Abstract
Osteosarcoma is one of the most common primary malignant bone tumors. The inhibitor of growth family of protein 5 has been identified as a tumor suppressor in many cancers. In this study, we confirmed the downregulation of the both inhibitor of growth family of protein 5 and messenger RNA levels in cancer tissues using Western blot and real-time polymerase chain reaction. In order to find the antitumor roles of inhibitor of growth family of protein 5, osteosarcoma cells, HOS, and MG63 were transfected with the plasmid pCDNA-3.1-inhibitor of growth family of protein 5. Overexpression of Inhibitor of growth family of protein 5 could induce apoptosis and inhibit cell proliferation in osteosarcoma cells. Furthermore, Western blot analysis showed that p-Smad2, p-Smad3, and Smad4 were increased in inhibitor of growth family of protein 5-expressing osteosarcoma cells. Our results indicated that overexpression of inhibitor of growth family of protein 5 in osteosarcoma cells induces apoptosis by activating the Smad pathway, thus proposing a promising role for inhibitor of growth family of protein 5 in treatment of patients with osteosarcoma.
Keywords: osteosarcoma, ING5, apoptosis, survival rate, the Smad pathway
Introduction
Osteosarcoma (OS), originating from the metaphysis of the long bones, is the most common primary malignant bone tumor among children and young adults.1 In recent years, great treatment strategies have been made in the OS; however, treatment efficacy and survival rate remain poor.2 Therefore, the investigation into the novel targets for the OS is warranted.
The inhibitor of growth family of proteins (ING1-ING5) has been identified as tumor suppressors.3 They played an important role in cell cycle, cell senescence, and apoptosis.4 Downregulation of ING5 has been found in many cancers, including colorectal cancer,5 gastric cancer,6 and breast cancer.7 Further studies demonstrated that ING5 is a cofactor of Tip60 for acetylation of p53 at K120, leading to activation of p53, and induces apoptosis in response to DNA damage.8–10 However, no reports showed the roles of ING5 in OS. In this study, we evaluated the correlation between ING5 expression and the prognosis of OS using clinical samples.
Furthermore, we assessed the antitumor roles of ING5 in OS cells. The Smad signaling pathway is a pivotal pathway in regulating a range of cellular functions in cancer cells, including proliferation, mobility, and apoptosis.11 Central mediators of the Smad pathways are Smad2 and Smad3. They are phosphorylated and activated through the formation of the phosphorylation of Smad2 (ser465/467) and Smad3 (ser423/425).11 P-Smad2 and P-Smad3 formed complexes with Smad4 that accumulate in the nucleus and regulate the progression of cancer cells.11 This is the first report to show the influence of ING5 on the Smad signaling pathway in OS cells.
Materials and Methods
Osteosarcoma Cells and Tissue Samples
The human OS cell lines, HOS and MG63, and the osteoblast cell line, NHOst, were obtained from the Cell Bank of Central South University. All cells were maintained in Dulbecco Modified Eagle’s Medium (DMEM; Gaithersburg, Maryland) supplemented with 10% (vol/vol) fetal bovine serum (FBS) and antibiotics (100 units/mL of penicillin and 100 mg/mL of streptomycin) at 37ºC in a 5% CO2 incubator. Osteosarcoma tissues and matched normal tissues were obtained from Jintan Hospital between May 2008 and June 2012, with the agreement of each patient. The procedure was approved by Jiangsu University Ethics Committee.
