between japonica and indica. Unlike host genes near unique LTR-RTs, no bin had significant gene expression differences (supplementary fig. 14, Supplementary Material online). Further, there was no correlation between the host gene expression and distance to nearest LTR-RT across 12 japonica tissue and 1 indica tissue data set (supplementary table 5, Supplementary Material online).
Fig. 9.
Boxplot of inverse hyperbolic sine transformed gene expression differences between indica and japonica genes, where the japonica gene has a LTR-RT nearby and indica gene does not. Gene expression differences were binned by distance to nearest LTR-RT in the japonica genome. Numbers above boxplot represent the sample size. *P < 0.05.
Selection against Highly Methylated LTR-RT
The number of shared and unique LTR-RTs was used to examine the host selective pressures that are limiting the activity of the LTR-RTs. The removal of LTR-RTs may involve various host factors such as recombination that physically removes the LTR-RT from the host genome (Devos et al. 2002), or through epigenetic repressive marks silencing LTR-RTs which then is removed from the host population through selection or drift. We divided the LTR-RTs into equally sized recombination rate or LTR-RT sequence PmC bins and calculated the proportion of shared LTR-RTs per bin. Assuming unique LTR-RTs are results of recent randomly transposing RTs, then a decrease in the proportion of shared LTR-RT across different bins can be used to infer the extent of host selection removing LTR-RTs insertions from the population.
Focusing on the japonica results, we divided LTR-RTs into three equally sized recombination rate bins and counted the number of shared and unique LTR-RTs per bin. With higher recombination rate, the proportion of shared LTR-RTs decreased (supplementary table 6, Supplementary Material online; Fisher’s exact test [FET] P = 8.85 × 10−6), suggesting recombination-mediated excisions are likely to remove LTR-RTs from the host genome. This result was consistent with Tian et al. (2009) which suggested host recombination is an important factor removing LTR-RTs from the O. sativa genome. The japonica LTR-RTs were then divided into three equally sized bins according to their PmC levels. Results showed that with increased methylation there was a decrease in the proportion of shared LTR-RTs (supplementary table 7, Supplementary Material online; FET P = 1.32 × 10−20), suggesting LTR-RTs that are highly methylated were likely to be removed from the host genome.
Since there was a positive correlation between host recombination rate and LTR-RT methylation (fig. 3), we examined if the host recombination-mediated excision was driving the removal of highly methylated LTR-RTs, by examining the proportion of shared LTR-RT across increasing recombination rates for each PmC bin. Results showed a trend of decreasing proportion of shared LTR-RTs with increased recombination rate per PmC bin, however none of it were significant (table 1; Low PmC bin FET P = 0.21; Mid PmC bin FET P = 0.67; High PmC bin FET P = 1.0). On the other hand, for each recombination rate bin, increased LTR-RT PmC led to a decreased proportion of shared LTR-RT and was significant (table 1; Low recombination rate bin FET P = 2.57 × 10−5; Mid recombination rate bin FET P = 2.90 × 10−5; High recombination rate bin FET P = 4.70 × 10−6), suggesting there were mechanism(s) independent of the host recombination that was removing highly methylated LTR-RTs from the host genome.
Table 1.
Counts of Shared and Unique LTR-RT Per Methylation and Recombination Rate Bins.
| Low PmC | ||||||||
|---|---|---|---|---|---|---|---|---|
|
Low Recombination Rate |
Mid Recombination Rate |
High Recombination Rate |
||||||
| Shared LTR-RT | Unique LTR-RT | %Shared LTR-RT | Shared LTR-RT | Unique LTR-RT | %Shared LTR-RT | Shared LTR-RT | Unique LTR-RT | %Share LTR-RT |
| 154 | 29 | 0.84 | 113 | 36 | 0.76 | 123 | 47 | 0.72 |
| Mid PmC | ||||||||
|
Low Recombination Rate |
Mid Recombination Rate |
High Recombination Rate |
||||||
| Shared LTR-RT | Unique LTR-RT | %Shared LTR-RT | Shared LTR-RT | Unique LTR-RT | %Shared LTR-RT | Shared LTR-RT | Unique LTR-RT | %Shared LTR-RT |
| 79 | 32 | 0.71 | 86 | 56 | 0.61 | 75 | 55 | 0.58 |
| High PmC | ||||||||
|
Low Recombination Rate |
Mid Recombination Rate |
High Recombination Rate |
||||||
| Shared LTR-RT | Unique LTR-RT | %Shared LTR-RT | Shared LTR-RT | Unique LTR-RT | %Shared LTR-RT | Shared LTR-RT | Unique LTR-RT | %Shared LTR-RT |
| 57 | 45 | 0.56 | 75 | 83 | 0.47 | 117 | 140 | 0.46 |
Note.—Methylation levels were divided into three bins: Low PmC = 0–0.344; Mid PmC = 0.344–0.425; High PmC = 0.425–0.659. Recombination rates were divided into three bins: Low recombination rate = 0–1.72 cM/Mb; Mid recombination rate = 1.72–4.28 cM/Mb; High recombination rate = 4.28–12.68 cM/Mb.
Using logistic regression, we specifically modeled the association between LTR-RT PmC and their recombination rate to the absence or presence of a LTR-RT in a genome. Unique LTR-RTs that are only observed in one genome and not the other were encoded as zero, and shared LTR-RTs that are present in both genomes were encoded as one. Chi-square goodness-of-fit test indicated the logistic regression including an interaction term between LTR-RT PmC and recombination rate was not significantly better fit than a model that did not include the interaction term (χ2 test P = 1.0). With logistic regression, LTR-RT PmC was a significant negative predictor of shared LTR-RT status (β = −5.38 and P = 3.17 × 10−16), whereas recombination rate was not significant (β = −0.063 and P = 0.097), consistent with the binning results.
Since LTR-RTs can amplify in recent bursts, the logistic regression result may have been influenced by increased number of highly methylated unique LTR-RTs, and not due to a decrease in methylated shared LTR-RTs. Further, the chromosomal environment may bias the preference for shared or unique LTR-RTs. Pericentromeric regions had significantly lower proportion of unique LTR-RTs than nonpericentromeric regions (supplementary table 8, Supplementary Material online; FET P = 3.3 × 10−5), however this can either be due to increased number of unique LTR-RTs in the nonpericentromeric regions from recent LTR-RT burst or due to increased shared LTR-RTs in pericentromeric regions from the inefficient selection removing the LTR-RT elements. Thus, we conducted a logistic regression removing LTR-RTs that had a divergence time of <100,000 years to remove recently amplified LTR-RTs, and LTR-RTs that were located in the pericentromeric regions. A chi-square goodness-of-fit test showed no significant improvement in a model that included an interaction term between LTR-RT PmC and recombination rate (chi-square test P = 1.0). The logistic regression still indicated LTR-RT PmC was a significant negative predictor of shared LTR-RT status (β = −4.87 and P = 3.30 × 10−5) while recombination rate was not (β = 0.001 and P = 1.0).
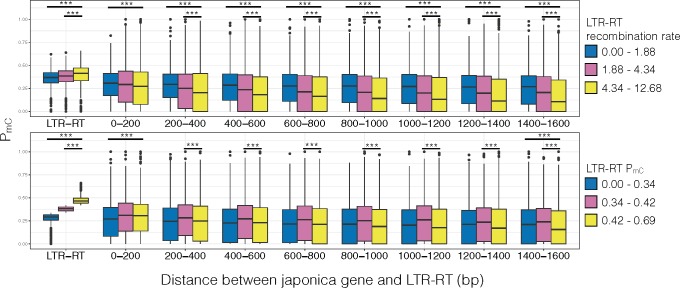
We then examined if there were any differences in methylation spreading for the LTR-RTs classified into three different recombination rate and PmC level bins, and whether the differences in spreading were contributing to the selection and removal of LTR-RTs. Results showed that LTR-RTs in the highest recombination rate or highest PmC bins had the lowest PmC levels across its surrounding region (fig. 10).
