Abstract
Purpose
Indoleamine 2,3 dioxygenase 1 (IDO1) mediates potent immunosuppression in multiple preclinical models of cancer. However, the basis for elevated IDO1 expression in human cancer, including the most common primary malignant brain tumor in adults, glioblastoma (GBM), is poorly understood. The major objective of this study is to address this gap in our understanding of how IDO1 expression contributes to the biology of GBM, and whether its level of expression is a determinant of GBM patient outcome.
Experimental Design
Patient-resected GBM, the cancer genome atlas, human T cell:GBM co-cultures, as well as nu/nu, NOD-scid and humanized (NSG-SGM3-BLT) mice engrafted human GBM, form the basis of our investigation.
Results
In situ hybridization for IDO1 revealed transcript expression throughout patient-resected GBM, whereas immunohistochemical IDO1 positivity was highly variable. Multivariate statistical analysis revealed that higher levels of IDO1 transcript predict a poor patient prognosis (P=0.0076). GBM IDO1 mRNA levels positively correlated with increased gene expression for markers of cytolytic and regulatory T cells, in addition to decreased patient survival. Humanized mice intracranially-engrafted human GBM revealed an IFNγ-associated T cell-mediated increase of intratumoral IDO1.
Conclusions
Our data demonstrate that high intratumoral IDO1 mRNA levels correlate with a poor GBM patient prognosis. It also confirms the positive correlation between increased GBM IDO1 levels and human-infiltrating T cells. Collectively, this study suggests that future efforts aimed at increasing T cell-mediated effects against GBM, should consider combinatorial approaches that co-inhibit potential T cell-mediated IDO1 enhancement during therapy.
Keywords: IDO, immunotherapy, immunosuppression, immune checkpoint, glioma, Treg
INTRODUCTION
Glioblastoma (GBM, astrocytoma, WHO grade IV) is the most common and aggressive primary central nervous system (CNS) cancer in adults (1). Numerous efforts have been made to identify prognostic biomarkers for GBM patients, resulting in the determination of O-6-methylguanine-DNA methyltransferase (MGMT) gene promoter methylation (2, 3), mutant isocitrate dehydrogenase 1 and 2 (mIDH1/2) (4), as well as chromosome 1p/19q co-deletion, as important indicators of tumor malignancy and/or response to specific therapies (5, 6). Novel immunotherapies, which have caused the reconsideration of patient management among multiple malignancies including melanoma (7, 8) and lung cancer (9, 10), have been largely unsuccessful in treating GBM. A caveat associated with this lack of success is the relative dearth of knowledge regarding biomarkers that could be used to guide patient selection for specific immunotherapies.
Indoleamine 2,3-dioxygense 1 (IDO1) is an interferon-inducible tryptophan (Trp) catabolic enzyme (11) that facilitates immunosuppression in cancer (12) through regulatory T cell (Treg; CD3+CD4+CD25+FoxP3+)-mediated suppression of cytolytic CD8+ effector T cells (Tc), as demonstrated in preclinical GBM models (13). The protein level and/or activity of IDO1 has been correlated with prognosis in several tumor types (14–19). In GBM, however, interpretations of the prognostic significance of IDO1 expression have been inconsistent, with immunohistochemical analysis revealing cellular positivity ranging from 8% to 96%, respectively (20, 21). Additionally, while we previously demonstrated that the upregulation of IDO1 mRNA levels inversely correlate with glioma patient survival (22), microarray-based expression analysis did not prognostically stratify GBM patient survival, possibly due to an inability to quantify full length IDO1 mRNA transcript as a result of oligo-based hybridization technology.
Here, we have compared mRNA- vs. antibody-based IDO1 detection in GBM surgical specimens, and determined a correlation between high IDO1 mRNA levels and decreased patient survival for grade II, III or IV (GBM) gliomas. Increased IDO1 mRNA was associated with a commensurate increase in the expression for cytolytic T cell markers, CD3E and CD8A, suggesting a T cell-associated influence on intratumoral IDO1 expression.
METHODS
The cancer genome atlas (TCGA) sample description
The TCGA data for all the cancer types analyzed in current study were accessed from the UCSC Xena browser (http://xena.ucsc.edu/). mRNA expression data represented by RNASeq (Illumina Hi-seq platform) includes RSEM normalized level 3 data present in TCGA as of April 13th, 2017. DNA methylation data and exon expression RNASeq data were extracted from the same TCGA dataset. Glioma patient data were also acquired from the Molecular Brain Neoplasia Data (REMBRANDT) database (https://wiki.cancerimagingarchive.net/display/Public/REMBRANDT) as of April 13th, 2017. TCGA GBM gene expression data by AffyU133a array were also acquired from the UCSC Xena browser.
Patient samples
Peripheral blood from GBM patients were collected at the Northwestern Brain Tumor Institute (NBTI) Tissue Bank. PBMCs were isolated using Ficoll-Paque (GE Healthcare) density gradient separation and stored at liquid nitrogen tank for coculture experiments. Snap-frozen tissue and formalin-fixed, paraffin-embedded (FFPE) tissue from surgically-removed GBM were also collected at the NBTI Tissue Bank. All tumors were diagnosed according to WHO diagnostic criteria by Dr. Craig Horbinski. Detailed information for patient tissue samples used in gene expression analysis and in situ RNA hybridization /immunohistochemistry are provided in Supplementary Table 1.
Animal and tissue preparation
Immunocompetent, humanized mice (NSG-SGM3-BLT) were established by implantation of human fetal liver and thymus fragments as well as haematopoietic stem cells into immunodeficient NOD.Cg-Prkdcscid IL2rgtm1Wjl Tg (CMV-IL3, CSF2, KITLG)1Eav/MloySzJ (NSG-SGM3) mice, NOD.CB17-Prkdcscid/J (NOD-scid) mice were obtained from The Jackson Laboratory and CrTac:NCr-Foxn1nu mice were obtained from Taconic. Mice were maintained under specific pathogen–free conditions in the Northwestern University Center for Comparative Medicine. For T cell depletion experiments, 200 µg anti-human CD4 (clone OKT-4; BioXCell) and/or 200 µg anti-human CD8 (clone OKT-8; BioXCell) was delivered/mouse by intraperitoneal (i.p.) injection 3 days prior to tumor cell engraftment and maintained every 3 days until experimental endpoints. Mouse IgG2b (clone MPC-11, BioXCell) and mouse IgG2a (clone C1.18.4, BioXCell) were administrated in the same dose and approach as isotype control. For orthotopic brain tumor mouse modeling, 3 × 105 human U87, PDX12 or PDX43 GBM cells were intracranially-engrafted similar to our previous studies (22). U87 cells were acquired from ATCC and engrafted at < 10 total passages while PDX tumor cells were provided by the laboratory of Dr. C. David James, PhD, from continuously propagated GBM subcutaneously engrafted in nude mice. Human GBM was not tested for mycoplasma prior to analysis. Mice were euthanized at the indicated time point(s). Brain tumor, non-tumor brain tissue isolated from the contralateral hemisphere, cervical lymph node and spleen were collected, dissected and washed in ice-cold phosphate-buffered saline (PBS), frozen in liquid nitrogen and stored at −80°C until analysis or processed for other techniques. Patient-derived GBM xenografts (PDX) were prepared as previously reported (23, 24). Procedures for all mouse experiments were reviewed and approved by the Institutional Animal Care and Use Committee at Northwestern University and are in compliance with national and institutional guidelines.
