Abstract
Stem cell leukemia/lymphoma syndrome (SCLL) is driven by constitutive activation of chimeric FGFR1 kinases generated by chromosome translocations. We have shown that FGFR inhibitors significantly suppress leukemia and lymphoma development in vivo, and cell viability in vitro. Since resistance to targeted therapies is a major reason for relapse, we developed FGFR1-overexpressing mouse and human cell lines that are resistant to the specific FGFR inhibitors AZD4547 and BGJ398, as well as non-specific inhibitors, such as ponatinib, TKI258 and E3810. Two mutually exclusive mechanisms for resistance were demonstrated; an activating V561M mutation in the FGFR1 kinase domain and mutational inactivation of PTEN resulting in increased PI3K/AKT activity. Ectopic expression of PTEN in the PTEN-mutant cells resensitizes them to FGFR inhibitors. Treatment of resistant cells with BGJ398, in combination with the BEZ235 PI3K inhibitor, shows an additive effect on growth in vitro and prolongs survival in xenograft models in vivo. These studies provide the first direct evidence for both the involvement of the FGFR1 V561M mutation and PTEN inactivation in the development of resistance in leukemias overexpressing chimeric FGFR1. These studies also provide a potential strategy to treat leukemias and lymphomas driven by FGFR1 activation that become resistant to FGFR1 inhibitors.
INTRODUCTION
The fibroblast growth factor receptors (FGFR1–4) play an important role in normal embryonic development, regulating cellular proliferation, survival, migration and differentiation in a variety of cell types, including the hematopoietic system.1, 2 Abnormalities of FGF receptors, however, are seen in nearly all common types of cancers where either gene amplification, or activating mutation and rearrangements, have been found in subsets of lung, bladder and breast cancers as well as leukemias and lymphomas.3, 4 The only neoplasm in which FGFR1 is consistently activated, however, is the stem cell leukemia/lymphoma (SCLL) syndrome,5 which is characterized by chromosome translocations that lead to a constitutively activated, ligand independent, chimeric, FGFR1 kinase.6, 7 These patients develop a myeloproliferative disorder and may concurrently develop either T- or B- cell lymphomas, and 80% of patients eventually progress to AML.5 We and others have shown that these various leukemias are all dependent on FGFR1 for their survival, since treatment with an FGFR1 inhibitor such as ponatinib or BGJ398 leads to suppression of FGFR1 activation and reduced cell viability in vitro and in vivo.8–10 Recently, a patient with BCR-FGFR1 driven SCLL treated with ponatinib showed partial morphological remission after 12 weeks treatment and markedly reduced disease burden following bone marrow transplantation, demonstrating the importance of FGFR1 kinase activity in a clinical setting.11 As a result of our development of mouse models of SCLL8, 12–15 several cell lines expressing different chimeric kinases have been developed and have proved useful in evaluating the effectiveness of various anti-FGFR1 drug regimens.8, 9
To date, several different FGFR inhibitors have been evaluated in SCLL models including PKC412,16 E381017 and TKI258,18, 19 which are relatively broad-spectrum kinase inhibitors. Ponatinib, which was originally developed to target the BCR-ABL, T315I mutation,20 was shown to also inhibit FGFR1 at low concentrations and is particularly effective against FGFR1 driven neoplasms8 but also inhibits other kinases such as KIT and FLT3. More recently, however, more specific FGFR inhibitors such as ADZ4547,21 JNJ4275649322 and BGJ39823 have been developed. We have evaluated the relative efficiency of these different FGFR1 inhibitors in suppressing growth of leukemic cells lines expressing chimeric FGFR1 kinases and demonstrated that they are highly effective in suppressing in vitro growth at nanomolar concentrations,9 and have demonstrated the effectiveness of BGJ398 against SCLL syndromic and de novo FGFR1 overexpressing AML in vivo, paving the way for clinical trials using FGFR1 inhibitors in subtypes of AML overexpressing FGFR1.
One of the ultimate consequences of approaches targeting a single protein, however, is the almost inevitable development of resistance to that inhibitor, through a variety of mechanisms. It is important, therefore, to identify potential mechanisms of resistance, so that alternative therapeutic strategies are available once resistance arises. To address this possibility, we have now derived cell lines, carrying various constitutively activated FGFR1 kinases seen in SCLL, that are resistant to FGFR1 inhibitors, and investigated the mechanisms of the resistance. Two different, mutually exclusive mechanisms were identified, the first involves mutation of the FGFR1 tyrosine kinase domain and the second involves mutational inactivation of the PTEN gene, which resulted in consequential upregulation of PI3K kinase. Targeting PI3K in these resistant cells lines led to decreased viability of leukemic cells and, in combination with FGFR1 inhibitors, showed an additive effect on growth inhibition. Re-expressing PTEN in the PTEN-mutant cell lines reverted them to FGFR1 inhibitor sensitivity. In vivo, the combined treatment with drugs inhibiting both FGFR1 and PI3K led to suppression of leukemogenesis and significantly prolonged survival. These studies suggest a possible future application of this drug combination for resistant, FGFR1-driven leukemias.
MATERIALS AND METHODS
Establishment of resistant cell lines
All cell lines were grown in RPMI 1640 medium supplemented with 5% fetal calf serum (Hyclone, Logan, UT) and 1% penicillin-streptomycin. FGFR-inhibitor resistant leukemia sub-lines were established by exposing parental cells to a regimen of progressively increasing concentrations of ponatinib. The authenticity of the KG1 cell line was confirmed by western blot demonstration of the presence the 60kD FGFR1OP1-FGFR1 kinase as described previously12.
