Abstract
The enzymes that determine protein O-GlcNAcylation, O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA), act on key transcriptional and epigenetic regulators, and both are abundantly expressed in the brain. However, little is known about how alterations in O-GlcNAc cycling affect human embryonic stem cell (hESC) neural differentiation. Here, we studied the effects of perturbing O-GlcNAcylation during neural induction of hESCs using the metabolic inhibitor of OGT, peracetylated 5-thio-N-acetylglucosamine (Ac4-5SGlcNAc). Treatment of hESCs with Ac4-5SGlcNAc during induction limited protein O-GlcNAcylation and also caused a dramatic decrease in global levels of UDP-GlcNAc. Concomitantly, a subpopulation of neural progenitor cells (NPCs) acquired an immature neuronal morphology and expressed early neuronal markers such as β-III tubulin (TUJ1) and microtubule associated protein 2 (MAP2), phenotypes that took longer to manifest in the absence of OGT inhibition. These data suggest that chemical inhibition of OGT and perturbation of protein O-GlcNAcylation accelerate the differentiation of hESCs along the neuronal lineage, thus providing further insight into the dynamic molecular mechanisms involved in neuronal development.
Graphical Abstract
O-GlcNAcylation of serine or threonine residues is a widespread modification of nuclear, cytoplasmic, and mitochondrial proteins that is dynamic and reversible on the time scale of hours or even seconds.1–4 Unlike phosphorylation, which is regulated by hundreds of kinases and phosphatases, only one O-GlcNAc transferase (OGT) installs the GlcNAc residue, and a single O-GlcNAcase (OGA) removes it. UDP-GlcNAc, the nucleotide sugar donor for OGT, is produced via the hexosamine biosynthetic pathway (HBP), and its levels are highly responsive to intracellular concentrations of glucose, amino acids, and lipids.5 This sensitivity allows O-GlcNAcylation to respond to changes in nutrient availability and the cell’s metabolic state.6 Moreover, maintaining a proper balance of O-GlcNAc in the cell is important, as the dysregulation of O-GlcNAcylation underlies several disease states.7
A number of studies point to a role for O-GlcNAcylation in stem cell maintenance and differentiation. The complete absence of OGT is lethal to mouse ESCs and embryos,8 and experiments with conditional alleles reveal that OGT is required in a tissue-specific manner at later stages of development.9 O-GlcNAcylation has been shown to both enhance and suppress activity of proteins important for embryonic stem cell (ESC) pluripotency and differentiation.10–15 Jang and colleagues reported that a loss of OGT reduced proliferation and self-renewal of ESCs.11 Conversely, high O-GlcNAcylation decreased differentiation of these cells. The study also showed that O-GlcNAcylation of pluripotency factors OCT4 and SOX2 was necessary for maintaining ESC pluripotency. In contrast, a recent study by Myers et al. showed that O-GlcNAcylation of SOX2 at a specific serine residue inhibited stem cell pluripotency and maintenance, suggesting a new mechanism by which O-GlcNAc regulates SOX2 in response to developmental cues.15
O-GlcNAcylation seems to be particularly important in brain development. Many proteins important for neuronal cell signaling, synaptic plasticity, learning, and memory are O-GlcNAc-modified.4,16–19 Indeed, studies of brain-specific OGT knockout mice point to a role for O-GlcNAc in neuronal function and neurodegeneration.9,20–22 Liu et al. reported higher levels of O-GlcNAc, OGT, and OGA in neurons compared to non-neuronal cells in the rat brain.23 Maintaining high levels of O-GlcNAcylation prevents ectodermal differentiation of mouse ESCs,13 impairs axonal branching,24 and inhibits proteasome function.25 A recent study using human embryonic stem cells (hESCs) found that excess O-GlcNAc decreased the expression of neural markers PAX6 and SOX1.26 However, the authors did not examine the effect of decreasing O-GlcNAc during hESC differentiation.
Here, we characterize O-GlcNAc cycling during neural induction of hESCs and the effect of chemical inhibition of OGT on the differentiation process. We found that O-GlcNAc levels oscillate during neural differentiation both with and without OGT inhibition by Ac4-5SGlcNAc. Upon treatment with the inhibitor, we also observed that neural progenitor cells (NPCs) gained morphology reminiscent of immature neurons, acquiring the neuronal markers β-III tubulin (TUJ1) and microtubule-associated protein 2 (MAP2) on a faster time scale than without OGT inhibition. Gene expression analysis revealed that genes involved in neuronal differentiation and forebrain development were significantly upregulated in OGT-inhibited cells. These findings improve our mechanistic understanding of neural differentiation and may provide a faster route to generate neurons from hESCs.
RESULTS AND DISCUSSION
Global O-GlcNAcylation Oscillates As hESCs Differentiate to NPCs
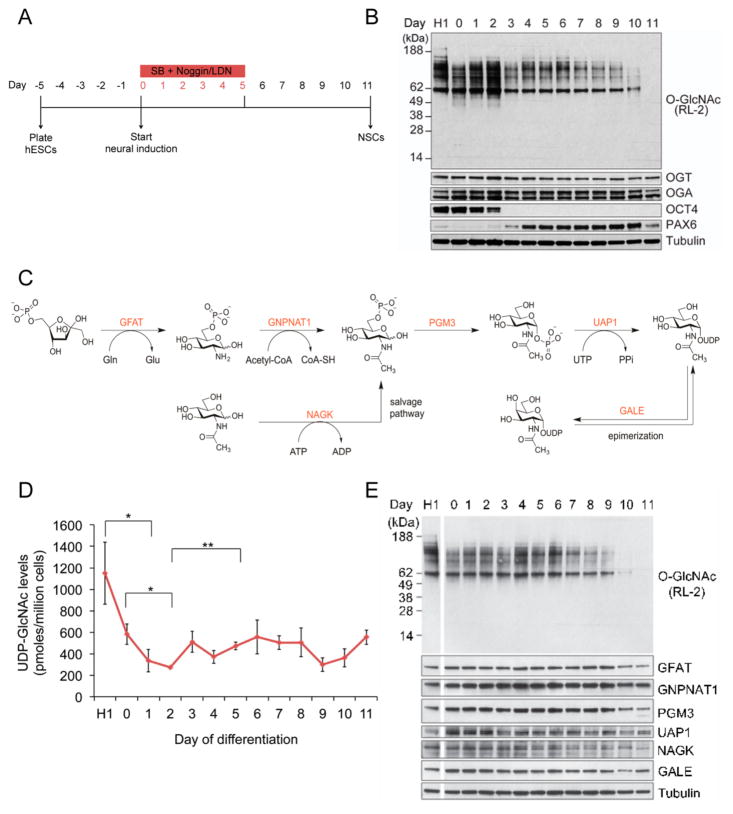
hESCs were differentiated into NPCs using the dual SMAD-inhibition protocol (Figure 1A).27 This protocol relies on the synergistic action of SB431542 (SB), which inhibits the Lefty/Activin/TGFβ pathways, and LDN-193189 (LDN) or Noggin, inhibitors of bone morphogenetic protein (BMP). During NPC formation, expression of the pluripotency marker OCT4 was undetectable by Western blot after day 2 of neural induction, while the emergence of transcription factor and neuroectodermal marker PAX6 was clearly observed by Western blot 4 days post induction (Figure 1B). This indicated a successful differentiation of the hESC line H1 to NPCs over 11 days, as reported previously.27 We then analyzed global O-GlcNAcylation at every day of neural differentiation by Western blot with an O-GlcNAc-specific antibody (RL2). Global O-GlcNAc levels oscillated during hESC neural induction, dramatically decreasing after day 9 of induction. The expression of both OGT and OGA also decreased toward the end of the neural induction protocol (Figure 1B), as has been seen in studies of O-GlcNAcylation in rat brain and mouse embryonic neural precursor cells.23,28 However, OGT and OGA protein expression did not oscillate similarly to O-GlcNAc levels. Together, these data suggest that a decrease in O-GlcNAcylation may be important for neural induction of hESCs and that the oscillation in the levels of O-GlcNAc is not due to changes in OGT and OGA abundance.