Glioma cell lines, coculture assays, immunohistochemistry, in situ mRNA hybridization, flow cytometry, Western blotting, real-time RT-PCR, HPLC, Gene Set Enrichment Analysis
Procedures described in the Supplementary Materials and Methods section.
Statistical analysis
The cutoff value for each gene expression level was determined with Cutoff Finder software (http://molpath.charite.de/cutoff/) using significance as the cutoff optimization method (25). Kaplan-Meier (KM) survival analysis was performed to estimate the survival distribution, while the log-rank test was used to assess the statistical significance of differences between the stratified survival groups using GraphPad Prism (version 6, GraphPad Software, Inc., La Jolla, CA). Renyi family of test statistics was computed via SAS software (version9.4, SAS Institute Inc., Cary, NC) to determine the survival difference between two groups given the presence of crossing hazard rates. Cox proportional hazards regression analyses were performed to assess the independent contribution of the mRNA signature and clinicopathologic variables to survival prediction using MedCalc (version 16.4.3, MedCalc Software bvba, Belgium). Pearson’s correlation was used to analyze the relationship between each two genes’ mRNA expression level. Comparisons between multiple groups were analyzed by One-way ANOVA using GraphPad Prism software. The correlation between IDO1 mRNA levels and different cell types (TAM, MDSC, Treg, Neutrophil, Tc) were examined by Canonical Correlation analysis, where each cell type was defined by a linear combination of the corresponding signature marker genes. One-way Repeated Measurement Analysis of Variance, followed by Tukey multiple comparison was conducted to determine difference between IDO1 exon expression levels. Differences were considered to be statistically significant when P < 0.05. Standard error of the mean (SEM) is presented as the error bar in all bar graphs, and mean ± SEM was utilized to describe the data throughout the text unless specifically noted.
RESULTS
Comparison of IDO1 expression by mRNA and protein detection methods in GBM
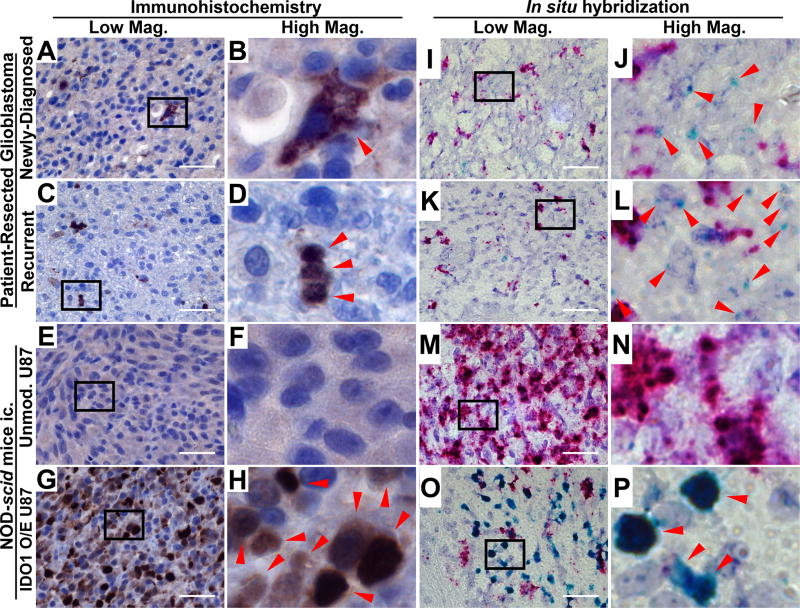
Previous studies have demonstrated highly variable IDO1 levels in GBM using immunohistochemical detection. In accordance with a recent independent report (26), minimal IDO1 protein expression was detected in both newly-diagnosed and recurrent surgical GBM specimens (Fig. 1A–D, Supp. Fig. 1A,B), as indicated with human IDO1 mAb (Clone D5J4E, Cell Signaling Technology). IDO1 expression was not detectable in unmodified human U87 tumor intracranially-engrafted (ic.) into NOD-scid mice (Fig. 1E,F; Supp. Fig. 1C), although it was evident in U87 GBM cells modified to overexpress human IDO1 cDNA and engrafted into NOD-scid mice (Fig. 1G,H; Supp. Fig. 1, 2). In contrast to the antibody-based detection properties, in situ hybridization readily detected IDO1 mRNA in both primary and recurrent patient GBM tissue samples (Fig. 1I–1L; Supp. Fig. 1E, F). Unmodified intracranial U87 GBM were negative for IDO1 mRNA (Fig. 1M, N; Supp. Fig. 1G), whereas U87 modified to express IDO1 cDNA were positive (Fig. 1O, P, Supp. Fig. 1H).
Figure 1. Immunohistochemical (IHC) IDO1 localization and in situ hybridization (ISH) for IDO1 in human glioblastoma (GBM).
(A–H) IHC detection with a human-specific IDO1 antibody was performed in (A, B) newly diagnosed primary patient-resected GBM, (C, D) recurrent patient-resected GBM or tumors isolated from immunodeficient NOD-scid mice intracranially-engrafted (ic.) (E, F) unmodified U87 cells and (G, H) U87 overexpressing human IDO1 conjugated to an HA tag (HA-IDO1 O/E). A positive signal is indicated by red arrows in the high magnification images (B, D, F, H). (I–P) ISH for human IDO1 mRNA was performed in the same tissues described for IHC and is indicated with red arrows in the high magnification images (J, L, N, P). The proliferation marker, Ki67, was used as a positive control and appears as a red signal. All lower magnification images were obtained under 64× objective lens with a scale bar representing 400 µm.