Fig. 10.
Japonica subpopulation levels of methylation for LTR-RTs and their surrounding region for bins divided by the LTR-RT recombination rate and methylation level. Comparisons were only made between the highest bin to the mid or lowest bin. ***P < 0.001.
Discussion
The evolutionary epigenomic consequences of TEs on surrounding host genes have been investigated in three model organisms, Arabidopsis thaliana (Hollister and Gaut 2009), Drosophila melanogaster, and D. simulans (Lee 2015; Lee and Karpen 2017). In both studies, the spread of repressive epigenetic marks (methylation in A. thaliana and histone di- and tri-methylation of H3 lysine 9 [H3K9me2/3] in Drosophila) from TEs into surrounding regions were associated with reduced host gene expression (Quadrana et al. 2016; Stuart et al. 2016). Importantly, TEs that had higher epigenetic repression and were closer to host genes had lower population frequencies, suggesting host selection against the spreading of repressive epigenetic marks can be another factor determining the TE frequencies. Although the model had strong evidence of support from both A. thaliana and Drosophila, its generality to other organisms is questionable. Compared with the majority of plant species, A. thaliana is atypical in terms of its small genome size and low TE content (The Arabidopsis Genome Initiative 2000; Nystedt et al. 2013; Wendel et al. 2016) suggesting the epigenetic response and TE population dynamic could be a unique evolutionary trait of A. thaliana. Drosophila, on the other hand, has lost the ability of DNA methylation (Feng et al. 2010; Zemach, McDaniel, et al. 2010), calling into question whether differences in TE epigenetic modification can also lead to differences in epigenetic repressive mark spreading. To test the deleterious epigenetic spreading model, in this study, we examined the host-TE epigenetic regulation in a different model organism, the Asian rice O. sativa.
The transposition–selection balance model has been the main hypothesis for modeling TE population dynamics, and its main prediction is that negative selection maintains a low frequency of TEs in the host population (Charlesworth and Langley 1989; Petrov et al. 2003; González et al. 2008; Lockton et al. 2008; Lee and Langley 2010). The transposition–selection balance model assumes a steady state of transposition rate for TEs; however evidence of bursts of TE transpositions (Kidwell 1983; SanMiguel et al. 1998; Bowen and McDonald 2001; Kofler et al. 2012; Lu et al. 2012; Cridland et al. 2013; Belyayev 2014) have also suggested alternative models such as the transposition-burst model (Bergman and Bensasson 2007; Blumenstiel et al. 2014). Our phylogenetic analysis of O. sativa LTR-RTs had indicated instances of transposition bursts in both copia- and gypsy-like elements, where there were groups separated by long internal branches but members within the groups had very short terminal branch lengths. Further, there was pervasive evidence of horizontal transfer of LTR-RTs between the japonica and indica subpopulation. Given the extensive evidence of admixture between the two subpopulation (Caicedo et al. 2007; Gao and Innan 2008; He et al. 2011; Huang et al. 2012; Choi et al. 2017), domestication-related hybridization between japonica and indica would have introduced new LTR-RTs to the recipient genome, which could have led to transposition bursts for certain LTR-RTs in the novel genomic environment (Engels 1992; Kofler et al. 2015). This suggests TE transposition bursts could be used to infer the timing of past domestication-related gene flow between japonica and indica.
Past studies have focused on the TE mediated physical disruptions on the host genome (Langley et al. 1988; Montgomery et al. 1991; Biémont and Cizeron 1999; Petrov et al. 2003; Wright et al. 2003; Boissinot et al. 2006; Mieczkowski et al. 2006; Hedges and Deininger 2007), whereas the deleterious effects arising from the epigenetic regulation of TEs (Lisch 2009; Diez et al. 2014) has been a relatively understudied area of host-TE evolution. Plants are ideal organisms to study TE mediated epigenetic influences due to their exceptional genomic architecture, where the functional coding sequences are scattered across a sea of repetitive DNA sequences (Kelly and Leitch 2011; Wendel et al. 2016). RTs have no ability to excise itself and mobilize to new locations (Feschotte et al. 2002), hence there would be strong selective pressures on the host to epigenetically silence the activity of those elements. Since repressive epigenetic marks are able to spread beyond TE sequences (Cokus et al. 2008; Ahmed et al. 2011; Eichten et al. 2012; Quadrana et al. 2016; Stuart et al. 2016), spreading of epigenetic modifications from TEs are expected to have a strong influence on the evolution of plant genomes.
Spreading of methylation is variable (Rebollo et al. 2011) and in plants it was first observed in maize where the degree of repressive epigenetic mark spreading was dependent on the RT family (Eichten et al. 2012). Here, we discovered in O. sativa, spreading is not only dependent on the RT family but also on the age of the inserted element, recombination rate, level of LTR-RT methylation, and the chromosomal location. Interestingly, rice and maize despite having a divergence time of 70 Ma (Wang et al. 2015) and substantially different repetitive genomic content (rice 35% vs. maize 85%) (Wendel et al. 2016), both had similar methylation spreading profiles for LTR-RTs. Specifically among the superfamilies, Gypsy elements had the highest level of spreading while LTR-RT elements near the centromere were likely to have higher levels of methylation spreading. This suggested there may be a common evolutionary mechanism between rice and maize (and potentially other plants) that maintains the methylation spreading. Here, highly spreading LTR-RT elements and families are limited to the gene-poor pericentromeric regions where they are likely to have less deleterious effects on surrounding genomic regions. Whether there is an active mechanism that facilitates this process or is an indirect product of natural selection may require further studies.
The spread of methylation was specifically initiated by the presence of a LTR-RT sequence and was not an insertion preference of LTR-RTs toward highly methylated epigenomic environments. Interestingly, the spreading of methylation marks was relatively short for newly transposed LTR-RTs, limited to ∼200 bp from the RT sequence. In O. sativa, younger LTR-RTs are closer to host genes (vonHoldt et al. 2012; this study), and uncontrolled epigenetic regulation of newly transposed LTR-RTs are likely to affect host gene expression if their methylation spreads to surrounding regions (Hollister and Gaut 2009). This may explain the short distance of spreading for the unique LTR-RTs. In addition, LTR-RTs that are likely to be under strong selection for its deleterious effects (i.e., elements in high recombining regions and consequently having higher chance of deleterious ectopic recombination between nonhomologous chromosomal positions or those that are strongly methylated) had significantly reduced levels of methylation spreading, suggesting there may be additional host factors or self-regulating factors from the RT sequence itself, limiting the methylation spreading in LTR-RTs that are already under strong selection. Possible genetic mechanisms from the host and TE that limits the spreading can be seen from the involvement of genes containing the Jumonji C protein domain in regulating the spread of heterochromatic marks in euchromatin boundaries (Tamaru 2010), and TE sequences containing insulator domain and function (Bell et al. 2001; Kuhn and Geyer 2003; Gaszner and Felsenfeld 2006; Bushey et al. 2008).
