Abstract
Enzymatic transformations of primary, canonical metabolites generate active biomolecules that regulate important cellular and physiological processes. Roles include regulation of histone demethylation in epigenetics, inflammation in tissue injury, insulin sensitivity, cancer cell invasion, stem cell pluripotency status, inhibition of nitric oxide signaling and others. Such modified compounds, defined as epimetabolites, have functions distinct from classic hormones as well as removed from generic anabolism and catabolism. Epimetabolites are discovered by untargeted metabolomics using liquid- or gas chromatography–high resolution mass spectrometry and structurally annotated by in-silico fragmentation prediction tools. Their specific biological functions are subsequently investigated by targeted metabolomics methods.
Graphical abstract
Introduction
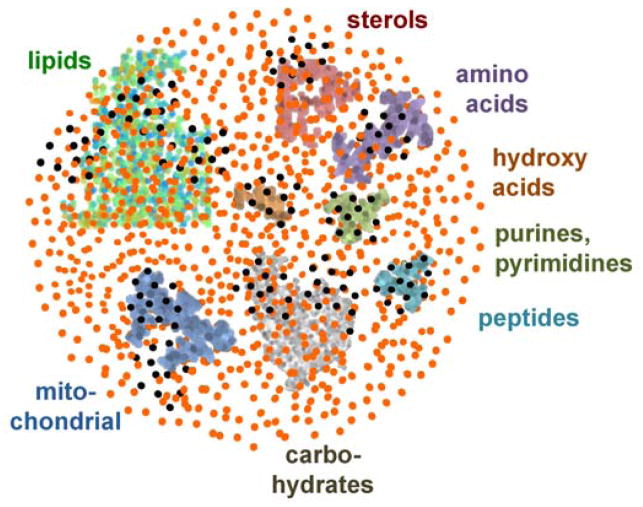
Metabolism intersects mechanisms in molecular biology with disease endpoints. While new discoveries are still being made in classic central metabolism, the dark matter of the metabolome has been largely ignored [1] (Figure 1). Metabolomics explores the impact of changes in our microbiome [2], repair mechanisms for damaged metabolites [3], the exposome [4•] and a range of fields of biologically active metabolites, from oncometabolites [5,6] to the sterol-mediated regulation of the activity of signaling cascades [7,8]. The idea of metabolites with regulatory functions goes well beyond classic biochemical feedback inhibition: metabolites act on distal modules of the molecular and organismal network that are impossible to be explained by the central dogma of unidirectional information flow from genotype to phenotype. This review will focus how targeted and untargeted mass spectrometry methods and software contributes to reveal biological functions of identified and novel, hitherto unknown metabolites.
Figure 1.
The dark matter of metabolism. Known biochemical modules (colored) are interspersed with epimetabolites (black), in addition to exposome compounds (orange) detected by untargeted mass spectrometry such as household chemicals, pharmaceuticals, food components, pesticides.
Beyond building and burning
Metabolomics has focused for too long on classic, well-defined pathways of primary metabolites that constitute the major highways in cellular anabolism or catabolism. This biochemical pathway-centric focus has been nurtured by a view that metabolites only rarely act as regulators, isolating well-researched fields as exceptions to the rule rather than as an overarching theme of metabolism in its own right. Examples include lipid mediators of inflammation [9•], or insulin sensitivity [10••], and regulation of histone demethylases by the oncometabolite 2-hydroxyglutarate (2HG) [11]. These compounds share commonalities that they are often low abundant, transient in nature, and removed from mainstream energy or polymer metabolism. In plants, secondary or specialized metabolites can be considered a synonym to epimetabolites. Specialized metabolites are readily recognized as active regulators and have dedicated and lengthy metabolic pathways for their production [12,13]. Bacteria are also known to use metabolites as regulators for gene expression through riboswitches [14,15]. However, these non-canonical metabolite roles are considered isolated examples and not evidence of a larger role for metabolites in vivo.
We are proposing the field of epimetabolites to provide an umbrella term for these non-canonical metabolite functions to fall into. We define an epimetabolite as a metabolite removed from its classical function in anabolism or catabolism. These non-canonical metabolites serve a functional role, including but not limited to, regulation, defense, communication, storage or transport functions. Epimetabolites often remain chemically similar to their canonical counterparts and may use simple modifications like methylation or acetylation that can be easily reversed (Figure 2). They may have once been formed by enzyme errors or chemical damage, but gained biological roles over time. Using untargeted metabolomics, new hypotheses can be generated by discovery of new epimetabolites. Once a new hypothesis is generated, targeted metabolomics methods can be created to accurately quantify them.