Patient survival and IDO1 expression in different grades of gliomas
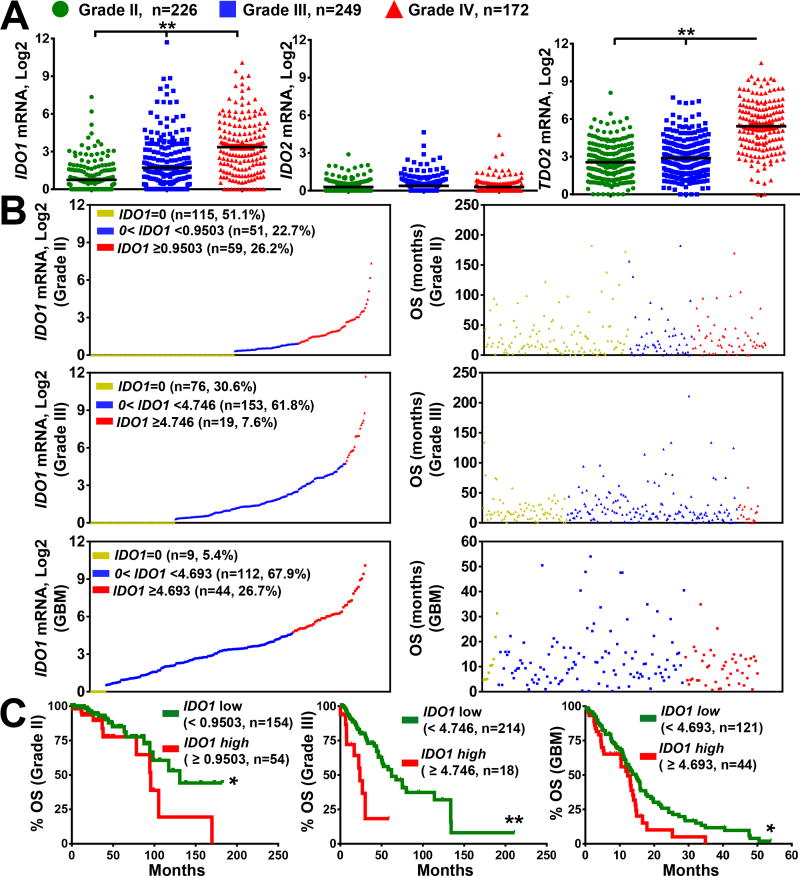
To follow-up our previous work, we evaluated the surgical specimen cohort using Hi-RNA sequencing technology for the mammalian tryptophan catabolic genes, IDO1 and TDO2, as well as the pseudogene, IDO2, for WHO grade II, grade III and grade IV (GBM) glioma, as accessed through the TCGA database. IDO1, IDO2 and TDO2 (tryptophan 2,3-dioxygenase) mRNA expression levels were quantified and correlated with overall survival (OS). IDO1 expression progressively increased with tumor grade (Fig. 2A; P < 0.0001). Interestingly, while a large subset of grade II and III glioma specimens were undetectable for IDO1 mRNA, 95% of GBM specimens possessed detectable IDO1 mRNA levels (Fig. 2B). In contrast, IDO2 expression did not change with tumor grade and the majority of samples analyzed were undetectable for gene transcript (Fig. 2A). In accordance with a previous report, TDO2 mRNA levels were significantly increased in grade IV glioma (27). Additionally, further analysis of IDO1 mRNA in GBM revealed differential exon expression for IDO1 (Supp. Fig. 3, Supp. Table 2), indicating the possibility of multiple IDO1 mRNA isoforms being expressed in GBM.
Figure 2. mRNA expression for tryptophan catabolic enzymes in human glioma and the association of IDO1 with overall survival in glioma patients.
(A) The mRNA expression levels for IDO1, IDO2 and TDO2 in grade II (green; n=226) and grade III (blue; n=249) and grade IV (GBM; red; n=172) analyzed from the cancer genome atlas RNA-Hi-Seq. Illumina database. Horizontal lines in the scatter plots represents mean ± SEM. (B) Relative expression of IDO1 mRNA expression (left column) in grade II (top), grade III (middle) and grade IV (bottom) glioma and the corresponding survival (right column). Each dot represents one patient sample that is displayed in 3 colored groups based on IDO1 expression level: undetectable IDO1 mRNA (yellow; IDO1=0); IDO1 mRNA < cutoff (blue) and IDO1 mRNA > cutoff (red). Sample size (n), frequency of the representative population, and cutoff values within each grade of glioma are annotated in the IDO1 mRNA distribution dot plot. (C) Kaplan-Meier (KM) survival analysis of grade II (left), grade III (center) and grade IV (right) glioma patients stratified by IDO1-low (green; below the cutoff) and IDO1-high (red; equal or above the cutoff) expression levels. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Based on the correlation between increasing glioma grade and increasing IDO1 mRNA, we examined whether high IDO1 expression correlates with patient survival using Kaplan-Meier (KM) analysis. Cutoff Finder was utilized to generate individual cutoff values within each glioma grade. IDO1 mRNA levels were stratified into IDO1-low and -high expressing groups based on the determined cutoff values. High IDO1 mRNA levels were significantly correlated with decreased patient survival across all 3 glioma patient tumor grades (Fig. 2C; P < 0.05). Because clinicopathologic parameters including age, tumor grade and mode of therapy can also contribute to the prognosis of glioma patients, multivariate COX regression analysis was performed to evaluate whether IDO1 mRNA levels can be utilized as an independent prognostic factor. The results indicate that, IDO1 mRNA expression functions as an independent prognostic factor in grade II and III glioma, as well as for GBM patients (Supp. Table 3,4, respectively; Table 1). Karnofsky Performance status data was missing in up to 50% of the analyzed specimens and were therefore not included in the multivariate analysis.
Table 1.
Multivariate analysis of IDO1 mRNA levels as an independent prognostic marker in GBM patients (n=148).
| Variables | Total No. of Patient Events |
Death | Survival (months) KM Analysis | Multivariate Cox Regression | |||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| No. | % | Median | 95% CI | P | HR | 95% CI | P | ||
| Age at diagnosis, year | |||||||||
| < 50 | 33 | 21 | 63.6 | 14.5 | [12.7, 21.9] | 0.146 | |||
| ≥ 50 | 115 | 81 | 70.4 | 13.0 | [10.4, 15.4] | ||||
|
| |||||||||
| Sex | |||||||||
| Male | 99 | 65 | 65.7 | 13.0 | [11.3, 15.1] | 0.814 | |||
| Female | 49 | 37 | 75.5 | 14.7 | [9.86, 17.9] | ||||
|
| |||||||||
| Tumor Subtypes | |||||||||
| Classical | 39 | 27 | 69.2 | 14.0 | [11.8, 16.1] | 0.656 | |||
| Mesenchymal | 51 | 35 | 68.6 | 11.3 | [10.3, 15.9] | ||||
| Neural | 25 | 20 | 80.0 | 14.9 | [5.39, 18.0] | ||||
| Proneural | 33 | 20 | 60.6 | 14.7 | [10.9, 22.2] | ||||
|
| |||||||||