Figure 1.
Oscillation of global O-GlcNAcylation during neural differentiation of hESCs. (A) Overview of the dual-SMAD inhibition protocol. Cells were grown to 90% confluency on days –2 and –1 before starting neural induction on day 0. (B) Whole cell lysates were immunoblotted for O-GlcNAc, OGT, OCT4, PAX6, and OGA. H1 refers to undifferentiated hESCs. (C) Hexosamine biosynthetic pathway (HBP) enzymes involved in UDP-GlcNAc synthesis. (D) Quantitation of UDP-GlcNAc levels on each day of neural differentiation by high performance anion exchange chromatography (HPAEC; n = 4; mean ± SEM; *P < 0.05, **P < 0.01). H1 refers to undifferentiated hESCs. (E) Western blot analysis of HBP enzymes during each day of neural differentiation. H1 refers to undifferentiated hESCs.
Protein O-GlcNAcylation is influenced by glucose flux through the HBP, which produces UDP-GlcNAc (Figure 1C). Levels of UDP-GlcNAc are positively correlated to cellular protein O-GlcNAcylation.29 To determine whether the availability of UDP-GlcNAc might be correlated with the observed difference in global O-GlcNAc levels between the hESCs and NPCs, we measured the levels of UDP-GlcNAc at every day of differentiation. Analysis of extracted UDP-GlcNAc using high performance anion exchange chromatography (HPAEC) revealed a similar fluctuation in the levels of the nucleotide sugar donor to global protein-associated O-GlcNAc in the early stages of neural induction (Figure 1D). Given that cellular differentiation is generally associated with proliferation and increased metabolic requirement,30 it is possible that the changes in UDP-GlcNAc concentration reflect a difference in glucose flux through the HBP. This result might suggest that protein O-GlcNAcylation is being modulated in part by the cellular concentration of UDP-GlcNAc. Notably, though, UDP-GlcNAc levels toward the end of neural differentiation did not parallel the dramatic decrease observed in global protein O-GlcNAcylation.
To determine if changes in the expression of the enzymes in the HBP were responsible for the observed changes in UDP-GlcNAc levels during neural induction, we analyzed the protein levels of every enzyme in the HBP by Western blotting. As shown in Figure 1E, the protein levels of HBP enzymes remained constant during the beginning of the neural induction, except for the enzymes UDP-N-acetylglucosamine pyrophosphorylase 1 (UAP1) and N-acetylglucosamine kinase (NAGK), whose expression seems to drop off as early as day 3 and 5, respectively. Interestingly, as we observed for OGT and OGA, the abundance of most enzymes in the HBP decreased toward the end of differentiation, including the enzymes glutamine fructose-6-phosphate amidotransferase (GFAT1) and phosphoglucomutase 3 (PGM3). Although GFAT1 is the rate-limiting enzyme of HBP, its decrease in expression does not correlate with the increase in UDP-GlcNAc levels on day 11. It has been shown that UDP-GlcNAc levels are also influenced by the expression of the enzyme glucosamine-phosphate N-acetyltransferase 1 (GNPNAT1), whose expression is mostly constant during the neural induction.31 Similarly, the expression of the enzyme UDP-galactose-4-epimerase (GALE) did not change during neural induction. The lack of correlation between HBP enzyme expression and O-GlcNAc levels could be due to consumption of UDP-GlcNAc in other glycosylation pathways.
Changes in the expression of glucose transporters, such as GLUT1 and GLUT4, have been shown to influence UDP-GlcNAc levels in mice.32 While we did not examine these proteins directly, the transcript levels of the transporters GLUT1 (SLC2A1) and GLUT3 (SLC2A3) increased as cells differentiated toward NPCs, whereas the transcript levels of GLUT4 (SLC2A4) decreased during neural induction (vide infra and Supporting Information Figure 1). These results correspond to an increase in glucose uptake32 and possibly account for the increase in UDP-GlcNAc levels at later days of differentiation. Furthermore, both GLUT1 and GLUT3 have been shown to be important in brain metabolism and development.33
Treatment with the OGT Inhibitor Ac4-5SGlcNAc Lowers Global O-GlcNAc Levels in Differentiating hESCs
To determine whether global O-GlcNAcylation influences the process of neuronal differentiation, we used the small molecule peracetylated 5-thio-N-acetylglucosamine (Ac4-5SGlcNAc) recently shown by Gloster et al. to disrupt the catalytic activity of OGT.34 Ac4-5SGlcNAc is converted within cells to UDP-5SGlcNAc, a nucleotide sugar analogue of UDP-GlcNAc that competes with the native substrate and inhibits protein O-GlcNAcylation by OGT. As well, Ac4-5SGlcNAc reduces the cellular pool of native UDP-GlcNAc, another mode by which it may suppress protein O-GlcNAcylation.
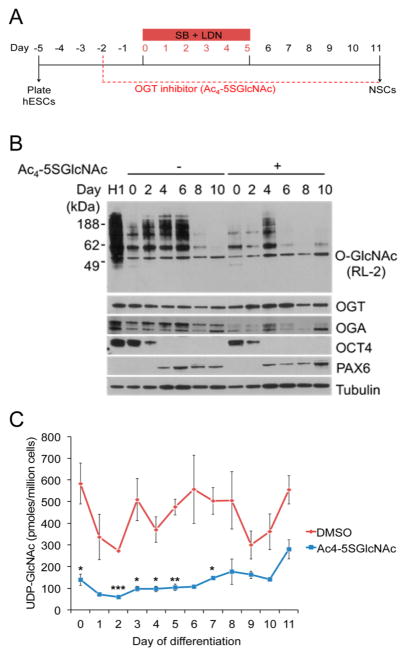
We first tested various inhibitor doses to identify concentrations of Ac4-5SGlcNAc that decrease global O-GlcNAcylation, as determined by Western blot with the anti-O-GlcNAc antibody. As shown in Supporting Information Figure 2A, treatment with 50 μM Ac4-5SGlcNAc for 24 h abrogated the majority of detectable protein O-GlcNAcylation. We also observed that treatment with Ac4-5SGlcNAc for 5 days led to a time-dependent decrease in O-GlcNAc for up to 3 days, after which time O-GlcNAc levels started to increase (Supporting Information Figure 2B). We also observed a decrease in OGA protein expression during the 5-day period. This may reflect a feedback mechanism in which reduction in OGT activity and protein O-GlcNAcylation leads to down-regulation of OGA expression.34,35 OGT inhibition by Ac4-5SGlcNAc did not affect hESC pluripotency, as measured by OCT4 expression (Supporting Information Figure 2B). Importantly, treatment of cells with the OGT inhibitor did not cause apparent changes in cell morphology and proliferation (Supporting Information Figure 2C).
We next analyzed global O-GlcNAcylation levels after treating neural-induced hESCs with 50 μM Ac4-5SGlcNAc (Figure 2A). Overall, O-GlcNAcylation levels were lower in inhibitor-treated cells, but the oscillation pattern characteristic of the neural differentiation process was maintained (Figure 2B). As above, perturbations in O-GlcNAc cycling induced a compensatory decrease in OGA and an increase in OGT protein levels. The expressions of OCT4 and PAX6 were not affected by the OGT inhibitor (Figure 2B). Furthermore, we observed lower UDP-GlcNAc levels when differentiating cells in the presence of Ac4-5SGlcNAc, as previously reported by Gloster et al. (Figure 2C).34 These results confirm that Ac4-5SGlcNAc is metabolized by the cells and inhibits protein O-GlcNAcylation. Also, the oscillatory pattern in both O-GlcNAc and UDP-GlcNAc levels during neural induction, even after OGT inhibition, indicates a possible regulatory mechanism utilized by hESCs to properly differentiate into NPCs.
Figure 2.