Figure 2.
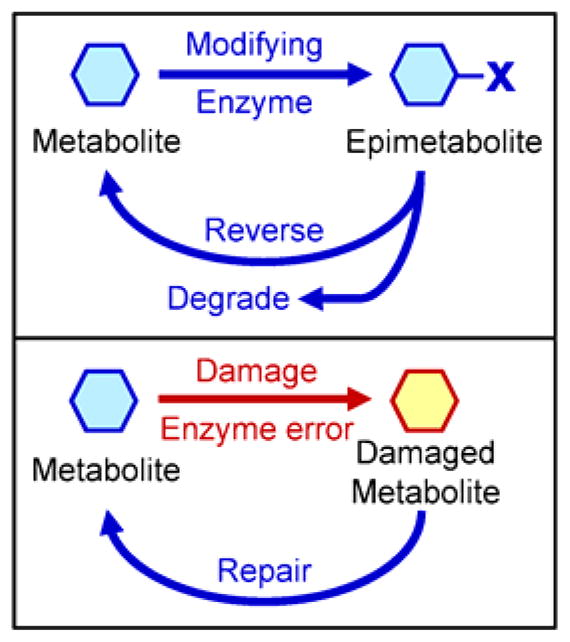
Origin of novel metabolites. Specific enzymes modify or repair metabolites without creating new pathways.
Metabolomics methods to target the biological role of epimetabolites
Researchers may be driven by a hypothesis of the involvement of a specific metabolite in biological context: validating such hypothesis is best achieved by targeting this compound by mass spectrometry at high selectivity and sensitivity. To obtain increased specificity, any physicochemical property of small molecules can be exploited to separate metabolites prior to reaching the detector such as differences in boiling point (gas chromatography, GC), lipophilicity (liquid chromatography, LC) electric surfaces charges and migration against a fluid (capillary electrophoresis, CE) or migration against a gas (ion mobility, IM). Hence, complex mixtures of metabolites can be efficiently separated prior to reaching the mass spectrometer. In addition, sensitivity is enhanced by fragmenting the intact molecules inside the mass spectrometer (MS/MS). By monitoring molecule-specific fragmentations, the signal-to-noise ratio of detection is greatly enhanced because noise molecules or co-eluting compounds that have, by chance, the same mass-to-charge ratio (m/z) and the same chromatographic retention time will unlikely also have the same MS/MS fragmentation. Especially in LC, signal-to-noise ratios are dominated by buffer and solvent clusters, making LC–MS/MS a very effective way to target epimetabolites. To accurately quantify targeted compounds, internal standards and calibration curves are used. Open access software packages for multitarget metabolomics analyses have been reviewed elsewhere [16,17], but lack thorough validation through independent round-robin tests (ring trials).
Commonly measured using targeted metabolomics, oxylipins are prime examples of epimetabolites with very potent regulatory activity. These compounds are released from phospholipid lipases and produced on demand by at least one oxidation step involving molecular oxygen. There are hundreds of different oxylipins in aerobic organisms, ranging from cyclized forms (such as prostaglandins and thromboxanes) to epoxides such as leucotrienes and monohydroxy fatty acids (HETEs). In animals, most oxylipins belong to the family of 20-carbon eicosanoids and have multiple physiological roles, including balancing pro-inflammatory and anti-inflammatory roles in tissue injury. The literature on the roles, regulation and biochemistry of oxylipins abound with hundreds of studies each year. Many metabolomic tools target these classes, with specific emphasis on sample preparation and accurate quantification in plasma by LC–MS/MS [18,19] and on multi-target methods that combine oxylipin profiling with other bioactive lipid classes such as endocannabinoids [20].
A novel class of lipid epimetabolites has recently been discovered, the fatty esters of monohydroxy fatty acids (FAHFAs) [10••]. Specifically, the FAHFA member palmitic acid-9-hydroxystearic acid, 9-PAHSA, was shown to correlate highly with insulin sensitivity and to be reduced in both adipose tissue and serum of insulin-resistant humans. In mice, administering 9-PAHSA improved glucose tolerance while stimulating GLP-1 and insulin secretion, giving mechanistic insights how this new endogenous epimetabolite might act. Once one member of an epimetabolite class has been discovered, detecting similar compounds through metabolomics becomes of high interest. An in-silico library of accurate mass MS/MS spectra was generated and experimentally validated, including the detection of previously unknown FAHFA metabolites, to guide the identification of FAHFAs in untargeted metabolomic screens [21].