Stable expression of human PTEN in resistant cells
The wild type, PTEN mouse ORF clone (Cat#MR206321) was purchased from OriGene (www.origene.com) and subsequently subcloned into the pLenti-C-Myc-DDK-IRES-tRFP vector (OriGene, plasmid #PS100080). The PTEN deletion cells were transduced with lentiviral particles carrying wild type PTEN using standard procedures. Cells stably transduced with exogenous PTEN (RFP positive) were isolated using fluorescence-activated cell sorting.
Molecular analyses and Western blot analysis
Total RNA was isolated using Trizol (Invitrogen, Waltham, MA) and retro-transcribed with a QuantiTect reverse transcription kit (Qiagen, Valencia, CA). Amplification was performed using standard PCR (New England Bio, MA) in combination with specific primers and conditions (available upon request). Proteins were isolated as described previously.8 Whole-cell lysates (30 µg) were separated using SDS-PAGE and immunoblotted with specific antibodies. The anti-phospho-FGFR1 and anti-FGFR1 antibodies were purchased from Abcam (Cambridge, MA) and Santa Cruz Biotechnology (Paso Robles, CA), respectively. All other antibodies were obtained from Cell Signaling (Danvers, MA).
Reverse phase protein array (RPPA)
RPPAs were constructed and analyzed as described previously.24 Briefly, denatured cellular lysates were arrayed on nitrocellulose membranes at various concentrations and probed with antibodies (n=167 for mouse and n=218 for human) that recognize most common signaling pathways. A complete list of antibodies used is available (www.bcm.edu). Signals were captured by tyramide dye deposition and a DAB colorimetric reaction, quantified and normalized to various control lysates and peptides on the array as described.24
Animal studies and drug regimens
NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice (obtained from the Jackson Laboratories), were maintained as a breeding colony. All experiments were conducted under Augusta University IACUC approved protocols. Female, 6–8 week old mice were used in all xenograft experiments. Mice were engrafted with 1–2 × 106 cells via tail vein injection. All mice were treated with either drug, or vehicle control (PEG300:acetic buffer = 1:1), orally using a gavage needle once per day. All treatments were performed 5 days per week for 4 weeks.
Ponatinib was provided by Ariad Pharmaceuticals Inc. (Cambridge, MA). PD173074 was purchased from Cayman Chemical, AZD4547 and BGJ398 from ChemieTek, JNJ-42756493 from Active Biochem and TKI258 from LC Laboratories. E3810 was provided by EOS pharmaceuticals. All drugs were dissolved in DMSO and stored at −80°C before use. For drug treatments, cells were seeded at 3,000–10,000 cells/well, depending on the cell line, in 96-well plates and incubated overnight. Cells were then treated with either DMSO (control) or different FGFR inhibitors as indicated in the results section at concentrations defined by the experiments. Cell viability was determined using CellTiter-Glo® luminescence cell viability kits (Promega) and a SpectraMax® M5e (Molecular Probes) luminescence plate reader as described previously.8
RESULTS
Generation of cell lines resistant to FGFR1 inhibitors
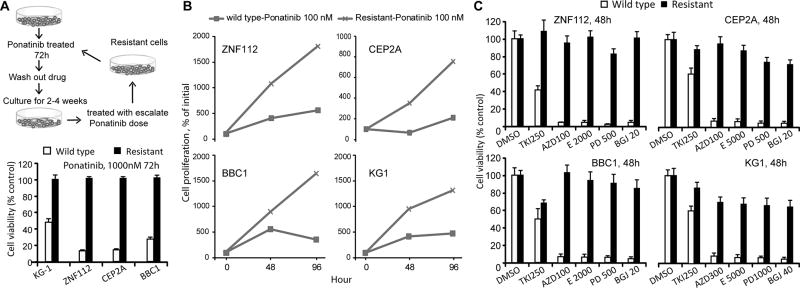
SCLL patients typically develop a myeloproliferative disorder that progresses to AML, but may also develop T-cell or B-cell lymphomas depending on the specific chimeric kinase present.5 We have established murine cell lines representative of all of these lineages. ZNF112 was derived from a T-lymphoma expressing the ZMYM2-FGFR1 chimeric kinase,12 BBC1 was derived from a B-cell lymphoma in a model for BCR-FGFR1 kinase15 and CEP2A was derived from an AML expressing the CNTRL-FGFR1 chimeric kinase.13 In addition, we have characterized a murine model of the FGFR1OP1-FGFR1 kinase25 present in the KG1 human cell line.26 Ponatinib (50–100 nM) suppresses FGFR1 activation in all of these cell lines with concomitant growth suppression,8 demonstrating that their survival is dependent on the constitutive activation of the chimeric kinases. Since development of resistance is one of the inevitable consequences of prolonged treatment with single drug regimens, we established derivative cell lines that were resistant to ponatinib through exposure of the parental cells to step-wise increases in concentration as illustrated in Figure 1A. After 2 years of this successive selection process, derivative cell lines were established which were resistant to at least ~1000 nM ponatinib (Figure 1A lower panel) as shown by cell proliferation assays (Figure 1B).
Figure 1. Generation of cell lines resistant to FGFR inhibitors.