Real-Time Polymerase Chain Reaction and reverse transcription PCR (RT-PCR)
Total RNA was isolated from cells and tissues using TRIzol reagent (Invitrogen Gibco, Carlsbad, California). RNA (1 μg) was reverse transcribed using TaKaRa Reverse Transcription Kit (TaKaRa, Dalian, China). The resultant complementary DNA was then used for polymerase chain reaction. The ING5 primers were 5′-GGGAGATGATTGGCTGTG-3′ (sense) and 5′-CCTTTGGGTT TCGTGGTA-3′ (antisense). The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers were 5′-AGAAGGCTGGGGCTCATTTG-3′ (sense) and 5′-CGATCCACACGGAGTACTTGC-3′ (antisense). Amplification of ING5 and GAPDH was performed with 1 cycle at 95ºC for 10 minutes and 40 cycles of 95ºC for 15 seconds and 60ºC for 60 seconds. Calculation of the relative expression of each transcript was performed using the 2−ΔΔCt method 12.
Immunohistochemistry
Tissues were fixed in formalin, embedded in paraffin, and cut into 4 μm thick sections. The sections were incubated with a polyclonal antibody against ING5 (1:200; ab3716, Abcam, Shanghai, China) or P-Smad3 (1:200; ab52903, Abcam) overnight at 4°C. Secondary antibodies were applied and incubated at room temperature for 30 minutes. Reaction products were visualized by incubation with 3,3′-diaminobenzidine and then counterstained with hematoxylin. Negative controls (NC) were achieved by omitting the primary antibody. Sections treated without primary antibodies were used as NCs. The positive percentage of counted cells was graded semiquantitatively according to a 4-tier scoring system: negative (−), 0∼5%; weakly positive (+), 6∼25%; moderately positive (++), 26∼50%; and strongly positive (+++), 51∼100%.
Plasmid and Transfection
Transfection of the plasmid pCDNA-3.1-ING5 into HOS and MG63 cells was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Plasmid pCDNA-3.1 was used as a control. Briefly, cells were plated in DMEM with 10% FBS and cultured until they achieved 70% to 80% confluence. Culture medium was then replaced with low-serum media (containing 0.5% FBS), and then cells were transfected with 5 μg/well of plasmid. For 48 hours after transfection, cells were used in the following studies.
Colony Formation Assay
Cells (5 × 103 cells /well) in 24-well culture plates were incubated for 14 days when the colonies were visible. Crystal violet staining was performed, and the number of colonies was counted.
Cell Apoptosis Assay
The apoptosis was determined using the Apoptosis Detection Kit (KeyGEN, Nanjing, China) based on the procedures provided by the manufacturer. Cells were stained with Annexin V-FITC and propidium iodide (PI) in the dark for 30 minutes. The cells were analyzed using a fluorescence-activated cell sorting flow cytometer (BD Biosciences, San Jose, California).
Western Blot
Total protein was extracted from cells and tissues using lysed buffer (20 mM Tris-HCl, 150 mM NaCl, 2 mM EDTA, 1% Triton-X100) containing a protease inhibitor cocktail (Sigma-Aldrich, Saint Louis, Missouri). Proteins (30 µg) were separated via 8% SDS-PAGE and transferred to PVDF membranes. The membranes were incubated with primary antibodies overnight (Table 1). Then, the membranes were incubated with a secondary horseradish peroxidase -conjugated antibody for 1 hour at room temperature. Bound antibody complexes were detected and visualized with enhanced chemiluminescence substrate (Thermo Scientific, Rockford, Illiniois). All immunoblots images were performed using Bio-Rad ChemiDoc instrument (Bio-Rad Laboratories, Inc, Shanghai, China). Relative band intensities were performed using Image J software, National Institutes of Health, USA.
Table 1.
The Antibodies Used in the Western Blot Analysis.
| Protein | Producer | Catalog number | Dilution |
|---|---|---|---|
| ING5 | Abcam | ab3716 | 1:200 |
| Smad 3 | Abcam | ab40854 | 1:100 |
| P-Smad 3 | Abcam | ab52903 | 1:100 |
| Smad 2 | Abcam | ab40855 | 1:100 |
| P-Smad 2 | Abcam | ab53100 | 1:100 |
| Smad 4 | Abcam | ab40759 | 1:100 |
| β-actin | Santa Cruz biotechnology | sc-130301 | 1:500 |
Abbreviation: ING5, inhibitor of growth family of protein 5.