Ac4-5SGlcNAc perturbation of O-GlcNAc and UDP-GlcNAc levels in hESCs undergoing neural differentiation. (A) Overview of neural differentiation protocol with OGT inhibitor added. (B) Western blot analysis of hESCs differentiated in the presence of OGT inhibitor (50 μM) showed a decrease in O-GlcNAc levels, and in OGA expression, while enhancing OGT expression; tubulin immunoblotting showed equal loading. (C) UDP-GlcNAc levels measured by HPAEC analysis during neural induction with DMSO (red line, as in Figure 1D) or Ac4-5SGlcNAc (blue line, concentration at 50 μM; n = 3–4; mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001).
OGT Inhibition Promotes Neural Differentiation of hESCs
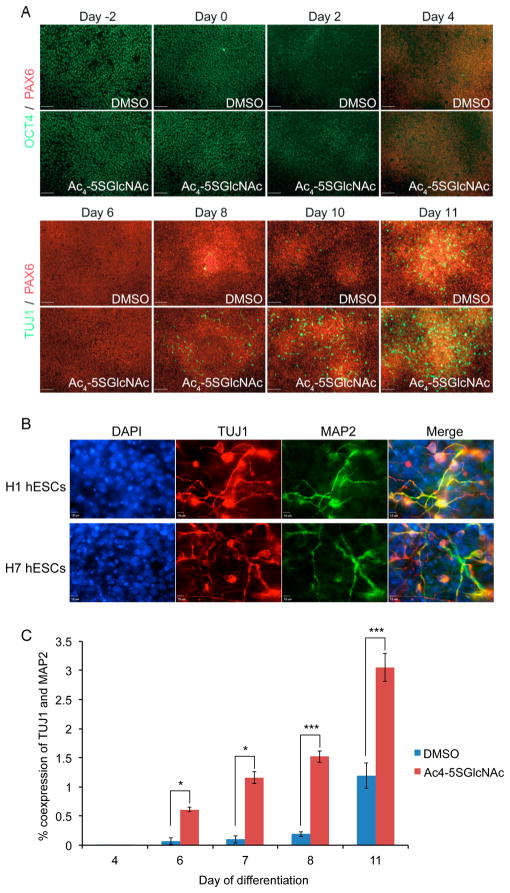
We next asked whether treatment of hESCs with Ac4-5SGlcNAc during neural induction caused perturbations in the differentiation process. Treatment of hESCs with 50 μM Ac4-5SGlcNAc for 48 h prior to initiating neural induction with SMAD inhibitors accelerated expression of the neuronal marker TUJ1 compared to noninhibited cells (Supporting Information Figure 3A). Thus, all further neural inductions were performed in the presence of OGT inhibitor from day –2 to 11, and LDN and SB from day 0 to 5. Compared to cells treated with DMSO, hESCs differentiated in the presence of Ac4-5SGlcNAc prematurely expressed TUJ1 as early as day 8, and immunofluorescence analysis confirmed that they displayed greater extension of neurites (Figure 3A, Supporting Information Figure 5A). Similarly, we observed colabeling of neuronal markers TUJ1 and MAP2 by day 11 of induction in the presence of Ac4-5SGlcNAc in multiple hESC lines: H1, H9, and H7 (Figure 3B and Supporting Information Figure 3B). This underscores the reproducibility of these effects of OGT inhibitor treatment during neural differentiation of hESCs. Similar results were obtained when cells were treated with a lower concentration of Ac4-5SGlcNAc (10 μM) during neural induction (Supporting Information Figure 3C).
Figure 3.
Neuronal differentiation accelerated by treatment with Ac4-5SGlcNAc. (A) Immunofluorescence shows an earlier appearance of neuronal marker, TUJ1, in Ac4-5SGlcNAc-inhibited hESCs, in comparison to DMSO-treated cells. Expression of neural progenitor marker, PAX6, stays relatively constant in DMSO- and Ac4-5SGlcNAc-treated cells. (B) Coexpression of neuronal markers TUJ1 and MAP2 is shown in two hESC lines, H1 and H7, via immunofluorescence, validating the early neuronal phenotype observed in cells differentiated under Ac4-5SGlcNAc treatment. (C) Flow cytometry analysis of H1 hESCs shows a higher percent of coexpression of TUJ1 and MAP2 in cells treated with Ac4-5SGlcNAc than cells treated with DMSO (day 4, 6, 7, n = 2; day 8, 11, n = 6; mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001).
We also analyzed TUJ1 and MAP2 expression by flow cytometry analysis. In line with the microscopy results, we observed a rapid increase in the coexpression of these markers as early as day 6 of neural differentiation in the presence of Ac4-5SGlcNAc, with a 10-fold difference in expression between Ac4-5SGlcNAc-treated and DMSO-treated cells (Figure 3C). Coexpression of TUJ1 and MAP2 in Ac4-5SGlcNAc-treated cells continued to increase until day 11, at which point 3% of Ac4-5SGlcNAc-treated cells were coexpressing both neuronal markers, compared to 1% of DMSO-treated cells. Notably, MAP2 staining has been shown to follow TUJ1 labeling and is found in brain regions where the cell bodies of more mature neurons are located.36 TUJ1 was found to be O-GlcNAc modified in retinal pigment epithelial cells and mouse brain.37,38 In addition, previous studies have shown that O-GlcNAcylation of tubulin proteins inhibits their polymerization and negatively regulates microtubule formation.39 These findings suggest that a decrease in O-GlcNAcylation of TUJ1 proteins might be involved in increased microtubule formation and neuronal growth.
Additionally, as shown above, we did not detect a difference in OCT4, SOX2, or PAX6 expression when cells were treated with Ac4-5SGlcNAc (Figures 2B, 3A and Supporting Information Figure 6). These results suggest that the accelerated neuronal induction produced by OGT inhibition is not due to a premature exit from the pluripotent state and entrance into the neural progenitor state controlled by the abundance of OCT4, SOX2, and PAX6. SOX2 is involved in hESC pluripotency and differentiation,40 and it is known to be O-GlcNAcylated.10,11,15 Thus, it would be interesting to investigate whether its O-GlcNAcylation state and its interaction with other transcriptional regulatory proteins is altered due to hESC treatment with Ac4-5SGlcNAc,15 and consequently, whether these alterations are involved in the mechanism that accelerates hESC neural induction.
A complicating feature of Ac4-5SGlcNAc is that it can affect glycosylation pathways beyond O-GlcNAcylation, possibly causing pleiotropic effects that convolute biological studies. Indeed, Walker and co-workers found Ac4-5SGlcNAc to affect glycosylation of proteins that populate the cell surface, presumably due to inhibition of other glycosyltransferases or depletion of nucleotide sugars.35 As well, Gloster et al. reported that cells convert Ac4-5SGlcNAc to UDP-5SGalNAc, which in turn decreased the endogenous UDP-GalNAc pool.34 As UDP-GalNAc is the sugar donor for polypeptide GalNAc transferases that initiate mucin-type O-glycosylation, this finding suggests that Ac4-5SGlcNAc treatment can affect this sector of the glycome as well.
To determine whether cell-surface glycosylation patterns of hESCs are grossly perturbed by Ac4-5SGlcNAc, we performed staining experiments with the fluorescent lectins Canavalia ensiformis agglutinin (ConA), Lens Culiniaris Agglutinin (LCA), Artocarpus integrifolia agglutinin (AIL or Jacalin), and Arachis hypogaea agglutinin (PNA). Jacalin and PNA are lectins that bind galactose/N-acetylgalactosamine residues in mucin O-glycans, whereas LCA and ConA are lectins that bind the high-mannose core region of N-glycans. hESCs treated with 50 μM Ac4-5SGlcNAc or DMSO vehicle for 4 days of neural induction stained similarly with Jacalin and LCA, as determined by flow cytometry (Supporting Information Figure 4). In contrast, we observed significant differences between inhibitor- and vehicle-treated cells when we stained with PNA and ConA, specifically on day –2 and day 0. However, we did not observe differences in cell surface staining by any of the lectins on day 2 of neural induction. Thus, while Ac4-5SGlcNAc may have pleiotropic effects on hESC glycosylation beyond protein O-GlcNAcylation, these appear minimal once neural induction is underway.
