Abstract
Unbiased genomic screening analyses have highlighted novel immunomodulatory properties of the active form of vitamin D, 1,25-dihydroxyvitamin D (1,25(OH)2D). However, clearer interpretation of the resulting gene expression data is limited by cell model specificity. The aim of the current study was to provide a broader perspective on common gene regulatory pathways associated with innate immune responses to 1,25(OH)2D, through systematic re-interrogation of existing gene expression databases from multiple related monocyte models (the THP-1 monocytic cell line (THP-1), monocyte-derived dendritic cells (DCs), and monocytes). Vitamin D receptor (VDR) expression is common to multiple immune cell types, and thus pathway analysis of gene expression using data from multiple related models provides an inclusive perspective on the immunomodulatory impact of vitamin D. A bioinformatic workflow incorporating pathway analysis using PathVisio and WikiPathways was utilised to compare each set of gene expression data based on pathway level context. Using this strategy, pathways related to the TCA cycle, oxidative phosphorylation and ATP synthesis and metabolism were shown to significantly regulated by 1,25(OH)2D in each of the repository models (Z-scores 3.52 – 8.22). Common regulation by 1,25(OH)2D was also observed for pathways associated with apoptosis and the regulation of apoptosis (Z-scores 2.49 – 3.81). In contrast to the primary culture DC and monocyte models, the THP-1 myelomonocytic cell line showed strong regulation of pathways associated with cell proliferation and DNA replication (Z-scores 6.1 – 12.6). In short, data presented here support a fundamental role for active 1,25(OH)2D as a pivotal regulator of immunometabolism.
Keywords: vitamin D, monocyte, dendritic cells, gene expression, pathway
Introduction
In recent years studies in vivo and in vitro have shown that vitamin D is able to influence biological responses that extend far beyond its classical effects on skeletal homeostasis. Prominent amongst these extra-skeletal effects is the interaction between vitamin D and the immune system, including regulation of both innate and adaptive immune responses (Adams and Hewison 2008; Hewison 2011; Wei and Christakos 2015). As a consequence of these observations, vitamin D-deficiency has been linked to increased risk of bacterial and viral infection (Lake and Adams 2011; Nnoaham and Clarke 2008), as well as inflammatory and autoimmune disease (Cantorna 2012; Jeffery, et al. 2016). The ability of supplementary or therapeutic vitamin D to prevent or improve these immune disorders is much less clear, and is the subject of randomized placebo control trials currently underway. Crucially, improved understanding of the mechanisms that underpin the immunomodulatory actions of vitamin D has greatly helped to improve the design and outcome of clinical trials. In particular, unbiased analysis of gene responses to vitamin D supplementation has uncovered previously unrecognised immune targets for vitamin D (Chun, et al. 2014; Liu, et al. 2006) that are now key markers of vitamin D function in supplementation trials.
The initial observation linking vitamin D and immune function was detection of the nuclear vitamin D receptor (VDR) in lymphocytic and myeloid cells (Bhalla, et al. 1983; Mangelsdorf, et al. 1984), indicating that these cells are able to respond to the active form of vitamin D, 1,25(OH)2D which binds with VDR. Further studies showed that cells from innate immune system such as monocytes/macrophages (Kreutz, et al. 1993) and dendritic cells (DC) (Hewison, et al. 2003) are also able to synthesise 1,25(OH)2D from the inactive precursor 25-hydroxyvitamin D (25D), the main circulating form of vitamin D. As the principal effect of dietary vitamin D supplementation is to raise serum levels of 25D, the ability of monocytes and DCs to convert 25D to 1,25(OH)2D provides a localised, autocrine, pathway by which enhanced 25D can influence both innate and adaptive immunity (Jeffery, et al. 2012; Liu et al. 2006). These observations placed monocytes and DCs at the centre of the immunomodulatory activity of vitamin D. Synthesis of 1,25(OH)2D by these cells has the potential to influence endogenous innate immune cell function in the form of enhanced antibacterial activity (Hewison 2011; Liu et al. 2006), and/or modulated antigen presentation (Hewison et al. 2003; Jeffery et al. 2012). Furthermore, 1,25(OH)2D generated by monocytes and/or DCs may also impact adaptive immune function by exert exogenous effects on T cells or B cells expressing VDR (Jeffery et al. 2012). Despite this, our understanding of the broader impact of vitamin D on cells from the innate immune system such as monocytes, macrophages and DCs remains far from clear.
Recent genome-wide expression studies using a monocyte-derived DC model highlighted the potential role of pathways associated with glucose metabolism, the TCA cycle and oxidative phosphorylation in mediating the effects of 1,25(OH)2D in promoting a tolerogenic DC phenotype (Ferreira, et al. 2015). The aim of the current study was to provide a broader analysis of monocyte/DC responses to 1,25(OH)2D using existing gene expression repositories for multiple models of innate immune function. A meta-analysis of various datasets has been performed using an integrative workflow of open-source bioinformatics tools in order to give a biological context to the genes that are significantly modulated by 1,25(OH)2D. Finally, a comparison between the immune cell types selected has been applied to highlight the common biological processes that are altered by vitamin D.
Materials & Methods
Workflow overview
A workflow was developed that utilises and integrates gene expression data from public repositories and places them in a biological context. Supplemental Figure 1 illustrates step by step the different tools and methods used in the analysis workflow. The first procedure consisted of open and free modules in ArrayAnalysis to i) perform a data quality check, ii) normalize the raw transcriptomic data and iii) perform a statistical analysis to obtain the parameters that show gene expression changes. The processed datasets were then applied to PathVisio v. 3.2.4 software (Kutmon, et al. 2015; van Iersel, et al. 2008) in order to perform statistical analyses that highlight the biological processes significantly altered as a consequence of gene expression changes and to visualize them in pathway diagrams from the WikiPathways pathway repository downloaded January 2017 (Kutmon, et al. 2016). Comparison of the pathways shown to be significantly altered between the different cell types using PathVisio and WikiPathways was also represented by heat maps diagrams. In another approach, genes shown to be regulated by 1,25(OH)2D in all three cell models were used to identify enriched biological processes by Gene Ontology (GO) analysis and the results were visualized in biological networks using ClueGO v 2.3.3 (Bindea, et al. 2009), an app of Cytoscape 3.4.0 (Smoot, et al. 2011) (Supplemental Figure 1). This workflow is designed in a user friendly fashion that does not require programming skills. In order to reproduce this workflow, it is recommended to use the latest versions of designated software. Furthermore, it is also important that gene annotation database and pathways repositories used in the analysis are regularly updated.
Transcriptomic datasets
Publically available transcriptomic data sets for 1,25(OH)2D-treated monocytes and dendritic cells (DC) were selected from ArrayExpress (https://www.ebi.ac.uk/arrayexpress/, (Kolesnikov, et al. 2015)) and Gene Expression Omnibus (GEO, http://ncbi.nlm.nih.gov/geo/, (Barrett and Edgar 2006)). Selection criteria were based on gene expression studies performed in human cells including monocytic cell lines, monocytes and monocyte-derived DCs treated with 1,25(OH)2D: THP-1 cells (GSE52819 (Verway, et al. 2013)), dendritic cells (GSE13762 (Széles, et al. 2009)) and human monocytes (GSE56490 (Ferreira et al. 2015)). The selection of the three studies was based on experimental design and quality control analysis of the raw data. In all studies data were derived from human immune cells treated with 1,25(OH)2D, or from control (vehicle-treated) cells (Supplemental Table 1). In addition to the three cell models outlined above, preliminary analyses were also carried out using other 1,25(OH)2D-treated cell data repositories: THP-1 cells (GSE60102 (Nurminen, et al. 2015)) and human monocytes (GSE46268 (Wheelwright, et al. 2014)). In addition the data for GSE13762 (Széles, et al. 2009) included both immature and mature DC, and both these models were include in the preliminary analysis. Finally, as a negative control, the preliminary pathway analysis included a 1,25(OH)2D-treated B cell dataset (GSE22523 (Lisse, et al. 2011). Because of missing raw data, incomplete gene annotation, incompatibility with ArrayAnalysis, and non-monocyte origin, some data from the preliminary analysis were not included in the main analysis of this study, but relevant findings related to the data were addressed in Results.
