Abstract
Genetic selection was used to explore the probability of finding enzymes in protein sequence space. Large degenerate libraries were prepared by replacing all secondary structure units in a dimeric, helical bundle chorismate mutase with simple binary-patterned modules based on a limited set of four polar and four nonpolar residues. Two-stage in vivo selection yielded catalytically active variants possessing biophysical and kinetic properties typical of the natural enzyme even though ≈80% of the protein originates from the simplified modules and >90% of the protein consists of only eight different amino acids. This study provides a quantitative assessment of the number of sequences compatible with a given fold and implicates previously unidentified residues needed to form a functional active site. Given the extremely low incidence of enzymes in completely unbiased libraries, strategies that combine chemical information with genetic selection, like the one used here, may be generally useful in designing novel protein scaffolds with tailored activities.
Despite recent progress on the de novo design of structurally defined proteins (1–4), creation of stable scaffolds with tailored enzymatic activities remains an unrealized challenge. Not only is our understanding of the relationship between sequence, structure, and function incomplete, but the requirement for catalysis imposes severe constraints on design. Misplacement of catalytic residues by even a few tenths of an angstrom can mean the difference between full activity and none at all.
Direct selection of catalysts from pools of fully randomized polypeptides is a conceivable alternative to de novo design, requiring no foreknowledge of structure or mechanism. An analogous approach has yielded RNA catalysts for a variety of chemical reactions (5). However, a 100-residue protein has 20100 (1.3 × 10130) possible sequences. Even a library with the mass of the Earth itself—5.98 × 1027 g—would comprise at most 3.3 × 1047 different sequences, or a miniscule fraction of such diversity. Unless protein catalysts are unexpectedly abundant and evenly distributed in sequence space, such a strategy will clearly be impractical.
Combination of these two approaches represents a potentially attainable middle ground. For example, basic structural information, such as the sequence preferences of helices and sheets or the tendency of hydrophobic residues to be buried in the protein interior, might be used to design focused libraries from which catalysts could be selected with reasonable probability. In fact, binary patterning of polar and nonpolar amino acids (6–8) has been used to generate four-helix bundle proteins (9) that exhibit some native-like properties, including protease resistance and cooperative unfolding (10, 11).
Here we show that a combinatorial approach that couples modular design and selection can successfully reproduce a known catalytic activity with an unnatural sequence based on a severely restricted set of building blocks. Specifically, we have constructed large binary-patterned libraries based on the AroQ-class chorismate mutase (CM) from Methanococcus jannaschii (MjCM′) (12, 13) and applied evolutionary strategies to evaluate their catalytic capabilities. Our results provide a quantitative assessment of the number of different sequences compatible with a catalytically active helical bundle fold and illustrate the importance of subtle interactions in the formation of a functional active site.
Materials and Methods
Reagents.
Restriction enzymes and Klenow fragment were from New England Biolabs. T4 DNA ligase was from Fermentas, Vilnius, Lithuania. HotStarTaq polymerase was from Qiagen, Basel, Switzerland. Protein concentration was determined with the Coomassie Plus Protein Assay Reagent from Pierce, using BSA as the calibration standard.
Oligonucleotides.
The H1 library was prepared with oligonucleotides RPH1CS-01 (99 bp, 5′-TATGGGAGATGGATACATATGHTSRAVRAVHTSHTSRAVHTSCGTRAVRAVHTSRAVRAVHTSRAVRAVRAVHTSHTSAAGCTTCGCCT- GCCCAAGCTT) and RPH1NS-02 (97 bp, 5′-AAATCGTTGGAATAGGAATTCCSADBTYBTYCTTSADBTYSADSADBTYBTYSADSADBTYBTYACGBTYSADSADAAGCTTGGGCAGGCGAAGCTT), where R = A or G; V = A, G, or C; H = A, C, or T; S = C or G; B = C, G, or T; Y = C or T; and D = A, G, or T. The HindIII sites that were used for library construction are underlined. For the H2/H3 library, RPH2CS-03 (81 bp, 5′-GGAATTCCGATCAACGACHTSRAVCGTGAARAVHTSHTSHTSRAVRAVHTSRAVRAVHTSHTSRAVGAGCACAACGTCGAC) and RPH3NS-04 (108 bp, 5′-GTGGTGCCGCTCGAGBTYBTYBTYSADSADBTYBTYCTGSADSADBTYBTYBTYBTYSADSAD- SADBTYSADSADBTYSADSADSADBTYBTYGTCGACGTTGTGCTC) were used. Primers for H1 amplification were: RPH1A1 (5′-CAACGTTATGTTCTTTACAA) and KCMBU1 (5′-CTGGCCTTTTGCTCACAT). Primers for H2/H3 amplification were: RPH2H3A1 (5′-TTGATGAGATTGACAATAAGATA) and RPH2H3A2 (5′-CAACTGTTGGGAAGGGCGAT). All oligonucleotides were synthesized by Microsynth (Balgach, Switzerland).