Targeting methylated epimetabolites
The name “epimetabolite” invokes the analogy to the term epigenetics, a broad class of modifications made outside of changes to gene sequences. Methylation plays and important role in epigenetic regulation and has mounting evidence as an important modification to form epimetabolites. Methylation of classic canonical pathway metabolites such as glycine, nicotinamide or arginine yields epimetabolites with profound cellular or physiological roles. The oncometabolite N-methylglycine, or sarcosine, stimulates invasion and aggressivity in prostate cancer cells, initially discovered through untargeted metabolomics [22]. Further studies show the addition of sarcosine but not glycine or alanine induced tumorgenic changes in in vivo prostate cancer models [23]. Another example of a methylated epimetabolite is 1-methylnicotinamde (1MNA), which acts as a methylation sink in naïve embryonic stem cells (ESC) preventing deposit of H2K27me3 marks. Increasing levels of 1MNA, and decreasing levels of S-adenosylmethionine (SAM), are essential to naïve ESC maintenance shown by differentiation of naïve ESC nicotinamide n-methyltransferase knock out line even in the presence of naïve state stabilizers. Targeted and untargeted metabolomics were able to distinguish the primed from naïve in both human and mouse ESCs [24••]. A third example of methylation of canonical metabolites leading to gained regulatory function is asymmetric and symmetric dimethyl arginine (ADMA and SDMA). ADMA and SDMA are produced by the repeated methylation of arginine residues by protein arginine methyltransferases and subsequent proteolysis of methylated proteins. Liberated ADMA, but not SDMA, then works as a competitive inhibitor for endothelial nitric oxide synthase. ADMA inhibition of eNOS can further promote uncoupling of eNOS and production of reactive oxygen and nitrogen species. A meta-analysis of 16 cohort studies involving more than 4,000 subjects showed that ADMA levels alone are significantly associated with an increased risk of coronary artery disease [25]. SDMA itself can be used as biomarker for renal insufficiency, as quantified by targeted LC–MS/MS in urine [26].
Targeting isomeric variants of epimetabolites
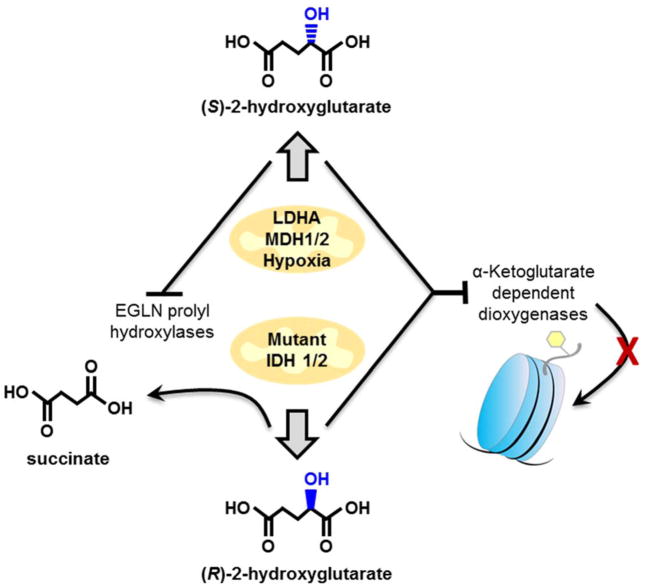
When targeting specific metabolites, separating isomers can be critical. Mass spectrometry is usually insufficient in this regard therefore most targeted metabolomic methods utilize chromatographic separation. As biologically relevant compounds are often stereo-specific, it becomes necessary to separate stereoisomers. This can be done with chiral columns and derivatization in LC or GC methods. One example is the well-established oncometabolite 2-hydroxyglutarate (2HG) [27], discovered by untargeted metabolomics [28], which is now known to have different bio-active roles depending upon stereo-conformation. 2HG can be produced by mutated or wild type enzymes in either R- or S-enantiomer forms respectively (Figure 3). In multiple cancer types, R-2HG is produced from mutations to either isocitrate dehydrogenase 1 or 2 (IDH1 or IDH2). Cells without IDH1 or IDH2 mutations can produce S-2HG, especially during times of hypoxic stress by either lactate dehydrogenase A or malate dehydrogenase 1 or 2 [29]. Both R- and S- forms of 2-HG are inhibitors of α-ketoglutarate dependent dioxgenases, which include notably, histone lysine demethylase 4C, an important epigenetic regulator [11]. While both enantiomeric forms of 2HG inhibit cellular demethylases, S-2HG is also an inhibitor for EGLN prolyl hydroxylases (involved in HIF-1α regulation) [30••] while R-2HG is a substrate for this reaction [29]. Accurate in vivo studies of the oncometabolite 2-hydroxyglutarate must therefore include targeted methods capable of enantiomeric separation of R- and S-2HG.