Pre-processing raw data and statistical data analysis in ArrayAnalysis
Unprocessed transcriptomics data were collected from the repositories and processed using the on-line workflow of ArrayAnalysis performed in January 2017 (https://www.arrayanalysis.org (Eijssen, et al. 2013)) to obtain quality control reports, normalized data and perform statistical analysis. The workflow uses an R script as a core module with functions from several Bioconductor libraries. Quality control showed that there was no need to exclude any samples in the three selected transcriptomic datasets (data not shown). Depending on the microarray technology used the normalization algorithm was selected. RMA method was used to normalize GSE52819 (Verway et al. 2013) and GSE56490 (Ferreira et al. 2015). GC-RMA method was applied to GSE13762 (Szeles et al. 2009). Statistical analysis of the normalized data to determine genes that are significantly altered (up/down-regulated), was carried out with a second module from ArrayAnalysis including the moderated t-test from the R/Bioconductor package limma (Ritchie, et al. 2009). The output is a table with the expression parameters for each gene showing fold change (FC), in a log scale) after 1,25(OH)2D treatment and the significance of changes by the Benjamini-Hochberg adjusted p-value. Based on statistical gene expression parameters the significantly up/down-regulated genes were identified using the criteria: absolute log2FC => 0.26 and adjusted p value < 0.05. The average expression (AE) was used to filter out lowly expressed genes and the cut-off was specific for each dataset based on the density plot of the intensities after normalization: THP-1 AE > 4.25; immature DC AE > 3.86; and monocytes AE > 9.5 (Table 1).
Table 1. Regulation of gene expression by 1,25(OH)2D in THP-1, dendritic cells and monocyte models.
Total number of genes measured for each gene expression repository and the number of genes with significantly altered expression. Genes with an absolute log fold change > 0.26 and adjusted p-value < 0.05 were considered as regulated. Based on the expression parameter log fold change genes are classified by up-regulated (log fold-change > 0.26) and down-regulated (log fold-change <−0.26).
| Dataset | Type of cell | Genes measured | Genes significantly altered |
Genes up-regulated | Genes down-regulated |
|---|---|---|---|---|---|
| Verway M et al. (2013) | THP-1 | 21,664 | 2,652 | 1,194 | 1,458 |
| Széles L et al. (2009) | Immature DC | 20,111 | 2,093 | 1,130 | 963 |
| Ferreira G et al (2014) | Monocytes | 21,664 | 1,248 | 525 | 723 |
Pathway statistics and analysis in PathVisio
PathVisio v. 3.2.4 (Kutmon, et al. 2015; van Iersel, et al. 2008), an open-source pathway creation and analysis tool, was used to contextualize significantly altered genes, and altered biological processes were visualised using the biological pathway repository WikiPathways (Kutmon, et al. 2016). Utilizing the BridgeDb identifier mapping feature released on 18 October 2016 (http://www.bridgedb.org/) (van Iersel, et al. 2010), PathVisio recognizes genes (probe) identifiers from the most used databases and microarray platforms such as NCBI and Affymetrix. This identifier mapping database (Homo sapiens Derby Ensembl 85 gene database) enables linking of statistical values from analyzed data to the corresponding gene boxes in the pathway diagrams of WikiPathways. In the analysis, the curated human pathways collection (released April 2017, http://data.wikipathways.org/20170410/gpml/) of WikiPathways was used. For pathway analysis, a criterion is chosen to select genes that are significantly altered within each dataset based on the expression difference parameter of fold change (on log2 scale) and the significance of that change represented as the adjusted p value (<0.05). PathVisio performs an overrepresentation analysis taking into account all genes measured (N), genes that satisfy the criterion (R), genes measured in the experiment that are present in the pathway (n) and genes in the pathway measured in the experiment that satisfy the criteria (r):
The Z-score is used to measure how significantly a subset of genes is altered in a certain pathway compared to the complete dataset. In our analysis, biological pathways that have a Z-score equal to or above 1.96 are considered significantly altered. The function heatmap.2 from the R library gplots (v.3.0.1) was used to create a heatmap that compared and hierarchically clustered pathways for each of the datasets based on Z-scores.
GO analysis with ClueGO in Cytoscape
GO analysis of the in common significantly altered genes (absolute log2FC >= 0.26 and adjusted p value < 0.05) in the three types of immune cells was performed to identify and visualize which GO biological process terms were significantly overrepresented. Within the commonly used open-source network analysis tool Cytoscape v. 3.4 (Smoot et al. 2011), the ClueGO app v 2.3.3 performs GO analysis to group a set of genes in GO terms minimizing redundancy and display their relationship in a network. Connections between the GO nodes containing a common sub-set of genes were calculated with kappa statistics based on the correlated genes that are grouped.
Results
Differentially expressed genes in 1,25(OH)2D-treated THP1 cells, dendritic cells and monocytes
A preliminary pathway analysis of multiple THP-1, monocyte and DC datasets revealed common regulation of several key cell pathways by 1,25(OH)2D in cells from the myeloid lineage: electron transport, oxidative phosphorylation, the TCA cycle, glycolysis and gluconeogenesis, and apoptosis. By contrast, the non-myeloid B cell gene expression dataset showed no similarity with the myeloid effects of 1,25(OH)2D, underlining the lineage-specific effects of 1,25(OH)2D (Supplemental Figure 2). From this initial pathway analysis it was also interesting to note that 1,25(OH)2D regulation of pathways associated with the cell cycle and cell proliferation were only observed in the myeloid leukemic cell line THP-1, and not the other non-neoplastic primary cell models used in the study (Supplemental Figure 2). Because of incomplete raw data or gene annotation, or incompatibility with ArrayAnalysis, further, more detailed, pathway analysis was restricted to raw datasets for GSE52819, GSE13762, GSE56490.
Pathway analysis reveals altered biological processes based on molecular changes in 1,25(OH)2D-treated immune cells
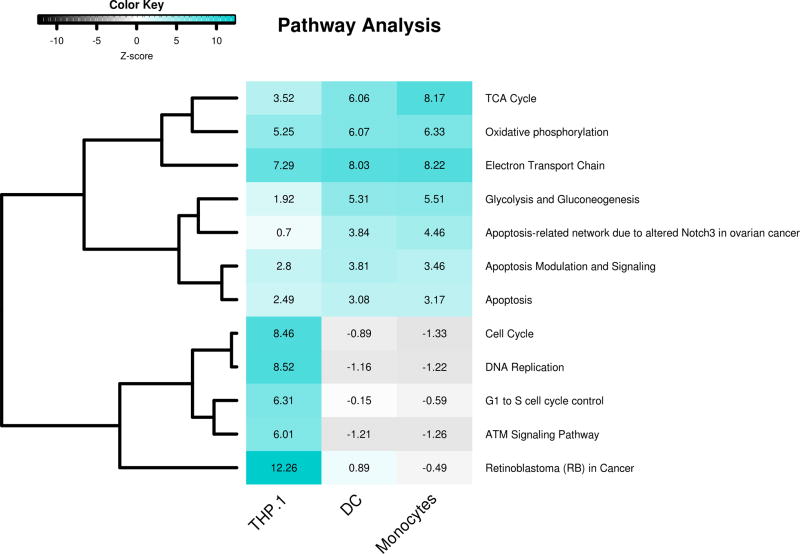
Pathway overrepresentation analysis performed on immune cell datasets GSE52819, GSE13762, and GSE56490 generated a list of biological processes regulated by 1,25(OH)2D in each of the three different innate immune cell models. For each cell type the significantly altered pathways were selected and their Z-scores plotted and clustered in a heatmap (Figure 1) showing Z-score comparisons for major affected pathways between the three cell models. Data indicate that 1,25(OH)2D-regulated pathways were either cell-specific or common to all three models. Pathways related to DNA replication, the cell cycle, and cancer (retinoblastoma) showed high Z-scores, and were more strongly regulated by 1,25(OH)2D in THP-1 cells, consistent with the proliferative leukemic nature of this cell line (25). By contrast, primary cells (DCs and monocytes) showed stronger 1,25(OH)2D-regulation of pathways associated with glycolysis/gluconeogenesis and the apoptosis-related network due to altered Notch3 in ovarian cancer pathway relative to THP-1 cells, albeit at lower Z-scores than observed for THP-1-specific pathways. Pathways associated with apoptosis and apoptosis modulation and signalling also showed lower Z-scores, but were equally induced by 1,25(OH)2D in THP-1 cells, DCs and monocytes. The highest Z-score for 1,25(OH)2D-regulated pathways common to all three cell models was for pathways related to the TCA cycle, oxidative phosphorylation and the electron transport chain.