Pursuant to these observations, we sought to determine whether intentional modifications to cell surface glycosylation reproduced the same neuronal phenotype seen under treatment with Ac4-5SGlcNAc. We differentiated hESCs in the presence of the O-glycosylation inhibitor benzyl-α-GalNAc (2 mM) or the enzyme α-mannosidase (0.05 U/ml) from day –2 to 8. These reagents have been previously shown to reduce the complexity of cell surface O- and N-glycans, respectively.41–43 However, neither treatment affected TUJ1 expression in the manner that we observed with Ac4-5SGlcNAc (Supporting Information Figure 5A). This observation supports the notion that Ac4-5SGlcNAc’s effect on neuronal differentiation kinetics is due to inhibition of O-GlcNAcylation rather than altered cell surface glycosylation. As a control experiment, differentiated hESCs treated with α-mannosidase were fixed on day 8 and probed against the lectins Arachis hypogaea agglutinin (PNA) and Canavalia ensiformis agglutinin (ConA) to test the efficacy of α-mannosidase treatment on cells (Supporting Information Figure 5B). Treatment of cells with α-mannosidase caused a reduction in mannose residues on the cell surface as shown by immunofluorescence imaging with a fluorescent ConA lectin. In contrast, the fluorescent signal of the PNA lectin did not decrease in α-mannosidase-treated cells, as compared to DMSO-treated cells. Overall, our results indicate that further characterization of Ac4-5SGlcNAc needs to be performed to better understand the metabolic and phenotypic effects of treating cells with this OGT inhibitor.
OGA Inhibition Does Not Prevent Neural Induction of hESCs
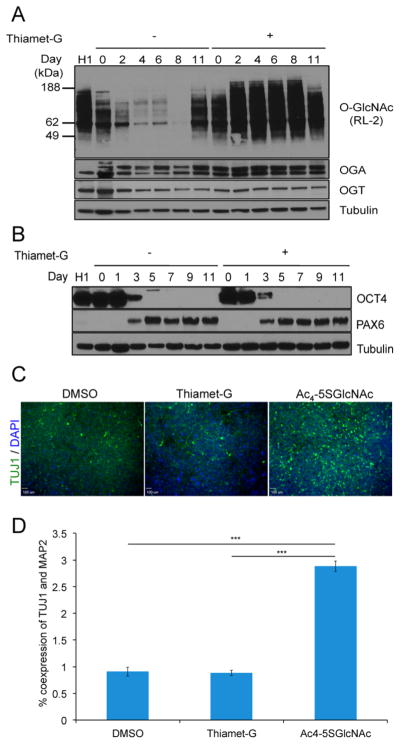
Maintaining high levels of O-GlcNAc in cells has been shown to inhibit neural differentiation of embryonic stem cells.13,26 Therefore, we examined whether elevating total O-GlcNAcylation levels in differentiating hESCs by treatment with the OGA inhibitor Thiamet-G (TMG) prevented cells from differentiating into NPCs.44 We induced cells from day –2 to 11 in the presence of 100 nM TMG and compared these to cells treated with either 50 μM Ac4-5SGlcNAc or DMSO. Treatment of cells with TMG during neural induction caused an increase in total O-GlcNAcylation (Figure 4A). Similarly, we observed an increase in OGA expression, whereas OGT expression decreased. As shown by Maury et al., OCT4 levels did not change due to TMG treatment (Figure 4B).26 However, contrary to the study by Maury et al., we did not observe a significant change in PAX6 expression upon treatment with TMG (although significantly higher concentrations were used in their study). As well, we did not observe a difference in the expression of TUJ1 between TMG-treated and DMSO-treated cells nor in the coexpression of MAP2 and TUJ1, as shown by immunofluorescence and flow cytometry, respectively (Figure 4C,D). These results suggest that treating differentiating hESCs with 100 nM TMG does not affect the hESC’s potential to become NPCs. These findings are consistent with a recent study in which oga disruption in the mouse brain was found to increase neurogenesis.45 Embryonic stem cells derived from these mice maintained the ability to differentiate in the presence of retinoic acid but failed to express the neuronal marker TUJ1, which further corroborates our findings with Ac4-5SGlcNAc. In contrast to what we observed, the differentiated mouse stem cells exhibited altered expression of pluripotency and differentiation markers, such as SOX2 and NESTIN.
Figure 4.
Thiamet-G increases global O-GlcNAcylation in differentiating hESCs but does not affect differentiation kinetics. (A) Western blot shows that O-GlcNAc levels increase with Thiamet-G treatment from day –2 to 11. (B) Western blot shows that treatment with Thiamet-G does not affect differentiation kinetics since no difference in OCT4 and PAX6 protein expression was observed in Thiamet-G-treated cells in comparison to DMSO-treated cells. (C) Immunofluorescence analysis of TUJ1 expression levels in differentiated hESCs treated with DMSO, Thiamet-G, or Ac4-5SGlcNAc from day –2 to 11. Thiamet-G treatment does not recapitulate the neuronal phenotype obtained in Ac4-5SGlcNAc-treated hESCs. (D) Percent coexpression of TUJ1 and MAP2 in cells treated with DMSO, Thiamet-G, or Ac4-5SGlcNAc for the entire duration of neural differentiation, as determined by flow cytometry analysis, confirms the results obtained by immunofluorescence (n = 6; mean ± SEM; ***P < 0.001).
OGT Inhibition Upregulates Neuronal Genes
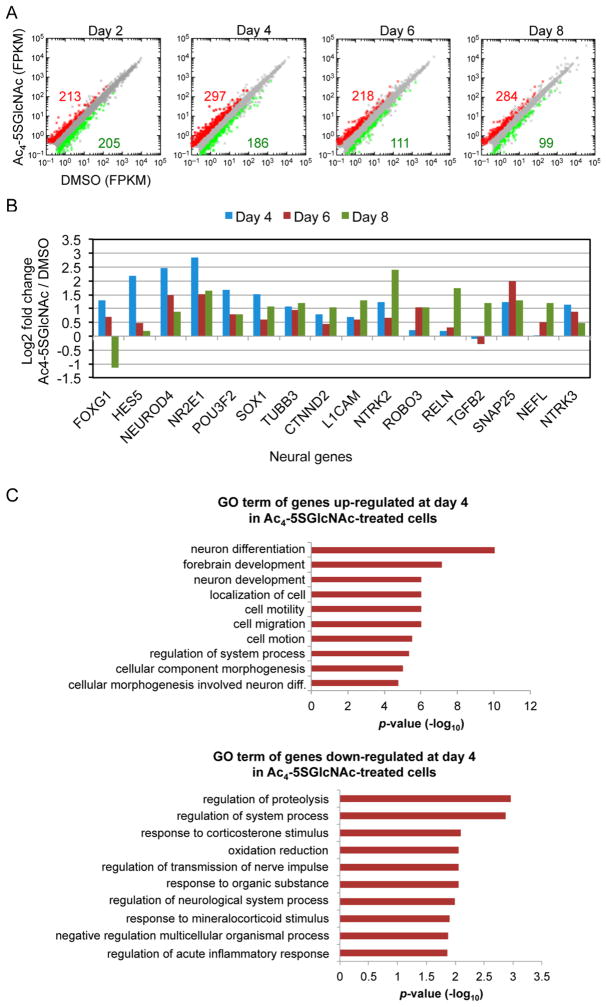
To obtain a more comprehensive view of the role of O-GlcNAc during neural differentiation, we performed RNA-seq analysis every 2 days during neural differentiation of DMSO- and Ac4-5SGlcNAc-treated hESCs. Strikingly, except for day 2, the number of genes that were upregulated due to OGT inhibition was always much higher than the number of genes that were downregulated, which suggests that O-GlcNAcylation could function as a repressor of the expression of neural genes (Figure 5A). Similarly, the marked difference between the number of upregulated and downregulated genes suggests that the gradual change in the transcriptome, as cells exit pluripotency and enter the neural progenitor stage, is controlled in part by protein O-GlcNAcylation. Day 4 corresponded to the day where the expression of a higher number of genes was affected due to OGT inhibition—483 genes total. Interestingly, day 4 coincides with a time point where cells lose expression of OCT4 and start expressing the neural progenitor marker PAX6. This may signify that cells are exiting the pluripotent state and moving down a path to become NPCs. These changes in transcription are consistent with studies in Drosophila where it has been shown that OGT is encoded by the Polycomb group (PcG) gene, which acts to repress gene expression.46–48 OGT has also been shown to interact with the Sin3-HDAC complex to promote gene silencing.49
Figure 5.