Library Construction.
For construction of the H1 and H2/H3 libraries, complementing oligonucleotide pairs (RPH1CS-01/RPH1NS-02, and RPH2CS-03/RPH3NS-04, respectively) were annealed and extended with Klenow fragment to make double-stranded DNA. The primer extension products were purified by agarose gel electrophoresis, eluted from DEAE membranes, and precipitated with isopropanol. The purified DNA was treated with restriction enzymes (NdeI/EcoRI and EcoRI/XhoI for the H1 and H2/H3 libraries, respectively), purified as above, and ligated with appropriate fragments (2,958 bp NdeI/EcoRI for H1 or 2,925 bp EcoRI/XhoI for H2/H3) of the respective acceptor vectors pKMCMT-λ or pKMCMT-λ2, containing the MjCM′ gene fragment encoding the rest of the protein (i.e., either H2/H3-WT or H1-WT). Vectors pKMCMT-λ and pKMCMT-λ2, which will be described in detail elsewhere, are derived from a variant of the trc-promoter expression plasmid pKMCMT-W (13) that contained the 282-bp NdeI/XhoI fragment encoding MjCM′ from pET-MjCM′-pATCH (14) and, additionally, a silent EcoRI restriction site in the segment encoding the L1 loop. Replacement of the 124-bp segment encoding the H1 helix of MjCM′ with the 964-bp NdeI/EcoRI fragment from phage λ yielded pKMCMT-λ. Replacement of the 158-bp segment encoding H2/L2/H3 with the 1,751-bp EcoRI/XhoI fragment from phage λ gave pKMCMT-λ2. The DNA overlap region used for construction of the H1 library subsequently was excised by HindIII digestion, and religation restored a single HindIII site encoding Lys-23–Leu-24.
For construction of the H1/H2/H3 library, the active H1 and H2/H3 variants were harvested by scraping the colonies from the selection plates and purifying the plasmid DNA. The randomized segments from the H1 and H2/H3 plasmid pools were amplified by 20 cycles of PCR using primer pairs RPH1A1/KCMBU1 and RPH2H3A1/RPH2H3A2, respectively. PCR products were restriction-digested (NdeI/EcoRI and EcoRI/XhoI for the H1 and H2/H3 fragments, respectively), purified as above, and ligated with the 2,800-bp NdeI/XhoI fragment of vector pKT-λ3. Vector pKT-λ3 is a derivative of pKMCMT-W (13) with the 964-bp NdeI/EcoRI and 1,751-bp EcoRI/XhoI fragments of λ phage replacing the entire 300-bp MjCM gene.
Selection.
The library plasmid pools were electroporated into the KA12/pKIMP-UAUC selection strain (15). Transformed cells were washed in minimal medium and plated onto M9c minimal media plates (lacking Tyr and Phe) (16). Plates were incubated at 25°C for either 9 (H1 and H2/H3 library experiments) or 13 days (H1/H2/H3 library experiments).
Protein Production and Purification.
Genes encoding wild-type (WT) MjCM′ and active variants were subcloned as 282-bp NdeI/XhoI fragments into plasmid pET-22b-pATCH and overexpressed in Escherichia coli strain KA13 (13). Overproduced protein was purified by affinity chromatography on Ni2+-NTA agarose and assayed directly for CM activity. Before detailed characterization, MjCM′ and clone H1/H2/H3–12 were additionally purified by FPLC on an Amersham Pharmacia Superose 12-HR 10/30 gel filtration column eluted with PBS (pH 7.5) at 4°C.
Protein Characterization.
Size-exclusion chromatography, CD measurements, and CM activity assays were performed as described (13, 16). Reactions with H1/H2/H3–12 were supplemented with BSA (0.1 mg/ml).
Sequence Analysis.
The sequences of ≥15 correct length clones from the H1 and H2/H3 libraries before and after selection were obtained. Roughly 20 unselected and 100 selected H1/H2/H3 variants were sequenced. Because the H1/H2/H3 library contained many duplicates of individual clones or their component parts after selection, only the sequences of unique H1 and H2/H3 regions were used for detailed comparisons with the corresponding segments from the H1 and H2/H3 libraries. Slight deviations from expected amino acid compositions at degenerate codon positions caused by biases in the synthetic oligonucleotides were neglected as they were present equally in unselected and selected clones.
Results
Design Strategy.