Statistical Analysis
Relationships between ING5 expression and the clinicopathological characteristics of the patients were analyzed by χ2 test. The Kaplan-Meier method was used to test the association between ING5 expression and overall survival. All statistical analyses were operated using SPSS version 17.0. P value <.05 was considered to be statistically significant. Each experiment was repeated for 3 times.
Results
The Levels of ING5 mRNA and Protein were Reduced in OS
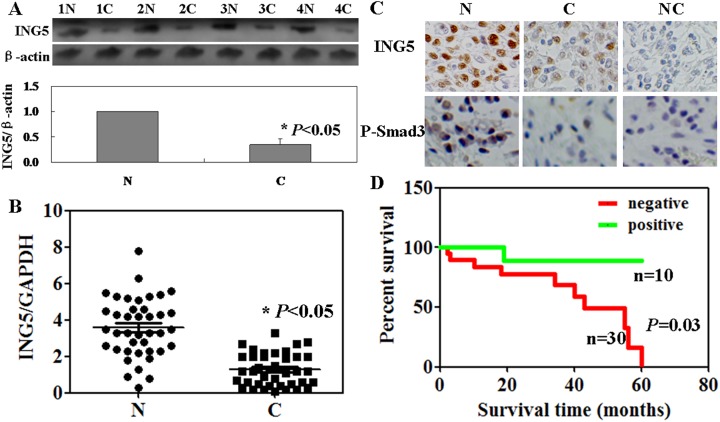
We compared ING5 protein and messenger RNA (mRNA) in cancer tissues from the patients with OS to matched normal tissues. As shown in Figure 1A and B, both the protein and the mRNA levels of ING5 in cancer tissues were lower than that in normal tissues (P < .05). Low ING5 protein expression was found in the cytoplasm of OS cells by immunohistochemistry (Figure 1C). As summarized in Table 2, expression of ING5 was inversely correlated with tumor size, lung metastasis, and local recurrence (P < .05) but not with sex, primary location, or histological type (P > .05). The cumulative survival rate of the patients with ING5 expression was obviously higher than the ones without its expression (P = .03, Figure 1D).
Figure 1.
Inhibitor of growth family of protein 5 (ING5) protein and messenger RNA (mRNA) levels in osteosarcoma (OS) cancer tissues using Western blot (A), real-time polymerase chain reaction (B), and immunohistochemisrty (C). (D) Kaplan-Meier curves relating to the cumulative survival rate of patients with OS. N, normal, C, cancer, NC, negative control.
Table 2.
ING5 Expression and Clinicopathological Parameters of Osteosarcoma.
| Clinicopathological features | n | ING5 expression | |||
|---|---|---|---|---|---|
| − | + | χ2 | P | ||
| Sex | 0.038 | .845 | |||
| Female | 13 | 10 | 3 | ||
| Male | 27 | 20 | 7 | ||
| Primary location | 1.888 | .864 | |||
| Humerus | 6 | 5 | 1 | ||
| Radius | 8 | 6 | 2 | ||
| Femur | 7 | 6 | 1 | ||
| Tibia | 5 | 3 | 2 | ||
| Fibula | 9 | 7 | 2 | ||
| Others | 5 | 3 | 2 | ||
| Histological type | 0.737 | .946 | |||
| Osteoblastic | 7 | 5 | 2 | ||
| Chondroblastic | 8 | 6 | 2 | ||
| Fibroblastic | 10 | 8 | 2 | ||
| Dilated blood vessels | 9 | 6 | 3 | ||
| Others | 6 | 5 | 1 | ||
| Surgery type | 0.135 | .713 | |||
| Amputation | 18 | 13 | 5 | ||
| Limb sparing | 22 | 17 | 5 | ||
| Lung metastasis | 15.68 | .00008 | |||
| - | 15 | 6 | 9 | ||
| + | 25 | 24 | 1 | ||
| Local recurrence | 10.15 | .001 | |||
| − | 12 | 5 | 7 | ||
| + | 28 | 25 | 3 | ||
| Tumor size | 8.889 | .002 | |||
| < 6 cm | 16 | 8 | 8 | ||
| ≥ 6 cm | 24 | 22 | 2 | ||
Abbreviations: ING5, inhibitor of growth family of protein 5; PR, positive rate, χ2 value: χ2 distribution.