| Chemotherapy | |||||||||
| Yes | 117 | 74 | 63.2 | 14.5 | [13.0, 16.1] | <0.0001 | |||
| No | 31 | 28 | 90.3 | 3.90 | [2.73, 7.63] | ||||
|
| |||||||||
| Radiation therapy | |||||||||
| Yes | 128 | 83 | 64.8 | 14.7 | [13.0, 15.9] | <0.0001 | 7.50 | [4.39, 12.80] | <0.0001 |
| No | 20 | 19 | 95.0 | 2.24 | [0.953, 3.75] | ||||
|
| |||||||||
| IDO1 | |||||||||
| Low | 106 | 71 | 67.0 | 14.9 | [11.7, 16.1] | 0.016 | 1.82 | [1.17, 2.81] | 0.0076 |
| High | 42 | 31 | 73.8 | 12.3 | [10.3, 14.0] | ||||
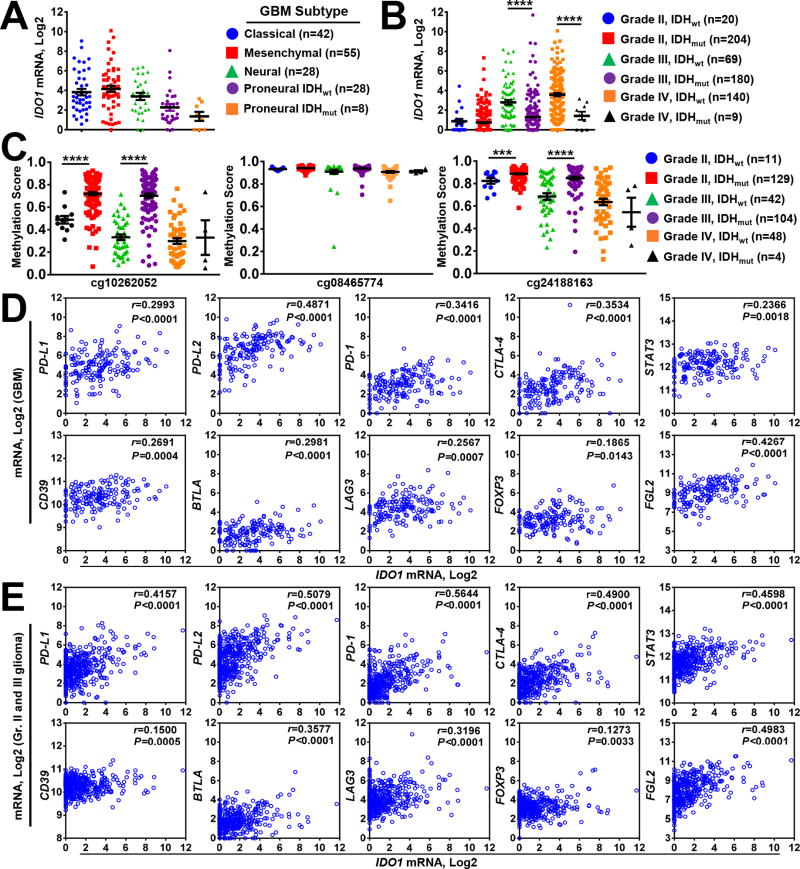
IDO1 expression with respect to GBM subtype and IDH1 mutation status
Given the commonly used transcriptome-based classification of GBM (28), we examined IDO1 mRNA expression with respect to GBM transcriptional subclasses. One-way ANOVA identified the proneural GBM subtype, among which isocitrate dehydrogenase 1 and 2 (mIDH1/2) mutation is most common, with significantly lower IDO1 mRNA levels (Fig. 3A; P < 0.05; Supp. Table 5). Decreased IDO1 expression was also evident in mIDH1/2-associated grade III and IV glioma specimens (Fig. 3B, P < 0.0001, Supp. Table 5). Given the established relationship between IDH1 mutation and cytosine hypermethylation in glioma (29, 30), we also analyzed genomic CpG motifs for DNA methylation throughout the human IDO1 gene locus. TCGA analysis of the DNA methylation status identified 3 CpG loci within the human IDO1 gene (locus 1: cg10262052, 1500bp of upstream transcription start site; loci 2 and 3: cg0846577, cg24188163, gene body). Methylation was increased at 2 of the 3 CpG loci in mIDH1/2 grade II and III glioma (Fig. 3C). In contrast, methylation patterns did not significantly change at any CpG locus among GBM specimens, confirming that, while lower glioma grade IDO1 expression is significantly affected by mIDH status, independent mechanisms appear to regulate IDO1 mRNA levels in GBM.
Figure 3. Correlation of GBM IDO1 mRNA levels with IDH mutation, DNA methylation and immunosuppressive gene expression.
(A) IDO1 mRNA levels were compared among classical (blue circle), mesenchymal (red square) and neural (green triangle) GBM subtypes analyzed from the cancer genome atlas RNA-Hi-Seq. Illumina database. The Proneural GBM subtype was further stratified into IDH1/2-wild-type (IDHwt; purple circle) and IDH1/2-mutant (IDHmut; orange square) samples. (B) IDO1 mRNA levels were compared among grade II (IDHwt; blue circle and IDHmut; red square), grade III (IDHwt; green triangle and IDHmut; purple circle), as well as grade IV (IDHwt; orange circle and IDHmut; black triangle) glioma. (C) DNA methylation analysis of cg10262052, cg08465774 and cg24188163 in grade II (IDHwt; blue circle and IDHmut; red square), grade III (IDHwt; green triangle and IDHmut; purple circle), as well as grade IV (IDHwt; orange circle and IDHmut; black triangle) glioma. Pearson’s correlation analysis for IDO1 mRNA with PD-L1, PD-L2, PD-1, CTLA-4, STAT3, CD39, BTLA, LAG3, FOXP3 and FGL2 in (D) GBM and (E) pooled grade II and III glioma. Each small circle in the plot represents a single patient data point. For all the scatter plots, horizontal lines represent mean ± SEM. ***, P < 0.001, ****, P < 0.0001.
Due to IDO1 activity acting as an immunosuppressant, we examined IDO1 expression with respect to the expression of genes that influence immune response. Using Pearson’s correlation analysis, we found significant relationships between mRNA expression for IDO1 and PD-L1 (r = 0.2993 and r = 0.4157), PD-L2 (r = 0.4871 and r = 0.5079), PD-1 (r = 0.3416 and r = 0.5644), CTLA-4 (r = 0.3534 and r = 0.4900), STAT3 (r = 0.2366 and r = 0.4598), CD39 (r = 0.2691 and r = 0.1500), BTLA (r = 0.2981 and r = 0.3577), LAG3 (r = 0.2567 and r = 0.3196), FOXP3 (r = 0.1865 and r = 0.1273) and FGL2 (r = 0.4267 and r = 0.4983) both in GBM and grade II/III glioma, respectively, (P < 0.0143 and P < 0.01, respectively) (Fig. 3D, E). These results suggest that increased IDO1 expression is most evident in tumors expressing additional immunosuppressive factors.
We also examined IDO1 expression with respect to immune cell infiltrates in tumor, including the markers: (i) CD14, HLA-DR, CD312, CD115, CD163, CD204, CD301, CD206 associated with tumor-associated macrophages (TAM), (ii) CD14, CD11b, CD33, Arg1 associated with myeloid-derived suppressor cells (MDSC), and (iii) CD11b, CD16, CD66b and ELANE associated with neutrophils. Correlation analyses showed that GBM-derived IDO1 mRNA levels positively correlate with the expression of all of the cell-type specific marker genes (Supp. Fig. 4A, B; P < 0.001).