Figure 1. Common altered pathways in 1,25(OH)2D-treated THP-1, DCs and monocytes.
Heat map representing Z-scores for pathways significantly altered by 1,25(OH)2D after performing a pathway analysis on datasets for THP-1, DCs and monocytes treated with 1,25(OH)2D. Z-scores >1.96 indicate that more genes are significantly altered in this pathway compared to the complete dataset.
GO analysis shows metabolic, immunological and apoptotic processes altered by 1,25D in THP-1 cells, dendritic cells and monocytes
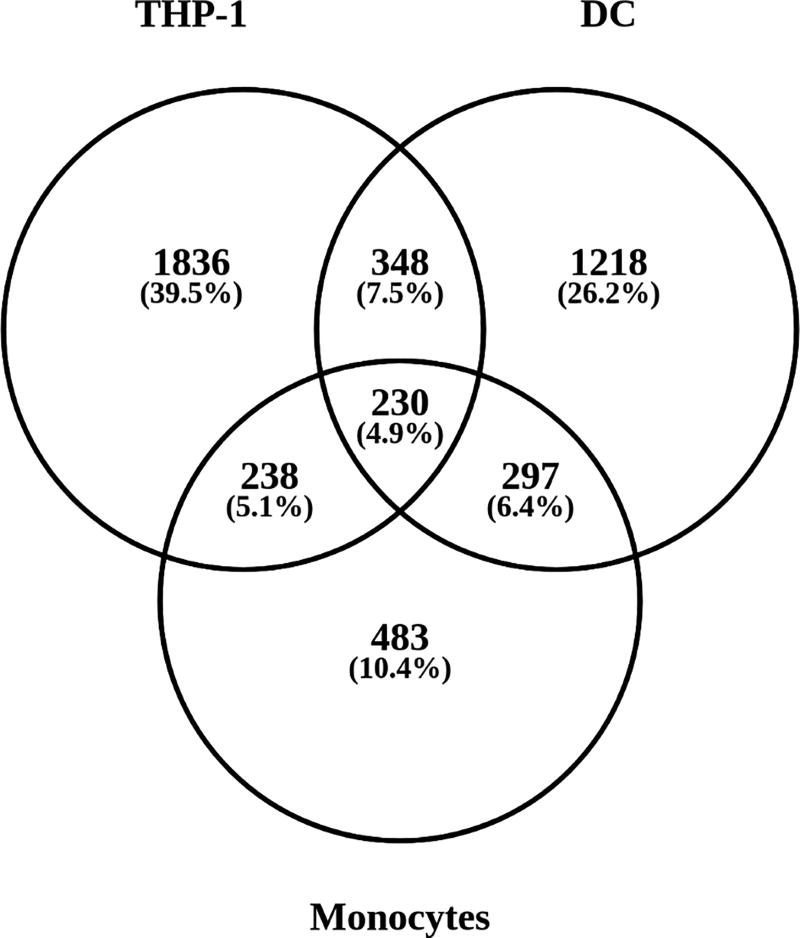
ArrayAnalysis processing of GSE52819, GSE13762, GSE56490 datasets was carried out to define the number of genes measured and number of genes significantly regulated by 1,25(OH)2D in each of the three cell models (Table 1). Further analysis of these data showed that a total of 230 genes were significantly regulated by 1,25(OH)2D in all three cell models (Figure 2), and these genes were selected for subsequent GO analysis.
Figure 2. 1,25(OH)2D-regulated genes in THP-1 cells, DCs and monocytes.
Venn diagrams showing numbers of common and cell-specific genes regulated by 1,25(OH)2D in THP-1 cells, DCs and monocytes. Genes with an absolute log2 fold change > 0.26 and adjusted p-value < 0.05 were considered as regulated.
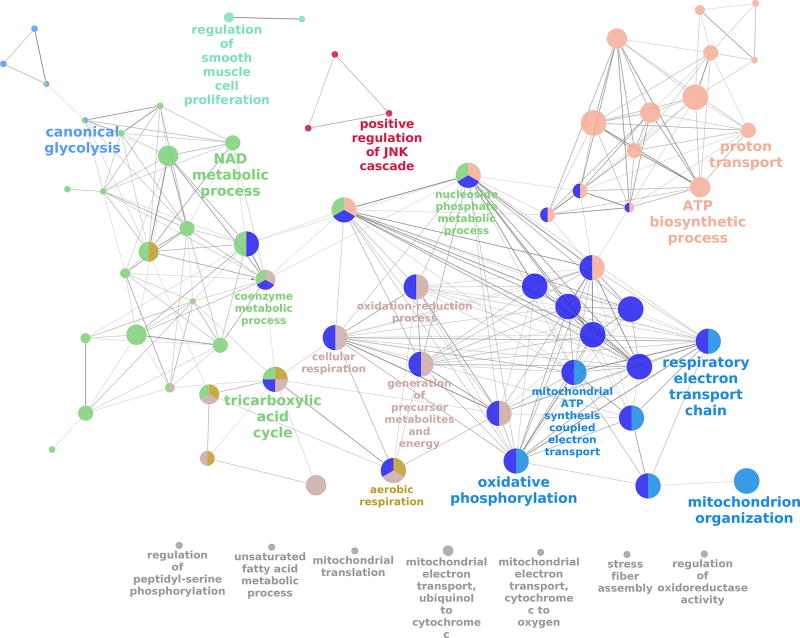
GO analysis using the list of 230 commonly altered genes was carried out using ClueGO in Cytoscape to display networks of overrepresented biological processes associated with this list. A total of 39 groups of GO terms resulted in the identification of three broad sub-networks common to each of the cell models (Figure 3 and Supplemental Figure 3A–3B).
Figure 3. ClueGO Gene ontology analysis of common 1,25(OH)2D-regulated bioenergetics genes in THP-1 cells, DCs and monocytes.
Cytoscape analysis showing overrepresented bioenergetic pathways for 1,25(OH)2D-treated THP-1 cells, DCs and monocytes. Colours represent the GO group, each node is a GO biological process and the edges shows the connectivity between each node based on the connection of the genes that contain. The size of the nodes depends on the amount of genes that are grouped. Nodes with no connections are coloured in grey.
The first of these networks was associated with bioenergetic GO-terms including the electron transport chain (GO:22904), oxidation-regulation process (GO:55114), oxidative phosphorylation (GO:6119), tricarboxylic acid cycle (GO:6099) and canonical glycolysis (GO:61621) (Figure 3). A second sub-network was based on immunological GO-terms such as responses to molecules of bacterial origin (GO:2237), cytokine production (GO:1816), inflammatory response (GO:6954) and myeloid cell differentiation (GO:30099) (Supplemental Figure 3A). The final network identified that groups of apoptotic processes such as intrinsic apoptotic signaling (GO:97193) and negative regulation of apoptotic signaling (GO:2001234) (Supplemental Figure 3B).
Visualization of 1,25(OH)2D induced changes in gene expression in THP-1 cells, DCs and monocytes on altered metabolic pathways
Pathways related to ATP metabolism showed high Z-scores (>1.96), indicating strong regulation by 1,25(OH)2D (Figure 2). To investigate the significant changes in gene expression in more depth the effect of 1,25(OH)2D on gene expression was visualized on the altered metabolic pathways discovered with pathway analysis, i.e., electron transport chain, oxidative phoshorylation and TCA cycle and glycolysis.