Scheme (A, above) for the development of resistance to ponatinib where, after 2 years of selection, four different cell lines overexpressing FGFR1 kinase were developed which are resistant to >1000 nM (below). At a 100 nM concentration, which significantly inhibits proliferation in the parental cells, the resistant cells still show active proliferation (B). Although resistance was induced following exposure to ponatinib, the same cells are also resistant to other FGFR1 inhibitors; TKI258 (TKI), AZD4547 (AZD), E3810 (E). PD173074 (PD) and BGJ398 (BGJ) at the concentrations (nM) indicated in (C).
To determine whether the resistance in these cell lines was specific to ponatinib, or led to a pan-resistance to FGFR1 inhibitors, we challenged the ponatinib resistant cell lines with the relatively more specific FGFR inhibitors, AZD4547 (2000nM) and BGJ398 (3000 nM), as well as two less specific FGFR inhibitors, TKI258 and E3810. As shown in Figure 1C, treatment of the cells expressing chimeric FGFR1 kinases with these FGFR inhibitors, at a concentration 10× their IC50, did not significantly inhibit cell proliferation when compared with the parental cells, suggesting common mechanisms for all inhibitors.
Mechanisms of FGFR1 inhibitor resistance involves FGFR1 mutations and PTEN deletion
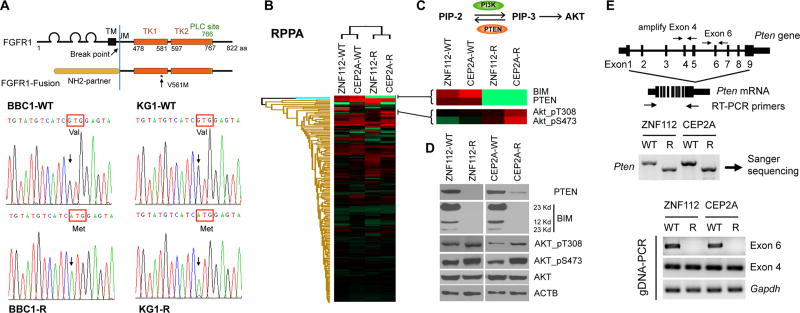
To establish the mechanism that underlies the resistance to FGFR1 inhibitors, we used Sanger sequencing of RT-PCR products derived from the tyrosine kinase domains of the chimeric FGFR1 kinases in the parental and resistant cells. All four lines showed the wild type sequence in the parental, drug sensitive cell lines (Figure 2A). In contrast, a single, homozygous G→A mutation in the kinase domain, leading to a V561M missense change, was identified in the BBC1 and KG1 resistant cell lines. ZNF112 and CEP2A cells did not show this mutation in the resistant cells.
Figure 2. Mechanisms of FGFR1 inhibitor resistance involves FGFR1 mutation and PTEN deletion.
Sanger sequencing of RT-PCR products derived from the kinase domains of the chimeric kinases in the parental (WT) and resistant cells (R) of BBC1 and KG1 cell lines (A). Heat map of relative protein levels identified using reverse phase protein array (RPPA) analysis using parental ZNF112 and CEP2A cells (WT) compared with their resistant (R) counterparts (B) shows significant down regulation of the PTEN and BIM proteins (C) with upregulation of phospho-AKT. These observations were confirmed using western blotting (D) where the phospho-activated protein is elevated compared with overall proteins levels. (E) Location of primers used to amplify individual exons (above) and the full length mRNA (below) of PTEN. The resistant clones show a smaller length RT-PCR product compared with the parental (WT) cells (below left). Sequencing of the smaller product showed loss of exon 6 (data not shown) which was confirmed using exon specific amplification (below right).
To investigate the resistance mechanism in ZNF112 and CEP2A cells, we next performed an unbiased analysis of differential protein changes in the resistant cells compared with the parental cells, using reverse phase protein array (RPPA) analysis. This analysis surveys the levels of 167 mouse proteins involved in common signaling pathways.24 Although hierarchical cluster analysis of relative levels of proteins showed that the overall pattern was very similar between the parental cells and their resistant derivatives (Figure 2B), the PTEN and BIM proteins, while unaffected in BBC1 and KG1 resistant cells (data not shown), were significantly down-regulated in the ZNF112 and CEP2A resistant cells compared with the respective parental cells (Figure 2C). Consistent with down-regulation of PTEN, phosphorylation levels of AKT at amino acids T308 and S473 were enhanced in the resistant cells (Figure 2D). Western blot analysis using anti-PTEN or anti-BIM antibodies confirmed that levels of these proteins were remarkably reduced or lost (Figure 2D) in the resistant clones. To investigate the mechanism of PTEN suppression in the resistant ZNF112R and CEP2AR cells lines (Figure 2E), RT-PCR analysis of the full-length transcript clearly showed a smaller PTEN mRNA. Sanger sequencing of these RT-PCR products demonstrated homozygous deletions of exon 6 in PTEN in both cases (Figure 2E), which results in a predicted premature stop-codon resulting in loss of the PTEN protein. RPPA analysis also identified a remarkable decrease of BIM expression levels in both resistant cells lines (Figure 2D). The BIM protein (also known as BCL2L11) belongs to the BCL2 family, and functions as an apoptotic activator. To investigate whether decrease of BIM can lead to resistance to apoptosis, we treated three mouse resistant cell lines with the Nutlin, Etopside and Cisplatin apoptosis-inducing drugs, as well as Taxel which does not induce apoptosis. All cells were treated with individual drug at their GI50 for 48 hours where apoptosis levels were reduced in the resistant cells compared with the parental cells but not following treatment with Taxel (Supplementary Figure 1). Thus, decrease of BIM expression may also contribute to resistance to FGFR1 inhibitors in SCLL cells.