Regulation of IDO1 expression in glioblastoma
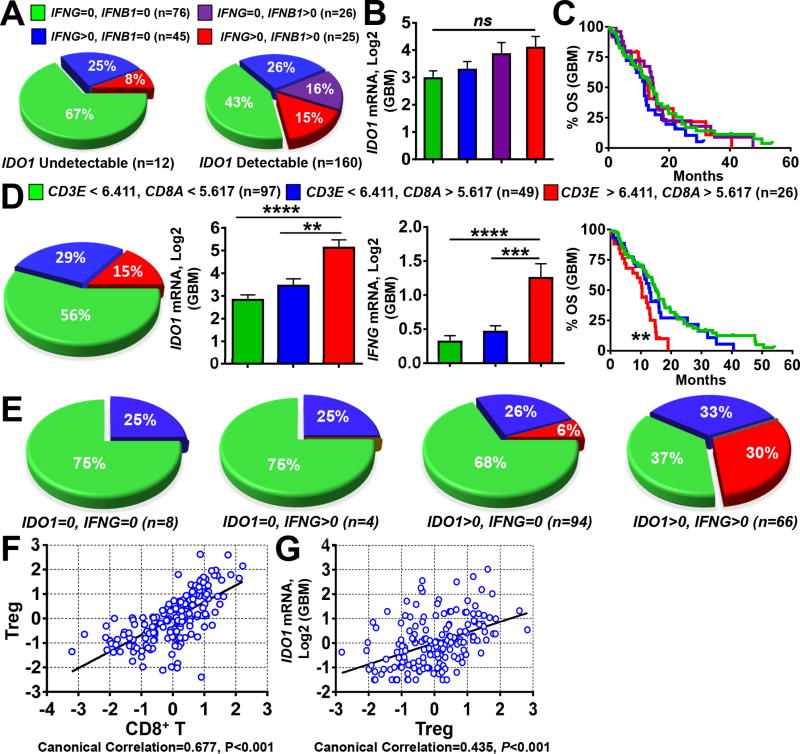
In many tissues, IDO1 expression is undetectable (20), but rapidly induced and made detectable by pro-inflammatory stimuli (31). Due to the multiple interferon-stimulated response elements (ISRE) in the promoter region of IDO1, treatment of in vitro-cultured human GBM cells with the T cell effector pro-inflammatory cytokine, IFNγ (IFNG), leads to robust IDO1 mRNA and protein expression (32, 33). To determine whether this occurs in situ, we examined TCGA data that revealed 12 IDO1-undetectable patient-resected GBM specimens, with 4 samples co-expressing detectable IFNG levels (Fig. 4A). In contrast, of the 160 GBM specimens with detectable IDO1 expression, >50% (n=94) were absent for IFNG. Strikingly, 43% (n=69) of GBM specimens were absent for both IFNG and IFNΒ. When the 172 total GBM samples were further stratified into IFNG/IFNΒ-expressing, versus -non-expressing groups, no difference was found among IDO1 mRNA levels (Fig. 4B), nor did this stratification yield any correlation with GBM patient survival (Fig. 4C).
Figure 4. Gene expression correlation analysis between IDO1, IFNG and T cells in human glioblastoma (GBM).
(A) Frequency analysis of GBM specimens with undetectable (left) or detectable (right) IDO1 expression and stratified for the co-absence of IFNG and IFNB1 (green), absence of IFNG and expression of IFNB1 (purple), expression of IFNG and absence of IFNB1 (blue) and co-expression for both IFNG and IFNB1 (red). (B) Comparison of IDO1 mRNA levels among GBM specimens co-absent for IFNG and IFNB1 (green), absent of IFNG and expression of IFNB1 (purple), expression of IFNG and absent of IFNB1 (blue) and co-expression for both IFNG and IFNB1 (red). Sample size (n) of each classification group is same as in (A). (C) KM analysis of GBM patients based on the stratification of IFNG and IFNB1 as performed in (A, B). (D) Classification of 172 GBM specimens based on the expression levels of CD3E and CD8A using the calculated cutoff values (left panel, pie chart) and IDO1 mRNA expression (central panel, bar graph) as well as KM analysis (right panel, survival curves) based on this classification. (E) 172 GBM samples were first classified into 4 groups based on IDO1 and IFNG expression status (undetectable = 0 or detectable > 0), then a frequency analysis was further performed using the same CD3E and CD8A-based stratification as in (A) within each of the four groups. Sample size (n) is the same as in (D). Canonical correlation analysis of (F) CD8+ T cell marker genes, CD3E and CD8A, with those of regulatory T cells (Treg) cells, CD3E, CD4, CD25 and FOXP3, as well as (G) Treg marker genes with IDO1 mRNA expression. Each blue spot represents a GBM patient data point. A regression line was fitted to the dot plot. *: P < 0.05, **: P < 0.01, ***: P < 0.001, ****: P < 0.0001.
TCGA analysis for human T cell-specific surface marker, CD3E, as well as Tc-associated surface marker, CD8A, was next assessed among the 172 patient-resected GBM samples and its correlation with IDO1, IFNG and patient survival. Sample stratification identified that 56% of GBM possess low CD3E and CD8A mRNA levels, 29% consist of low CD3E with high CD8A mRNA levels and 15% contain high CD3E and high CD8A mRNA levels (Fig. 4D). Notably, high CD3E and CD8A mRNA levels were correlated with higher IDO1 and IFNG mRNA levels, as well as decreased patient survival (P < 0.01). These results suggest that greater T cell infiltration of tumor is associated with higher IDO1 and IFNG expression, as well as lower overall survival of GBM patients.
Based on the well-characterized association of activated CD8+ T cells and IFNγ expression, we investigated potential associations between the expression of IFNG, CD3E and CD8A mRNA. Frequency analysis indicated that GBM specimens lacking in IDO1 expression also lacked detectable CD3E and CD8A T cell signatures (n=12), irrespective of tumor IFNG expression (Fig. 4E). Conversely, GBM specimens with high CD3E and CD8A transcript levels expressed detectable IDO1, with a higher frequency of specimens co-expressing IFNG (n=20). We also examined the association between markers for CD8+ T cells and Treg (Fig. 4F), as well as IDO1 mRNA and Treg (Fig. 4G), confirming a positive relationship among both correlative groups.
T cells directly regulate IDO1 expression in human GBM
To further explore associations between T cells and IDO1 expression in GBM, humanized immunocompetent mice (hNSG; NSG-SGM3-BLT) were intracranially-engrafted with human patient-derived GBM xenografts (PDX) 12 or 43 and treated with or without T cell depleting antibodies. Analysis of isolated GBM tumors, draining cervical lymph nodes (cLN) and spleen revealed the presence of both CD4+ and CD8+ human T cells that were significantly decreased when mice were co-administered T cell-depleting antibodies (Fig. 5A,B). The analysis of humanized immunocompetent mice engrafted U87 GBM revealed a predominantly human leukocyte composition in mouse spleen that was reflected in both the myeloid and lymphoid immune cell compartments (Supp. Fig. 5A). Similar to mice engrafted PDX GBM, analysis of U87 tumors confirmed the presence of human T cells in the GBM (Supp. Fig. 5B) as well as in the cLN and spleen (Supp. Fig. 5B), with a notable absence of human T cells in the contralateral brain hemisphere without tumor. Additionally, intratumoral CD4+ and CD8+ T cells were detectable among the CD3+ GBM-infiltrating T cells (Supp. Fig. 5C), with an increased frequency of CD3+CD4+ T cells in association with markers indicative of immunosuppressive Treg, as compared to the cLN (Supp. Fig. 5D). CD3−CD45+ human myeloid cells were also enriched in brain tumors, as compared to peripheral secondary lymphoid organs. Treatment with CD4 or CD8 mAbs decreased U87 GBM-infiltrating T cell levels (Supp. Fig. 6), confirming the availability of multiple GBM models for investigating the interactions between human brain tumor and human immune cells, in situ.