Figure 3.
The different effects of the enantiomeric forms of 2-hydroxyglutarate. Targeted methods must carefully distinguish chiral stereoisomers to unravel the different functions of the oncometabolites (R)- and (S)-2-hydroxyglutarate that impact histone methylations, or may be substrate to proline hydroxylases.
How to discover and identify new epimetabolites through untargeted metabolomics
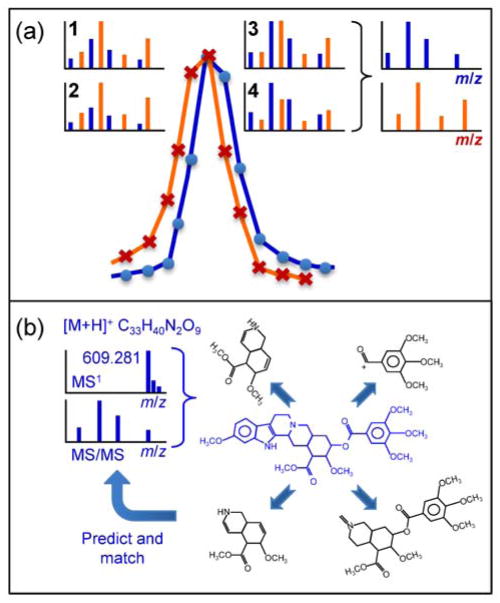
As defined above, epimetabolites are chemically modified versions of mainstream compounds that have defined biological roles. While targeted metabolomics methods for these compounds can aid in elucidating their biological roles, it limits the information that can be obtained from samples. However, targeted approaches can be combined with untargeted screening to create a “targeted-plus” method for quantification of metabolites of interest and simultaneous collection of untargeted data [31]. Two steps are involved in discovering genuine metabolites: first, unbiased detection and structural identification of compounds and their statistical association with biological endpoints are collected to yield a starting hypothesis on their potential cellular roles. Second, targeted subsequent studies, as outlined above, give deeper insights into the specifics of their biological functions. Untargeted metabolomics is frequently employed for finding such novel biomarkers, using either GC–MS or LC–MS methods. Typically, multiple extraction and chromatography methods are combined for an untargeted study to capture the greatest diversity of small molecules in a sample. Methods are optimized for coverage and reproducibility. Untargeted metabolomics produces semi-quantitative data on hundreds to thousands of compounds during a single run that are reported by relative intensities. Data processing with classic software such as MZmine 2 [32] and XCMS is still popular, while new subroutines for XCMS have been developed to reduce bias in peak detections [33]. Recently, improved software for peak detection, adduct identification and automated MS/MS deconvolution has been released, MS-DIAL [34••]. MS-DIAL works for both classic data-dependent MS/MS fragmentation experiments as well as data-independent fragmentation studies (Figure 4a). The software also includes large mass spectral libraries such as LipidBlast [35] for compound identifications by MS/MS matching, in addition to scoring deviations from predicted retention times. However, for an overwhelming number of metabolites detected by untargeted metabolomics, no satisfactory MS/MS match can be found.
Figure 4.
(a) Mass spectral deconvolution in untargeted metabolomics. In both GC–MS and data-independent LC–MS/MS, molecule fragments overlap for co-eluting metabolites. Following all MS/MS events (1–4) in MS-DIAL enables disentangling precursor and fragment ions to obtain pure MS1 and MS/MS spectra. (b) Predicting elemental composition and MS fragmentation by in-silico tools. From accurate mass, isotope and MS/MS data, elemental formulas are calculated. List of isomer structures are downloaded from databases, MS/MS spectra are predicted, and structures are ranked by highest matching scores.
For the remaining unknown peaks of interest (a combined feature of a specific m/z value at a specific retention time), the hard work starts: the structural annotation of those features. First, the analytical nature of this compounds needs to be defined, for example, as protonated molecular ion [M+H]+ or as one of many other adducts that are regularly detected in LC–MS runs [36]. Only afterwards can one match the accurate mass and isotope distribution information to calculate the most probable elemental composition, the start for annotating the unknown feature by lists of possible isomers. For calculating chemical formulas (Figure 4b), accurate masses are needed with accuracies better than 2 ppm, enabled by instruments that have high mass resolving power (10,000–450,000 FWHM) and relatively wide dynamic linear ranges (3.5–5 orders of magnitude). However, it has previously been shown that accurate mass information alone is insufficient to yield unambiguous elemental formulas, even at <1 ppm mass accuracy [37]. However, if MS/MS fragmentation data are available, the correct elemental formulas are retrieved at the top position in more than 98% of the cases validated for over 5,000 test compounds using the MS-FINDER software [38] or the Sirius 3 algorithm [39].