ING5 Displays Antitumor Activity in OS Cell Lines
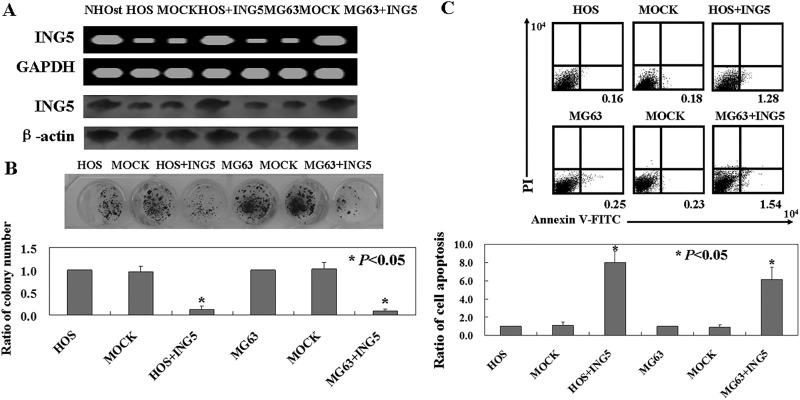
The levels of ING5 mRNA and protein were lower in the OS cells, HOS, and MG63 compared to the osteoblast cell line, NHOst (Figure 2A). In order to study the antitumor roles of ING5 in OS cells, pCDNA-3.1-ING5 was transfected into HOS and MG63 cells. The levels of ING5 mRNA and protein increased upon transfection in the 2 cells (Figure 2A). The colony formation assay showed that the proliferation rates of ING5-expressing OS cells were decreased compared to untransfected ones (P < .05; Figure 2B). Apoptotic cells were detected using Annexin V-FITC/PI double staining. The percentage of apoptosis in ING5-expressing cells was 6 to 8 times higher than that of untransfected ones (P < .05; Figure 2C).
Figure 2.
The antitumor roles of inhibitor of growth family of protein 5 (ING5) in osteosarcoma (OS) cells. (A) ING5 messenger RNA (mRNA) and protein levels in OS cells using reverse transcription PCR (RT-PCR) and Western blot, respectively. (B) The proliferation ratio of the cells was assayed using the soft agar assay. (C) Apoptotic cells were assayed by Annexin-V/FITC and propidium iodide (PI) double staining. HOS indicates untreated HOS cells; MOCK, HOS cells transfected with pCDNA-3.1; HOS+ING5, HOS cells transfected with pCDNA-3.1-ING5; MG63, untreated MG63 cells; MOCK, MG63 cells transfected with pCDNA-3.1; MG63+ING5, MG63 cells transfected with pCDNA-3.1-ING5.