1,25(OH)2D and the electron transport chain
In the human electron transport chain pathway (Waagmeester et al 2017 http://wikipathways.org/index.php/Pathway:WP111) significantly up-regulated or down-regulated genes (adj. p-value < 0.05) in the three immune cells treated with 1,25(OH)2D compared to untreated cells were collectively visualized. Overall, the electron transport chain was activated by 1,25(OH)2D in the three immune cells. Biological data in Figure 4 shows in all three cell types a significant increase in expression by 1,25(OH)2D of genes associated with electron transport chain complexes I (NDUFA2, NDUFA3, NDUFA8, NDUFB9, NDUFA10, NDUFB5, NDUFAB1, NDUFS3, NDUFV1, NDUFS1), complex II (SDHB), complex III (UQRC1-2), complex IV (COX4l1, COX5A) and complex V (ATP5A1, ATP5B, ATP5C1, ATP5G1, ATP5L). Interestingly, the gene expression of the uncoupling protein 2 (UCP2) is significantly (adj. p-value < 0.05) down-regulated in THP-1 cells and monocytes by 1,25(OH)2D, but unchanged in dendritic cells.
Figure 4. Effect of 1,25(OH)2D on genes associated with the electron transport chain.
Visualization of changes in gene expression after 1,25(OH)2D treatment in THP-1 cells, DCs and monocytes. Log2 fold changes are shown as a gradient from blue (downregulated) to red (upregulated) over white (unchanged). Adjusted p-values < 0.05 are shown in green.
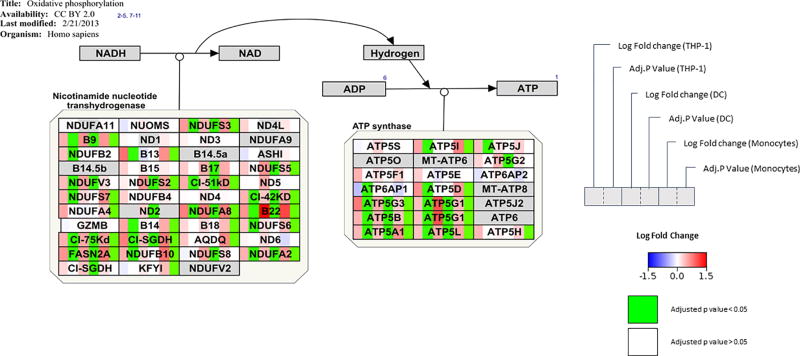
1,25(OH)2D and oxidative phosphorylation
In the oxidative phosphorylation pathway (Dalquist et al 2016 http://wikipathways.org/index.php/Pathway:WP623) all significantly changed genes were up-regulated, demonstrating that this process is activated by 1,25(OH)2D in THP-1 cells, DCs and monocytes. Biological data in Figure 5 shows that specifically genes related to ATP synthase (ATPG3, ATP5G1, ATP5B, ATP5G1, ATP5A1, ATP5L) and nicotinamide nucleotide transhydrogenase (NDUFA3, NDUFS3, NDUFV1, NDUFA10, NDUFA8, NUDFB9 NDUFS1, NDUFB5, NDUFAB1, NDUFA2) are significantly (adj. p-value < 0.05) up-regulated by 1,25(OH)2D in each of the cell models.
Figure 5. Effect of 1,25(OH)2D on genes associated with oxidative phosphorylation.
Visualization of changes in gene expression after 1,25(OH)2D treatment in THP-1 cells, DCs and monocytes. Log2 fold changes are shown as a gradient from blue (downregulated) to red (upregulated) over white (unchanged). Adjusted p-values < 0.05 are shown in green.
1,25(OH)2D and the TCA cycle and glycolysis
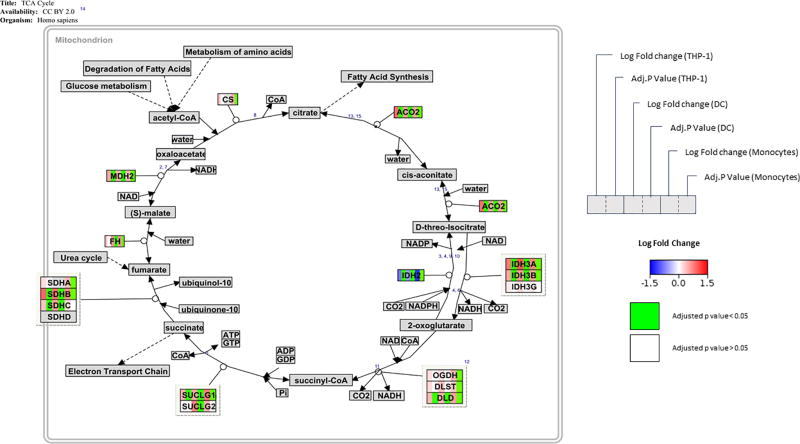
In the TCA cycle (Dalquist et al 2016 http://wikipathways.org/index.php/Pathway:WP78), the process that produces energy by oxidation of acetyl-CoA, 1,25(OH)2D up-regulates genes involved in the conversion of acetyl-CoA in carbohydrate and chemical energy. 1,25(OH)2D increased the expression of ACO2, IDH3, IDH4B, DLD, SUCLG1, SDHB and MDH2 and only the expression of IDH2, which functions in the opposite direction of the cycle, was significantly (adj. p-value < 0.05) down-regulated (Figure 6). Glucose metabolism was also affected by 1,25(OH)2D, with enzymes such as HK2, PFKM and FBP1 being significantly (adj. p-value < 0.05) up-regulated after treatment (Supplemental Figure 4).
Figure 6. Effect of 1,25(OH)2D on genes associated with the TCA cycle.
Visualization of changes in gene expression after 1,25(OH)2D treatment in THP-1 cells, DCs and monocytes. Log2 fold changes are shown as a gradient from blue (downregulated) to red (upregulated) over white (unchanged). Adjusted p-values < 0.05 are shown in green.
1,25(OH)2D and cell proliferation
Analysis of both THP-1 cell datasets (18, 19) revealed that 1,25(OH)2D had a significant effect on genes associated with the cell cycle (Conklin et al 2017 https://www.wikipathways.org/instance/WP179_r93002)(Figure 7A). This included suppression of complexes involved in phase transitions and checkpoint signalling such as BUB1/BUB3, p34cdc2/cyclin B, Chk1/Chk2, and cell cyclin-dependent kinases (CDKs) such as CDK1, CDK2, and associated cyclins A1, A2, B1, B2 and E1, E2. THP-1 cells treated with 1,25(OH)2D also showed decreased expression of genes associated with DNA replication (Koren et al 2017 https://www.wikipathways.org/instance/WP466_r94886)(Figure 7B). This included suppression of subunits of the maintenance protein complex (MCM), needed to initiate and elongate the replication fork in the DNA replication. 1,25(OH)2D also suppressed expression of genes that participate in progression of the cell cycle such as components of the assembly of the pre-replicative complex. This includes subunits of the component of the origin recognition complex (ORC): ORCL 1, 3 and 6. Genes that participate in the assembly of pre-replicative DNA complexes were also down-regulated by 1,25(OH)2D. These include: CDC6, and CDT1/GMNN complex. Finally, components of the DNA polymerase (POLa2, POLa, POLe2), primase (PRIM1, PRIM2) and subunits of the replication factor C (RFC1-5) were also down-regulated by 1,25(OH)2D in THP-1 cells (Supplemental Figure 5A–B). In contrast to effects in leukemic THP-1 cells, pathways associated with cell proliferation and cell cycling were unaffected by 1,25(OH)2D in primary cultures of monocytes and dendritic cells (Supplemental Figure 2).
Figure 7. Effect of 1,25(OH)2D on the expression of genes associated with cell proliferation in THP-1 cells.
WikiPathways representation of A) cell cycle; B) DNA replication pathways. Visualization of changes in gene expression after 1,25(OH)2D treatment in THP-1 cells. Log2 fold changes are shown as a gradient from blue (downregulated) to red (upregulated) over white (unchanged). Adjusted p-values < 0.05 are shown in green.