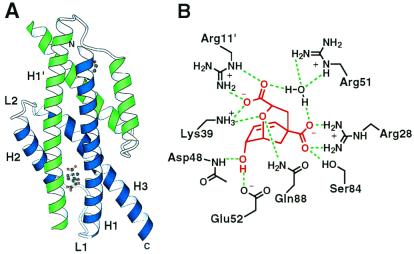
AroQ CMs catalyze the Claisen rearrangement of chorismate to prephenate with rate accelerations >106 (17). The structurally characterized prototype from E. coli (EcCM) (18) is a homodimeric helical bundle protein, whose intricately intertwined polypeptide chains fold as three helices (labeled H1, H2, and H3) separated by two loops (L1 and L2) (Fig. 1A). The two identical active sites are constructed from residues from both chains. Six highly conserved polar residues (Arg-11, Arg-28, Lys-39, Arg-51, Glu-52, and Gln-88; Fig. 1B) contribute hydrogen bonding and electrostatic interactions that are important for substrate binding and catalysis (19, 20). The side chains of several other, less conserved and largely aliphatic amino acids provide additional hydrophobic interactions (residues 14, 35, 55, 81, and 85) or hydrogen bonds (residue 84) to the binding pocket.
Figure 1.
AroQ structure and active site. (A) The homodimeric enzyme is shown with a transition state analog inhibitor bound at the active sites (18); the two identical polypeptide chains are colored blue and green for clarity. (B) An array of polar active site residues (black) provides extensive hydrogen bonding and electrostatic interactions with bound inhibitor (red). Residues 11, 28, 39, 51, 52, and 88 were held constant in the randomization experiments.
Because the chorismate rearrangement is essential for the biosynthesis of tyrosine (Tyr) and phenylalanine (Phe) in bacteria, genetic selection can be used to isolate proteins with CM activity from combinatorial libraries (15, 16, 21–23). As summarized in Figs. 2 and 3, our experimental strategy for constructing binary patterned CMs involved systematic replacement of all of the secondary structural elements in the protein with modules of random sequence. We chose the monofunctional MjCM′ AroQ protein as a design template because of its small size and thermostability (13). By extensively varying everything but the short L1 and L2 loops and the six highly conserved active site residues in this protein (Figs. 1B and 2), we expected to gain quantitative insight into the robustness of the helical bundle fold in forming active enzymes. To bias the libraries toward helical structures, subsets of either polar or nonpolar amino acids were assigned to each position in the individual modules according to the polarity of the corresponding MjCM′ residue (Fig. 2). Only eight of the 20 standard amino acids were permitted by design, thus ensuring that libraries contained only novel alternatives to naturally occurring CMs. Four polar amino acids—asparagine (Asn), aspartic acid (Asp), glutamic acid (Glu), and lysine (Lys)— and four apolar amino acids—isoleucine (Ile), leucine (Leu), methionine (Met), and Phe—constituted the two sets of allowable building blocks. At the genetic level, these residues can be specified by two degenerate codons. The RAV codon (R = A or G; V = A, G, or C) encodes Asn, Asp, Glu, and Lys, whereas the HTS codon (H = A, C, or T; S = C or G) encodes Ile, Leu, Met, and Phe. Using equimolar concentrations of the indicated bases at all degenerate positions during DNA synthesis results in a 3-fold bias for Leu in the nonpolar subset and a 2-fold bias for Glu and Lys in the polar subset. Overrepresentation of Leu, Glu, and Lys, which occur frequently in α-helices (24), should favor formation of native-like structures.
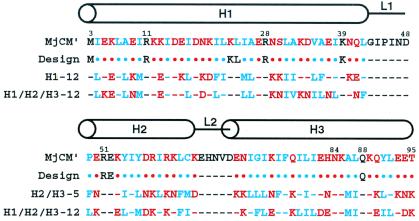
Figure 2.
Library design and sequences of selected variants. The MjCM′ sequence comprises the N-terminal 93 aa of AroQf from M. jannaschii plus an appended Leu-Glu-(His)6 tag (not shown); residue numbers and secondary structural elements were assigned by comparison with EcCM (18). Binary patterned helical modules were designed according to the observed distribution of hydrophilic and hydrophobic residues in the MjCM′ H1, H2, and H3 helices. Large degenerate libraries were obtained by providing mixtures of Asn, Asp, Glu, and Lys at polar (red) positions and mixtures of Ile, Leu, Met, and Phe at apolar (blue) positions. The starting methionine, highly conserved active site amino acids, and loop residues (black) were held constant. Lys-23 and Leu-24 in the H1 helix were also retained for construction purposes (see Fig. 3). Sequences of representative clones (H1–12, H2/H3–5, and H1/H2/H3–12) obtained by selection from the libraries are shown below the imposed binary pattern; dashes indicate residues that are identical to their MjCM′ counterpart.