ING5 Activates the Smad Signaling Pathway
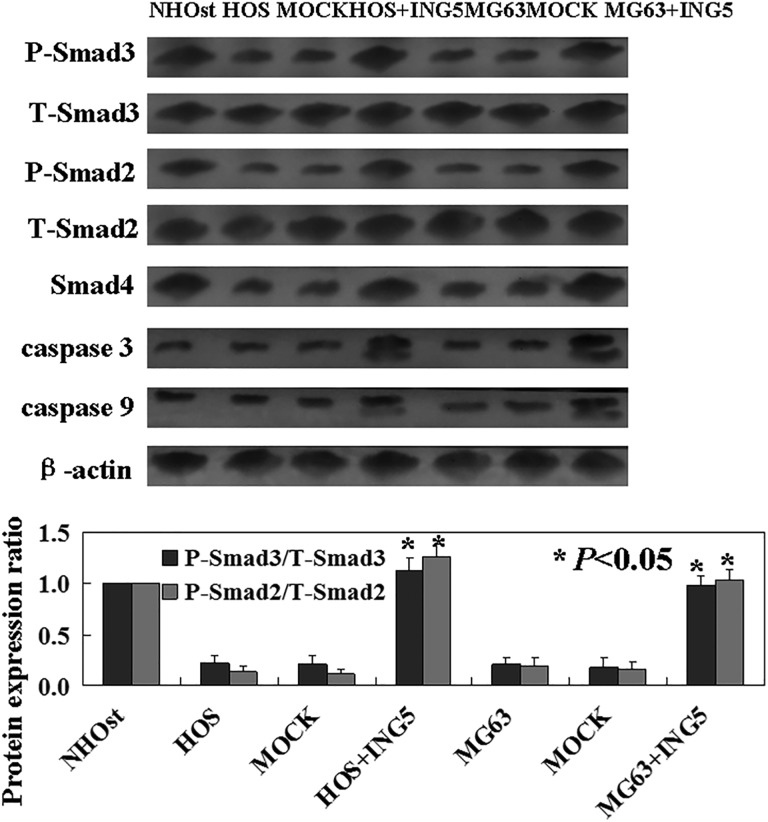
To confirm potential mechanisms following exogenous expression of ING5 in OS cells, we performed Western blot analysis using antibodies that recognize the Smad family members. Western blot analysis showed that p-Smad2, p-Smad3, and Smad4 were increased in ING5-expressing cells (Figure 3). Total protein levels of Smad2 and Smad3 remained unchanged (Figure 3). Followed with activation of Smad pathway, caspase 3 and caspase 9 were increased (Figure 3).
Figure 3.
Western blot was used to detect the potential mechanisms of inhibitor of growth family of protein 5 (ING5) in osteosarcoma (OS) cells. Levels of p-Smad2, p-Smad3, t-Smad2 (total Smad2), t-Smad3 (total Smad3), and Smad4 were detected in ING5-expressing OS cells.
Discussion
In the recent years, ING5 has received more attention due to its low expression in tumors.5–7 Consistent with previous results, here in we found that both ING5 protein and mRNA were downregulated in OS cancer tissues compared to the matched normal tissues. In the study of Gou et al,13 they found that ING5 expression was negatively linked to tumor size and lymph node metastasis of gastric cancer. Consistent with their results, we also confirmed that expression of ING5 was negatively associated with tumor size and lung metastasis.
In lung cancer cells, overexpression of ING5 induced G2 phase arrest and suppressed proliferation.14 In this study, we also found that overexpression of ING5 induced apoptosis and inhibited proliferation of OS cells. Furthermore, we found that p-Smad2, p-Smad3, and Smad4 were increased in ING5-expressing OS cells. It indicated that the activation of the Smad signaling pathway in ING5-expressing OS cells. To our knowledge, no previous studies showed the roles of ING5 in the Smad signaling pathway. However, another ING family member, ING2, mediates the ability of Smad pathway to activate transcription and inhibit cell proliferation.15 The ING proteins (ING1-5) are characterized by a high homology in their C-terminal domain, which contains a nuclear localization sequence and a Plant HomeoDomain. In our study, we confirmed ING5 could regulate the Smad signaling pathway.
In conclusion, our results demonstrate, for the first time that overexpression of ING5 in OS cells induces apoptosis by activating the Smad pathways, thus proposing a promising role of ING5 for patients’ with OS treatment.