From elemental formulas to correct epimetabolite structure annotations
Next, all isomer structures of these potential novel epimetabolites need to be searched by their calculated elemental formulas in metabolome databases. A good start is still the Human Metabolome Database (HMDB) [40] but of course, if an epimetabolite indeed has never been reported before, it cannot be retrieved from such resources. Therefore, the scope of possibly existing metabolites has been increased by assuming substrate ambiguity of enzymes, leading to the release of the Metabolic In-silico Network Expansion database (MINE) [41•] that includes more than 571,000 hypothetical compounds, including many metabolites that are derived from simple methylations, acetylations, hydroxylations or other single-reaction modifications that would signify the discovery of an epimetabolite. Albeit, none of these virtual metabolites has a validated MS/MS spectrum, making it difficult to rank the best structure to the experimental MS/MS data. Several research groups have generated tools to predict MS/MS spectra from molecular structures, using chemical bond energies in the improved MetFrag tool [42], known dissociation rules in MS2Analyzer [43], hydrogen bond rearrangements in MS-FINDER [38], fragmentation tree calculations in CSI:FingerID [44] or machine learning strategies in CFM-ID [45•]. These tools are tested in regular competitions, the CASMI contests [46]. At current, the glass is half full, at best as none of these tools yield better than 50% correct structure annotations within the top-5 hits in such blinded tests. There is much room for improvement, and each predicted epimetabolite must still be validated by confirmation using an authentic, synthesized chemical standard. Other approaches include using nuclear magnetic resonance (NMR) which can provide additional insight into unknown structure identification [47].
From identification of potential epimetabolites to defining biological roles
After data processing and identification, known metabolites can be linked to new biological changes, providing new hypotheses to study. For example, the link between microbiome metabolism of phosphatidylcholine and cardiovascular disease was initially discovered using untargeted metabolomics [48]. The epimetabolites discovered as predictors of cardiovascular disease, trimethylamine N-oxide (TMAO) and its precursor, γ-butyrobetaine are now appreciated to be important as proatherogenic actors, inducing the development of distinct microbial communities when added to the diet [49]. The recent identification of 4-phosphoerythronate and 2-phospho-L-lactate as side products of mammalian glyceraldehyde 3-phosphate dehydrogenase and pyruvate kinase respectively, illustrate the ability of epimetabolites to regulate classical pathways. 2-Phospho-L-lactate inhibits glycolysis and 4-phosphoerythronate inhibits flux to pentose phosphate pathway. These side products are dephosphorylated by phosphoglycolate phosphatase, now considered to be a metabolite repair enzyme [50]. Other untargeted epimetabolite discoveries may lack specific mechanisms, but have proven to be specific biomarkers for disease states. Diacetylspermine (DAS) has been discovered through untargeted HILIC–QTOF MS/MS as validated marker for non-small-cell lung cancer [51], yet a clear mechanism has yet to be reported. Using untargeted metabolomics, DAS has also been reported to be associated with biofilm formation in colon cancer progression [52]. These examples highlight the difficulties in metabolomics, from untargeted discovery and structure identifications to biological validations.
Conclusions
Metabolites are not mere outputs of genetic networks, but actively participate in many aspects of cellular regulation. The number of regulatory metabolites (beyond classic feed-back enzyme inhibition) has expanded to a level that justifies defining a new umbrella classification, epimetabolites. Advances in analytical chemistry have made fast, selective, sensitive and affordable detection of such epimetabolites possible. Hypotheses are generated by finding new epimetabolites through untargeted metabolomics while their biological roles are subsequently validated in targeted metabolomics studies.
Highlights.