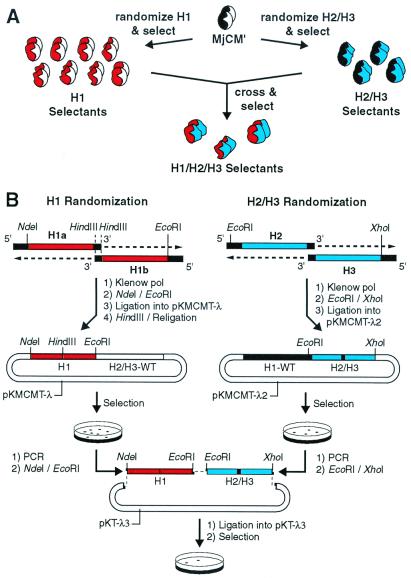
Figure 3.
Construction of binary patterned CMs. (A) The general experimental strategy involved initially selecting functional enzymes from protein libraries in which only the H1 (red) or H2/H3 (blue) helices were replaced with randomized modules created according to the binary patterning scheme of Fig. 2. In a second stage, selected H1 and H2/H3 modules were combined combinatorially, and functional H1/H2/H3 enzymes were identified by genetic selection. (B) The H1 and H2/H3 binary patterned libraries were constructed at the genetic level from synthetic oligonucleotides in which polar and nonpolar residues were designated by specific degenerate codons (see Materials and Methods). Briefly, appropriately randomized oligonucleotide pairs were annealed at complementary overlap sites, extended with Klenow polymerase fragment, and cloned into acceptor vectors containing the complementary WT gene segment. Functional H1 and H2/H3 modules from the libraries were identified by genetic selection. PCR amplification of the encoding gene segments and subsequent three-fragment ligation with the acceptor vector pKT-λ3 yielded the H1/H2/H3 library plasmid pool, encoding CMs in which all secondary structural elements (≈80% of the protein) are derived from the binary patterned helical modules. Active clones were again identified by genetic selection.
Partial Randomization.
Initially, randomized modules corresponding to the H1 and H2/H3 helices were combined individually with complementary WT MjCM′ segments (Fig. 3A). Both libraries were constructed from two ≈100-base-long oligonucleotides with appropriately randomized regions and ligated into vectors containing the complementary WT MjCM′ gene fragment (Fig. 3B). The segment encoding the L2 loop provided a convenient DNA overlap for construction of the H2/H3 library, but construction of the H1 library required that two additional residues (Lys-23 and Leu-24) in the middle of the dimer-spanning helix be held constant (Fig. 3B). The E. coli strain KA12/pKIMP-UAUC (15) was transformed with the resulting plasmid pools to give >107 transformants (Table 1). Sequencing of unselected clones from both libraries showed that approximately two-thirds of the genes had insertions or deletions that probably arose during oligonucleotide synthesis or library construction (25), but one-third had reading frames of the correct length. The randomized regions of the final H1 and H2/H3 libraries constituted 37% and 42% of the entire protein, respectively.
Table 1.
Library statistics
| Library | Total no. of library clones | Fraction of correct length clones | Total no. of complementing clones | Complementation rate |
|---|---|---|---|---|
| H1 | 3.0 × 107 | ≈30% | 1,980 | 1 in 4,500 |
| H2/H3 | 3.8 × 107 | ≈30% | 654 | 1 in 17,500 |
| H1/H2/H3 | 7.4 × 107 | 100% | 7,430 | 1 in 10,000 |
The propensity of the partially randomized proteins to fold into catalytically active structures was assessed by complementation of the endogenous CM deficiency of KA12/pKIMP-UAUC (15). Without CM activity, this strain is unable to synthesize Tyr and Phe and hence cannot grow on minimal medium. When a plasmid encoding a functional CM is introduced into the auxotroph, it regains prototrophy. Roughly 1 in 104 of the correct length members of each library was found to be active in this system (Table 1). In other words, ≈0.01% of the theoretically possible binary-patterned sequences yield folded, functional enzymes.
Genes of active clones from each library were subcloned into an overexpression vector and purified enzymes were assayed for CM activity. Kinetic data presented in Table 2 show that enzymes from both libraries are quite active, with kcat and Km values within a factor of 30 of MjCM′ (see Fig. 2 for sequences). In general, however, unnatural H2/H3 variants were found to be less active than unnatural H1 variants, suggesting that the H2/H3 helices are less tolerant to variation. Binary patterning is obviously useful for catalyst design, but the rarity of active enzymes underscores the need for efficient selection.
Table 2.
Characterization of CM variants
| CM variant | kcat, s−1 | Km, μM | % Identity to MjCM′ in randomized region |
|---|---|---|---|
| MjCM′* | 5.7 | 41 | — |
| H1-12 | 2.3 | 120 | 35 |
| H2/H3-5 | 0.20 | 550 | 26 |
| H1/H2/H3-12 | 0.38 | 1,700 | 33 |
The values for MjCM′ were taken from ref. 13.