Abbreviations
- DMEM
Dulbecco’s Modified Eagle’s Medium
- FBS
fetal bovine serum
- ING5
inhibitor of growth family of protein 5
- mRNA
messenger RNA
- NC
negative control
- PI
propidium iodide
- OS
osteosarcoma.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
References
- 1. Gianferante DM, Mirabello L, Savage SA. Germline and somatic genetics of osteosarcoma— connecting aetiology, biology and therapy. Nat Rev Endocrinol. 2017;13(8):480–491. [DOI] [PubMed] [Google Scholar]
- 2. Lin YH, Jewell BE, Gingold J, et al. Osteosarcoma: molecular pathogenesis and iPSC modeling. Trends Mol Med. 2017;23(8):737–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Soliman MA, Riabowol K. After a decade of study-ING, a PHD for a versatile family of proteins. Trends Biochem Sci. 2007;32(11):509–519. [DOI] [PubMed] [Google Scholar]
- 4. Campos EI, Chin MY, Kuo WH, Li G. Biological functions of the ING family tumor suppressors. Cell Mol Life Sci. 2004;61(19-20):2597–2613. [DOI] [PubMed] [Google Scholar]
- 5. Zheng HC, Xia P, Xu XY, Li G. The nuclear to cytoplasmic shift of ING5 protein during colorectal carcinogenesis with their distinct links to pathologic behaviors of carcinomas. Hum Pathol. 2011;42(3):424–433. [DOI] [PubMed] [Google Scholar]
- 6. Xing YN, Yang X, Xu XY, Takahashi H, Takano Y. The altered expression of ING5 protein is involved in gastric carcinogenesis and subsequent progression. Hum Pathol. 2011;42(1):25–35. [DOI] [PubMed] [Google Scholar]
- 7. Zhao QY, Ju F, Wang ZH, Ma XZ, Zhao H. ING5 inhibits epithelialmesenchymal transition in breast cancer by suppressing PI3K/Akt pathway. Int J Clin Exp Med. 2015;8(9):15498–15505. [PMC free article] [PubMed] [Google Scholar]
- 8. Shiseki M, Nagashima M, Pedeux RM, et al. p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res. 2003;63(10):2373–2378. [PubMed] [Google Scholar]
- 9. Jafarnejad SM, Li G. Regulation of p53 by ING family members in suppression of tumor initiation and progression. Cancer Metastasis Rev. 2012;31(1-2):55–73. [DOI] [PubMed] [Google Scholar]
- 10. Zhang F, Bäumer N, Rode M, et al. The inhibitor of growth protein 5 (ING5) depends on INCA1 as a co-factor for its antiproliferative effects. PLoS One. 2011;6(7):e21505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu JF, Nie XC, Shao YC, Su WH, Ma HY, Xu XY. Bleomycin suppresses the proliferation and the mobility of human gastric cancer cells through the smad signaling pathway. Cell Physiol Biochem. 2016;40(6):1401–1409. [DOI] [PubMed] [Google Scholar]
- 12. Xu R, Wei S, Zhou G, et al. Multiplex TaqMan locked nucleic acid real-time PCR for the differential identification of various meat and meat products. Meat Sci. 2017;137:41–46. [DOI] [PubMed] [Google Scholar]
- 13. Gou WF, Shen DF, Yang XF, et al. ING5 suppresses proliferation, apoptosis, migration and invasion, and induces autophagy and differentiation of gastric cancer cells: a good marker for carcinogenesis and subsequent progression. Oncotarget. 2015;6(23):19552–19579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jain P, Karthikeyan C, Moorthy NS, Waiker DK, Jain AK, Trivedi P. Human CDC2-like kinase 1 (CLK1): a novel target for Alzheimer’s disease. Curr Drug Targets. 2014;15(5):539–550. [DOI] [PubMed] [Google Scholar]
- 15. Zhang R, Jin J, Shi J, Hou Y. INGs are potential drug targets for cancer. J Cancer Res Clin Oncol. 2017;143(2):189–197. [DOI] [PubMed] [Google Scholar]