LTR-RTs are removed from the host genome by homologous and nonhomologous recombination-mediated mechanisms (Roeder et al. 1980; Devos et al. 2002), and is a universal mechanism across various angiosperms for removing RTs (Vitte and Bennetzen 2006). In O. sativa, there is an inverse relationship between the host recombination rate and both LTR-RT density and LTR-RT fragment sizes (Tian et al. 2009), indicating the important role of recombination for physically removing LTR-RTs. Paradoxically, however, regions of high recombination rate are expected to also have an increased probability of deleterious ectopic recombination between highly identical LTR-RTs in nonhomologous genomic locations (Langley et al. 1988). Here, a potential trade-off could occur between the host recombination rate and its role in removing LTR-RTs. We discovered a positive correlation between recombination rate and LTR-RT methylation, indicating host epigenetic factors may play an additional role in regulating RTs in high recombination rate regions. For example, in A. thaliana, centromeric and pericentromeric regions are hypermethylated and suppressed for recombination (Gaut et al. 2007; Cokus et al. 2008; Lisch 2009), whereas the repression occurs directly in cis on the methylated sequences (Mirouze et al. 2012). Thus, increased methylation across LTR-RT sequences may have a role in suppressing recombination. Consistent with these previous observations, we found no significant effect of increased recombination rate on removing LTR-RTs across the three methylation bins and based on logistic regression results. This suggested the suppression of recombination from increased methylation, also suppresses recombination-mediated excision mechanisms that physically removes LTR-RTs from the genome. This was surprising, especially for the highly methylated LTR-RTs, given that these were likely to be deleterious based on our results showing these elements as likely to be recent transpositions and closer to host genes. Hence, the increased LTR-RT methylation in high recombination regions may relate to suppressing LTR-RT mediated deleterious ectopic recombination from happening.
It was unclear then, what mechanism(s) drive the preferential removal of highly methylated LTR-RT. Since methylation spreading was reduced for highly methylated LTR-RTs, variation in methylation spreading could not explain the reduced proportion of highly methylated shared LTR-RTs. We note our annotation of LTR-RTs was aimed at identifying somewhat intact elements, which would bias ourselves to more recent LTR-RTs. Many LTR-RTs that have been partially deleted or degraded will be missing from our analysis. Thus, there may be crosstalk between host recombination and LTR-RT epigenetic marks, which then effectively removes highly methylated LTR-RTs via recombination-mediated excision mechanisms. On the other hand, it is also possible with higher recombination rate there is less of a Hill–Robertson effect (Hill and Robertson 1966), and the higher methylation reflects the efficient selection from host epigenetic factors silencing LTR-RT activities (Dolgin and Charlesworth 2008). Here, the host recombination has a role in decoupling the linkage between the deleterious LTR-RT with the surrounding genes, and selection would be more efficient in removing the deleterious recombinants.
Consistent with observations in maize (Gent et al. 2013; Li et al. 2015), both japonica and indica LTR-RTs closer to host genes had higher CHH site methylation. This suggested the stricter heterochromatin–euchromatin boundaries enforced by the CHH methylation might restrict the silencing more strongly on LTR-RTs that are nearer host genes. This may be why only host genes that had LTR-RTs inserted within its intron had significantly reduced levels of gene expression. Evidence from the highly active DNA transposon mPing family found that the majority of TEs inserted near host genes had no significant effect on host gene expression under controlled conditions (Naito et al. 2009), further suggesting TEs may have minimal influence on nearby host gene expression.
This, however, contrasted previous studies in plants, where TE methylation and nearby host gene expression were negatively correlated (Hollister and Gaut 2009; Eichten et al. 2012; Wang et al. 2013; Diez et al. 2014). In O. sativa, Zhang et al. (2015) found weak but significantly positive correlation between host gene expression and distance to nearest TE, suggesting TEs have negative effect on nearby host gene expression. However, our study was mainly focused on class I LTR-RTs while Zhang et al. (2015) reported results from both class I RTs and class II DNA transposons. Given that higher proportion of class II, DNA transposons are found nearer host genes (Bureau and Wessler 1994; Zhang et al. 2015; Wicker et al. 2016), DNA transposons may have methylation spreading with stronger effects on nearby host gene expression. In addition, host genes with TEs inserted within the gene had the largest reduction in gene expression (Zhang et al. 2015), suggesting TEs inserted within the host gene has the strongest effect on its expression.
On the other hand, Zhang et al. (2015) had measured gene expression from a single genome, comparing expression from genes with and without a TE nearby. Hence, it is equally possible that the positive correlation between host gene expression and distance to nearby TE can be interpreted as an insertion bias of TEs to transpose near host genes with low gene expression. We note, however, our study was mainly based on a single tissue sample (6-week-old mature leaf) and comparing gene expression between two individuals that have or do not have an LTR-RT nearby. Thus, there is an issue of power that has affected our gene expression analysis, and the lack of methylation spreading caused gene expression effect may be due to a sampling bias. Here, a necessary future work would be to examine the population frequency of each TE and infer the strength of selection against each element. This would then be compared with the epigenome of multiple individuals to differentiate whether host selection on the levels of TE mediated epigenetic marks shape the TE frequency, or if there is a transposition bias of TEs favoring specific host epigenetic environment (Lee and Karpen 2017).
In this study, we have shown significant evidence of LTR-RT originating methylation spreading and various factors that determine the amount of spreading. These spreading can be deleterious if it affects the host gene expression and will be selected against (Hollister and Gaut 2009; Lee 2015). In O. sativa, however, there was no significant effect of LTR-RT originating methylation spreading on host gene expression, except for those that are inserted within the intron. This suggested in O. sativa, LTR-RT originating methylation spreading might not have strong deleterious consequences on host gene expression. In fact, a recent study by Lee and Karpen (2017) have found that whether the TE originating spread of epigenetic repressive marks causes an excess of host genes with reduced gene expression, was dependent on the host genetic background. Further, proximity of a TE insertion does not always lead to a reduction in host gene expression (Lisch 2013) while methylation is known to be influenced by environmental conditions (Dowen et al. 2012; Dubin et al. 2015; Secco et al. 2015; Kawakatsu et al. 2016). Thus, if there were any deleterious effect of TE originating methylation spreading in O. sativa, it would be complex depending on both genetic and environmental conditions. As plants are immovable from their surrounding environment, environmental factors may play an important role in the TE-host dynamic of a plant (Naito et al. 2009; Galindo-González et al. 2017). Thus, examining the environmental effects would be crucial for future plant studies of host epigenetic impact on TE evolution.
Materials and Methods
Analyzed Data Set
Reference genomes for O. sativa ssp. japonica and O. sativa ssp. indica were downloaded from Ensembl Plants release 30 (http://plants.ensembl.org/; last accessed November 4, 2017). The japonica genome was sequenced from the nipponbare cultivar (International Rice Genome Sequencing Project 2005) and the indica genome was sequenced from the 93-11 cultivar (Yu et al. 2002).
Methylomic and transcriptomic data for the two O. sativa subpopulation were obtained from Chodavarapu et al. (2012). The data were generated from stage matched 6-week-old leaf tissues originating from the same cultivars used as the reference genome in this study. Additional functional genomic data from various tissue samples were analyzed only for japonica, as it was the only subpopulation that had matching transcriptomic and methylomic data from the nipponbare genome. Methylation and transcriptome data from five additional tissue and developmental time point (embryo, endosperm, seedling root, seedling shoot, and 2- to 3-month-old leaf) were obtained from Zemach, Kim, et al. (2010) and Zemach, McDaniel, et al. (2010), and additional transcriptome data from 11 additional tissue and developmental time point (anther, calli, early inflorescence, emerging inflorescence, embryo 25 days after pollination, endosperm 25 days after pollination, 20-day leaf, pistil, seed 5 day after pollination, seed 10 day after pollination, and seedling 2 week) were obtained from Davidson et al. (2012).
LTR-RT De Novo Annotation and Analysis
For the japonica and indica genomes LTR-RTs were annotated using the program LTRharvest (Ellinghaus et al. 2008). Parameters for LTRharvest were adapted from Copetti et al. (2015) which annotated repetitive DNA content across 11 unpublished high-quality Oryza genomes (Jacquemin et al. 2013). LTR-RT sequence features and protein domains were annotated with LTRdigest (Steinbiss et al. 2009). Using the protein models from PFAM (Finn et al. 2014), we annotated the integrase domain (rve, PF00665.24), RNase H domain (PF00075.22), reverse transcriptase domain (RVT1, PF00078.25), and the retrotransposon gag protein domain (gag, PF03732.15) for each candidate LTR-RT sequences.