Ectopic expression of PTEN can re-sensitize the resistant cells to FGFR1 inhibitors
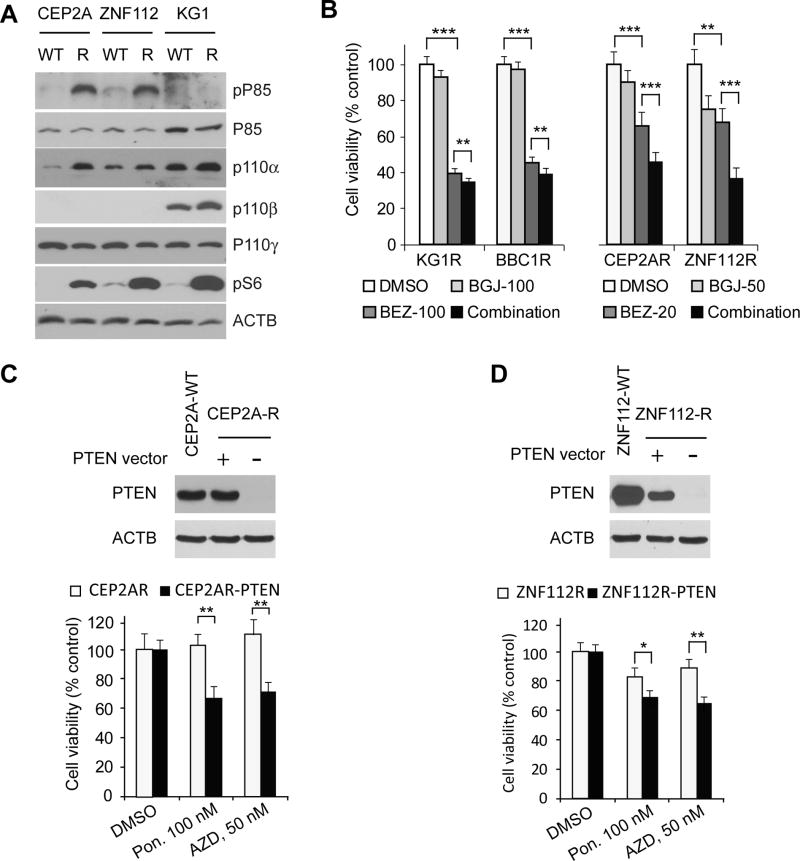
Since a consequence of PTEN loss is activation of its downstream target, PI3K, we used western blotting to analyze members of the PI3K signaling cascade. As shown in Figure 3A, the PI3K signaling pathway was activated in the most resistant cells compared with the parental cells, as shown by increased phosphorylation levels of either p85, p110α or p110β in the resistant cells (but p85 phosphorylation levels were comparable between resistant and parental KG1, and p110α levels were comparable between resistant and parental ZNF112 cells). Consistently, S6 phosphorylation levels were also significantly upregulated in the resistant cells compared with the parental cells (Figure 3A). Activation of PI3K signaling was less pronounced in BBC1 cells (data not shown) which is consistent with the variant nature of this particular rearrangement (see discussion). PI3K activation in KG1 resistant cells (without PTEN deletion) is also increased compared with wild type KG1 cells (Figure 3A).
Figure 3. Ectopic expression of PTEN can re-sensitize the resistant cells to FGFR1 inhibitors.
Analysis of protein expression levels downstream of PI3K (A) shows increased activation of p110α, p110β (the p110β antibody is specific for the human antigen) and pS6 in the resistant cells. Treatment of the resistant cells individually with either BGJ398 or BEZ235 show reduced cell viability, which is enhanced when both drugs are used in combination, ** p < 0.01, *** p < 0.001 (B). Re-expression of PTEN in resistant cells (C) leads to a recovery of sensitivity to ponatinib (Pon) and AZD4547 (AZD) in both CEP2A and ZNF112 cells (D).
Given the upregulation of PI3K signaling in the resistant cells, we reasoned that resistant cells treated with PI3K inhibitors may lead to growth inhibition. Indeed, in vitro drug treatment shows that the BEZ235 PI3K inhibitor, can significantly (p < 0.001) reduce proliferation in the resistant cells regardless of the specific chimeric kinase being expressed (Figure 3B). Furthermore, analysis of cell proliferation following combined treatment with BEZ235 and BGJ398 showed an additive effect (Figure 3B). To functionally characterize the effect further, we ectopically expressed PTEN in the CEP2AR and ZNF112R resistant cells (Figure 3C and 3D, top panel). When both the resistant derivative cells expressing exogenous PTEN were treated with ponatinib or AZD4547 for 48h, the resistant cells were resensitized to FGFR inhibitors (Figure 3C and 3D). Together, these results indicate that constitutive activation of the PI3K signaling pathway, resulting from PTEN deletion, is a significant mechanism leading to resistance to FGFR inhibitors in this cell system.