Epimetabolites are modified classic metabolites with new roles
Example roles include regulation of tumorigenesis, inflammation and pluripotency
New epimetabolites are discovered by untargeted metabolomics
Structure annotation by in-silico prediction software
Acknowledgments
This work was funded by grants NIH U24DK097154, NIH HL113452 and NSF MCB 1611846.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.da Silva RR, Dorrestein PC, Quinn RA. Illuminating the dark matter in metabolomics. Proceedings of the National Academy of Sciences. 2015;112:12549–12550. doi: 10.1073/pnas.1516878112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Mahony SM, Clarke G, Borre YE, Dinan TG, Cryan JF. Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behavioural brain research. 2015;277:32–48. doi: 10.1016/j.bbr.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 3.Linster CL, Van Schaftingen E, Hanson AD. Metabolite damage and its repair or pre-emption. Nature chemical biology. 2013;9:72–80. doi: 10.1038/nchembio.1141. [DOI] [PubMed] [Google Scholar]
- 4•.Rappaport SM, Barupal DK, Wishart D, Vineis P, Scalbert A. The blood exposome and its role in discovering causes of disease. Environmental Health Perspectives (Online) 2014;122:769. doi: 10.1289/ehp.1308015. This paper is a seminal contribution due to its comprehensive investigation of exposome compounds present in human blood, and ranges of concentrations that tightly overlap with levels of endogenous metabolites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santagata S, Eberlin LS, Norton I, Calligaris D, Feldman DR, Ide JL, Liu X, Wiley JS, Vestal ML, Ramkissoon SH. Intraoperative mass spectrometry mapping of an onco-metabolite to guide brain tumor surgery. Proceedings of the National Academy of Sciences. 2014;111:11121–11126. doi: 10.1073/pnas.1404724111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nowicki S, Gottlieb E. Oncometabolites: tailoring our genes. The FEBS Journal. 2015;282:2796–2805. doi: 10.1111/febs.13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Gianoulis TA, Yip KY, Gerstein M, Snyder M. Extensive in vivo metabolite-protein interactions revealed by large-scale systematic analyses. Cell. 2010;143:639–650. doi: 10.1016/j.cell.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Meara TR, Veri AO, Polvi EJ, Li X, Valaei SF, Diezmann S, Cowen LE. Mapping the Hsp90 Genetic Network Reveals Ergosterol Biosynthesis and Phosphatidylinositol-4-Kinase Signaling as Core Circuitry Governing Cellular Stress. PLoS Genet. 2016;12:e1006142. doi: 10.1371/journal.pgen.1006142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9•.Serhan CN. Pro-resolving lipid mediators are leads for resolution physiology. Nature. 2014;510:92–101. doi: 10.1038/nature13479. This review provides a very good overview over the roles and regulation of oxylipins and resolvins in inflammation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10••.Yore MM, Syed I, Moraes-Vieira PM, Zhang T, Herman MA, Homan EA, Patel RT, Lee J, Chen S, Peroni OD. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell. 2014;159:318–332. doi: 10.1016/j.cell.2014.09.035. The authors here report on the discovery and validation of FAHFA lipids and their direct involvement in insulin resistance and sensitivity, with a prospect of using this new class of lipid epimetabolites for treatment of type 2 diabetes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su X, Wellen KE, Rabinowitz JD. Metabolic control of methylation and acetylation. Current opinion in chemical biology. 2016;30:52–60. doi: 10.1016/j.cbpa.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laursen T, Moller BL, Bassard JE. Plasticity of specialized metabolism as mediated by dynamic metabolons. Trends in Plant Science. 2015;20:20–32. doi: 10.1016/j.tplants.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Cheynier V, Comte G, Davies KM, Lattanzio V, Martens S. Plant phenolics: Recent advances on their biosynthesis, genetics, and ecophysiology. Plant Physiology and Biochemistry. 2013;72:1–20. doi: 10.1016/j.plaphy.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Johnson JE, Reyes FE, Polaski JT, Batey RT. B-12 cofactors directly stabilize an mRNA regulatory switch. Nature. 2012;492:133. doi: 10.1038/nature11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henkin TM. Riboswitch RNAs: using RNA to sense cellular metabolism. Genes & Development. 2008;22:3383–3390. doi: 10.1101/gad.1747308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorrochategui E, Jaumot J, Lacorte S, Tauler R. Data analysis strategies for targeted and untargeted LC-MS metabolomic studies: Overview and workflow. TrAC Trends in Analytical Chemistry. 2016;82:425–442. [Google Scholar]
- 17.Misra BB, der Hooft JJJ. Updates in metabolomics tools and resources: 2014–2015. Electrophoresis. 2016;37:86–110. doi: 10.1002/elps.201500417. [DOI] [PubMed] [Google Scholar]