Increased neural gene expression in Ac4-5SGlcNAc-treated cells as early as day 4 of differentiation. (A) RNA-seq analysis of Ac4-5SGlcNAc-treated cells versus DMSO-treated cells at different days of neural differentiation. Scatter plots represent FPKM values for genes expressed in DMSO- and Ac4-5SGlcNAc-treated cells. Red and green dots are genes whose expression is affected more than or less than 2-fold with p values < 0.05, respectively. Gray dots represent genes not changing within the selected cutoff. (B) Selected neural genes from RNA-seq analysis are illustrated as a log2 fold change between Ac4-5SGlcNAc-treated cells and DMSO-treated cells on days 4, 6, and 8 of differentiation. (C) The top 10 gene ontology (GO) term Biological Process categories obtained from RNA-seq analysis of Ac4-5SGlcNAc-treated and DMSO-treated cells, ranked by p value. Top: GO term analysis of genes upregulated by treatment of Ac4-5SGlcNAc on day 4 of neural differentiation. Bottom: GO term analysis of genes downregulated by treatment of Ac4-5SGlcNAc on day 4 of neural differentiation.
Consistent with the induction of neural development in OGT-inhibited cells, selected neuronal genes showed an increase in expression (Figure 5B). For instance, TUBB3, NEUROD4, RELN, TGFB2, and L1CAM transcripts were all upregulated in Ac4-5SGlcNAc-treated cells. Enhanced expression of the TUBB3 gene, which encodes for TUJ1, in Ac4-5SGlcNAc-treated cells confirmed our observation of enhanced protein expression as determined by flow cytometry and immunofluorescence microscopy (Figure 3A–C). The decrease in expression of FOXG1 and HES5 genes by day 8 suggests that the Ac4-5SGlcNAc-treated NPCs are exiting their proliferative state. Gene ontology (GO) term analysis of transcripts on day 4 of differentiation showed that neuron differentiation, forebrain development, and neuron development were significantly enriched categories among upregulated genes in Ac4-5SGlcNAc-treated cells compared to DMSO-treated cells (Figure 5C). These data demonstrate that a critical time in the differentiation process occurs at least 48 h after induction and that transcriptional changes occur earlier than previously observed at the protein level.
By contrast, transcripts that were downregulated in Ac4-5SGlcNAc-treated cells were enriched in categories such as regulation of proteostasis and redox status. Notably, decreased protein O-GlcNAcylation in neuronal cells has been shown to elevate autophagic flux, reduce levels of toxic protein aggregates, and provide protection from neurodegeneration.50 Modulation of autophagic flux and proteotoxicity has also been observed in Caenorhabditis elegans O-GlcNAc cycling mutants.51 Thus, we hypothesize that inhibition of OGT in our study could lead to increased mitochondrial activity and autophagic flux, which are known to benefit neuronal health.52 Continued analysis of key metabolic transitions occurring within the first several days of neural induction may lead to improved understanding of the role that O-GlcNAc plays as a potential regulatory switch from the pluripotent state to the progenitor state, and finally the transition to the postmitotic neuronal fate.
CONCLUSION
The results presented in this study further corroborate O-GlcNAcylation as a key regulator in the process of embryonic stem cell differentiation to neurons. Our findings that Ac4-5SGlcNAc induces expression of neuronal genes as early as day 4 of differentiation and initiates increased axonal branching by day 8 of differentiation suggests a novel mechanism by which in vitro neuronal differentiation can be accelerated. From a practical standpoint, this finding may prove useful for the development of stem-cell-based therapies for neurodegenerative conditions. In the future, it will be important to study whether the effects of OGT inhibition during neural differentiation of hESCs could be reproduced with more specialized neurons, as well as in adult neurogenesis. In addition, these results may contribute new insights into other functions of OGT and O-GlcNAc in the development of the human brain.
METHODS
Cell Culture
Human embryonic stem cell (hESC) lines H1, H7, and H9 (WA01, WA07, WA09; WiCell) were cultured with hESC-qualified basement membrane matrix Matrigel (BD Biosciences). Cells were grown under standard conditions (37 °C, 5% CO2) in TeSR2 media (STEMCELL Technologies Inc.). Media were exchanged daily after the first 48 h of cell culture, and cells were passaged every 3 to 5 days using 1 mg mL–1 Dispase (Gibco) and mechanical lifting. Karyotype analysis was routinely performed and indicated that all samples were diploid and had no chromosomal abnormalities.
Neural Induction
hESC lines were differentiated into neural progenitors as reported in ref 27. Briefly, cells were dissociated using Accutase (STEMCELL Technologies Inc.) and plated on Matrigel (BD Biosciences)-treated dishes at a density of 40 000–50 000 cells/cm2 in the presence of MEF-conditioned hESC medium containing 10 ng/mL FGF-2 (R&D Systems) and 10 μM ROCK inhibitor (Y-27632; Tocris). MEF-conditioned hESC medium was obtained by culturing mytomycin-C-treated mouse embryonic fibroblasts (MEF; Millipore PMEF-CF) in DMEM/F12, 20% (v/v) Knockout Serum Replacement, 1 mM GlutaMAX, 100 μM MEM nonessential amino acids, 0.1 mM β-mercaptoethanol (Invitrogen), and 6 ng/mL FGF-2. After 24 h of cell growth, media were harvested and sterile-filtered. The ROCK inhibitor was withdrawn the day after plating, and hESCs were allowed to expand for 3–4 days or until they were 80–90% confluent. Neural differentiation was initiated using KSR medium, which contained Knockout DMEM (Invitrogen), 15% (v/v) Knockout Serum Replacement, 1 mM GlutaMAX, 100 μM MEM nonessential amino acids, and 0.1 mM β-mercaptoethanol. To inhibit SMAD signaling, 100 nM LDN-193189 (Stemgent) or 200 ng/mL Noggin (33-NG-050, R&D Systems) and 10 μM SB431542 (Tocris) were added on days 0–5 of induction. Cells were fed daily, and N2 medium was added in increasing 25% increments every other day starting on day 4 and leading to 100% N2 on day 10. N2 media consisted of 1:1 DMEM/F12 powder (Gibco/Invitrogen) resuspended in distilled water, glucose, sodium bicarbonate, putrescine, progesterone, sodium selenite, insulin (Sigma), and apo-transferrin (Kamada Ltd.).
Chemical Inhibitors
Synthesis of Ac4-5SGlcNAc was carried out as described previously.34 Thiamet-G was purchased from Cayman Chemical. hESCs undergoing neural differentiation were treated with a 10 or 50 μM final concentration of OGT inhibitor, starting 2 days before the neural induction began. The inhibitor was added to the media daily throughout the neural induction protocol. To determine the effects of OGA inhibition during neural induction, cells were treated at different times during induction with 100 nM Thiamet-G. DMSO (Sigma) was used as a control for all of the inhibitions.