Simultaneously targeting FGFR and PI3K signaling pathways can overcome the resistance of FGFR1 inhibitors in vivo
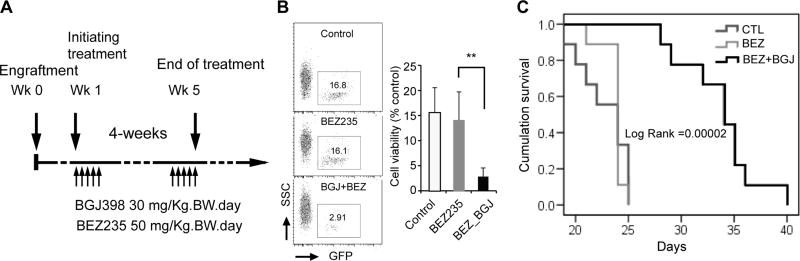
Loss of PTEN function leads to activation of the PI3K signaling pathway, and this was particularly evident in ZNF112R cells. Therefore, to investigate whether targeting PI3K signaling in these cells would suppress growth in vivo, we engrafted murine leukemia ZNF112R cells into NSGS mice and, one week post-engraftment, the mice were randomized into three groups (N = 10); vehicle (PEG300) control, BEZ235 (50 mg/kg body weight, previously shown to have no adverse effects27–29 at this dosage) and, based on our previous studies,9 combined BEZ235/BGJ398 (each 30 mg/kg body weight). The mice were treated on a 5-day on, 2-day off regimen for 4 consecutive weeks as illustrated in Figure 4A. The retrovirus used to transform murine stem cells carries a GFP marker expressed from an IRES downstream of the fusion kinases, which can be used to specifically identify leukemic cells.30 Two weeks post-treatment, blood samples were collected from the tail veins and analyzed using flow cytometry for the presence of GFP. As shown in Figure 4B, although there was a reduction in the percentage of GFP+ cells following treatment with BEZ235, albeit not significant, the percentage of GFP+ cells was significantly decreased in the group treated with the BGJ398-BEZ235 combination. This observation is consistent with the in vitro studies shown in figure 3B. Kaplan-Meier survival analysis further showed that the cohort treated with the drug combination showed a significantly prolonged survival compared with groups treated with either vehicle or BEZ235 alone (Figure 4C).
Figure 4. Simultaneously targeting FGFR1 and PI3K signaling pathways can overcome resistance to FGFR inhibitors in vivo.
Scheme outlining the drug treatment regimen (A). Flow cytometric analysis of peripheral blood samples from mice engrafted with GFP+ ZNF112R cells following treatment either (1) vehicle control (PEG300), (2) BEZ235 alone or (3) BEZ235 (BEZ) in combination with BGJ398 (BGJ) (B) demonstrates an additive effect with the drug combination. In vivo xenograft studies show that, while BEZ235 treatment alone does not significantly prolong survival in ZNF112 xenotransplanted mice compared with vehicle control (CTL) treated mice. Mice treated with the drug combination show significantly prolonged survival (C).
DISCUSSION
The main reason for failure of targeted cancer therapies, using single agents, is the emergence of resistant clones. Ideally, therefore, a second-line strategy for treatment should be identified that is available immediately to treat the resistant clone as determined by the mechanism of resistance. Treating FGFR1-driven neoplasms associated with SCLL with FGFR inhibitors is still in developmental stages, with only one report to date showing suppression of leukemia development, in a single patient.11 While our focus has been on the relatively rare SCLL syndrome, where FGFR1 is the consistent driver of the neoplasm, FGFR overexpression has also been identified in de novo AML9 as well as subgroups of other (solid) tumor types, where FGFR inhibitors have also proved effective.31 With increasing numbers of small molecule FGFR inhibitors entering clinical trials, it was timely to investigate alternative strategies for treating emerging resistance.
In this report we describe two mutually exclusive mechanisms underlying resistance to FGFR inhibitors with the first demonstration that a homozygous V561M mutation is definitively associated with the development of resistance in 50% of the resistant cell lines analyzed. Previous studies, notably in childhood T-ALL, have presented correlative associations between the presence of the V561M mutation with poor survival, but no mechanistic studies confirming the association.32 The demonstration that the V561 mutation leads to constitutive activation of FGFR1 was provided through chemical biology approaches. Proteins expressed in bacterial systems were crystalized and the association dynamics for ADZ4547 and E3810 in wild type and mutant proteins determined.33 Furthermore, the FGFR V561M mutation was also reported to induce strong resistance to PD17307434 and BGJ398.35 These studies showed the V561M mutation is a significant FGFR1 activating event. Molecular modeling from in vitro binding assays, however, suggested that both E3810 and AZD4547 showed reduced affinity to the V561M mutation, identifying a possible mechanism for the inability of AZD4547 to suppress activation of the mutant FGFR1 kinase. These results support the observation that resistance selection through an acquired V561M mutation is the underlying mechanism of resistance in mutant SCLL cell lines. Studies in transient transfection of the V561M-FGFR1 into COS7 cells36 or 293T cells34 further demonstrated increased FGFR1 autophosphorylation. In these same studies, when the V561M mutation was ectopically expressed in model cell lines, they became less sensitive to pan-kinase inhibitors. Similarly, in squamous cell lung cancer, ectopic expression of the V561M mutant FGFR1 in FGFR1 overexpressing cells abolished sensitivity to PD173074.31 These observations support the conclusion that the mutations that were selected for in the BBC1 and KG1 cell lines are responsible for the resistance to specific FGFR kinase inhibitors.