We then searched for LTR-RTs that overlapped with O. sativa gene coordinates. These were likely to be false positive LTR-RT annotations that were called from pairs of solo LTRs left over from recombination-mediated excision of LTR-RTs (Bennetzen et al. 2005). LTR-RTs that overlapped exons of an O. sativa gene were excluded but any LTR-RTs that were completely within the intron sequence of an O. sativa gene was included for downstream analysis (see supplementary fig. 1, Supplementary Material online, for examples). In order to classify each LTR-RT into superfamily (Wicker et al. 2007), we used the RepeatClassifier program from the RepeatModeler ver. 1.0.8 suite (http://www.repeatmasker.org; last accessed November 4, 2017), and searched against Repbase release 20170127 library (Bao et al. 2015).
In O. sativa, genes are positioned along a recombination gradient where heterochromatic or low recombination regions are expected to have less genic sequences (Tian et al. 2009; Flowers et al. 2012). We focused on LTR-RTs that were located on the assembled 12 pseudomolecules of each reference genome, since unassembled scaffolds are likely to be from unmappable highly repetitive heterochromatic regions that are devoid of O. sativa genes. Gene annotations for each subpopulation genomes were obtained from Ensembl Plants release 30. Distance between LTR-RT and O. sativa gene was measured as the number of basepairs that separated between the LTR and the nearest exon of an O. sativa gene. LTR-RTs that were inserted within an intron of an O. sativa gene were assigned a distance of zero.
Phylogenetic relationship of the japonica and indica LTR-RTs was inferred using the rve gene. Multi-sequence alignment was conducted using MAFFT on all 3,613 rve genes. The alignment was then used by RAxML ver. 8.2.10 to conduct 100 bootstrap analyses to search for the best fitting maximum-likelihood tree. Phylogenetic tree was illustrated with iTOL ver. 3 (Letunic and Bork 2016).
Classification of LTR-RT into Specific Families
Annotated LTR-RTs were further classified into specific families using the 242 consensus sequences of LTR-RTs from the RetrOryza database (Chaparro et al. 2007). We used nucleotide blast (blastn) from the blast ver. 2.2.31 suite (Camacho et al. 2009) to search the RetrOryza LTR-RT sequences to each other to remove redundant consensus sequences. We followed the “95-80-98” rule (Flutre et al. 2011): two consensus sequences are considered identical if it covers 98% of its length with at least 80 bp and over 95% identity. Since blast can identify multiple overlapping high scoring pairs (HSPs) between a query and target sequence, identity between the query and target sequence was calculated by averaging the percent identity of all identified HSPs. By this rule, we found dendrobat_osj was rn_561-394, osr19 was rn_219-129, osr20 was rn_89-81, osr22 was rn_453-234, and rn_44-26 was rn_44-393. We then searched our annotated LTR-RTs to the redundancy removed RetrOryza database, and a LTR-RT was identified using the “80-80-80” rule (Wicker et al. 2007): two TEs belong to the same family if they were 80% identical over at least 80 bp and 80% of their length.
LTR-RT Insertion Time Estimation
Insertion time for each LTR-RT was estimated using the approach of SanMiguel et al. (1998). The pair of LTR sequences for each LTR-RT was aligned to each other using the program MAFFT ver. 7.154 b (Katoh and Standley 2013) with the L-INS-i algorithm. DNA divergence between the sequence was estimated with the baseml program from PAML ver. 4.8 (Yang 2007) using the Kimura-2-parameter base substitution model (Kimura 1980). Divergence time (i.e., insertion time) between the pair of LTR sequence was then calculated by dividing the DNA divergence to twice the O. sativa substitution rate (Ma and Bennetzen 2004).
Oryza sativa Recombination Rate Estimation
Recombination rate was estimated using 3267 genetic markers generated from the genetic map study of Rice Genome Project (Harushima et al. 1998; http://rgp.dna.affrc.go.jp/E/publicdata/geneticmap2000/index.html; last accessed November 4, 2017). We mapped the physical location of those genetic markers against the japonica genome using the megablast algorithm from the blast ver. 2.2.31 suite (Camacho et al. 2009). We selected genetic markers with blast results where coverage were >90% (qcovhsp > 90), percent of identical matches that were >90% (pident > 90), and markers that were located in the correct chromosome. Several markers had sequences from both 5′ and 3′ region. Both of them were considered redundant and only one end was used to map its physical position. The middle physical position of each genetic marker was used to represent its physical map position. The genetic (cM) and physical map (bp) of each marker were then loaded onto the MareyMap package (Rezvoy et al. 2007) to estimate the recombination rate (cM/Mb). The genetic versus physical map were plotted for each chromosome and visually inspected to remove anomalous genetic markers, leading to a total of 1381 markers being used (supplementary table 9, Supplementary Material online). For a given physical position of the chromosome, we used the approach of Muyle et al. (2011) to fit a loess function curve to estimate the recombination rate. Any physical position that had a negative recombination rate estimation was assumed to have a recombination rate of zero.
Estimating Pericentromeric Regions
Using the recombination rate estimates from above, we estimated the pericentromeric region for the japonica genome. For each of the 12 pseudomolecules starting from genomic position 500,000 bp, we estimated the recombination rate of the midpoint position in 1-Mb nonoverlapping sliding windows. Window with the lowest recombination rate was assumed to be where the centromere was located and the surrounding 2-Mb up- and downstream was assumed to be the pericentromeric region. Estimated pericentromeric region for the japonica genome can be found in supplementary table 10, Supplementary Material online. We note our estimated pericentromeric region largely overlaps those that were found independently by Tian et al. (2009).
Methylomic Data Analysis
Initially, raw bisulfite sequencing (BS-seq) FASTQ reads were under quality control using the program trim galore! Ver. 0.4.3 (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/; last accessed November 4, 20) with default parameters. BS-seq reads were then mapped to the reference genomes using the program bismark ver. 0.16.3 (Krueger and Andrews 2011) designed specifically for BS-seq read mapping and methylation calling. Each BS-seq library was mapped to the corresponding O. sativa subpopulation genome the BS-seq data was generated from.
Each cytosine site was determined whether if it was a methylated cytosine (mC) using a binomial test (Lister et al. 2008; Greaves et al. 2012). P value for each cytosine site was determined using the binomial distribution where m is the number of mC reads, x is the total number of mC and unmethylated cytosine (uC) reads, and ɛ is the error rate. Error rate was determined by aligning the BS-seq read to the chloroplast genome of each subpopulation. Since the chloroplast does not have evidence of methylation (Cokus et al. 2008), any reads with evidence of methylation would be an error (Lister et al. 2008). We used bismark to align the BS-seq reads to the respective chloroplast genomes (genbank accession number AY522330 for nipponbare and AY522329 for 93-11) and the error rates were determined by counting the total number mC reads divided by the total number of mC and uC reads. Error rates for each sample can be found in supplementary table 11, Supplementary Material online. Cytosine sites with P value <0.001 were then considered as sites with significant evidence of methylation.
For a given genomic region, its level of DNA methylation was determined by calculating the proportion of cytosine sites with significant evidence of methylation. This was calculated by dividing the total number of mC sites to the total number of mC and uC sites. Consistent with vonHoldt et al. (2012), only LTR-RTs that had at least 100 cytosines sites with at least 2× coverage were analyzed. We also analyzed the spreading of methylation around LTR-RT sequences. Upstream and downstream regions of LTR-RTs were divided into 200-bp bins, and each bin was analyzed if >50% of its cytosine had mC or uC calls.