Cell Surface Glycan Perturbation
hESCs undergoing neural differentiation were treated either with 2 mM of the O-glycosylation inhibitor, benzyl-α-GalNAc (Sigma), or 0.05 U/mL of α-mannosidase (Sigma) from day –2 to 8. Concentrations were determined based on previous literature.40–42
Western Blot Analysis
Whole cell protein extracts were lysed by probe sonication in buffer consisting of 50 mM Tris-HCl at pH 8.0, 150 mM NaCl, 1% Triton X-100 or Tween, and 2 mM EDTA (including protease and phosphatase inhibitors (Roche) and 1 μM Thiamet-G). Samples were separated by SDS-PAGE (4–12% Bio-Rad), transferred to nitrocellulose membranes, and blocked with 5% milk (LabScientific, inc.) or BSA (Sigma) in PBS with Tween 20 (Sigma). The following primary antibodies were used to detect the indicated proteins: anti-OGT (DM-17, Sigma and sc-32921, Santa Cruz Biotech), anti-OCT4 (MAB4401, Millipore), anti-O-GlcNAc (RL2, Thermo Scientific), anti-OGA (SAB4200267, Sigma), anti-PAX6 (PRB-278P, Covance), antitubulin (T5168, Sigma), anti-GFAT1 (3818, Cell Signaling), anti-GNPNAT1 (SAB2701548, Sigma), anti-PGM3 (sc-100410, Santa Cruz Biotech), anti-UAP1 (GTX103592, GeneTex), anti-NAGK (N6788, Sigma), and anti-GALE (ab155997, abcam). The following horseradish peroxidase-linked secondary antibodies were used for detection at a dilution ratio of 1:5000 goat antirabbit-HRP-conjugated secondary antibody (4010–05, Southern Biotech) and goat antimouse-HRP-conjugated secondary antibody (1010–05, Southern Biotech).
Immunofluorescence Microscopy
Cells were fixed with 4% (v/v) paraformaldehyde (Thermo Scientific) for 40 min, rinsed three times with 1× PBS for 5 min, and blocked and permeabilized for at least 1 h with 5% (v/v) FBS plus 0.3% (v/v) Triton X-100 in PBS. Cells were stained with the following primary antibodies: TUJ1 (MAB1637, Millipore), MAP-2 (MAB3418, Millipore), PAX6 (PRB-278P, Covance), and OCT4 (MAB4401, Millipore). Cells were rinsed three times with 1× PBS and were incubated with Alexa Fluor conjugated 488-conjugated goat-antimouse secondary antibody and Alexa Fluor 594-conjugated goat-antirabbit secondary antibody (Invitrogen) for 2 h at room temperature. After, cells were rinsed three times with 1× PBS, and DAPI (Invitrogen) was added for 5 min before analysis using an Olympus IX71 fluorescent microscope. Control cells were stained with mouse and rabbit IgG isotypes (PP54, PP64, Millipore).
Flow Cytometry
Colonies were dissociated with Accutase and rinsed twice with 1× PBS with 0.5% (w/v) BSA. Cells were counted using a hemocytometer, and roughly 1 × 106 cells per sample were used for analysis. All cells were fixed with 4% (v/v) paraformaldehyde for 40 min at 4 °C, washed twice with PBS, permeabilized using 0.1% (v/v) Triton X-100 in PBS, and blocked using 0.5% (w/v) BSA in PBS. Cells were stained with conjugated antibodies for 20 min in the dark on ice. Primary conjugated antibodies were the following: MAP2-PE (FCMAB318PE, Millipore), TUJ1 Alexa Fluor 488 (560381, BD Pharmingen), MAP2B Alexa Fluor 647 (560382, BD Pharmingen), OCT3/4 Alexa Fluor 647 (560329, BD Pharmingen), and SOX2 Alexa Fluor 488 (561593, BD Pharmingen). Flow cytometry (BD FACs Calibur from BD Biosciences) was performed, and data were analyzed using FlowJo software v9.8.1 (TreeStar Inc.). Total cell population was identified and gated based on characteristic voltage pulse area measurement of forward scatter (FSC: particle size) and side scatter (SSC: granularity/complexity of the particle) of the laser light.
Lectin Analysis
Cells were harvested and fixed in 4% paraformaldehyde at 4 °C for 40 min in the dark. Cells were then labeled with five fluorescein isothiocyanate (FITC)-labeled lectins (Vector Laboratories, Inc., Burlingame, CA, USA): Arachis hypogaea agglutinin (PNA), Canavalia ensiformis agglutinin (ConA), Artocarpus integrifolia agglutinin (AIL or Jacalin), and Lens culinaris agglutinin (LCH). All samples were mixed with 50 μg/mL of their designated lectins and incubated for 30 min at room temperature in the dark. The lectin-labeled cell samples were washed three times by centrifuging the samples for 5 min at 300g and then resuspending the resultant pellets in 1× PBS with 5% BSA. For controls, 50 μg/mL of each individual lectin was incubated in 100 mg mL–1 of hapten sugars overnight at 4 °C in the dark. After incubation, fixed cell samples were labeled with these sugar-treated lectins and used as controls. The following sugars (monosaccharides) were used as inhibitors: D-galactose (for PNA and Jacalin), N-acetyl-D-galactosamine (for Jacalin), and methyl α-D-mannopyranoside and D-mannose (for ConA and LCA). All samples were analyzed using a BD Accuri C6 flow cytometer, and data were acquired with BD Accuri C6 software (BD Biosciences). Data were analyzed using FlowJo software v9.8.1 (Treestar, Ashland, OR, USA) and gated as described above.
Cell Proliferation
The effect of Ac4-5SGlcNAc on cell proliferation was measured using a calorimetric (MTT) kit for cell survival and proliferation (CT02, Millipore) following the manufacturers’ instructions.
High Performance Anion Exchange Chromatography (HPAEC)
Cells were induced to neural progenitors and harvested as described above. Typically –3 × 106 cells were harvested per sample. Cells were then lysed with 80% “super-cold” methanol (on dry ice). Lysate was centrifuged at 2000g for 15 min. The supernatant was dried by rotary evaporation for 4–5 h. A metabolite pellet was resuspended in 40 mM sodium phosphate buffer (pH 7.4; 40 μL per million cells) and filtered through an Amicon Ultra centrifugal filter unit (Millipore, 10 000 MWCO). Filtrates were analyzed by HPAEC (ICS-3000 system, Dionex) with CarboPacPA1 (Dionex) with a pulsed amperometry detector (PAD) and UV-detector in-line.53,54 Typically, 20 μL of metabolite was injected into the sample loading loop before the sample enters a guard column (Dionex, 4 × 50 mm) and then an analytical column (Dionex, 4 × 250 mm). The eluents used were 1.0 mM NaOH (C) and 1.0 M NaOAc and 1.0 mM NaOH (D). Low-carbonate NaOH (50% in water) was obtained from Fisher Scientific (SS254–1), and NaOAc was from Sigma (71183). HPAEC was run with a flow rate = 1 mL/min, and the following gradient elution was performed: T0 min = 95% C, T5 = 85% C, T15 = 70% C, T20 = 60% C, T45 = 60% C, T50 = 0% C, T60 = 0% E2, T65 = 95%, T75 = 95%. UDP-GlcNAc standards (50 μM, 25 uM, 10 μM, and 2.5 μM) were also analyzed on the same day as cellular samples. UDP-GlcNAc peak areas were calculated by integration and converted to picomoles of UDP-GlcNAc by analyzing the peak areas of the UDP-GlcNAc standards and generating a comparable standard curve. Data were normalized to cell number.
RNA-Sequencing and Data Analysis
Total RNA was isolated from cells using the TRIzol Reagent (Invitrogen) and RNeasy Mini kit (Qiagen). RNA quality was assessed using the Agilent 2100 Bioanalyzer. One microgram of each sample was used to isolate mRNA and prepare the library following the protocol of the Illumina TruSeq RNA sample prep kit v2 (RS-122–2001). Prepared libraries were sequenced on an Illumina HiSeq 2500 sequencer Rapid Run mode at the QB3 Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley. Sequenced reads (27 million) were mapped to the human RefSeq (hg19) using TopHat55 with default parameters. Differentially expressed genes were identified using Cuffdiff.56 Biological duplicates for each condition were subjected to the analysis, and the p value cutof rf of 0.05 was applied to select significant differences in gene expression for scatter plots and for GO term analysis using DAVID Bioinformatics Resources 6.7.57
Statistical Analysis
All data are expressed as mean ± SEM. The n represents the number of biological replicates. P values were calculated using the unpaired two-tailed Student’s t test.