1,25(OH)2D and apoptosis
In the GO analysis, a network that relates apoptosis GO terms revealed three GO core groups: 1) negative regulation of apoptotic signaling; 2) intrinsic apoptotic signalling; 3) regulation of intrinsic apoptotic signaling pathway (Supplemental Figure 3B). These groups include genes that were up-regulated by 1,25(OH)2D in all three cells types such as: NDUFS3, TNFSF10, TRAP1, YBX3, NCK2, CEBPB, FXN, CYCS and NCK2. Conversely, CD74, DAPK1, ITGAV and FNIP2 were down-regulated by 1,25(OH)2D. Biological data in Supplemental Figures 6A and 6B show significant effects of 1,25(OH)2D on pathways associated with apoptosis. The receptor TNFTSF10D that participates negatively in apoptosis is up-regulated by 1,25(OH)2D in all three cell types, whilst the family of apoptotic caspases (2,3,4,6 and 7) and the apoptotic peptidase activating factor 1 (APAF1) are suppressed. Finally, it is interesting to note that JAK2 and MAP3K5 are down-regulated by 1,25(OH)2D in THP-1 cells but up-regulated in DCs and monocytes.
Discussion
Bioinformatics, or applying informatics to study biology, seeks to provide an unbiased and statistically robust insight into the molecular mechanisms that impact human health and disease. Large-scale projects such as the Human Genome Project (Roberts, et al. 2001) and ENCODE (Consortium, et al. 2007) have provided access to expansive repositories of data that have greatly helped to shed light on genomic function. However, utilisation of these data repositories to answer specific biological questions remains limited. This is particularly true for studies of vitamin D, despite detailed genome-wide analysis of VDR-chromatin interactions (Pike, et al. 2017). In most cases, analysis of the genomic responses to 1,25(OH)2D-VDR complexes has been cell-specific, incorporating both classical calciotropic (Meyer, et al. 2014), and extra-skeletal (Neme, et al. 2016) effects of vitamin D. Recently, a more integrated approach to the bioinformatic interrogation of 1,25(OH)2D-VDR datasets has been described, utilising VDR chromatin immunoprecipitation-sequencing (ChIP-Seq) datasets with National Human Genome Research Institute (NHGRI) Genome-Wide Association Study (GWAS) Single Nucleotide Polymorphism (SNP) datasets (Singh 2017). This strategy of combining publicly available datasets presents a distinct set of challenges, notably in establishing statistical rigour and appropriate analytical workflows, but nevertheless provides a novel perspective on the role of vitamin D and the VDR in human health. Notably, integration of ChIP-seq-GWAS datasets emphasized an important role for VDR in regulating of target genes associated with immunomodulation (Singh 2017). The current study proposes an alternative bioinformatic approach to further interrogate the immunomodulatory function of 1,25(OH)2D-VDR. Utilising pathway analysis workflows to assess multiple RNA expression datasets for monocyte-derived cell models allowed to the identification of common cellular pathways that are regulated by 1,25(OH)2D, revealing an important new immunometabolic function for vitamin D. The analysis pipeline described here offers an alternative statistical approach to conventional software packages. The most notable advantage of the workflow presented here is that it is open source and user-friendly for the bioscience community, and enables reproducible and straightforward methodology that can process data from different gene expression analysis platforms.
Transcriptomic analysis of gene-regulatory responses has played a pivotal role in defining the innate immune functions of vitamin D (Chun et al. 2014). Notable studies have utilized specific monocyte (Liu et al. 2006), and dendritic cell (Ferreira et al. 2015) models to identify previously unrecognized molecular targets for active 1,25(OH)2D, which have, in turn, revealed antibacterial (Adams, et al. 2009; Bacchetta, et al. 2014; Chun, et al. 2015), and metabolic regulatory (Vanherwegen, et al. 2017) functions for vitamin D in these cells. Collation of data from these different studies provides an alternative strategy for analysis of the immunomodulatory function of vitamin D, by incorporating groups of regulated genes into cellular pathway analyses. Using this strategy it was possible to identify common cellular processes that are regulated by 1,25(OH)2D, as well as those that are more cell lineage-specific. The major advantage of this workflow is that it contextualizes the experimental data at the biological process level using interactive pathways. Based on criteria, the pathway statistics approach of PathVisio defines the genes that are significantly altered in the dataset and highlights the pathways of the WikiPathways repository that are altered after treatment with 1,25(OH)2D. In our pathway analysis we used the manually curated and up-to-date human pathway collection of WikiPathways containing a broad spectrum of biological processes, including well-described metabolic processes as well as signalling and gene regulatory processes. Pathway analysis not only highlights the altered biological processes based on changes in gene expression but it enables the investigation of the relationship between genes and different datasets in great detail. This makes pathway analysis a suitable approach for an integrative and in depth analysis of large-scale transcriptomic data.
The most striking observation from the current analysis is that in all three cell models 1,25(OH)2D is strongly associated with changes in cellular metabolism, oxidative phosphorylation and energy generation. Oxidative phosphorylation has been shown to play an important role in promoting a tolerogenic phenotype in immune cells (Michalek, et al. 2011; Vats, et al. 2006). Previous studies have reported effects of 1,25(OH)2D on mitochondrial functionality and physiology, and pathways related to glucose metabolism in monocyte-derived DCs (Ferreira et al. 2015). In a similar fashion, serum 25(OH)D status has been linked to markers of bioenergetic pathways in human peripheral blood mononuclear cells (Calton, et al. 2016), and active 1,25(OH)2D has been shown to increase production of ATP and ROS, as well as altering mitochondrial functionality and morphology by increasing its membrane potential and total mass of mitochondria in differentiating monocytes (Ferreira et al. 2015). It was therefore notable in the current study that the most significantly altered pathways common to all three innate immune cell types were those associated with mitochondrial function: TCA cycle, electron transport chain and oxidative phosphorylation. Collectively these observations underline an important role for 1,25(OH)2D as a positive regulator of mitochondrial metabolism and bioenergetic pathways in immune cells. Interestingly, this effect of 1,25(OH)2D appears to be consistent not only across the three myeloid cell models studied in detail in Figure 1, but is also observed within different DC sub-types. In the current study we chose an immature DC (iDC) model which focused purely on the effect of 1,25(OH)2D, although this was for a longer time period (5 days) than the other models. Nevertheless, further analysis showed that the pathway effects of 1,25(OH)2D on iDC over 5 days were similar to those observed in immune-activated mature DC (mDC) treated with 1,25(OH)2D for 12 hours (Supplemental Figure 2).
Pathways associated with glycolysis and gluconeogenesis, such as the TCA cycle, were also significantly regulated by 1,25(OH)2D in all three cell types studied. Catabolism of glucose leads to the formation of pyruvate, a biomolecule that is then metabolized in the TCA cycle. At present, relatively little is known about the impact of vitamin D on the TCA cycle, other than clinical studies showing association between vitamin D deficiency and dysregulation of glucose metabolism like diabetes (Seida, et al. 2014). The present study demonstrates that several genes involved in the glucose metabolism are significantly regulated by vitamin D (Supplemental Figure 4). Comparison of the THP-1, DC, and monocyte datasets showed that genes for enzymes such as hexokinase, phosphofructokinase and fructose-bisphosphatase are commonly up-regulated after 1,25(OH)2D treatment. In contrast to oxidative phosphorylation and electron transport, the glycolysis/gluconeogenesis effects of 1,25(OH)2D were less consistent with some genes showing opposite patterns of regulation in different cell types. It is nevertheless clear that the regulation of glucose metabolism is a key facet of 1,25(OH)2D regulation of myeloid cells.