Transcriptomic Data Analysis
The raw RNA sequencing (RNA-seq) FASTQ reads were under quality control using trim galore! with default parameters. RNA-seq reads were then aligned to the subpopulation genomes using the program HISAT2 ver. 2.0.4 (Kim et al. 2015). To estimate gene expression values, the alignment files were then analyzed with HTSeq ver. 0.6.1 (Anders et al. 2015) to calculate RPKM values for each gene. To normalize the variation existing between different samples, we applied the trimmed mean of M value (TMM) method (Robinson and Oshlack 2010) from the edgeR ver. 3.18.0 package (Robinson et al. 2010) on each samples’ gene expression values. We aligned each RNA-seq data from japonica and indica to a single subpopulation genome, resulting in two alignment files for each subpopulation genome. This was necessary for downstream gene expression comparison analysis (see Results for detail), and was done so that any gene of a given subpopulation genome would have gene expression values from both japonica and indica samples. Genes with zero RPKM were not analyzed, as we could not differentiate whether it was due to no expression or low undetectable gene expression.
Comparative Genomic Analysis
To analyze orthologous regions between the subpopulation genomes, we used the approach of Choi et al. (2017) to align the genomes to each other. Briefly, this was done by using either the nipponbare genome or the 93-11 genome as the reference and aligned a query genome using LASTZ ver. 1.03.73 (Harris 2007). Alignment blocks were then chained together using the UCSC Kent utilities (Kent et al. 2003; https://github.com/ENCODE-DCC/kentUtils; last accessed November 4, 2017) and alignment chain with the highest alignment score was selected to represent the orthologous region.
We used the genome alignments to determine whether a LTR-RT insertion in a given genome (hereon termed as reference genome) would also be inserted in the orthologous position of the aligned genome (hereon termed as the query genome). For each LTR-RT discovered in the reference genome, we examined its orthologous 1-kb up- and downstream flanking regions in the query genome. We made sure both flanking orthologous regions were on the same chromosome and the query genome alignment covered at least 50% of the 1-kb region of the reference genome.
Distance between the flanking region of the LTR-RT in the reference genome and its orthologous position in the query genome were then calculated. Since this distance represents the LTR-RT size in the reference genome, we then examined its orthologous position in the query genome to see if the same LTR-RT existed in the query genome. If this distance in the query genome was <10% of the reference genome, we assumed the LTR-RT of the reference genome was missing in the orthologous position in the query genome. If the distance in the query genome was >10% but less than twice the size of the LTR-RT in the reference genome, we assumed that LTR-RT of the reference genome existed in the orthologous position of the query genome.
Statistical Analysis and Multiple Testing Corrections
For each reported median, we estimated the bootstrap confidence interval (BCI) of the median, by resampling the data with replacement and calculated the median of that bootstrapped sample. This was done 10,000 times to estimate the 95% BCI of the median.
A logistic regression analysis was conducted using the shared and unique LTR-RT as a binary dependent variable, whereas the LTR-RT methylation level (ML) and LTR-RT recombination rate (RR) as the independent variable:
Chi-square goodness of fit test was conducted to compare the fit between the two models.
All statistical tests with P values in this study (chi-square goodness-of-fit test, Fisher’s exact test, logistic regression, Mann–Whitney U test, Spearman’s rho correlation, and Wilcoxon signed-rank test) were pooled together to correct for multiple hypothesis testing. We used the p.adjust function from the program R (R Core Team 2016) to implement the Benjamini and Yekutieli (2001) method. We note all reported P values in this study are multiple hypothesis corrected P values.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Science Foundation Plant Genome Research Program (IOS-1546218), the Zegar Family Foundation (A16-0051), and the NYU Abu Dhabi Research Institute (G1205) to M.D.P. We appreciate the New York University—High Performance Computing for providing computational resources and support. We thank Grace Yuh Chwen Lee and Zoe Joly Lopez for providing critical comments and improving the manuscript.
References
- Ahmed I, Sarazin A, Bowler C, Colot V, Quesneville H.. 2011. Genome-wide evidence for local DNA methylation spreading from small RNA-targeted sequences in Arabidopsis. Nucleic Acids Res 3916:6919–6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W.. 2015. HTSeq – a Python framework to work with high-throughput sequencing data. Bioinformatics 312:166–169.http://dx.doi.org/10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J.. 2007. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 3185851:761–764.http://dx.doi.org/10.1126/science.1146484 [DOI] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O.. 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6:11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrón MG, Fiston-Lavier A-S, Petrov DA, González J.. 2014. Population genomics of transposable elements in Drosophila. Annu Rev Genet. 48:561–581. [DOI] [PubMed] [Google Scholar]
- Baucom RS, Estill JC, Leebens-Mack J, Bennetzen JL.. 2008. Natural selection on gene function drives the evolution of LTR retrotransposon families in the rice genome. Genome Res. 192:243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G.. 2001. Insulators and boundaries: versatile regulatory elements in the eukaryotic genome. Science 2915503:447..http://dx.doi.org/10.1126/science.291.5503.447 [DOI] [PubMed] [Google Scholar]
- Belyayev A. 2014. Bursts of transposable elements as an evolutionary driving force. J Evol Biol. 2712:2573–2584.http://dx.doi.org/10.1111/jeb.12513 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Yekutieli D.. 2001. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 294:1165–1188. [Google Scholar]
- Bennetzen JL, Ma J, Devos KM.. 2005. Mechanisms of recent genome size variation in flowering plants. Ann Bot. 951:127–132.http://dx.doi.org/10.1093/aob/mci008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman CM, Bensasson D.. 2007. Recent LTR retrotransposon insertion contrasts with waves of non-LTR insertion since speciation in Drosophila melanogaster. Proc Natl Acad Sci U S A. 10427:11340–11345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biémont C, Cizeron G.. 1999. Distribution of transposable elements in Drosophila species. Genetica 1051:43–62. [DOI] [PubMed] [Google Scholar]
- Biémont C, Vieira C.. 2006. Genetics: junk DNA as an evolutionary force. Nature 4437111:521–524. [DOI] [PubMed] [Google Scholar]
- Blumenstiel JP. 2011. Evolutionary dynamics of transposable elements in a small RNA world. Trends Genet. 271:23–31.http://dx.doi.org/10.1016/j.tig.2010.10.003 [DOI] [PubMed] [Google Scholar]
- Blumenstiel JP, Chen X, He M, Bergman CM.. 2014. An age-of-allele test of neutrality for transposable element insertions. Genetics 1962:523..http://dx.doi.org/10.1534/genetics.113.158147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S, Davis J, Entezam A, Petrov D, Furano AV.. 2006. Fitness cost of LINE-1 (L1) activity in humans. Proc Natl Acad Sci U S A. 10325:9590–9594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousios A, Gaut BS.. 2016. Mechanistic and evolutionary questions about epigenetic conflicts between transposable elements and their plant hosts. Curr Opin Plant Biol. 30:123–133.http://dx.doi.org/10.1016/j.pbi.2016.02.009 [DOI] [PubMed] [Google Scholar]