The second consistent observation in resistant FGFR1-driven leukemias was the loss of PTEN function in 50% of the cell lines, through an identical internal deletion of exon 6, leading to a premature stop codon. As a result, the PI3K/AKT/mTOR pathway was upregulated, and targeting PI3K in the resistant clones suppressed growth. Of note, however, we also observed that resistant cells became less sensitive to either the BEZ235 or BAY80-6946 PI3K inhibitors, as shown in Supplementary Figure 2. The decrease in sensitivity to PI3K inhibitors in resistant cells can be due to up-regulation of PI3K signaling pathway (Fig 3A) as a result of PTEN loss. We have also shown that increased concentrations of FGFR1 inhibitors leads to mild suppression of FGFR1 activation in the resistant cells. However, when targeting PI3K was combined with inhibition of FGFR1, the effects were synergistic. Although PTEN deletion as a cause of resistance in leukemias has not been reported before, it has been shown in breast cancer, for example, where metastatic tumors following treatment with a PI3K inhibitor, BYL719, showed various exon deletions in PTEN which was assumed to be the cause of the resistance.37 In vitro suppression of PTEN in breast cancer cell lines sensitive to BYL719 increased their resistance to the drug supporting this conclusion. Loss of PTEN has also been reported as a mechanism of resistance to EGFR inhibitors in lung cancer.38 Of note, in our studies, the PI3K/AKT pathway was also upregulated in KG1 resistant cells that expressed wild type PTEN. Although PTEN protein levels are comparable between the KG1 resistant and the parental cells (data not shown), the PI3K signaling pathway is obviously activated. The mechanism by which PI3K is activated in the KG1 resistance cells in the absence of a PTEN deletion, however, is not known. Since FGFR1 activates PI3K, it is likely that the persistent expression of mutant FGFR1 kinase in these cells may be responsible to some extent for the increased PI3K signaling.
The SCLL syndrome is driven by activation of various chimeric FGFR1 kinases where there is evidence that the different chimeric FGFR1 genes associated with the activation lead to different outcomes and disease course5. Despite minor differences, such as the presence of eosinophilia in some cases, or a preference for the development of T-lymphoma or B-lymphoma between the subsets of common rearrangements, the BCR-FGFR1 gene stands out in that this is typically a more aggressive disease. We have also seen this in our model systems where disease onset is disproportionately early amongst the mouse syngeneic models14 as well as in human xenograft models using transformed CD34+ stem cells.39 The BCR-FGFR1 rearrangement is also distinct amongst the SCLL rearrangements in that the BCR component of the chimeric gene encodes for a serine-threonine kinase which may modify and enhance the phenotypes generated as a result of its expression. This was demonstrated previously where mutation of a critical phosphorylation site in BCR led to a predominantly T-cell disease, compared with the B-lymphomas typically associated with the wild type rearrangement.40. It is possible, therefore, that the BCR component of this chimeric kinase may contribute to modifying the consequences of disregulation of the PI3K pathway and so different alternative therapies may be indicated.
Although we have defined two mechanisms of resistance to FGFR1 inhibitors, it is possible that other mechanisms may arise that will need alternative strategies to treat these resistant clones. Extensive molecular analyses of SCLL models in vivo and in vitro have suggested some potential targets,13–15, 25, 30, 41 such as FLT3, MYC and NOTCH1. Although AML is the primary consequence of constitutive activation of FGFR1 in SCLL, B- and T-cell lymphomas also develop.5 While the common driver event is the upregulation of FGFR1 in the various associated neoplasms, our gene expression analyses have defined other potential targets that could be incorporated into a multi-drug treatment of the syndromic neoplasms seen in SCLL. In ZNF198-FGFR1 expressing T-lymphomas, for example, upregulation of NOTCH signaling was a consistent observation and gamma secretase inhibitors were effective in suppressing leukemogenesis,30 suggesting a possible alternative drug combination for T-lymphomas arising in SCLL. Alternatively, BCL2 is also upregulated in SCLL tumors12 which provides another potential target to treat resistant cells.
In summary, we have identified two important mechanisms by which sub-clones of leukemic cells can develop resistance to FGFR inhibitors during long-term targeted therapy both of which lead to upregulation of PI3K signaling. It is possible, therefore, that simultaneously targeting both FGFR and PI3K may prove to be an effective way of treating clones resistant to FGFR inhibitors in future clinical trials, not only for SCLL related leukemias but other tumor types that are addicted to FGFR family kinases.42
Supplementary Material
Novelty and Impact.
Chromosome translocations in stem cell leukemia and lymphoma syndrome lead to constitutive over expression of FGFR1. These primary leukemias are sensitive to FGFR1 inhibitors but patients are at risk to developing resistance to mono therapies. This report describes unique mechanisms of FGFR1 mutation and PTEN deletion by which resistance arises in these leukemias and suggests second line approaches to treating resistant clones using personalized, alternative therapeutic strategies depending on the specific mechanism of resistance.
Acknowledgments
This work was supported by a grant from the National Cancer Institute of the National Institutes Health (CA076167).
Footnotes
CONFLICT INTEREST
The authors declare no conflict of interest.