Combinatorial Randomization.
Analogous to the way the immune system combines Ig heavy and light chains to generate diverse antibody structures, the selected H1 and H2/H3 segments were crossed to create CMs in which all secondary structural elements (≈80% of the protein) are derived from the simplified, binary-patterned modules (Fig. 3A). The randomized portions of all active clones in the H1 and H2/H3 libraries (Table 1) were amplified by PCR, and an H1/H2/H3 library was created by three-fragment ligation with an appropriate acceptor vector (Fig. 3B). Transformation of KA12/pKIMP-UAUC with the resulting plasmid pool yielded 7.4 × 107 transformants, ensuring adequate coverage of all possible combinations of active H1 and H2/H3 segments from the initial libraries (1,980 × 654 = 1.29 × 106). Surprisingly, only about 1 of every 104 transformants complements the CM deficiency. Most H1/H2/H3 combinations do not yield active enzymes, even though all of the preselected segments are functional in a native context. The WT MjCM′ H1 and H2/H3 helices are obviously more effective than their binary-patterned counterparts in templating correctly folded enzymes, presumably because they already have been optimized for stability (13) and are better able to accommodate imperfections in the rest of the protein.
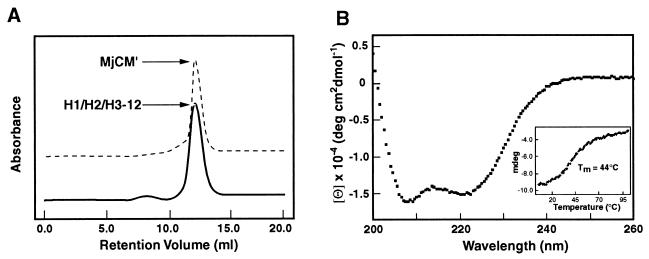
One fast-growing clone that was isolated multiple times from the H1/H2/H3 library (H1/H2/H3–12, Fig. 2) was characterized in detail. Its size-exclusion chromatographic profile (Fig. 4A) and CD spectrum (Fig. 4B) are essentially the same as those for MjCM′, suggesting that H1/H2/H3–12 is a compact helical homodimer. It undergoes cooperative thermal denaturation (Fig. 4B Inset) with the midpoint of the unfolding transition (Tm) at ≈44°C [for comparison, Tm = 88°C and 63°C for MjCM′ and EcCM, respectively (13)]. H1/H2/H3–12 also possesses considerable catalytic activity, with a kcat only 15-fold lower than MjCM′ and a 40-fold higher Km value (Table 2). The selected enzyme thus captures the key biophysical and functional properties of its natural counterpart, even though >90% of the protein consists of only eight different amino acids.
Figure 4.
Characterization of a binary patterned CM. (A) Size-exclusion chromatography traces of H1/H2/H3–12 (solid line) and MjCM′ (dashed line), showing similar retention times. MjCM′ was previously shown to be homodimeric by analytical ultracentrifugation (13). (B) The CD spectrum of H1/H2/H3–12. The molar ellipticities at 208 and 222 nm are comparable to those of WT MjCM′ (data not shown) and show that the protein is highly helical. (Inset) The cooperative thermal denaturation of H1/H2/H3–12.
Sequence Analysis.
The wealth of sequence data that emerges from these experiments is an invaluable resource for gaining deeper insight into the factors that influence protein folding, function, and evolution. Multiple clones from all three libraries were consequently analyzed and compared for salient patterns. First, no duplicates were found in the H1 and H2/H3 libraries or in the unselected H1/H2/H3 library, whereas several appeared among selected H1/H2/H3 clones. Some H1 and H2/H3 segments are also used repeatedly to construct functional H1/H2/H3 variants, albeit in different combinations. Second, sequence identity to WT MjCM′ generally increases after selection, although a direct correlation between sequence identity and activity is not observed. Third, Ile is overrepresented in active H1/H2/H3 clones. It is also relatively abundant in MjCM′ (13) and may be important for proper packing of the hydrophobic core. Fourth, spontaneous mutations are randomly distributed in all libraries at a low frequency (1 in every ≈400 nts). In the H1/H2/H3 library, however, an unprogrammed valine at position 32 was selected in multiple independent clones, presumably because it packs better than the larger hydrophobic amino acids in the starting set of building blocks. Valine is accessible by a single point mutation, whereas the WT Ala32 would require two base changes. Here we see evolution at work.