- 18.Ostermann AI, Willenberg I, Schebb NH. Comparison of sample preparation methods for the quantitative analysis of eicosanoids and other oxylipins in plasma by means of LC-MS/MS. Analytical and bioanalytical chemistry. 2015;407:1403–1414. doi: 10.1007/s00216-014-8377-4. [DOI] [PubMed] [Google Scholar]
- 19.Willenberg I, Ostermann AI, Schebb NH. Targeted metabolomics of the arachidonic acid cascade: current state and challenges of LC–MS analysis of oxylipins. Analytical and bioanalytical chemistry. 2015;407:2675–2683. doi: 10.1007/s00216-014-8369-4. [DOI] [PubMed] [Google Scholar]
- 20.Gouveia-Figueira S, Nording ML. Validation of a tandem mass spectrometry method using combined extraction of 37 oxylipins and 14 endocannabinoid-related compounds including prostamides from biological matrices. Prostaglandins & other lipid mediators. 2015;121:110–121. doi: 10.1016/j.prostaglandins.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Ma Y, Kind T, Vaniya A, Gennity I, Fahrmann JF, Fiehn O. An in silico MS/MS library for automatic annotation of novel FAHFA lipids. Journal of cheminformatics. 2015;7:1. doi: 10.1186/s13321-015-0104-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 23.Khan AP, Rajendiran TM, Ateeq B, Asangani IA, Athanikar JN, Yocum AK, Mehra R, Siddiqui J, Palapattu G, Wei JT, et al. The role of sarcosine metabolism in prostate cancer progression. Neoplasia. 2013;15:491. doi: 10.1593/neo.13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Sperber H, Mathieu J, Wang Y, Ferreccio A, Hesson J, Xu Z, Fischer KA, Devi A, Detraux D, Gu H. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nature cell biology. 2015;17:1523–1535. doi: 10.1038/ncb3264. This report unequivocally demonstrates that controlling the levels of the epimetabolite methyl-nicotinamide are required and sufficient to mediate the epigenetic status of pluripotent stem cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xuan C, Tian Q-W, Li H, Zhang B-B, He G-W, Lun L-M. Levels of asymmetric dimethylarginine (ADMA), an endogenous nitric oxide synthase inhibitor, and risk of coronary artery disease: A meta-analysis based on 4713 participants. European journal of preventive cardiology. 2015 doi: 10.1177/2047487315586094. 2047487315586094. [DOI] [PubMed] [Google Scholar]
- 26.Martens-Lobenhoffer J, Bode-Böger SM. Amino acid N-acetylation: Metabolic elimination of symmetric dimethylarginine as symmetric N α-acetyldimethylarginine, determined in human plasma and urine by LC–MS/MS. Journal of Chromatography B. 2015;975:59–64. doi: 10.1016/j.jchromb.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 27.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2010;465:966–966. doi: 10.1038/nature09132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Intlekofer AM, Dematteo RG, Venneti S, Finley LWS, Lu C, Judkins AR, Rustenburg AS, Grinaway PB, Chodera JD, Cross JR. Hypoxia induces production of L-2-hydroxyglutarate. Cell metabolism. 2015;22:304–311. doi: 10.1016/j.cmet.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30••.Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, Losman JA, Joensuu P, Bergmann U, Gross S. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–488. doi: 10.1038/nature10898. The authors show an enantiomeric difference of the effect of R/S-2-hydroxyglutarate controlling important aspects of cellular metabolism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cajka T, Fiehn O. Toward merging untargeted and targeted methods in mass spectrometry-based metabolomics and lipidomics. Analytical chemistry. 2015;88:524–545. doi: 10.1021/acs.analchem.5b04491. [DOI] [PubMed] [Google Scholar]
- 32.Pluskal T, Castillo S, Villar-Briones A, Orešič M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC bioinformatics. 2010;11:1. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Libiseller G, Dvorzak M, Kleb U, Gander E, Eisenberg T, Madeo F, Neumann S, Trausinger G, Sinner F, Pieber T. IPO: a tool for automated optimization of XCMS parameters. BMC bioinformatics. 2015;16:1. doi: 10.1186/s12859-015-0562-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34••.Tsugawa H, Cajka T, Kind T, Ma Y, Higgins B, Ikeda K, Kanazawa M, VanderGheynst J, Fiehn O, Arita M. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nature methods. 2015;12:523–526. doi: 10.1038/nmeth.3393. This is the recommended, freely available software for processing untargeted metabolomics data. The paper shows the utility for removing false positive peaks and compound identification, including large-scale data analyses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kind T, Liu K-H, Lee DY, DeFelice B, Meissen JK, Fiehn O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nature methods. 2013;10:755–758. doi: 10.1038/nmeth.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vinaixa M, Schymanski EL, Neumann S, Navarro M, Salek RM, Yanes O. Mass spectral databases for LC/MS-and GC/MS-based metabolomics: State of the field and future prospects. TrAC Trends in Analytical Chemistry. 2016;78:23–35. [Google Scholar]