- Bowen NJ, McDonald JF.. 2001. Drosophila euchromatic LTR retrotransposons are much younger than the host species in which they reside. Genome Res. 119:1527–1540.http://dx.doi.org/10.1101/gr.164201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookfield JFY. 1996. Models of the spread of non-autonomous selfish transposable elements when transposition and fitness are coupled. Genet Res. 6703:199..http://dx.doi.org/10.1017/S0016672300033681 [Google Scholar]
- Bureau TE, Wessler SR.. 1994. Mobile inverted-repeat elements of the Tourist family are associated with the genes of many cereal grasses. Proc Natl Acad Sci U S A. 914:1411–1415.http://dx.doi.org/10.1073/pnas.91.4.1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey AM, Dorman ER, Corces VG.. 2008. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell 321:1–9.http://dx.doi.org/10.1016/j.molcel.2008.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo AL, Williamson SH, Hernandez RD, Boyko A, Fledel-Alon A, York TL, Polato NR, Olsen KM, Nielsen R, McCouch SR.. 2007. Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet. 39:e163.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL.. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro C, Guyot R, Zuccolo A, Piégu B, Panaud O.. 2007. RetrOryza: a database of the rice LTR-retrotransposons. Nucleic Acids Res. 35(Database issue):D66–D70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. 1991. Transposable elements in natural populations with a mixture of selected and neutral insertion sites. Genet Res. 572:127..http://dx.doi.org/10.1017/S0016672300029190 [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D.. 1983. The population dynamics of transposable elements. Genet Res. 4201:1..http://dx.doi.org/10.1017/S0016672300021455 [Google Scholar]
- Charlesworth B, Langley CH.. 1986. The evolution of self-regulated transposition of transposable elements. Genetics 1122:359–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Langley CH.. 1989. The population genetics of Drosophila transposable elements. Annu Rev Genet. 23:251–287.http://dx.doi.org/10.1146/annurev.ge.23.120189.001343 [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B.. 1995. Transposable elements in inbreeding and outbreeding populations. Genetics 1401:415–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Ding B, Simon SA, Lopez D, Jia Y, Wang G-L, Meyers BC, Jacobsen SE, Pellegrini M.. 2012. Transcriptome and methylome interactions in rice hybrids. Proc Natl Acad Sci U S A. 10930:12040–12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JY, Platts AE, Fuller DQ, Hsing Y-I, Wing RA, Purugganan MD.. 2017. The rice paradox: multiple origins but single domestication in Asian rice. Mol Biol Evol. 344:969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE.. 2008. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 4527184:215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copetti D, Zhang J, El Baidouri M, Gao D, Wang J, Barghini E, Cossu RM, Angelova A, Maldonado L CE, Roffler S, et al. 2015. RiTE database: a resource database for genus-wide rice genomics and evolutionary biology. BMC Genomics 16:538.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridland JM, Macdonald SJ, Long AD, Thornton KR.. 2013. Abundance and distribution of transposable elements in two Drosophila QTL mapping resources. Mol Biol Evol. 3010:2311–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RM, Gowda M, Moghe G, Lin H, Vaillancourt B, Shiu S-H, Jiang N, Robin Buell C.. 2012. Comparative transcriptomics of three Poaceae species reveals patterns of gene expression evolution. Plant J. 713:492–502. [DOI] [PubMed] [Google Scholar]
- Devos KM, Brown JKM, Bennetzen JL.. 2002. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 127:1075–1079.http://dx.doi.org/10.1101/gr.132102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez CM, Roessler K, Gaut BS.. 2014. Epigenetics and plant genome evolution. Curr Opin Plant Biol. 18:1–8.http://dx.doi.org/10.1016/j.pbi.2013.11.017 [DOI] [PubMed] [Google Scholar]
- Dolgin ES, Charlesworth B.. 2008. The effects of recombination rate on the distribution and abundance of transposable elements. Genetics 1784:2169–2177.http://dx.doi.org/10.1534/genetics.107.082743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen RH, Pelizzola M, Schmitz RJ, Lister R, Dowen JM, Nery JR, Dixon JE, Ecker JR.. 2012. Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci U S A. 10932:E2183–E2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin MJ, Zhang P, Meng D, Remigereau M-S, Osborne EJ, Paolo Casale F, Drewe P, Kahles A, Jean G, Vilhjálmsson B, et al. 2015. DNA methylation in Arabidopsis has a genetic basis and shows evidence of local adaptation. Elife 4:e05255.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten SR, Ellis NA, Makarevitch I, Yeh C-T, Gent JI, Guo L, McGinnis KM, Zhang X, Schnable PS, Vaughn MW, et al. 2012. Spreading of heterochromatin is limited to specific families of maize retrotransposons. PLoS Genet. 812:e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D, Kurtz S, Willhoeft U.. 2008. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics 9:18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels WR. 1992. The origin of P elements in Drosophila melanogaster. BioEssays 1410:681–686.http://dx.doi.org/10.1002/bies.950141007 [DOI] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen P-Y, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. 2010. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A. 10719:8689–8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Jiang N, Wessler SR.. 2002. Plant transposable elements: where genetics meets genomics. Nat Rev Genet. 35:329–341.http://dx.doi.org/10.1038/nrg793 [DOI] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. 2014. Pfam: the protein families database. Nucleic Acids Res. 42:D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers JM, Molina J, Rubinstein S, Huang P, Schaal BA, Purugganan MD.. 2012. Natural selection in gene-dense regions shapes the genomic pattern of polymorphism in wild and domesticated rice. Mol Biol Evol. 292:675–687. [DOI] [PubMed] [Google Scholar]
- Flutre T, Duprat E, Feuillet C, Quesneville H, Xu Y.. 2011. Considering transposable element diversification in de novo annotation approaches. PLoS One 61:e16526.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo-González L, Mhiri C, Deyholos MK, Grandbastien M-A.. 2017. LTR-retrotransposons in plants: engines of evolution. Gene 626:14–25. [DOI] [PubMed] [Google Scholar]
- Gao L, Innan H.. 2008. Nonindependent domestication of the two rice subspecies, Oryza sativa ssp. indica and ssp. japonica, demonstrated by multilocus microsatellites. Genetics 1792:965–976.http://dx.doi.org/10.1534/genetics.106.068072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, McCarthy EM, Ganko EW, McDonald JF.. 2004. Evolutionary history of Oryza sativa LTR retrotransposons: a preliminary survey of the rice genome sequences. BMC Genomics 51:18..http://dx.doi.org/10.1186/1471-2164-5-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G.. 2006. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 79:703–713.http://dx.doi.org/10.1038/nrg1925 [DOI] [PubMed] [Google Scholar]
- Gaut BS, Wright SI, Rizzon C, Dvorak J, Anderson LK.. 2007. Recombination: an underappreciated factor in the evolution of plant genomes. Nat Rev Genet. 81:77–84. [DOI] [PubMed] [Google Scholar]
- Gent JI, Ellis NA, Guo L, Harkess AE, Yao Y, Zhang X, Dawe RK.. 2013. CHH islands: de novo DNA methylation in near-gene chromatin regulation in maize. Genome Res. 234:628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González J, Lenkov K, Lipatov M, Macpherson JM, Petrov DA.. 2008. High rate of recent transposable element-induced adaptation in Drosophila melanogaster. PLoS Biol. 610:e251.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves IK, Groszmann M, Ying H, Taylor JM, Peacock WJ, Dennis ES.. 2012. Trans chromosomal methylation in Arabidopsis hybrids. Proc Natl Acad Sci U S A. 1099:3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS. 2007. Improved pairwise alignment of genomic DNA [Ph.D. thesis]. Pennsylvania State University.
