Abstract
Esophageal squamous cell carcinoma (ESCC) is highly prevailing in Asia and it is ranked in the most aggressive squamous cell carcinomas. High-frequency loss of heterozygosity occurred in chromosome 14q11.2 in many tumors including ESCC, suggesting that one or more tumor-suppressor genes might exist within this region. In this study, we identified the tumor-suppressing role of DHRS2 (short-chain dehydrogenase/reductase family, member 2) at 14q11.2 in ESCCs. Downregulation of DHRS2 occurred in 30.8% of primary ESCC tumor tissues vs paired non-tumorous tissues. DHRS2 downregulation was associated significantly with ESCC invasion, lymph nodes metastasis and clinical staging (P<0.001). Survival analysis revealed that DHRS2 downregulation was significantly associated with worse outcome of patients with ESCC. In vitro and in vivo studies indicated that both DHRS2 variants could suppress cell proliferation and cell motility. Moreover, we demonstrated that DHRS2 could reduce reactive oxygen species and decrease nicotinamide adenine dinucleotide phosphate (oxidized/reduced), increase p53 stability and decrease Rb phosphorylation; it also decreased p38 mitogen-activated protein kinase phosphorylation and matrix metalloproteinase 2. In summary, these findings demonstrated that DHRS2 had an important part in ESCC development and progression.
Introduction
Esophageal squamous cell carcinoma (ESCC) is ranked in the most common cancers in the world, with an estimated 456 000 new cases annually.1 Esophageal cancer has two ESCC histological types: ESCC and esophageal adenocarcinoma. ESCC is a dominating histological type and prevalent in certain areas, especially in the northern China. Although advanced therapeutics in ESCC had been achieved recently, the 5-year survival rate is only 15–25% because of the late diagnosis. Like most solid tumors, chromosomal alteration is frequently observed in ESCC. For example, loss of 14q11.2 is one of the most common chromosomal changes identified by comparative genomic hybridization and high-resolution deletion mapping in many cancers, including ESCC,2 gastrointestinal tumors,3, 4 nasopharyngeal carcinoma,5 mesothelioma,6 suggesting that tumor-suppressor genes might exist in frequently deleted regions. As a gene located in the chromosome 14q11.2,7 DHRS2 (short-chain dehydrogenase/reductase family, member 2) was first acquired from a human hepatocarcinoma complementary DNA library.8 It codes for an enzyme that is a member of the short-chain dehydrogenase/reductase (SDR) family.
SDR enzyme family is characterized by some common sequence motifs: a glycine consensus of NAD/NADP cofactor-binding domain, amino acids as catalytic domain and highly conservative amino-acid sequence scattered among the sequences.9 Human SDR enzymes function actively in signaling molecules metabolism and intermediary and xenobiotic metabolism.10, 11, 12 Through their affection on regulatory signals, SDR enzymes take significant effect in controlling normal cell functions and some SDR enzymes have been associated with some diseases of human,11, 12 including metabolic disorders and tumors.13, 14
DHRS2 was reported to bind to mouse double minute 2 homolog (MDM2) and result in the weakening of MDM2-intermediated p53 degradation in osteosarcoma cell line.15 It is also activated by c-Myb and ETV5.16, 17 It was reported that DHRS2 expression correlated with estrogen receptor status in breast cancer,15, 18 and introduction of adenovirus harboring DHRS2 could suppress renal cancer cell growth.19 However, whether DHRS2 involved in ESCC cancer development and progression remained unclear. In this study, expression of DHRS2 was compared between ESCC primary tumor tissues and adjacent non-tumorous tissues. The clinical significance of DHRS2 in ESCC patients was explored. The tumor inhibition roles of DHRS2 in ESCC were identified by functional studies. The underlying mechanism of tumor inhibition of DHRS2 was also addressed.