Packing interactions, more generally, are likely to be crucial for the proper assembly of functional AroQ dimers. The unusually long H1-H1′ antiparallel coiled-coil in EcCM, for instance, is characterized by a standard heptad repeat of hydrophobic amino acids at the α (residues 7, 14, 21, 28, and 35) and δ (residues 10, 17, 24, 31, and 38) positions of each helix (18). A strong bias against phenylalanine at these positions and a preference for hydrophobic aliphatic residues, albeit not necessarily the MjCM′ WT residue, is evident in both the H1 and H1/H2/H3 libraries after selection. Residues 14 and 35, which also provide a portion of the hydrophobic surface of the substrate binding pocket, appear to have been subject to the greatest selection pressure (see below). Buried residues that contribute to tight packing of the H1-H1′ coiled-coil against the H2-H3 and H2′-H3′ segments similarly show a preference for aliphatic residues. At position 77, however, phenylalanine is highly enriched: 87% of active H1/H2/H3 clones have phenylalanine at this position compared with 68% of the H2/H3 library and only 21% of unselected clones. A similar, if less dramatic, trend is evident at positions 62 and 73. These phenylalanines may favor formation of a native-like dimer by providing a small hydrophobic core for nucleating folding of the H2-H3 segment and allowing it to pack against the H1-H1′ coiled-coil.
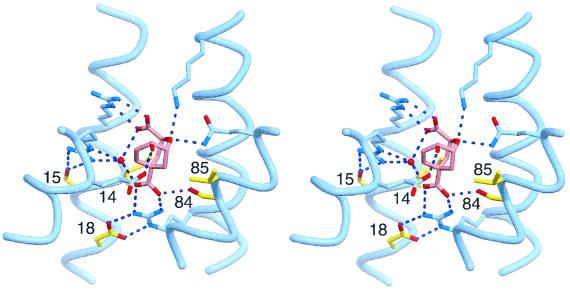
While most positions in the protein appear to be relatively tolerant to substitution, some are quite restrictive, implicating them in critical interactions needed for generating catalytically active binding pockets. For example, an Asn-Lys dyad is highly conserved at positions 84 and 85, and most active H1/H2/H3 clones have Ile-14, Asp-15, and Asp-18 (Fig. 2). These preferences are not evident in unselected clones, but can be rationalized by examination of the EcCM structure (18) (Fig. 5). The homologues of Asn-84, Lys-85, and the aforementioned Ile-14 interact directly with bound ligand, providing either hydrogen bonding (residue 84) or van der Waals (residues 14 and 85) interactions. They constitute a portion of the active site that was allowed to vary in the randomization experiments and their fixation during selection suggests that specific amino acids are required at these positions to form a functional binding pocket. Two other amino acids that contribute hydrophobic interactions to the active site, residues 35 and 55, show a strong bias toward Ile (>85% of the H1/H2/H3 clones), which mirrors the presence of hydrophobic β-branched amino acids at the corresponding positions in the WT MjCM′ sequence.
Figure 5.
Stereoview of the EcCM active site with bound transition state analog (pink) (18), showing the positions of the catalytic residues (blue) and the first and second sphere residues (yellow) that had highly restricted sequence requirements in active clones from the MjCM′ H1/H2/H3 library. The amino acid at position 84 forms a hydrogen bond with the tertiary carboxylate of the bound inhibitor (pink), while residues 14 and 85 are in van der Waals contact with the ligand. Amino acids at positions 15 and 18 help position active site residues Arg-51 and Arg-28, respectively. Image was created by using molscript (39) and RASTER3D (40).
Interestingly, the strongly selected residues Asp-15 and Asp-18 do not contact the bound ligand directly, but participate instead in second-sphere interactions with the active site. The corresponding EcCM residues interact with the side chains of the catalytically important residues Arg-51 and Arg-28 and may help position them for effective catalysis. In this case, inclusion of Arg-51 and Arg-28 as invariant residues in the libraries appears to have influenced the course of selection.
Discussion
This study demonstrates the feasibility of creating structurally complex and catalytically active enzymes by assembling randomized modules that are constructed from a limited set of building blocks and biased toward helical secondary structure by binary patterning. The binary distribution of hydrophilic/hydrophobic residues is inherent in the genetic code (NAN/NTN), and our results support suggestions (9, 26, 27) that modern enzymes could have evolved from primitive precursors constructed from a relatively small number of polar and nonpolar amino acids. There is, nevertheless, a low probability of finding catalysts, even when both position and identity of all critical active site residues are determined in advance. This finding contrasts with the ease of obtaining folded helical proteins through binary patterning (9), underscoring the exacting demands that catalysis places on protein design.