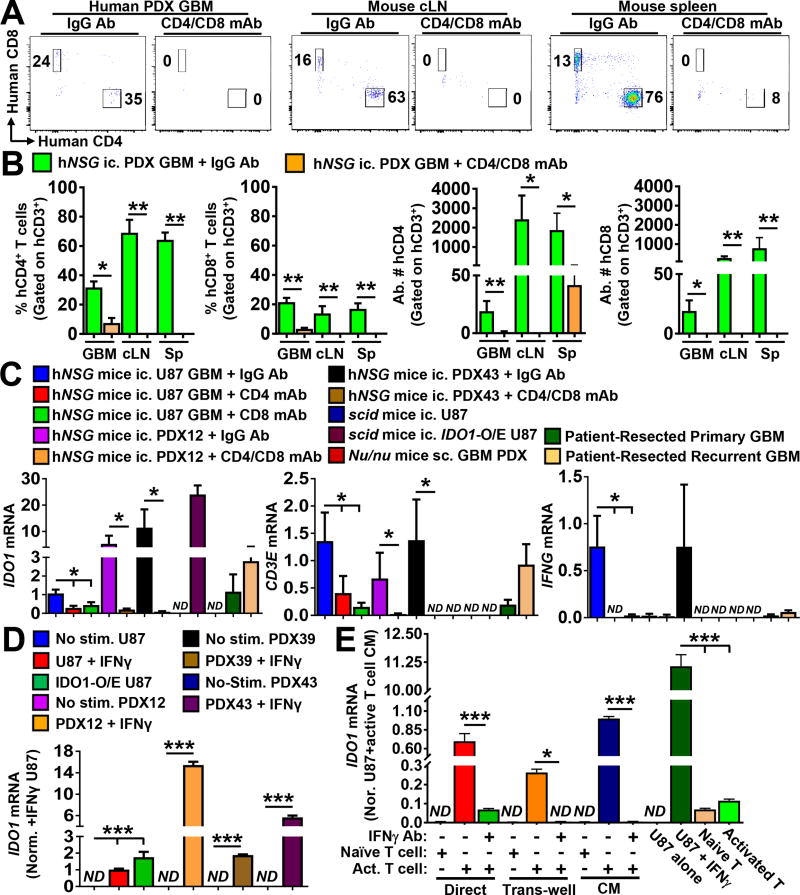
Figure 5. Activated human T cells directly increase IDO1 expression in human glioblastoma (GBM).
Humanized mice reconstituted with human immune cells (NSG-SGM3-BLT) were intracranially-engrafted patient-derived human GBM xenografts 12 or 43 (PDX12 or PDX43, respectively). (A) Representative flow plots of intracranial PDX GBM tumors, draining cervical lymph nodes (cLN) or spleen isolated at the time of symptomatic onset are shown for mice treated with IgG antibodies (Ab) or CD4 and CD8 T cell-depleting monoclonal antibodies (mAb). Data are representative of one mouse from each treatment group and tissue type. (B) Quantification of flow cytometric data of the frequency and absolute number for CD4+ and CD8+ T cells in the GBM, draining cLN and spleen of IgG Ab control- or CD4/CD8 mAb-treated humanized mice engrafted intracranial PDX12 or PDX43 GBM (n=5/group). (C) mRNA expression levels for human IDO1, CD3E or IFNG was quantified and compared among isolated intracranial human GBM (n=4–9/group). Unmodified U87, U87 modified to constitutively express human IDO1 cDNA (IDO1-O/E U87), PDX12, PDX39, PDX43, or patient-resected primary and recurrent GBM were included in the comparison. Intracranially (ic.)- or subcutaneously (sc.)-engrafted tumors were resected from T cell-deficient mice (Nu/nu or scid) or humanized mice with or without treatment of T cell depleting antibodies between 14 – 21 days post-intracranial injection. (D) In vitro expression analysis of human IDO1 mRNA in different GBM cells with or without addition of human interferon-gamma (IFNγ). Data represent pooled data from four independent experiments. (E) Detection of human IDO1 mRNA in the human GBM -T cell co-culture system, in vitro. CD3+ human T cells were isolated under positive selection from GBM patient PBMCs for the co-culture experiment. Human IDO1 mRNA levels were analyzed in U87 GBM cells under different treatment conditions, or naïve and activated T cells were analyzed in isolation and measured by real-time RT-PCR. Data were compiled from three independent experiments. hNSG: immunocompetent humanized mice (NSG-SGM3-BLT); SCID: NOD-scid mice; Nu/nu: nude mice; ND: not detectable. *: P < 0.05, **: P < 0.01, ***: P < 0.001.
Having validated the presence of human T cells in humanized mice with intracranial human PDX or U87 GBM, we further explored the hypothesis that T cells directly increase IDO1 expression in human glioma. Results from mice depleted for CD4+ and CD8+ T cells confirm that immune cell neutralization does not affect tumor growth (Supp. Fig. 7), and that T cell depletion decreases tumor IDO1 mRNA levels (P < 0.001, respectively) (Fig. 5C). Similarly, human IDO1 mRNA is undetectable in unmodified GBM isolated from T cell-deficient mice, but present in GBM tumors engineered to express human IDO1 cDNA (IDO1 O/E U87) and engrafted into mice without human T cells, as well as in surgically-resected GBM patient tumor. Expression for the T cell-specific marker, CD3E, as well as the T cell effector cytokine, IFNG, is readily detectable in normal hNSG mice, but is absent or decreased in the majority of GBM engrafted into mice depleted or deficient for human T cells. The analysis of subcutaneously-propagated human PDX GBM further confirmed the lack of IDO1 expression in T cell-deficient nude mice, although the transcript is potently induced after stimulation of PDX GBM cells with human IFNγ, ex vivo (Fig. 5D). In vitro GBM patient T cell:U87 GBM cell co-cultures confirmed that IDO1 is induced by activated T lymphocytes in an IFNγ-dependent manner (Fig. 5E; P < 0.001; Supp. Fig. 8B). Taken together, these data support the hypothesis that, GBM-infiltrating T cells directly increase immunosuppressive IDO1 expression in human GBM.
DISCUSSION
IDO1 is recognized as an important mediator of immunosuppression in cancer (12, 22, 34, 35). To address the potential usefulness of IDO1 expression as a prognostic tool for glioma patients, we compared the sensitivity of protein- and mRNA-based detection methods for IDO1 in human GBM. Our data indicate that the quantification of tumor IDO1 mRNA expression yields high prognostic value for GBM patient outcome. This also proved to be the case for grade II and grade III glioma. mRNA results also showed that IDO1 expression: (i) increases with glioma grade; (ii) is distinct among GBM molecular subtypes; (iii) is decreased in IDH mutant when compared to IDH wild-type tumors; (iv) is correlated with the expression of other immunosuppressive mediators; and (vii) intratumorally increases in association with increased expression for Tc and Treg markers.
To evaluate relationships between IDO1 expression and tumor-infiltrating T lymphocytes, our use of humanized immunocompetent (NSG-SGM3-BLT) mice intracranially-engrafted with human GBM showed that tumor IDO1 expression is influenced by infiltrating CD4+ and CD8+ T cells. In contrast to the general assumption that IFNγ is the primary regulator of IDO1 expression, our analysis of 172 patient-resected GBM revealed that 59% of tumor specimens are undetectable for IFNG (Fig. 4A). Coincidently, PDX12 tumor IDO1 expression increased in a T cell-dependent manner that was not associated with a commensurate increase in IFNγ levels, in situ (Fig. 5C). These findings confirm the existence of additional mechanisms responsible for IDO1 gene expression in GBM and is the subject of an ongoing investigation by our group. Therefore, we could conclude that, IFNG is sufficient, but not required, for increasing IDO1 levels in human GBM.