- Harushima Y, Yano M, Shomura A, Sato M, Shimano T, Kuboki Y, Yamamoto T, Lin SY, Antonio BA, Parco A, et al. 1998. A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 1481:479–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Zhai W, Wen H, Tang T, Wang Y, Lu X, Greenberg AJ, Hudson RR, Wu C-I, Shi S.. 2011. Two evolutionary histories in the genome of rice: the roles of domestication genes. PLoS Genet. 76:e1002100.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges DJ, Deininger PL.. 2007. Inviting instability: transposable elements, double-strand breaks, and the maintenance of genome integrity. Mutat Res Mol Mech Mutagen 616(1–2):46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A.. 1966. The effect of linkage on limits to artificial selection. Genet Res. 83:269–294.http://dx.doi.org/10.1017/S0016672300010156 [PubMed] [Google Scholar]
- Hoen DR, Hickey G, Bourque G, Casacuberta J, Cordaux R, Feschotte C, Fiston-Lavier A-S, Hua-Van A, Hubley R, Kapusta A, et al. 2015. A call for benchmarking transposable element annotation methods. Mob DNA 61:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister JD, Gaut BS.. 2009. Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 198:1419–1428.http://dx.doi.org/10.1101/gr.091678.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Kurata N, Wei X, Wang Z-X, Wang A, Zhao Q, Zhao Y, Liu K, Lu H, Li W, et al. 2012. A map of rice genome variation reveals the origin of cultivated rice. Nature 4907421:497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project. 2005. The map-based sequence of the rice genome. Nature 436:793–800.http://dx.doi.org/10.1038/nature03895 [DOI] [PubMed] [Google Scholar]
- Jacquemin J, Bhatia D, Singh K, Wing RA.. 2013. The International Oryza Map Alignment Project: development of a genus-wide comparative genomics platform to help solve the 9 billion-people question. Curr Opin Plant Biol. 162:147–156.http://dx.doi.org/10.1016/j.pbi.2013.02.014 [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 304:772–780.http://dx.doi.org/10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Huang SC, Jupe F, Sasaki E, Schmitz RJ, Urich MA, Castanon R, Nery JR, Barragan C, He Y, et al. 2016. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 1662:492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly LJ, Leitch IJ.. 2011. Exploring giant plant genomes with next-generation sequencing technology. Chromosom Res. 197:939–953.http://dx.doi.org/10.1007/s10577-011-9246-z [DOI] [PubMed] [Google Scholar]
- Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D.. 2003. Evolution’s cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc Natl Acad Sci U S A. 10020:11484–11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG. 1983. Hybrid dysgenesis in Drosophila melanogaster: factors affecting chromosomal contamination in the P-M system. Genetics 1042:317–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL.. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 124:357–360.http://dx.doi.org/10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 162:111–120.http://dx.doi.org/10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- Kofler R, Betancourt AJ, Schlötterer C.. 2012. Sequencing of pooled DNA samples (Pool-Seq) uncovers complex dynamics of transposable element insertions in Drosophila melanogaster. PLoS Genet. 81:e1002487.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Hill T, Nolte V, Betancourt AJ, Schlötterer C.. 2015. The recent invasion of natural Drosophila simulans populations by the P-element. Proc Natl Acad Sci U S A. 11221:6659–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR.. 2011. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2711:1571–1572.http://dx.doi.org/10.1093/bioinformatics/btr167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn EJ, Geyer PK.. 2003. Genomic insulators: connecting properties to mechanism. Curr Opin Cell Biol. 153:259–265.http://dx.doi.org/10.1016/S0955-0674(03)00039-5 [DOI] [PubMed] [Google Scholar]
- Kumar A, Bennetzen JL.. 1999. Plant Retrotransposons. Annu Rev Genet. 33:479–532.http://dx.doi.org/10.1146/annurev.genet.33.1.479 [DOI] [PubMed] [Google Scholar]
- Langley CH, Montgomery E, Hudson R, Kaplan N, Charlesworth B.. 1988. On the role of unequal exchange in the containment of transposable element copy number. Genet Res. 523:223.. [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE.. 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 113:204–220.http://dx.doi.org/10.1038/nrg2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Rouzic A, Deceliere G.. 2005. Models of the population genetics of transposable elements. Genet Res. 853:171..http://dx.doi.org/10.1017/S0016672305007585 [DOI] [PubMed] [Google Scholar]
- Lee YCG. 2015. The role of piRNA-mediated epigenetic silencing in the population dynamics of transposable elements in Drosophila melanogaster. PLoS Genet. 116:e1005269.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YCG, Karpen GH.. 2017. Pervasive epigenetic effects of Drosophila euchromatic transposable elements impact their evolution. Elife 6:e25762.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YCG, Langley CH.. 2010. Transposable elements in natural populations of Drosophila melanogaster. Philos Trans R Soc Lond B Biol Sci. 3651544:1219.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YCG, Langley CH.. 2012. Long-term and short-term evolutionary impacts of transposable elements on Drosophila. Genetics 1924:1411..http://dx.doi.org/10.1534/genetics.112.145714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat E. 2010. Identifying repeats and transposable elements in sequenced genomes: how to find your way through the dense forest of programs. Heredity (Edinb) 1046:520–533.http://dx.doi.org/10.1038/hdy.2009.165 [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P.. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1):W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Gent JI, Zynda G, Song J, Makarevitch I, Hirsch CD, Hirsch CN, Dawe RK, Madzima TF, McGinnis KM, et al. 2015. RNA-directed DNA methylation enforces boundaries between heterochromatin and euchromatin in the maize genome. Proc Natl Acad Sci U S A. 11247:14728–14733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang X, He K, Ma Y, Su N, He H, Stolc V, Tongprasit W, Jin W, Jiang J, et al. 2008. High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell 202:259–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhu J, Hu F, Ge S, Ye M, Xiang H, Zhang G, Zheng X, Zhang H, Zhang S, et al. 2012. Single-base resolution maps of cultivated and wild rice methylomes and regulatory roles of DNA methylation in plant gene expression. BMC Genomics 13:300.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, Gendrel A-V, Black M, Vaughn MW, Dedhia N, Richard McCombie W, Lavine K, Mittal V, May B, Kasschau KD, et al. 2004. Role of transposable elements in heterochromatin and epigenetic control. Nature 4306998:471–476. [DOI] [PubMed] [Google Scholar]
- Lisch D. 2009. Epigenetic regulation of transposable elements in plants. Annu Rev Plant Biol. 60:43–66.http://dx.doi.org/10.1146/annurev.arplant.59.032607.092744 [DOI] [PubMed] [Google Scholar]
- Lisch D. 2013. How important are transposons for plant evolution? Nat Rev Genet. 141:49–61. [DOI] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR, Aufsatz W, Mette MF, Matzke AJ, et al. 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 1333:523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockton S, Ross-Ibarra J, Gaut BS.. 2008. Demography and weak selection drive patterns of transposable element diversity in natural populations of Arabidopsis lyrata. Proc Natl Acad Sci U S A. 10537:13965–13970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Chen J, Zhang Y, Hu Q, Su W, Kuang H.. 2012. Miniature inverted-repeat transposable elements (MITEs) have been accumulated through amplification bursts and play important roles in gene expression and species diversity in Oryza sativa. Mol Biol Evol. 293:1005–1017.http://dx.doi.org/10.1093/molbev/msr282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Bennetzen JL.. 2004. Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci U S A. 10134:12404–12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Devos KM, Bennetzen JL.. 2004. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 145:860–869.http://dx.doi.org/10.1101/gr.1466204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ.. 2009. Small RNAs as guardians of the genome. Cell 1364:656–668.http://dx.doi.org/10.1016/j.cell.2009.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ.. 2009. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 213:367–376. [DOI] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA.. 2014. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet. 156:394–408.http://dx.doi.org/10.1038/nrg3683 [DOI] [PubMed] [Google Scholar]