- 37.Kind T, Fiehn O. Metabolomic database annotations via query of elemental compositions: mass accuracy is insufficient even at less than 1 ppm. BMC bioinformatics. 2006;7:1. doi: 10.1186/1471-2105-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsugawa H, Kind T, Nakabayashi R, Yukihira D, Tanaka W, Cajka T, Saito K, Fiehn O, Arita M. Hydrogen rearrangement rules: computational MS/MS fragmentation and structure elucidation using MS-FINDER software. Analytical Chemistry. 2016 doi: 10.1021/acs.analchem.6b00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dührkop K, Böcker S. Fragmentation trees reloaded. International Conference on Research in Computational Molecular Biology; Springer; 2015. pp. 65–79. [Google Scholar]
- 40.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E. HMDB 3.0—the human metabolome database in 2013. Nucleic acids research. 2012:gks1065. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41•.Jeffryes JG, Colastani RL, Elbadawi-Sidhu M, Kind T, Niehaus TD, Broadbelt LJ, Hanson AD, Fiehn O, Tyo KEJ, Henry CS. MINEs. open access databases of computationally predicted enzyme promiscuity products for untargeted metabolomics. Journal of cheminformatics. 2015;7:1. doi: 10.1186/s13321-015-0087-1. Substrate promiscuity of enzymes is known for a long time, but no database captures such information in a comprehensive manner. The authors here show how the database was constructed and used and how closely the predicted compounds resemble known natural products. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruttkies C, Schymanski EL, Wolf S, Hollender J, Neumann S. MetFrag relaunched: incorporating strategies beyond in silico fragmentation. Journal of cheminformatics. 2016;8:1. doi: 10.1186/s13321-016-0115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma Y, Kind T, Yang D, Leon C, Fiehn O. MS2Analyzer: a software for small molecule substructure annotations from accurate tandem mass spectra. Analytical chemistry. 2014;86:10724–10731. doi: 10.1021/ac502818e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dührkop K, Shen H, Meusel M, Rousu J, Böcker S. Searching molecular structure databases with tandem mass spectra using CSI: FingerID. Proceedings of the National Academy of Sciences. 2015;112:12580–12585. doi: 10.1073/pnas.1509788112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45•.Allen F, Pon A, Wilson M, Greiner R, Wishart D. CFM-ID: a web server for annotation, spectrum prediction and metabolite identification from tandem mass spectra. Nucleic acids research. 2014;42:W94–W99. doi: 10.1093/nar/gku436. At current, the MS/MS prediction capabilities of this software CFM-ID appears to slightly outperform alternative programs. This paper outlines the algorithms and assumptions behind it. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schymanski EL, Gerlich M, Ruttkies C, Neumann S. Solving CASMI 2013 with MetFrag, MetFusion and MOLGEN-MS/MS. Mass Spectrometry. 2014:3. doi: 10.5702/massspectrometry.S0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bingol K, Bruschweiler R. Knowns and unknowns in metabolomics identified by multidimensional NMR and hybrid MS/NMR methods. Current Opinion in Biotechnology. 2016;43:17–24. doi: 10.1016/j.copbio.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, DuGar B, Feldstein AE, Britt EB, Fu X, Chung Y-M. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koeth RA, Levison BS, Culley MK, Buffa JA, Wang Z, Gregory JC, Org E, Wu Y, Li L, Smith JD. γ-Butyrobetaine is a proatherogenic intermediate in gut microbial metabolism of L-carnitine to TMAO. Cell metabolism. 2014;20:799–812. doi: 10.1016/j.cmet.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Collard F, Baldin F, Gerin I, Bolsee J, Noel G, Graff J, Veiga-da-Cunha M, Stroobant V, Vertommen D, Houddane A, et al. A conserved phosphatase destroys toxic glycolytic side products in mammals and yeast. Nature Chemical Biology. 2016;12:601. doi: 10.1038/nchembio.2104. [DOI] [PubMed] [Google Scholar]
- 51.Wikoff WR, Hanash S, DeFelice B, Miyamoto S, Barnett M, Zhao Y, Goodman G, Feng Z, Gandara D, Fiehn O. Diacetylspermine is a novel prediagnostic serum biomarker for non–small-cell lung cancer and has additive performance with pro-surfactant protein B. Journal of Clinical Oncology. 2015 doi: 10.1200/JCO.2015.61.7779. JCO.2015.2061.7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson CH, Dejea CM, Edler D, Hoang LT, Santidrian AF, Felding BH, Ivanisevic J, Cho K, Wick EC, Hechenbleikner EM. Metabolism links bacterial biofilms and colon carcinogenesis. Cell metabolism. 2015;21:891–897. doi: 10.1016/j.cmet.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]