Supplementary Material
Acknowledgments
We thank Nathan Yee for assisting with Figure 1C and Ioannis Mountziaris, Douglas Fox, Jason Tsai, and Phung Gip for helpful discussions. This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 Instrumentation Grants S10RR029668 and S10RR027303. L.M.A. was supported by a National Institutes of Health F31 Predoctoral Fellowship. I.W.B. was supported by the California Institute for Regenerative Medicine Bridges to Stem Cell Research Award. N.G.R. was supported by a National Science Foundation Fellowship. T.Y. was supported by a Jane Coffin Childs Fund Fellowship. N.D.P. was supported by a NIH F30AG040909 fellowship. This work was funded by National Institutes of Health grant R01 CA200423 to C.R.B. and Cancer Prevention and Research Institute of Texas grant RP110080 to J.J.K.
Footnotes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschem-bio.7b00232.
Supporting Figures 1–6 (PDF)
Data tables (XLSX)
References
- 1.Kearse KP, Hart GW. Lymphocyte activation induces rapid changes in nuclear and cytoplasmic glycoproteins. Proc Natl Acad Sci U S A. 1991;88:1701–1705. doi: 10.1073/pnas.88.5.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kneass ZT, Marchase RB. Neutrophils Exhibit Rapid Agonist-induced Increases in Protein-associated O-GlcNAc. J Biol Chem. 2004;279:45759–45765. doi: 10.1074/jbc.M407911200. [DOI] [PubMed] [Google Scholar]
- 3.Yang X, Ongusaha PP, Miles PD, Havstad JC, Zhang F, So WV, Kudlow J, Michell RH, Olefsky JM, Field SJ, Evans RM. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature. 2008;451:964–969. doi: 10.1038/nature06668. [DOI] [PubMed] [Google Scholar]
- 4.Rexach JE, Clark PM, Mason DE, Neve RL, Peters EC, Hsieh-Wilson LC. Dynamic O-GlcNAc modification regulates CREB-mediated gene expression and memory formation. Nat Chem Biol. 2012;8:253–261. doi: 10.1038/nchembio.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Love DC, Ghosh S, Mondoux MA, Fukushige T, Wang P, Wilson MA, Iser WB, Wolkow CA, Krause MW, Hanover JA. Dynamic O-GlcNAc cycling at promoters of Caenorhabditis elegans genes regulating longevity, stress, and immunity. Proc Natl Acad Sci U S A. 2010;107:7413–7418. doi: 10.1073/pnas.0911857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross Talk Between O-GlcNAcylation and Phosphorylation: Roles in Signaling, Transcription, and Chronic Disease. Annu Rev Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bond MR, Hanover JA. O-GlcNAc cycling: a link between metabolism and chronic disease. Annu Rev Nutr. 2013;33:205–229. doi: 10.1146/annurev-nutr-071812-161240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shafi R, Iyer SP, Ellies LG, O’Donnell N, Marek KW, Chui D, Hart GW, Marth JD. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci U S A. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Donnell N, Zachara NE, Hart GW, Marth JD. Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol Cell Biol. 2004;24:1680–1690. doi: 10.1128/MCB.24.4.1680-1690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myers SA, Panning B, Burlingame AL. Polycomb repressive complex 2 is necessary for the normal site-specific O-GlcNAc distribution in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2011;108:9490–9495. doi: 10.1073/pnas.1019289108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jang H, Kim TW, Yoon S, Choi SY, Kang TW, Kim SY, Kwon YW, Cho EJ, Youn HD. O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell Stem Cell. 2012;11:62–74. doi: 10.1016/j.stem.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Shi FT, Kim H, Lu W, He Q, Liu D, Goodell MA, Wan M, Songyang Z. Ten-eleven translocation 1 (Tet1) is regulated by O-linked N-acetylglucosamine transferase (Ogt) for target gene repression in mouse embryonic stem cells. J Biol Chem. 2013;288:20776–20784. doi: 10.1074/jbc.M113.460386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Speakman CM, Domke TC, Wongpaiboonwattana W, Sanders K, Mudaliar M, van Aalten DM, Barton GJ, Stavridis MP. Elevated O-GlcNAc levels activate epigenetically repressed genes and delay mouse ES cell differentiation without affecting naive to primed cell transition. Stem Cells. 2014;32:2605–2615. doi: 10.1002/stem.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeon JH, Suh HN, Kim MO, Ryu JM, Han HJ. Glucosamine-Induced OGT Activation Mediates Glucose Production Through Cleaved Notch1 and FoxO1, Which Coordinately Contributed to the Regulation of Maintenance of Self-Renewal in Mouse Embryonic Stem Cells. Stem Cells Dev. 2014;23:2067–2079. doi: 10.1089/scd.2013.0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myers SA, Peddada S, Chatterjee N, Friedrich T, Tomoda K, Krings G, Thomas S, Maynard J, Broeker M, Thomson M, Pollard K, Yamanaka S, Burlingame AL, Panning B. SOX2 O-GlcNAcylation alters its protein-protein interactions and genomic occupancy to modulate gene expression in pluripotent cells. eLife. 2016;5:e10647. doi: 10.7554/eLife.10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffith LS, Mathes M, Schmitz B. β-Amyloid precursor protein is modified with O-linked N-acetylglucosamine. J Neurosci Res. 1995;41:270–278. doi: 10.1002/jnr.490410214. [DOI] [PubMed] [Google Scholar]
- 17.Liu F, Iqbal K, Grundke-Iqbal I, Hart GW, Gong CX. O-GlcNAcylation regulates phosphorylation of tau: A mechanism involved in Alzheimer’s disease. Proc Natl Acad Sci U S A. 2004;101:10804–10809. doi: 10.1073/pnas.0400348101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. Exploring the O-GlcNAc proteome: direct identification of O-GlcNAc-modified proteins from the brain. Proc Natl Acad Sci U S A. 2004;101:13132–13137. doi: 10.1073/pnas.0403471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vosseller K, Trinidad JC, Chalkley RJ, Specht CG, Thalhammer A, Lynn AJ, Snedecor JO, Guan S, Medzihradszky KF, Maltby DA, Schoepfer R, Burlingame AL. O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol Cell Proteomics. 2006;5:923–934. doi: 10.1074/mcp.T500040-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Wang AC, Jensen EH, Rexach JE, Vinters HV, Hsieh-Wilson LC. Loss of O-GlcNAc glycosylation in forebrain excitatory neurons induces neurodegeneration. Proc Natl Acad Sci U S A. 2016;113:15120–15125. doi: 10.1073/pnas.1606899113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lagerlöf O, Slocomb JE, Hong I, Aponte Y, Blackshaw S, Hart GW, Huganir RL. The nutrient sensor OGT in PVN neurons regulates feeding. Science. 2016;351:1293–1296. doi: 10.1126/science.aad5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lagerlöf O, Hart GW, Huganir RL. O-GlcNAc transferase regulates excitatory synapse maturity. Proc Natl Acad Sci U S A. 2017;114:1684–1689. doi: 10.1073/pnas.1621367114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Li X, Yu Y, Shi J, Liang Z, Run X, Li Y, Dai CL, Grundke-Iqbal I, Iqbal K, Liu F, Gong CX. Developmental Regulation of Protein O-GlcNAcylation, O-GlcNAc Transferase, and O-GlcNAcase in Mammalian Brain. PLoS One. 2012;7:e43724. doi: 10.1371/journal.pone.0043724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Francisco H, Kollins K, Varghis N, Vocadlo D, Vosseller K, Gallo G. O-GLcNAc post-translational modifications regulate the entry of neurons into an axon branching program. Dev Neurobiol. 2009;69:162–173. doi: 10.1002/dneu.20695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu K, Paterson AJ, Zhang F, McAndrew J, Fukuchi K, Wyss JM, Peng L, Hu Y, Kudlow JE. Accumulation of protein O-GlcNAc modification inhibits proteasomes in the brain and coincides with neuronal apoptosis in brain areas with high O-GlcNAc metabolism. J Neurochem. 2004;89:1044–1055. doi: 10.1111/j.1471-4159.2004.02389.x. [DOI] [PubMed] [Google Scholar]