Besides common targets for vitamin D in innate immune cells, analysis of multiple repositories also highlighted pathways that were cell model-specific. In THP-1 cells only, 1,25(OH)2D potently regulated genes involved in progression of cell cycle and DNA replication, consistent with the neoplastic origin of these cells. The pathways with high Z-score include Retinoblastoma in cancer, G1 to S cell cycle control, DNA replication and cell cycle, are similar to those reported previously for prostate cancer cells treated with 1,25(OH)2D (Kutmon, et al. 2015a). This suggests that, in addition to its common metabolic innate immune targets, 1,25(OH)2D also has common antiproliferative targets in neoplastic cell types, including suppression of key proteins that participate in progression of the cell cycle and regulation of DNA replication (Kriebitzsch, et al. 2009). Interestingly, pathway analysis of 1,25(OH)2D in prostate cancer cells (Kutmon et al. 2015a) did not reveal any significant regulation of the metabolic pathways as identified for monocytes and DCs in the current study. Thus, in the same way that suppression of cell cycle/DNA replication genes appears to be a cancer cell-specific effect of 1,25(OH)2D, genes associated with oxidative phosphorylation and energy metabolism appear to be innate immune cell-specific. Previous studies using THP-1 cells have also included analysis of the temporal variations in response to treatment with 1,25(OH)2D (Seuter, et al. 2016). The datasets in this particular study did not conform to requirements for the comparative pathway analysis used in our current study. However, we were able to carry out a preliminary pathway analysis of these data (Supplemental Figure 7). Here it was notable that metabolic and cell cycle effects of 1,25(OH)2D were only observed after 24 hours of treatment, suggesting that pathway regulation by vitamin D is highly time-dependent.
The third major group of pathways shown to be regulated by 1,25(OH)2D in THP-1, monocytes and DCs were those associated with programmed cell death (apoptosis). Previous studies have reported pro-apoptotic effects of 1,25(OH)2D in primary cultures of monocytes, with this effect being mediated via interference with CD40 responses (Almerighi, et al. 2009). Conversely, in the HL-60 leukemic cell line 1,25(OH)2D was reported to promote resistance to apoptosis (Mosieniak, et al. 2006). Furthermore, studies from our group have shown that antisense knockdown of VDR in another leukemic cell line, U937, promoted cell apoptosis (Hewison, et al. 1996). Data from the current study further support a role for 1,25(OH)2D in regulating apoptosis of innate immune cells: common genes in the three datasets compared are commonly up-regulated (TNFTSF10D) and down-regulated (caspases family proteins and APAF1).
Conclusions
Understanding the entire set of results of high-throughput gene expression studies has often been challenging, caused by the complicated process of data analysis, and the limited biological interpretation that could be extracted from the large datasets generated through bench experiments. As a consequence, genome-wide analyses have tended to focus on changes in expression for specific genes (Chun et al. 2014; Liu et al. 2006), or small groups of genes associated with a specific pathway (Ferreira et al. 2015). The aim of the current study was to provide, for the first time, an unbiased analysis of the function of vitamin D in immune cells at a broader pathway level. Our resulting data demonstrate the added value of implementing a workflow with different bioinformatic tools that allows us to analyze gene expression data in a rapid, automated, and reproducible fashion, to highlight pathways that are significantly altered by 1,25(OH)2D. It is possible to visualize and further interrogate these biological diagrams to provide a detailed description of the molecular effects of 1,25(OH)2D in different types of immune cells. Finally, comparison of different datasets has allowed us to identify pathways, notably those associated with cell metabolism, that are common to multiple different types of innate immune cells. This approach provides new insights into the immunomodulatory actions of vitamin D, but also has important implications for the many other physiological responses linked to vitamin D in recent studies.
Supplementary Material
Raw data were normalised and statistically analysed using ArrayAnalysis. Pathway analysis of normalised data was then carried out using PathVisio and WikiPathways. Commonly altered genes in different cell models were also analysed by gene ontology using Cytoscape and ClueGO.
Heat map representing Z-scores for pathways significantly altered by 1,25(OH)2D after performing a pathway analysis on datasets from immune cells treated with 1,25(OH)2D. Z-scores >1.96 indicate that more genes are significantly altered in this pathway compared to the complete dataset.
Nodes representing GO biological processes are displayed in three differentiated networks named as the biological processes that are overrepresented: bioenergetic (showed in figure 3) immune processes (A) and apoptosis (B). The colours represent the GO group, each node is a biologic process and the edges shows the connectivity between each node based on the connection of the genes that contain. Nodes with no connections are coloured in grey.
Visualization of changes in gene expression after 1,25(OH)2D treatment in THP-1 cells, DCs and monocytes. Log2 fold changes are shown as a gradient from blue (downregulated) to red (upregulated) over white (unchanged). Adjusted p-values < 0.05 are shown in green.
WikiPathways representation of A) G1 to S cell cycle control; B) Retinoblastoma in Cancer pathways identified by Pathvisio using gene expression data from THP-1 cells treated with 1,25(OH)2D. Log2 fold changes are shown as a gradient from blue (downregulated) to red (upregulated) over white (unchanged). Adjusted p-values < 0.05 are shown in green.
WikiPathways representation of A) apoptosis; B) apoptosis modulation by HSP70. Visualization of changes in gene expression after 1,25(OH)2D treatment in THP-1 cells, DCs and monocytes. Log2 fold changes are shown as a gradient from blue (downregulated) to red (upregulated) over white (unchanged). Adjusted p-values < 0.05 are shown in green.
To evaluate temporal effects of 1,25(OH)2D comparison of pathway analysis was carried out for THP-1 cell array analyses at 24 hours (Nurminem et al (GSE60102)), and 4 hours (Heikkinen et al (GSE27270)) and an RNA-seq study where THP-1 cells were treated with 1,25(OH)2D for 2.5, 4 and 24 hours (Seuter et al. (GSE36323)). Electron transport chain, oxidative phosphorylation and TCA cycle are only significantly altered (Z>1.96) in THP-1 cells treated with 1,25(OH)2D for 24 hours. Furthermore, cell cycle pathways related are only significantly altered (Z>1.96) in THP-1 cells of 1,25(OH)2D 24 hours treatment cells from Nurminem and Vermay datasets.
Acknowledgments
AMG is supported by a PhD Studentship from the Maastricht-Birmingham PhD Programme. MH is supported by a Royal Society Wolfson Merit Award (WM130118) and National Institutes of Health (AR063910). MK is supported by the Dutch Province of Limburg.
References
- Adams JS, Hewison M. Unexpected actions of vitamin D: new perspectives on the regulation of innate and adaptive immunity. Nat Clin Pract Endocrinol Metab. 2008;4:80–90. doi: 10.1038/ncpendmet0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams JS, Ren S, Liu PT, Chun RF, Lagishetty V, Gombart AF, Borregaard N, Modlin RL, Hewison M. Vitamin d-directed rheostatic regulation of monocyte antibacterial responses. J Immunol. 2009;182:4289–4295. doi: 10.4049/jimmunol.0803736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almerighi C, Sinistro A, Cavazza A, Ciaprini C, Rocchi G, Bergamini A. 1Alpha,25-dihydroxyvitamin D3 inhibits CD40L-induced pro-inflammatory and immunomodulatory activity in human monocytes. Cytokine. 2009;45:190–197. doi: 10.1016/j.cyto.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Bacchetta J, Chun RF, Gales B, Zaritsky JJ, Leroy S, Wesseling-Perry K, Boregaard N, Rastogi A, Salusky IB, Hewison M. Antibacterial responses by peritoneal macrophages are enhanced following vitamin D supplementation. PLoS One. 2014;9:e116530. doi: 10.1371/journal.pone.0116530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Edgar R. Mining microarray data at NCBI's Gene Expression Omnibus (GEO)*. Methods Mol Biol. 2006;338:175–190. doi: 10.1385/1-59745-097-9:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla AK, Amento EP, Clemens TL, Holick MF, Krane SM. Specific high-affinity receptors for 1,25-dihydroxyvitamin D3 in human peripheral blood mononuclear cells: presence in monocytes and induction in T lymphocytes following activation. J Clin Endocrinol Metab. 1983;57:1308–1310. doi: 10.1210/jcem-57-6-1308. [DOI] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calton EK, Keane KN, Soares MJ, Rowlands J, Newsholme P. Prevailing vitamin D status influences mitochondrial and glycolytic bioenergetics in peripheral blood mononuclear cells obtained from adults. Redox Biol. 2016;10:243–250. doi: 10.1016/j.redox.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MJ. Bioinformatic approaches to interrogating vitamin D receptor signaling. Mol Cell Endocrinol. 2017 doi: 10.1016/j.mce.2017.03.011. [DOI] [PubMed] [Google Scholar]
- Cantorna MT. Vitamin D, multiple sclerosis and inflammatory bowel disease. Arch Biochem Biophys. 2012;523:103–106. doi: 10.1016/j.abb.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun RF, Liu NQ, Lee T, Schall JI, Denburg MR, Rutstein RM, Adams JS, Zemel BS, Stallings VA, Hewison M. Vitamin D supplementation and antibacterial immune responses in adolescents and young adults with HIV/AIDS. J Steroid Biochem Mol Biol. 2015;148:290–297. doi: 10.1016/j.jsbmb.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun RF, Liu PT, Modlin RL, Adams JS, Hewison M. Impact of vitamin D on immune function: lessons learned from genome-wide analysis. Front Physiol. 2014;5:151. doi: 10.3389/fphys.2014.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin B, Pico A, Campillo Sach I, Kutmon M, Coort S, Yahaya B, Chichester C, Josip, Hanspers K, van Iersel M, Salomonis N, Dmitriev S, Kelder T. Cell cycle (Homo sapiens) 2017 www.wikipathways.org/instance/WP179_r93002.