Extrapolating from our data and from modest sequence constraints on interhelical turns (23, 28–30), we can estimate that if every position in the protein had been randomized, a library of ≈1024 members would have been needed to obtain AroQ mutases. This estimate is based on the experimentally observed frequencies for the binary-patterned helical modules and the assumption that only a single amino acid is tolerated at each highly conserved position in the active site (i.e., Arg-11, Arg-28, Lys-39, Arg-51, Glu-52, and Gln-88) Previous studies (23, 28–30) suggest further that 1–10% of all possible turn segments will yield active catalysts. Finally, based on the low incidence of active clones observed in the H1/H2/H3 library, a templating/assembly effect of 104 for organizing the H1 and H2/H3 segments is included; this factor could turn out to be larger for assembly of helices and loops that have not been optimized by a preselection step. The required library size can thus be calculated as follows: 4,500 (binary patterned H1) × 17,500 (binary patterned H2/H3) × 206 (randomized active site residues) × 102 (fully randomized L1) × 102 (fully randomized L2) × 104 (templating effects) = 5 × 1023.
The size of such a library is many orders of magnitude larger than that needed to identify noncatalytic ATP-binding proteins from random sequences (31). Although the estimated frequency of catalysts in protein sequence space will be contingent on the choice of building blocks and structural motif, on the difficulty of the chemical reaction, and on the level of catalytic activity needed for selection, construction of a moderately active enzyme also appears to be substantially more difficult than obtaining a ribozyme. For instance, it has been found that ≈1 in 1013 RNA molecules from a pool of random sequences promote a template-directed ligation with RNA substrates (32), despite an even more limited set of building blocks (33) and without any information biasing the population toward a particular scaffold. The diversity of side chains may provide proteins with intrinsically greater maximum catalytic potential than oligonucleotides, but coordinating the many interactions required to form complex secondary and tertiary structures while preventing misfolding may be inherently more difficult with amino acids than with nucleic acids that can exploit relatively simple hydrophobic base stacking and base-pairing motifs for folding. This observation is intriguing in the context of the RNA-world hypothesis, which postulates that RNA preceded proteins as the principal agent of catalysis (34).
Our estimate of the low frequency of protein catalysts in sequence space indicates that it will not be possible to isolate enzymes from unbiased random libraries in a single step. The required library sizes far exceed what is currently accessible by experiment, even with in vitro methods (31, 35). Instead, as in natural evolution, the design of new enzymes will require incremental strategies in which, for instance, a suitable scaffold is first generated, binding and catalytic groups are subsequently added, and the ensemble is optimized in an iterative fashion. Our two-stage approach to binary-patterned mutases and work on the redesign of existing enzymes (36–38) demonstrate the power of stepwise and modular procedures for directing the course of evolution. By iteratively combining combinatorial mutagenesis and selection with intelligent design, it may also prove possible to create novel protein scaffolds, unknown in nature, and to endow them with tailored catalytic activities.
Acknowledgments
We thank Stephanie Küng for experimental assistance and Kinya Hotta for assistance with figure preparation. This work was supported by the Eidgenössische Technische Hochschule (Zürich), the Schweizerischer Nationalfonds, and Novartis Pharma.
Abbreviations
- CM
chorismate mutase
- MjCM′
reengineered CM from Methanococcus jannaschii
- EcCM
Escherichia coli CM
- WT
wild type.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hill R B, Raleigh D P, Lombardi A, DeGrado W F. Acc Chem Res. 2000;33:745–754. doi: 10.1021/ar970004h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harbury P B, Plecs J J, Tidor B, Alber T, Kim P S. Science. 1998;282:1462–1467. doi: 10.1126/science.282.5393.1462. [DOI] [PubMed] [Google Scholar]
- 3.Dahiyat B I, Mayo S L. Science. 1997;278:82–87. doi: 10.1126/science.278.5335.82. [DOI] [PubMed] [Google Scholar]
- 4.Cordes M H J, Davidson A R, Sauer R T. Curr Opin Struct Biol. 1996;6:3–10. doi: 10.1016/s0959-440x(96)80088-1. [DOI] [PubMed] [Google Scholar]