- 26.Maury JJ, Chan KK, Zheng L, Bardor M, Choo AB. Excess of O-linked N-acetylglucosamine modifies human pluripotent stem cell differentiation. Stem Cell Res. 2013;11:926–937. doi: 10.1016/j.scr.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 27.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yanagisawa M, Yu RK. O-linked beta-N-acetylglucosaminylation in mouse embryonic neural precursor cells. J Neurosci Res. 2009;87:3535–3545. doi: 10.1002/jnr.22170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wells L, Vosseller K, Hart GW. Glycosylation of Nucleocytoplasmic Proteins: Signal Transduction and O-GlcNAc. Science. 2001;291:2376–2378. doi: 10.1126/science.1058714. [DOI] [PubMed] [Google Scholar]
- 30.Agathocleous M, Harris WA. Metabolism in physiological cell proliferation and differentiation. Trends Cell Biol. 2013;23:484–492. doi: 10.1016/j.tcb.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 31.Boehmelt G, Wakeham A, Elia A, Sasaki T, Plyte S, Potter J, Yang Y, Tsang E, Ruland J, Iscove NN, Dennis JW, Mak TW. Decreased UDP-GlcNAc levels abrogate proliferation control in EMeg32-deficient cells. EMBO J. 2000;19:5092–5104. doi: 10.1093/emboj/19.19.5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buse MG, Robinson KA, Marshall BA, Mueckler M. Differential effects of GLUT1 or GLUT4 overexpression on hexosamine biosynthesis by muscles of transgenic mice. J Biol Chem. 1996;271:23197–23202. doi: 10.1074/jbc.271.38.23197. [DOI] [PubMed] [Google Scholar]
- 33.Gerhart DZ, Broderius MA, Borson ND, Drewes LR. Neurons and microvessels express the brain glucose transporter protein GLUT3. Proc Natl Acad Sci U S A. 1992;89:733–737. doi: 10.1073/pnas.89.2.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gloster TM, Zandberg WF, Heinonen JE, Shen DL, Deng L, Vocadlo DJ. Hijacking a biosynthetic pathway yields a glycosyltransferase inhibitor within cells. Nat Chem Biol. 2011;7:174–181. doi: 10.1038/nchembio.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ortiz-Meoz RF, Jiang J, Lazarus MB, Orman M, Janetzko J, Fan C, Duveau DY, Tan ZW, Thomas CJ, Walker S. A small molecule that inhibits OGT activity in cells. ACS Chem Biol. 2015;10:1392–1397. doi: 10.1021/acschembio.5b00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menezes JR, Luskin MB. Expression of neuron-specific tubulin defines a novel population in the proliferative layers of the developing telencephalon. J Neurosci. 1994;14:5399–5416. doi: 10.1523/JNEUROSCI.14-09-05399.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung EJ, Chun JN, Jung SA, Cho JW, Lee JH. TGF-beta-stimulated aberrant expression of class III beta-tubulin via the ERK signaling pathway in cultured retinal pigment epithelial cells. Biochem Biophys Res Commun. 2011;415:367–372. doi: 10.1016/j.bbrc.2011.10.074. [DOI] [PubMed] [Google Scholar]
- 38.Clark PM, Dweck JF, Mason DE, Hart CR, Buck SB, Peters EC, Agnew BJ, Hsieh-Wilson LC. Direct In-Gel Fluorescence Detection and Cellular Imaging of O-GlcNAc-Modified Proteins. J Am Chem Soc. 2008;130:11576–11577. doi: 10.1021/ja8030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ji S, Kang JG, Park SY, Lee J, Oh YJ, Cho JW. O-GlcNAcylation of tubulin inhibits its polymerization. Amino Acids. 2011;40:809–818. doi: 10.1007/s00726-010-0698-9. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z, Oron E, Nelson B, Razis S, Ivanova N. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell. 2012;10:440–454. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 41.Paszkiewicz-Gadek A, Porowska H, Lemancewicz D, Wolczynski S, Gindzienski A. The influence of N- and O-glycosylation inhibitors on the glycosylation profile of cellular membrane proteins and adhesive properties of carcinoma cell lines. Int J Mol Med. 2006;17:669–674. [PubMed] [Google Scholar]
- 42.Arcaro KF, Wang J, Otis CN, Zuckerman BM. Beta-galactosidase and alpha-mannosidase inhibit formation of multicellular nodules in breast cancer cell cultures. Anticancer Res. 2004;24:139–144. [PubMed] [Google Scholar]
- 43.Persichetti E, Klein K, Paciotti S, Lecointe K, Balducci C, Franken S, Duvet S, Matzner U, Roberti R, Hartmann D, Gieselmann V, Beccari T. Lysosomal di-N-acetylchito-biase-deficient mouse tissues accumulate Man2GlcNAc2 and Man3-GlcNAc2. Biochim Biophys Acta, Mol Basis Dis. 2012;1822:1137–1146. doi: 10.1016/j.bbadis.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 44.Yuzwa SA, Macauley MS, Heinonen JE, Shan X, Dennis RJ, He Y, Whitworth GE, Stubbs KA, McEachern EJ, Davies GJ, Vocadlo DJ. A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat Chem Biol. 2008;4:483–490. doi: 10.1038/nchembio.96. [DOI] [PubMed] [Google Scholar]
- 45.Olivier-Van Stichelen S, Wang P, Comly M, Love DC, Hanover JA. Nutrient-driven O-GlcNAc cycling impacts neurodevelopmental timing and metabolism. J Biol Chem. 2017;292:6076. doi: 10.1074/jbc.M116.774042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gambetta MC, Oktaba K, Müller J. Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science. 2009;325:93–96. doi: 10.1126/science.1169727. [DOI] [PubMed] [Google Scholar]
- 47.Sinclair DA, Syrzycka M, Macauley MS, Rastgardani T, Komljenovic I, Vocadlo DJ, Brock HW, Honda BM. Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc) Proc Natl Acad Sci U S A. 2009;106:13427–13432. doi: 10.1073/pnas.0904638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu TW, Myschyshyn M, Sinclair DA, Cecioni S, Beja K, Honda BM, Morin RD, Vocadlo DJ. Genome-wide chemical mapping of O-GlcNAcylated proteins in Drosophila melanogaster. Nat Chem Biol. 2016;13:161–167. doi: 10.1038/nchembio.2247. [DOI] [PubMed] [Google Scholar]
- 49.Yang X, Zhang F, Kudlow JE. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell. 2002;110:69–80. doi: 10.1016/s0092-8674(02)00810-3. [DOI] [PubMed] [Google Scholar]
- 50.Kumar A, Singh PK, Parihar R, Dwivedi V, Lakhotia SC, Ganesh S. Decreased O-linked GlcNAcylation protects from cytotoxicity mediated by huntingtin exon1 protein fragment. J Biol Chem. 2014;289:13543–13553. doi: 10.1074/jbc.M114.553321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang P, Lazarus BD, Forsythe ME, Love DC, Krause MW, Hanover JA. O-GlcNAc cycling mutants modulate proteotoxicity in Caenorhabditis elegans models of human neurodegenerative diseases. Proc Natl Acad Sci U S A. 2012;109:17669–17674. doi: 10.1073/pnas.1205748109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Batlevi Y, La Spada AR. Mitochondrial autophagy in neural function, neurodegenerative disease, neuron cell death, and aging. Neurobiol Dis. 2011;43:46–51. doi: 10.1016/j.nbd.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomiya N, Ailor E, Lawrence SM, Betenbaugh MJ, Lee YC. Determination of nucleotides and sugar nucleotides involved in protein glycosylation by high-performance anion-exchange chromatography: sugar nucleotide contents in cultured insect cells and mammalian cells. Anal Biochem. 2001;293:129–137. doi: 10.1006/abio.2001.5091. [DOI] [PubMed] [Google Scholar]
- 54.Yu SH, Boyce M, Wands AM, Bond MR, Bertozzi CR, Kohler JJ. Metabolic labeling enables selective photocrosslinking of O-GlcNAc-modified proteins to their binding partners. Proc Natl Acad Sci U S A. 2012;109:4834–4839. doi: 10.1073/pnas.1114356109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.