- Maumus F, Quesneville H.. 2014. Ancestral repeats have shaped epigenome and genome composition for millions of years in Arabidopsis thaliana. Nat Commun. 5:4104.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy EM, Liu J, Lizhi G, McDonald JF.. 2002. Long terminal repeat retrotransposons of Oryza sativa. Genome Biol. 310:RESEARCH0053.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski PA, Lemoine FJ, Petes TD.. 2006. Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae. DNA Repair (Amst) 5(9–10): 1010–1020. [DOI] [PubMed] [Google Scholar]
- Mirouze M, Lieberman-Lazarovich M, Aversano R, Bucher E, Nicolet J, Reinders J, Paszkowski J.. 2012. Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc Natl Acad Sci U S A. 10915:5880–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery E, Charlesworth B, Langley CH.. 1987. A test for the role of natural selection in the stabilization of transposable element copy number in a population of Drosophila melanogaster. Genet Res. 4901:31–41.http://dx.doi.org/10.1017/S0016672300026707 [DOI] [PubMed] [Google Scholar]
- Montgomery EA, Huang SM, Langley CH, Judd BH.. 1991. Chromosome rearrangement by ectopic recombination in Drosophila melanogaster: genome structure and evolution. Genetics 1294:1085–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan MT. 2001. Transposable element number in mixed mating populations. Genet Res. 773:261–275. [DOI] [PubMed] [Google Scholar]
- Murtagh F, Legendre P.. 2014. Ward’s hierarchical agglomerative clustering method: which algorithms implement Ward’s criterion? J Classif. 313:274–295. [Google Scholar]
- Muyle A, Serres-Giardi L, Ressayre A, Escobar J, Glémin S.. 2011. GC-biased gene conversion and selection affect GC content in the Oryza genus (rice). Mol Biol Evol. 289:2695–2706. [DOI] [PubMed] [Google Scholar]
- Naito K, Zhang F, Tsukiyama T, Saito H, Hancock CN, Richardson AO, Okumoto Y, Tanisaka T, Wessler SR.. 2009. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 4617267:1130–1134. [DOI] [PubMed] [Google Scholar]
- Nuzhdin SV. 1999. Sure facts, speculations, and open questions about the evolution of transposable element copy number. Genetica 107(1–3):129–137.http://dx.doi.org/10.1023/A:1003957323876 [PubMed] [Google Scholar]
- Nuzhdin SV, Pasyukova EG, Mackay TFC.. 1996. Positive association between copia transposition rate and copy number in Drosophila melanogaster. Proc R Soc Lond B Biol Sci. 2631372:823.. [DOI] [PubMed] [Google Scholar]
- Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin Y-C, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, et al. 2013. The Norway spruce genome sequence and conifer genome evolution. Nature 4977451:579–584. [DOI] [PubMed] [Google Scholar]
- Petrov DA, Aminetzach YT, Davis JC, Bensasson D, Hirsh AE.. 2003. Size matters: non-LTR retrotransposable elements and ectopic recombination in Drosophila. Mol Biol Evol. 206:880–892. [DOI] [PubMed] [Google Scholar]
- Quadrana L, Bortolini Silveira A, Mayhew GF, LeBlanc C, Martienssen RA, Jeddeloh JA, Colot V.. 2016. The Arabidopsis thaliana mobilome and its impact at the species level. Elife 5:e15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2016. R: a language and environment for statistical computing. Vienna. [Google Scholar]
- Rebollo R, Karimi MM, Bilenky M, Gagnier L, Miceli-Royer K, Zhang Y, Goyal P, Keane TM, Jones S, Hirst M, et al. 2011. Retrotransposon-induced heterochromatin spreading in the mouse revealed by insertional polymorphisms. PLoS Genet. 79:e1002301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezvoy C, Charif D, Gueguen L, Marais GAB.. 2007. MareyMap: an R-based tool with graphical interface for estimating recombination rates. Bioinformatics 2316:2188–2189.http://dx.doi.org/10.1093/bioinformatics/btm315 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 261:139–140.http://dx.doi.org/10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A.. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 113:R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder GS, Farabaugh PJ, Chaleff DT, Fink GR.. 1980. The origins of gene instability in yeast. Science 2094463:1375–1380.http://dx.doi.org/10.1126/science.6251544 [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL.. 1998. The paleontology of intergene retrotransposons of maize. Nat Genet. 201:43–45. [DOI] [PubMed] [Google Scholar]
- Secco D, Wang C, Shou H, Schultz MD, Chiarenza S, Nussaume L, Ecker JR, Whelan J, Lister R.. 2015. Stress induced gene expression drives transient DNA methylation changes at adjacent repetitive elements. Elife 4:e09343.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbiss S, Willhoeft U, Gremme G, Kurtz S.. 2009. Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res. 3721:7002–7013.http://dx.doi.org/10.1093/nar/gkp759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Eichten SR, Cahn J, Karpievitch YV, Borevitz JO, Lister R.. 2016. Population scale mapping of transposable element diversity reveals links to gene regulation and epigenomic variation. Elife 5:e20777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H. 2010. Confining euchromatin/heterochromatin territory: jumonji crosses the line. Genes Dev. 2414:1465–1478.http://dx.doi.org/10.1101/gad.1941010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815.http://dx.doi.org/10.1038/35048692 [DOI] [PubMed] [Google Scholar]
- Tian Z, Rizzon C, Du J, Zhu L, Bennetzen JL, Jackson SA, Gaut BS, Ma J.. 2009. Do genetic recombination and gene density shape the pattern of DNA elimination in rice long terminal repeat retrotransposons? Genome Res. 1912:2221–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitte C, Bennetzen JL.. 2006. Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc Natl Acad Sci U S A. 10347:17638–17643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitte C, Panaud O.. 2003. Formation of Solo-LTRs through unequal homologous recombination counterbalances amplifications of LTR retrotransposons in rice Oryza sativa L. Mol Biol Evol. 204:528–540.http://dx.doi.org/10.1093/molbev/msg055 [DOI] [PubMed] [Google Scholar]
- Vitte C, Panaud O, Quesneville H.. 2007. LTR retrotransposons in rice (Oryza sativa, L.): recent burst amplifications followed by rapid DNA loss. BMC Genomics 8:218.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vonHoldt BM, Takuno S, Gaut BS.. 2012. Recent retrotransposon insertions are methylated and phylogenetically clustered in Japonica rice (Oryza sativa spp. japonica). Mol Biol Evol. 2910:3193–3203.http://dx.doi.org/10.1093/molbev/mss129 [DOI] [PubMed] [Google Scholar]
- Wang X, Wang J, Jin D, Guo H, Lee T-H, Liu T, Paterson AH.. 2015. Genome alignment spanning major poaceae lineages reveals heterogeneous evolutionary rates and alters inferred dates for key evolutionary events. Mol Plant 86:885–898. [DOI] [PubMed] [Google Scholar]
- Wang X, Weigel D, Smith LM.. 2013. Transposon variants and their effects on gene expression in Arabidopsis. PLoS Genet. 92:e1003255.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF, Jackson SA, Meyers BC, Wing RA.. 2016. Evolution of plant genome architecture. Genome Biol. 17:37.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al. 2007. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 812:973–982. [DOI] [PubMed] [Google Scholar]
- Wicker T, Yu Y, Haberer G, Mayer KFX, Marri PR, Rounsley S, Chen M, Zuccolo A, Panaud O, Wing RA, et al. 2016. DNA transposon activity is associated with increased mutation rates in genes of rice and other grasses. Nat Commun. 7:12790.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Agrawal N, Bureau TE.. 2003. Effects of recombination rate and gene density on transposable element distributions in Arabidopsis thaliana. Genome Res. 138:1897–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Schoen DJ.. 1999. Transposon dynamics and the breeding system. Genetica 107(1–3):139–148. [PubMed] [Google Scholar]
- Xu L, Zhang Y, Su Y, Liu L, Yang J, Zhu Y, Li C.. 2010. Structure and evolution of full-length LTR retrotransposons in rice genome. Plant Syst Evol. 287(1–2):19–28. [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 248:1586–1591.http://dx.doi.org/10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK-S, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al. 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 2965565:79–92. [DOI] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Silva P, Rodrigues JA, Dotson B, Brooks MD, Zilberman D.. 2010. Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci U S A. 10743:18729–18734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D.. 2010. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 3285980:916–919.http://dx.doi.org/10.1126/science.1186366 [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu Y, Xia E-H, Yao Q-Y, Liu X-D, Gao L-Z.. 2015. Autotetraploid rice methylome analysis reveals methylation variation of transposable elements and their effects on gene expression. Proc Natl Acad Sci U S A. 11250:E7022–E7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shiu S, Cal A, Borevitz JO, Borevitz JO.. 2008. Global analysis of genetic, epigenetic and transcriptional polymorphisms in Arabidopsis thaliana using whole genome tiling arrays. PLoS Genet. 43:e1000032.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.