- Consortium EP, Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlquist K, Pico A, Hanspers K, Willighagen E, Chichester C, Slenter D, Jagers F, Kutmon M. Electron Transport Chain (Homo sapiens) 2016 www.wikipathways.org/instance/WP111_r95118.
- Dahlquist K, Stobbe M, Pico A, Hanspers K, van Iersel M, Kelder T, Digles D, Willighagen E, Nijveen H, Fidelman N, Andrews R, II, Houten S. TCA Cycle (Homo sapiens) 2016 www.wikipathways.org/instance/WP78_r90661.
- Eijssen LM, Jaillard M, Adriaens ME, Gaj S, de Groot PJ, Muller M, Evelo CT. User-friendly solutions for microarray quality control and pre-processing on ArrayAnalysis.org. Nucleic Acids Res. 2013;41:W71–76. doi: 10.1093/nar/gkt293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira GB, Vanherwegen AS, Eelen G, Gutierrez AC, Van Lommel L, Marchal K, Verlinden L, Verstuyf A, Nogueira T, Georgiadou M, et al. Vitamin D3 Induces Tolerance in Human Dendritic Cells by Activation of Intracellular Metabolic Pathways. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Hewison M. Antibacterial effects of vitamin D. Nat Rev Endocrinol. 2011;7:337–345. doi: 10.1038/nrendo.2010.226. [DOI] [PubMed] [Google Scholar]
- Hewison M, Dabrowski M, Vadher S, Faulkner L, Cockerill FJ, Brickell PM, O'Riordan JL, Katz DR. Antisense inhibition of vitamin D receptor expression induces apoptosis in monoblastoid U937 cells. J Immunol. 1996;156:4391–4400. [PubMed] [Google Scholar]
- Hewison M, Freeman L, Hughes SV, Evans KN, Bland R, Eliopoulos AG, Kilby MD, Moss PA, Chakraverty R. Differential regulation of vitamin D receptor and its ligand in human monocyte-derived dendritic cells. J Immunol. 2003;170:5382–5390. doi: 10.4049/jimmunol.170.11.5382. [DOI] [PubMed] [Google Scholar]
- Hochreiter S, Clevert DA, Obermayer K. A new summarization method for Affymetrix probe level data. Bioinformatics. 2006;22:943–949. doi: 10.1093/bioinformatics/btl033. [DOI] [PubMed] [Google Scholar]
- Jeffery LE, Raza K, Hewison M. Vitamin D in rheumatoid arthritis-towards clinical application. Nat Rev Rheumatol. 2016;12:201–210. doi: 10.1038/nrrheum.2015.140. [DOI] [PubMed] [Google Scholar]
- Jeffery LE, Wood AM, Qureshi OS, Hou TZ, Gardner D, Briggs Z, Kaur S, Raza K, Sansom DM. Availability of 25-Hydroxyvitamin D3 to APCs Controls the Balance between Regulatory and Inflammatory T Cell Responses. J Immunol. 2012;189:5155–5164. doi: 10.4049/jimmunol.1200786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikov N, Hastings E, Keays M, Melnichuk O, Tang YA, Williams E, Dylag M, Kurbatova N, Brandizi M, Burdett T, et al. ArrayExpress update--simplifying data submissions. Nucleic Acids Res. 2015;43:D1113–1116. doi: 10.1093/nar/gku1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren D, Kutmon M, Kelder T, Pico A, Hanspers K, Dahlquist K, Roudbari Z. DNA replication (Homos sapiens) 2017 https://www.wikipathways.org/instance/WP466_r94886.
- Kreutz M, Andreesen R, Krause SW, Szabo A, Ritz E, Reichel H. 1,25-dihydroxyvitamin D3 production and vitamin D3 receptor expression are developmentally regulated during differentiation of human monocytes into macrophages. Blood. 1993;82:1300–1307. [PubMed] [Google Scholar]
- Kriebitzsch C, Verlinden L, Eelen G, Tan BK, Van Camp M, Bouillon R, Verstuyf A. The impact of 1,25(OH)2D3 and its structural analogs on gene expression in cancer cells--a microarray approach. Anticancer Res. 2009;29:3471–3483. [PubMed] [Google Scholar]
- Kutmon M, Coort SL, de Nooijer K, Lemmens C, Evelo CT. Integrative network-based analysis of mRNA and microRNA expression in 1,25-dihydroxyvitamin D3-treated cancer cells. Genes Nutr. 2015a;10:484. doi: 10.1007/s12263-015-0484-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutmon M, Riutta A, Nunes N, Hanspers K, Willighagen EL, Bohler A, Melius J, Waagmeester A, Sinha SR, Miller R, et al. WikiPathways: capturing the full diversity of pathway knowledge. Nucleic Acids Res. 2016;44:D488–494. doi: 10.1093/nar/gkv1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutmon M, van Iersel MP, Bohler A, Kelder T, Nunes N, Pico AR, Evelo CT. PathVisio 3: an extendable pathway analysis toolbox. PLoS Comput Biol. 2015b;11:e1004085. doi: 10.1371/journal.pcbi.1004085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake JE, Adams JS. Vitamin D in HIV-Infected Patients. Curr HIV/AIDS Rep. 2011;8:133–141. doi: 10.1007/s11904-011-0082-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SM, Du P, Huber W, Kibbe WA. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2008;36:e11. doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisse TS, Liu T, Irmler M, Beckers J, Chen H, Adams JS, Hewison M. Gene targeting by the vitamin D response element binding protein reveals a role for vitamin D in osteoblast mTOR signaling. FASEB J. 2011;25:937–947. doi: 10.1096/fj.10-172577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PT, Stenger S, Li H, Wenzel L, Tan BH, Krutzik SR, Ochoa MT, Schauber J, Wu K, Meinken C, et al. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science. 2006;311:1770–1773. doi: 10.1126/science.1123933. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Koeffler HP, Donaldson CA, Pike JW, Haussler MR. 1,25-Dihydroxyvitamin D3-induced differentiation in a human promyelocytic leukemia cell line (HL-60): receptor-mediated maturation to macrophage-like cells. J Cell Biol. 1984;98:391–398. doi: 10.1083/jcb.98.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MB, Benkusky NA, Lee CH, Pike JW. Genomic determinants of gene regulation by 1,25-dihydroxyvitamin D3 during osteoblast-lineage cell differentiation. J Biol Chem. 2014;289:19539–19554. doi: 10.1074/jbc.M114.578104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, Sullivan SA, Nichols AG, Rathmell JC. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millenaar FF, Okyere J, May ST, van Zanten M, Voesenek LA, Peeters AJ. How to decide? Different methods of calculating gene expression from short oligonucleotide array data will give different results. BMC Bioinformatics. 2006;7:137. doi: 10.1186/1471-2105-7-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosieniak G, Sliwinska M, Piwocka K, Sikora E. Curcumin abolishes apoptosis resistance of calcitriol-differentiated HL-60 cells. FEBS Lett. 2006;580:4653–4660. doi: 10.1016/j.febslet.2006.07.038. [DOI] [PubMed] [Google Scholar]
- Neme A, Nurminen V, Seuter S, Carlberg C. The vitamin D-dependent transcriptome of human monocytes. J Steroid Biochem Mol Biol. 2016;164:180–187. doi: 10.1016/j.jsbmb.2015.10.018. [DOI] [PubMed] [Google Scholar]
- Nnoaham KE, Clarke A. Low serum vitamin D levels and tuberculosis: a systematic review and meta-analysis. Int J Epidemiol. 2008;37:113–119. doi: 10.1093/ije/dym247. [DOI] [PubMed] [Google Scholar]