We found significant correlations between GBM IDO1 levels, decreased patient survival, and increased marker expression for Tc and Treg (Fig. 4D, E, F, G). This finding aligns with our previous observations in syngeneic, immunocompetent, intracranial mouse GBM models which showed that: (i) tumor cell IDO1 facilitates local Treg accumulation; (ii) Tc and Treg coincidently infiltrate IDO1-expressing tumors; and (iii) tumor cell IDO1 expression decreases animal subject survival (13, 36). Supplementary Fig. 9 presents a model showing tumor cell - T cell interactions and supports the hypothesis that GBM-infiltrating Tc facilitate IFNγ-dependent increases in tumor cell IDO1, followed by the intratumoral accumulation of immunosuppressive Treg. This model highlights the negative repercussions of inflammatory enhancement in human GBM and is supported by the data in Fig. 3A, B, showing the inverse association between the presence of mIDH and decreased intratumoral IDO1 levels. Given that mIDH suppresses CD8+ Tc cell accumulation in glioma (37), in addition to the favorable prognosis it carries for GBM patient survival (38), it is tempting to speculate that the mIDH suppression of GBM-infiltrating Tc increases patient survival by virtue of the additional suppression of IDO1 and Treg accumulation in those patient tumors.
Our observations suggest that future immunotherapeutic strategies incorporating IDO1 inhibition into a GBM patient treatment regimen may be more effective in treating classical, mesenchymal and neural GBM subtypes, when compared to the lower IDO1-expressing proneural GBM and glioma grades II and III. These data further infer that GBM patients with high baseline tumor-infiltrating T cell levels, or those who enroll on T cell enhancing therapeutic treatments, may maximally benefit from the addition of an IDO1 inhibitor. Our data also indicate a gene expression correlation between IDO1 and other mediators of immunosuppression (Fig. 3D, E), highlighting the potential for enhancing anti-tumor effects when combining IDO1 inhibition with a blockade of multiple immune checkpoint targets.
In conclusion, we show for the first time that IDO1 mRNA levels are a useful, sensitive and prognostic predictor of grade II – IV glioma patient survival. We also introduce a new, enzymatically active modified U87 human GBM cell line that expresses an C-terminus HA tag conjugated to human IDO1 (Supp. Fig. 2). We further present novel data showing differential IDO1 exon expression among patient-resected GBM (Fig. 3; Supp. Table 2), highlighting complex transcriptional mechanisms that are likely associated with the expression of multiple isoforms. Analysis of IDO1 expression in GBM isolated from humanized mice engrafted PDX43 and U87, versus PDX12 GBM, provide new comparative platforms for investigating IFNγ-independent mechanisms of IDO1 regulation by tumor-infiltrating T cells. Given the past preclinical strategies demonstrating the survival benefit of pharmacologic IDO1 blockade in preclinical brain tumor models (39), in addition to clinical trials evaluating IDO1 targeting strategies in glioma patients (NCT02052648, NCT02764151), the data collectively suggest that future treatment approaches designed to enhance T cell-mediated antitumor immunity may maximally benefit from the further addition of an IDO1 inhibitor.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Glioblastoma (GBM) is the most common primary malignant brain tumor in adults with a dismal prognosis. Increased IDO1 expression is associated with decreased survival among glioma patients, but the significance of IDO1 expression exclusively in GBM patients, has yet to demonstrated. This study reveals that, high IDO1 mRNA levels, as assessed using the Hi-RNA-Sequencing Illumina platform, are consistently associated with decreased GBM patient survival. It also shows that, infiltrating human T cells directly increase IDO1 expression in GBM. Given the growing number of clinical trials aimed at immunotherapeutically enhancing T cell functions in GBM, these data suggest that the further inhibition with an IDO1 inhibitor, may increase the number of individuals that respond favorably in the clinic.
Acknowledgments
Funding: This work was supported by NIH grants R00 NS082381 (D.A.W.) and R01 NS097851-01 (D.A.W.), the Cancer Research Institute – Clinic and Laboratory Integration Program (D.A.W.), the Robert H. Lurie Comprehensive Cancer Center – Zell Scholar Program of the Zell Family Foundation Gift (D.A.W.) and Northwestern Brain Tumor Institute.
We thank the Northwestern University Center for Advanced Microscopy (generously supported by NCI CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center) and the Northwestern University Flow Cytometry Core Facility (generously supported by Cancer Center Support Grant, NCI CA060553) for their technical support of microscope image acquisition and flow cytometry, respectively. Human tissues and histologic analyses were supported by the Northwestern Nervous System Tumor Bank. We thank Mr. Michael Gallagher for his illustration expertise that led to the creation and rendering of Figure 9.
Footnotes
Conflict of Interest Disclosure: None.
References
- 1.Adamson C, Kanu OO, Mehta AI, Di C, Lin N, Mattox AK, et al. Glioblastoma multiforme: a review of where we have been and where we are going. Expert Opin Investig Drugs. 2009;18:1061–83. doi: 10.1517/13543780903052764. [DOI] [PubMed] [Google Scholar]
- 2.Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–4. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 3.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 4.SongTao Q, Lei Y, Si G, YanQing D, HuiXia H, XueLin Z, et al. IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Sci. 2012;103:269–73. doi: 10.1111/j.1349-7006.2011.02134.x. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, Su H-k, Zhao H-f, Chen Z-p, To S-sT. Progress in the application of molecular biomarkers in gliomas. Biochemical and biophysical research communications. 2015;465:1–4. doi: 10.1016/j.bbrc.2015.07.148. [DOI] [PubMed] [Google Scholar]
- 6.Aldape K, Zadeh G, Mansouri S, Reifenberger G, von Deimling A. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129:829–48. doi: 10.1007/s00401-015-1432-1. [DOI] [PubMed] [Google Scholar]
- 7.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. The New England journal of medicine. 2010;363:711–23. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369:122–33. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rizvi NA, Mazieres J, Planchard D, Stinchcombe TE, Dy GK, Antonia SJ, et al. Activity and safety of nivolumab, an anti-PD-1 immune checkpoint inhibitor, for patients with advanced, refractory squamous non-small-cell lung cancer (CheckMate 063): a phase 2, single-arm trial. Lancet Oncol. 2015;16:257–65. doi: 10.1016/S1470-2045(15)70054-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antonia SJ, Lopez-Martin JA, Bendell J, Ott PA, Taylor M, Eder JP, et al. Nivolumab alone and nivolumab plus ipilimumab in recurrent small-cell lung cancer (CheckMate 032): a multicentre, open-label, phase 1/2 trial. Lancet Oncol. 2016;17:883–95. doi: 10.1016/S1470-2045(16)30098-5. [DOI] [PubMed] [Google Scholar]