References
- 1.Dorey K, Amaya E. FGF signalling: diverse roles during early vertebrate embryogenesis. Development. 2010;137:3731–3742. doi: 10.1242/dev.037689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine & growth factor reviews. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nature reviews Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 4.Wesche J, Haglund K, Haugsten EM. Fibroblast growth factors and their receptors in cancer. The Biochemical journal. 2011;437:199–213. doi: 10.1042/BJ20101603. [DOI] [PubMed] [Google Scholar]
- 5.Jackson CC, Medeiros LJ, Miranda RN. 8p11 myeloproliferative syndrome: a review. Hum Pathol. 2010;41:461–476. doi: 10.1016/j.humpath.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Baumann H, Kunapuli P, Tracy E, Cowell JK. The oncogenic fusion protein-tyrosine kinase ZNF198/fibroblast growth factor receptor-1 has signaling function comparable with interleukin-6 cytokine receptors. The Journal of biological chemistry. 2003;278:16198–16208. doi: 10.1074/jbc.M300018200. [DOI] [PubMed] [Google Scholar]
- 7.Still IH, Cowell JK. The t(8;13) atypical myeloproliferative disorder: further analysis of the ZNF198 gene and lack of evidence for multiple genes disrupted on chromosome 13. Blood. 1998;92:1456–1458. [PubMed] [Google Scholar]
- 8.Ren M, Qin H, Ren R, Cowell JK. Ponatinib suppresses the development of myeloid and lymphoid malignancies associated with FGFR1 abnormalities. Leukemia. 2013;27:32–40. doi: 10.1038/leu.2012.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Q, Bhole A, Qin H, Karp J, Malek S, Cowell JK, et al. Targeting FGFR1 to suppress leukemogenesis in syndromic and de novo AML in murine models. Oncotarget. 2016 doi: 10.18632/oncotarget.10438. e-pub ahead of print 6 July 2016; doi:2010.18632/oncotarget.10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lierman E, Smits S, Cools J, Dewaele B, Debiec-Rychter M, Vandenberghe P. Ponatinib is active against imatinib-resistant mutants of FIP1L1-PDGFRA and KIT, and against FGFR1-derived fusion kinases. Leukemia. 2012;26:1693–1695. doi: 10.1038/leu.2012.8. [DOI] [PubMed] [Google Scholar]
- 11.Khodadoust MS, Luo B, Medeiros BC, Johnson RC, Ewalt MD, Schalkwyk AS, et al. Clinical activity of ponatinib in a patient with FGFR1-rearranged mixed-phenotype acute leukemia. Leukemia. 2016;30:947–950. doi: 10.1038/leu.2015.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren M, Li X, Cowell JK. Genetic fingerprinting of the development and progression of T-cell lymphoma in a murine model of atypical myeloproliferative disorder initiated by the ZNF198-fibroblast growth factor receptor-1 chimeric tyrosine kinase. Blood. 2009;114:1576–1584. doi: 10.1182/blood-2009-03-212704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren M, Qin H, Kitamura E, Cowell JK. Dysregulated signaling pathways in the development of CNTRL-FGFR1-induced myeloid and lymphoid malignancies associated with FGFR1 in human and mouse models. Blood. 2013;122:1007–1016. doi: 10.1182/blood-2013-03-489823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren M, Qin H, Wu Q, Savage NM, George TI, Cowell JK. Development of ZMYM2-FGFR1 driven AML in human CD34+ cells in immunocompromised mice. International journal of cancer. 2016;139:836–840. doi: 10.1002/ijc.30100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ren M, Tidwell JA, Sharma S, Cowell JK. Acute progression of BCR-FGFR1 induced murine B-lympho/myeloproliferative disorder suggests involvement of lineages at the pro-B cell stage. PLoS One. 2012;7:e38265. doi: 10.1371/journal.pone.0038265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Deangelo DJ, Kutok JL, Williams IR, Lee BH, Wadleigh M, et al. PKC412 inhibits the zinc finger 198-fibroblast growth factor receptor 1 fusion tyrosine kinase and is active in treatment of stem cell myeloproliferative disorder. Proc Natl Acad Sci U S A. 2004;101:14479–14484. doi: 10.1073/pnas.0404438101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bello E, Colella G, Scarlato V, Oliva P, Berndt A, Valbusa G, et al. E-3810 is a potent dual inhibitor of VEGFR and FGFR that exerts antitumor activity in multiple preclinical models. Cancer Res. 2011;71:1396–1405. doi: 10.1158/0008-5472.CAN-10-2700. [DOI] [PubMed] [Google Scholar]
- 18.Chase A, Grand FH, Cross NC. Activity of TKI258 against primary cells and cell lines with FGFR1 fusion genes associated with the 8p11 myeloproliferative syndrome. Blood. 2007;110:3729–3734. doi: 10.1182/blood-2007-02-074286. [DOI] [PubMed] [Google Scholar]
- 19.Wasag B, Lierman E, Meeus P, Cools J, Vandenberghe P. The kinase inhibitor TKI258 is active against the novel CUX1-FGFR1 fusion detected in a patient with T-lymphoblastic leukemia/lymphoma and t(7;8)(q22;p11) Haematologica. 2011;96:922–926. doi: 10.3324/haematol.2010.036558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Hare T, Shakespeare WC, Zhu X, Eide CA, Rivera VM, Wang F, et al. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009;16:401–412. doi: 10.1016/j.ccr.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gavine PR, Mooney L, Kilgour E, Thomas AP, Al-Kadhimi K, Beck S, et al. AZD4547: an orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res. 2012;72:2045–2056. doi: 10.1158/0008-5472.CAN-11-3034. [DOI] [PubMed] [Google Scholar]