- Nurminen V, Neme A, Ryynanen J, Heikkinen S, Seuter S, Carlberg C. The transcriptional regulator BCL6 participates in the secondary gene regulatory response to vitamin D. Biochim Biophys Acta. 2015;1849:300–308. doi: 10.1016/j.bbagrm.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Pike JW, Meyer MB, Lee SM, Onal M, Benkusky NA. The vitamin D receptor: contemporary genomic approaches reveal new basic and translational insights. J Clin Invest. 2017;127:1146–1154. doi: 10.1172/JCI88887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Carvalho BS, Hetrick KN, Tavare S, Irizarry RA. R/Bioconductor software for Illumina's Infinium whole-genome genotyping BeadChips. Bioinformatics. 2009;25:2621–2623. doi: 10.1093/bioinformatics/btp470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts L, Davenport RJ, Pennisi E, Marshall E. A history of the Human Genome Project. Science. 2001;291:1195. doi: 10.1126/science.291.5507.1195. [DOI] [PubMed] [Google Scholar]
- Seida JC, Mitri J, Colmers IN, Majumdar SR, Davidson MB, Edwards AL, Hanley DA, Pittas AG, Tjosvold L, Johnson JA. Clinical review: Effect of vitamin D3 supplementation on improving glucose homeostasis and preventing diabetes: a systematic review and meta-analysis. J Clin Endocrinol Metab. 2014;99:3551–3560. doi: 10.1210/jc.2014-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seuter SA, Neme A, Carlberg C. Epigenome-wide effects of vitamin D and their impact on the transcriptome of human monocytes involve CTCF. Nucleic Acids Res. 2016;44:4090–4104. doi: 10.1093/nar/gkv1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh PK, van den Berg PR, Long MD, Vreugdenhil A, Grieshober L, Ochs-Balcom HM, Wang J, Delcambre S, Heikkinen S, Carlberg C, Campbell MJ, Sucheston-Campbell LE. Integration of VDR genome wide binding and GWAS genetic variation data reveals co-occurrence of VDR and NF-κB binding that is linked to immune phenotypes. BMC Genomics. 2017;18:132. doi: 10.1186/s12864-017-3481-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeles L, Keresztes G, Torocsik D, Balajthy Z, Krenacs L, Poliska S, Steinmeyer A, Zuegel U, Pruenster M, Rot A, et al. 1,25-dihydroxyvitamin D3 is an autonomous regulator of the transcriptional changes leading to a tolerogenic dendritic cell phenotype. J Immunol. 2009;182:2074–2083. doi: 10.4049/jimmunol.0803345. [DOI] [PubMed] [Google Scholar]
- van Iersel MP, Kelder T, Pico AR, Hanspers K, Coort S, Conklin BR, Evelo C. Presenting and exploring biological pathways with PathVisio. BMC Bioinformatics. 2008;9:399. doi: 10.1186/1471-2105-9-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Iersel MP, Pico AR, Kelder T, Gao J, Ho I, Hanspers K, Conklin BR, Evelo CT. The BridgeDb framework: standardized access to gene, protein and metabolite identifier mapping services. BMC Bioinformatics. 2010;11:5. doi: 10.1186/1471-2105-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanherwegen AS, Gysemans C, Mathieu C. Vitamin D endocrinology on the cross-road between immunity and metabolism. Mol Cell Endocrinol. 2017 doi: 10.1016/j.mce.2017.04.018. [DOI] [PubMed] [Google Scholar]
- Vats D, Mukundan L, Odegaard JI, Zhang L, Smith KL, Morel CR, Wagner RA, Greaves DR, Murray PJ, Chawla A. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab. 2006;4:13–24. doi: 10.1016/j.cmet.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verway M, Bouttier M, Wang TT, Carrier M, Calderon M, An BS, Devemy E, McIntosh F, Divangahi M, Behr MA, et al. Vitamin D induces interleukin-1beta expression: paracrine macrophage epithelial signaling controls M. tuberculosis infection. PLoS Pathog. 2013;9:e1003407. doi: 10.1371/journal.ppat.1003407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waagmeester A, Hanspers K, Kutmon M, Roudbari Z, Pico A, Kuchinsky A, van Iersel M. Oxidative phosphorylation (Homo sapiens) 2017 www.wikipathways.org/instance/WP623_r86899.
- Wei R, Christakos S. Mechanisms Underlying the Regulation of Innate and Adaptive Immunity by Vitamin D. Nutrients. 2015;7:8251–8260. doi: 10.3390/nu7105392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelwright M, Kim EW, Inkeles MS, De Leon A, Pellegrini M, Krutzik SR, Liu PT. All-trans retinoic acid-triggered antimicrobial activity against Mycobacterium tuberculosis is dependent on NPC2. J Immunol. 2014;192:2280–2290. doi: 10.4049/jimmunol.1301686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman C, Jensen LJ, Jarmer H, Berka R, Gautier L, Nielser HB, Saxild HH, Nielsen C, Brunak S, Knudsen S. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-9-research0048. research0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Raw data were normalised and statistically analysed using ArrayAnalysis. Pathway analysis of normalised data was then carried out using PathVisio and WikiPathways. Commonly altered genes in different cell models were also analysed by gene ontology using Cytoscape and ClueGO.
Heat map representing Z-scores for pathways significantly altered by 1,25(OH)2D after performing a pathway analysis on datasets from immune cells treated with 1,25(OH)2D. Z-scores >1.96 indicate that more genes are significantly altered in this pathway compared to the complete dataset.
Nodes representing GO biological processes are displayed in three differentiated networks named as the biological processes that are overrepresented: bioenergetic (showed in figure 3) immune processes (A) and apoptosis (B). The colours represent the GO group, each node is a biologic process and the edges shows the connectivity between each node based on the connection of the genes that contain. Nodes with no connections are coloured in grey.
Visualization of changes in gene expression after 1,25(OH)2D treatment in THP-1 cells, DCs and monocytes. Log2 fold changes are shown as a gradient from blue (downregulated) to red (upregulated) over white (unchanged). Adjusted p-values < 0.05 are shown in green.
WikiPathways representation of A) G1 to S cell cycle control; B) Retinoblastoma in Cancer pathways identified by Pathvisio using gene expression data from THP-1 cells treated with 1,25(OH)2D. Log2 fold changes are shown as a gradient from blue (downregulated) to red (upregulated) over white (unchanged). Adjusted p-values < 0.05 are shown in green.
WikiPathways representation of A) apoptosis; B) apoptosis modulation by HSP70. Visualization of changes in gene expression after 1,25(OH)2D treatment in THP-1 cells, DCs and monocytes. Log2 fold changes are shown as a gradient from blue (downregulated) to red (upregulated) over white (unchanged). Adjusted p-values < 0.05 are shown in green.
To evaluate temporal effects of 1,25(OH)2D comparison of pathway analysis was carried out for THP-1 cell array analyses at 24 hours (Nurminem et al (GSE60102)), and 4 hours (Heikkinen et al (GSE27270)) and an RNA-seq study where THP-1 cells were treated with 1,25(OH)2D for 2.5, 4 and 24 hours (Seuter et al. (GSE36323)). Electron transport chain, oxidative phosphorylation and TCA cycle are only significantly altered (Z>1.96) in THP-1 cells treated with 1,25(OH)2D for 24 hours. Furthermore, cell cycle pathways related are only significantly altered (Z>1.96) in THP-1 cells of 1,25(OH)2D 24 hours treatment cells from Nurminem and Vermay datasets.