- 11.Yasui H, Takai K, Yoshida R, Hayaishi O. Interferon enhances tryptophan metabolism by inducing pulmonary indoleamine 2,3-dioxygenase: its possible occurrence in cancer patients. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:6622–6. doi: 10.1073/pnas.83.17.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uyttenhove C, Pilotte L, Theate I, Stroobant V, Colau D, Parmentier N, et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med. 2003;9:1269–74. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 13.Wainwright DA, Balyasnikova IV, Chang AL, Ahmed AU, Moon KS, Auffinger B, et al. IDO expression in brain tumors increases the recruitment of regulatory T cells and negatively impacts survival. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:6110–21. doi: 10.1158/1078-0432.CCR-12-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inaba T, Ino K, Kajiyama H, Shibata K, Yamamoto E, Kondo S, et al. Indoleamine 2,3-dioxygenase expression predicts impaired survival of invasive cervical cancer patients treated with radical hysterectomy. Gynecologic oncology. 2010;117:423–8. doi: 10.1016/j.ygyno.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 15.Laimer K, Troester B, Kloss F, Schafer G, Obrist P, Perathoner A, et al. Expression and prognostic impact of indoleamine 2,3-dioxygenase in oral squamous cell carcinomas. Oral oncology. 2011;47:352–7. doi: 10.1016/j.oraloncology.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Speeckaert R, Vermaelen K, van Geel N, Autier P, Lambert J, Haspeslagh M, et al. Indoleamine 2,3-dioxygenase, a new prognostic marker in sentinel lymph nodes of melanoma patients. European journal of cancer (Oxford, England : 1990) 2012;48:2004–11. doi: 10.1016/j.ejca.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Jia Y, Wang H, Wang Y, Wang T, Wang M, Ma M, et al. Low expression of Bin1, along with high expression of IDO in tumor tissue and draining lymph nodes, are predictors of poor prognosis for esophageal squamous cell cancer patients. International journal of cancer Journal international du cancer. 2015;137:1095–106. doi: 10.1002/ijc.29481. [DOI] [PubMed] [Google Scholar]
- 18.Masaki A, Ishida T, Maeda Y, Suzuki S, Ito A, Takino H, et al. Prognostic Significance of Tryptophan Catabolism in Adult T-cell Leukemia/Lymphoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:2830–9. doi: 10.1158/1078-0432.CCR-14-2275. [DOI] [PubMed] [Google Scholar]
- 19.Ferdinande L, Decaestecker C, Verset L, Mathieu A, Moles Lopez X, Negulescu AM, et al. Clinicopathological significance of indoleamine 2,3-dioxygenase 1 expression in colorectal cancer. British journal of cancer. 2012;106:141–7. doi: 10.1038/bjc.2011.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Théate I, van Baren N, Pilotte L, Moulin P, Larrieu P, Renauld J-C, et al. Extensive Profiling of the Expression of the Indoleamine 2,3-Dioxygenase 1 Protein in Normal and Tumoral Human Tissues. Cancer Immunology Research. 2015;3:161–72. doi: 10.1158/2326-6066.CIR-14-0137. [DOI] [PubMed] [Google Scholar]
- 21.Mitsuka K, Kawataki T, Satoh E, Asahara T, Horikoshi T, Kinouchi H. Expression of indoleamine 2,3-dioxygenase and correlation with pathological malignancy in gliomas. Neurosurgery. 2013;72:1031–8. doi: 10.1227/NEU.0b013e31828cf945. discussion 8–9. [DOI] [PubMed] [Google Scholar]
- 22.Wainwright DA, Balyasnikova IV, Chang AL, Ahmed AU, Moon K-S, Auffinger B, et al. IDO Expression in Brain Tumors Increases the Recruitment of Regulatory T Cells and Negatively Impacts Survival. Clinical Cancer Research. 2012;18:6110–21. doi: 10.1158/1078-0432.CCR-12-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giannini C, Sarkaria JN, Saito A, Uhm JH, Galanis E, Carlson BL, et al. Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neuro-oncology. 2005;7:164–76. doi: 10.1215/S1152851704000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarkaria JN, Carlson BL, Schroeder MA, Grogan P, Brown PD, Giannini C, et al. Use of an orthotopic xenograft model for assessing the effect of epidermal growth factor receptor amplification on glioblastoma radiation response. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006;12:2264–71. doi: 10.1158/1078-0432.CCR-05-2510. [DOI] [PubMed] [Google Scholar]
- 25.Budczies J, Klauschen F, Sinn BV, Gyorffy B, Schmitt WD, Darb-Esfahani S, et al. Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PloS one. 2012;7:e51862. doi: 10.1371/journal.pone.0051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theate I, van Baren N, Pilotte L, Moulin P, Larrieu P, Renauld JC, et al. Extensive profiling of the expression of the indoleamine 2,3-dioxygenase 1 protein in normal and tumoral human tissues. Cancer immunology research. 2015;3:161–72. doi: 10.1158/2326-6066.CIR-14-0137. [DOI] [PubMed] [Google Scholar]
- 27.Opitz CA, Litzenburger UM, Sahm F, Ott M, Tritschler I, Trump S, et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478:197–203. doi: 10.1038/nature10491. [DOI] [PubMed] [Google Scholar]
- 28.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–8. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–83. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carlin JM, Borden EC, Sondel PM, Byrne GI. Biologic-response-modifier-induced indoleamine 2,3-dioxygenase activity in human peripheral blood mononuclear cell cultures. J Immunol. 1987;139:2414–8. [PubMed] [Google Scholar]
- 32.Taylor MW, Feng GS. Relationship between interferon-gamma, indoleamine 2,3-dioxygenase, and tryptophan catabolism. The FASEB Journal. 1991;5:2516–22. [PubMed] [Google Scholar]
- 33.Miyazaki T, Moritake K, Yamada K, Hara N, Osago H, Shibata T, et al. Indoleamine 2,3-dioxygenase as a new target for malignant glioma therapy. Laboratory investigation. J Neurosurg. 2009;111:230–7. doi: 10.3171/2008.10.JNS081141. [DOI] [PubMed] [Google Scholar]
- 34.Muller AJ, Sharma MD, Chandler PR, DuHadaway JB, Everhart ME, Johnson BA, et al. Chronic inflammation that facilitates tumor progression creates local immune suppression by inducing indoleamine 2,3 dioxygenase. Proceedings of the National Academy of Sciences. 2008;105:17073–8. doi: 10.1073/pnas.0806173105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muller AJ, DuHadaway JB, Chang MY, Ramalingam A, Sutanto-Ward E, Boulden J, et al. Non-hematopoietic expression of IDO is integrally required for inflammatory tumor promotion. Cancer immunology, immunotherapy : CII. 2010;59:1655–63. doi: 10.1007/s00262-010-0891-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhai L, Ladomersky E, Dostal CR, Lauing KL, Swoap K, Billingham LK, et al. Non-tumor cell IDO1 predominantly contributes to enzyme activity and response to CTLA-4/PD-L1 inhibition in mouse glioblastoma. Brain, Behavior and Immunity. 2017 doi: 10.1016/j.bbi.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohanbash G, Carrera DA, Shrivastav S, Ahn BJ, Jahan N, Mazor T, et al. Isocitrate dehydrogenase mutations suppress STAT1 and CD8+ T cell accumulation in gliomas. The Journal of clinical investigation. 2017;127:1425–37. doi: 10.1172/JCI90644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nobusawa S, Watanabe T, Kleihues P, Ohgaki H. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:6002–7. doi: 10.1158/1078-0432.CCR-09-0715. [DOI] [PubMed] [Google Scholar]
- 39.Wainwright DA, Chang AL, Dey M, Balyasnikova IV, Kim C, Tobias AL, et al. Durable therapeutic efficacy utilizing combinatorial blockade against IDO, CTLA-4 and PD-L1 in mice with brain tumors. Clinical Cancer Research. 2014 doi: 10.1158/1078-0432.CCR-14-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.