- 22.Angibaud PR, Mevellec L, Saxty G, Adelinet C, et al. Discovery of JNJ42756493, a potent fibroblast growth factor receptor (FGFR) inhibitor using a fragment based approach. Cancer Research (Abstract) 2014;74 [Google Scholar]
- 23.Guagnano V, Furet P, Spanka C, Bordas V, Le Douget M, Stamm C, et al. Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamin o]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase. Journal of medicinal chemistry. 2011;54:7066–7083. doi: 10.1021/jm2006222. [DOI] [PubMed] [Google Scholar]
- 24.Federici G, Gao X, Slawek J, Arodz T, Shitaye A, Wulfkuhle JD, et al. Systems analysis of the NCI-60 cancer cell lines by alignment of protein pathway activation modules with "-OMIC" data fields and therapeutic response signatures. Molecular cancer research : MCR. 2013;11:676–685. doi: 10.1158/1541-7786.MCR-12-0690. [DOI] [PubMed] [Google Scholar]
- 25.Qin H, Wu Q, Cowell JK, Ren M. FGFR1OP2-FGFR1 induced myeloid leukemia and T-cell lymphoma in a mouse model. Haematologica. 2016;101:e91–94. doi: 10.3324/haematol.2015.137695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furley AJ, Reeves BR, Mizutani S, Altass LJ, Watt SM, Jacob MC, et al. Divergent molecular phenotypes of KG1 and KG1a myeloid cell lines. Blood. 1986;68:1101–1107. [PubMed] [Google Scholar]
- 27.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nature medicine. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maira SM, Stauffer F, Brueggen J, Furet P, Schnell C, Fritsch C, et al. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol Cancer Ther. 2008;7:1851–1863. doi: 10.1158/1535-7163.MCT-08-0017. [DOI] [PubMed] [Google Scholar]
- 29.Sandhofer N, Metzeler KH, Rothenberg M, Herold T, Tiedt S, Groiss V, et al. Dual PI3K/mTOR inhibition shows antileukemic activity in MLL-rearranged acute myeloid leukemia. Leukemia. 2015;29:828–838. doi: 10.1038/leu.2014.305. [DOI] [PubMed] [Google Scholar]
- 30.Ren M, Cowell JK. Constitutive Notch pathway activation in murine ZMYM2-FGFR1-induced T-cell lymphomas associated with atypical myeloproliferative disease. Blood. 2011;117:6837–6847. doi: 10.1182/blood-2010-07-295725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Science translational medicine. 2010;2:62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutierrez A, Sanda T, Grebliunaite R, Carracedo A, Salmena L, Ahn Y, et al. High frequency of PTEN, PI3K, and AKT abnormalities in T-cell acute lymphoblastic leukemia. Blood. 2009;114:647–650. doi: 10.1182/blood-2009-02-206722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sohl CD, Ryan MR, Luo B, Frey KM, Anderson KS. Illuminating the molecular mechanisms of tyrosine kinase inhibitor resistance for the FGFR1 gatekeeper mutation: the Achilles' heel of targeted therapy. ACS chemical biology. 2015;10:1319–1329. doi: 10.1021/acschembio.5b00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou W, Hur W, McDermott U, Dutt A, Xian W, Ficarro SB, et al. A structure-guided approach to creating covalent FGFR inhibitors. Chemistry & biology. 2010;17:285–295. doi: 10.1016/j.chembiol.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan L, Wang J, Tanizaki J, Huang Z, Aref AR, Rusan M, et al. Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors. Proc Natl Acad Sci U S A. 2014;111:E4869–4877. doi: 10.1073/pnas.1403438111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blencke S, Zech B, Engkvist O, Greff Z, Orfi L, Horvath Z, et al. Characterization of a conserved structural determinant controlling protein kinase sensitivity to selective inhibitors. Chemistry & biology. 2004;11:691–701. doi: 10.1016/j.chembiol.2004.02.029. [DOI] [PubMed] [Google Scholar]
- 37.Juric D, Castel P, Griffith M, Griffith OL, Won HH, Ellis H, et al. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kalpha inhibitor. Nature. 2015;518:240–244. doi: 10.1038/nature13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sos ML, Koker M, Weir BA, Heynck S, Rabinovsky R, Zander T, et al. PTEN loss contributes to erlotinib resistance in EGFR-mutant lung cancer by activation of Akt and EGFR. Cancer Res. 2009;69:3256–3261. doi: 10.1158/0008-5472.CAN-08-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cowell JK, Qin H, Chang C-S, Kitamura E, Ren M. A model of BCR-FGFR1 driven human AML in immunocompromised mice. British Journal of Heamatology. 2015 doi: 10.1111/bjh.13877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roumiantsev S, Krause DS, Neumann CA, Dimitri CA, Asiedu F, Cross NC, et al. Distinct stem cell myeloproliferative/T lymphoma syndromes induced by ZNF198-FGFR1 and BCR-FGFR1 fusion genes from 8p11 translocations. Cancer Cell. 2004;5:287–298. doi: 10.1016/s1535-6108(04)00053-4. [DOI] [PubMed] [Google Scholar]
- 41.Ren M, Qin H, Ren R, Tidwell J, Cowell JK. Src activation plays an important key role in lymphomagenesis induced by FGFR1 fusion kinases. Cancer Res. 2011;71:7312–7322. doi: 10.1158/0008-5472.CAN-11-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pearson A, Smyth E, Babina IS, Herrera-Abreu MT, Tarazona N, Peckitt C, et al. High-Level Clonal FGFR Amplification and Response to FGFR Inhibition in a Translational Clinical Trial. Cancer Discov. 2016;6:838–851. doi: 10.1158/2159-8290.CD-15-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.