- 5.Wilson D S, Szostak J W. Annu Rev Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 6.West M W, Hecht M H. Protein Sci. 1995;4:2032–2039. doi: 10.1002/pro.5560041008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong H, Buckwalter B L, Shieh H M, Hecht M H. Proc Natl Acad Sci USA. 1995;92:6349–6353. doi: 10.1073/pnas.92.14.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowie J U, Clarke N D, Pabo C O, Sauer R T. Proteins Struct Funct Genet. 1990;7:257–264. doi: 10.1002/prot.340070307. [DOI] [PubMed] [Google Scholar]
- 9.Kamtekar S, Schiffer J M, Xiong H, Babik J M, Hecht M H. Science. 1993;262:1680–1685. doi: 10.1126/science.8259512. [DOI] [PubMed] [Google Scholar]
- 10.Roy S, Hecht M H. Biochemistry. 2000;39:4603–4607. doi: 10.1021/bi992328e. [DOI] [PubMed] [Google Scholar]
- 11.Roy S, Ratnaswamy G, Boice J A, Fairman R, McLendon G, Hecht M H. J Am Chem Soc. 1997;119:5302–5306. [Google Scholar]
- 12.Gu W, Williams D S, Aldrich H C, Xie G, Gabriel D W, Jensen R A. Microb Comp Genomics. 1997;2:141–158. doi: 10.1089/omi.1.1997.2.141. [DOI] [PubMed] [Google Scholar]
- 13.MacBeath G, Kast P, Hilvert D. Biochemistry. 1998;37:10062–10073. doi: 10.1021/bi980449t. [DOI] [PubMed] [Google Scholar]
- 14.MacBeath G, Kast P. BioTechniques. 1998;24:789–794. doi: 10.2144/98245st02. [DOI] [PubMed] [Google Scholar]
- 15.Kast P, Asif-Ullah M, Jiang N, Hilvert D. Proc Natl Acad Sci USA. 1996;93:5043–5048. doi: 10.1073/pnas.93.10.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gamper M, Hilvert D, Kast P. Biochemistry. 2000;39:14087–14094. doi: 10.1021/bi0016570. [DOI] [PubMed] [Google Scholar]
- 17.Haslam E. Shikimic Acid: Metabolism and Metabolites. New York: Wiley; 1993. [Google Scholar]
- 18.Lee A Y, Karplus P A, Ganem B, Clardy J. J Am Chem Soc. 1995;117:3627–3628. [Google Scholar]
- 19.Zhang S, Kongsaeree P, Clardy J, Wilson D B, Ganem B. Bioorg Med Chem. 1996;4:1015–1020. doi: 10.1016/0968-0896(96)00099-5. [DOI] [PubMed] [Google Scholar]
- 20.Liu D R, Cload S T, Pastor R M, Schultz P G. J Am Chem Soc. 1996;118:1789–1790. [Google Scholar]
- 21.MacBeath G, Kast P, Hilvert D. Protein Sci. 1998;7:1757–1767. doi: 10.1002/pro.5560070810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacBeath G, Kast P, Hilvert D. Science. 1998;279:1958–1961. doi: 10.1126/science.279.5358.1958. [DOI] [PubMed] [Google Scholar]
- 23.MacBeath G, Kast P, Hilvert D. Protein Sci. 1998;7:325–335. doi: 10.1002/pro.5560070212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Creighton T E. Proteins: Structures and Molecular Properties. New York: Freeman; 1993. [Google Scholar]
- 25.Hecker K H, Rill R L. BioTechniques. 1998;24:256–260. doi: 10.2144/98242st01. [DOI] [PubMed] [Google Scholar]
- 26.Crick F H C. J Mol Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 27.Wong J T-F. Proc Natl Acad Sci USA. 1975;72:1909–1912. doi: 10.1073/pnas.72.5.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castagnoli L, Vetriani C, Cesareni G. J Mol Biol. 1994;237:378–387. doi: 10.1006/jmbi.1994.1241. [DOI] [PubMed] [Google Scholar]
- 29.Brunet A P, Huang E S, Huffine M E, Loeb J E, Weltman R J, Hecht M H. Nature (London) 1993;364:355–358. doi: 10.1038/364355a0. [DOI] [PubMed] [Google Scholar]
- 30.Predki P F, Agrawal V, Brünger A T, Regan L. Nat Struct Biol. 1996;3:54–58. doi: 10.1038/nsb0196-54. [DOI] [PubMed] [Google Scholar]
- 31.Keefe A D, Szostak J W. Nature (London) 2001;410:715–718. doi: 10.1038/35070613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartel D P, Szostak J W. Science. 1993;261:1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- 33.Rogers J, Joyce G F. Nature (London) 1999;402:323–325. doi: 10.1038/46335. [DOI] [PubMed] [Google Scholar]
- 34.Gesteland R F, Cech T R, Atkins J F, editors. The RNA World. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. [Google Scholar]
- 35.Roberts R W. Curr Opin Chem Biol. 1999;3:268–273. doi: 10.1016/S1367-5931(99)80042-8. [DOI] [PubMed] [Google Scholar]
- 36.Yano T, Oue S, Kagamiyama H. Proc Natl Acad Sci USA. 1998;95:5511–5515. doi: 10.1073/pnas.95.10.5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altamirano M M, Blackburn J M, Aguayo C, Fersht A R. Nature (London) 2000;403:617–622. doi: 10.1038/35001001. [DOI] [PubMed] [Google Scholar]
- 38.Joo H, Lin Z, Arnold F H. Nature (London) 1999;399:670–673. doi: 10.1038/21395. [DOI] [PubMed] [Google Scholar]
- 39.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 40.Merrit E A, Murphy M E P. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]