Results
DHRS2 downregulation was significantly associated with worse prognosis of ESCC
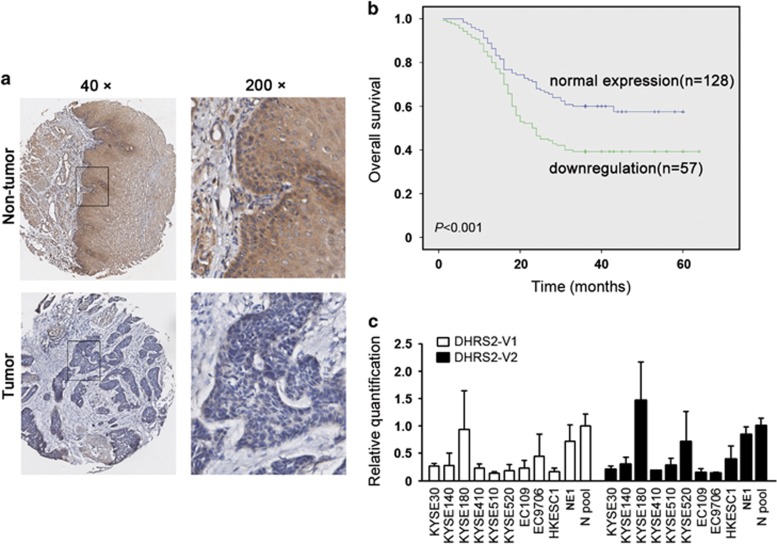
The protein level of DHRS2 was detected by immunohistochemistry staining using a tissue microarray and 73 pairs of tumor and non-tumorous specimens of ESCC. Informative data were obtained from 185 pairs of ESCCs (both tumor and adjacent non-tumor). Compared with the corresponding non-tumorous tissues, downregulation of DHRS2 was detected in 57/185 (30.8%) informative ESCC tumor tissues (Figure 1a). Association analysis revealed that DHRS2 downregulation was correlated with tumor invasion, lymph nodes metastasis and clinical staging (P<0.001, Table 1). Survival analysis revealed that the downregulation of DHRS2 was significantly associated with worse outcome of patients with ESCC (Kaplan–Meier method, P<0.001, Figure 1b). DHRS2 downregulation and other clinicopathologic features (tumor invasion, lymph nodes metastasis and staging) were analyzed by Cox regression analysis. The results revealed that it was not an independent prognosis factor for overall survival (Table 2). As the antibody targeting the common amino-acid sequence coded by DHRS2-V1 (variant 1, NM_182908.4) and DHRS2-V2 (variant 2, NM_005794.3), we next examined the RNA level of the variants in ESCC cell lines. Compared with the non-tumorous tissues pool and NE1 (an immortalized human esophageal epithelial cell line), the RNA levels of DHRS2-V1 and V2 decreased in most ESCC cell lines (Figure 1c). The DHRS2-V2 was relatively higher than DHRS2-V1 in most ESCC cell lines (Supplementary Figure 1A).
Figure 1.
DHRS2 downregulation was significantly associated with poorer prognosis of ESCC. (a) The representative pictures of DHRS2 staining in ESCC tumor tissue and paired non-tumor tissue. The boxed regions were magnified and shown in the right panels. (b) Kaplan–Meier analysis showed that DHRS2 downregulation was significantly associated with ESCC patients’ poorer survival. (c) The relative quantification of DHRS2-V1 and DHRS2-V2 in ESCC cell lines, NE1 (an immortalized human esophageal epithelial cell line) and non-tumor esophageal tissues pool (three cases).
Table 1. Correlation analysis of DHRS2 downregulation with clinicopathologic characteristics of ESCC patients.
| Clinicopathologic characteristics | DHRS2 downregulation | P-value |
|---|---|---|
| Age (years) | 0.836 | |
| <60 | 29/92 (31.52%) | |
| >=60 | 28/93 (30.11%) | |
| Gender | 0.911 | |
| Male | 32/105 (30.48%) | |
| Female | 25/80 (31.25%) | |
| Tumor cell differetiation | 0.703 | |
| Well | 6/25 (24.00%) | |
| Moderate | 40/114 (35.09%) | |
| Poor | 11/46 (23.91%) | |
| Tumor invasion | <0.001 | |
| 0–1 | 10/74 (13.51%) | |
| 2 | 3/10 (30.00%) | |
| 3 | 44/101 (43.56%) | |
| Lymph node metastasis | <0.001 | |
| − | 30/131 (22.90%) | |
| + | 27/54 (50.00%) | |
| Staging | <0.001 | |
| 0, I | 9/74 (12.16%) | |
| II, III, IV | 48/111 (43.24%) |
Abbreviations: DHRS2, short-chain dehydrogenase/reductase family, member 2; ESCC, eesophageal squamous cell carcinoma. Bold values indicate statistically significant.
Table 2. Univariate and multivariate analysis of different prognostic variables in patients with ESCC.
| Clinical–pathological features |
Univariable analysis |
Multivariable analysis |
||
|---|---|---|---|---|
| HR (95% CL) | P-value | HR (95% CL) | P-value | |
| Age | 1.094 (0.725–1.650) | 0.670 | ||
| Gender | 1.009 (0.666–1.529) | 0.966 | ||
| Differetiation | 1.375 (0.991–1.910) | 0.057 | ||
| Tumor invasion | 2.562 (1.923–3.413) | <0.001 | 1.166 (0.674–2.018) | 0.582 |
| Lymph nodes metastasis | 5.063 (3.316–7.729) | <0.001 | 2.434 (1.545–3.835) | <0.001 |
| TNM stage | 8.017 (4.244–15.143) | <0.001 | 3.736 (1.067–13.078) | 0.039 |
| DHRS2 downregulation | 2.283 (1.504–3.466) | <0.001 | 1.307 (0.851–2.008) | 0.221 |
Abbreviations: CI, confidence interval; DHRS2, short-chain dehydrogenase/reductase family, member 2; ESCC, esophageal squamous cell carcinoma; HR, hazard ratio; TNM staging system (T, tumor; N, lymph node; M, metastasis). Bold values indicate statistically significant.
As deletion of 14q11.2 was often reported in many cancers, fluorescence in situ hybridization (FISH) was used to examine the DNA copy number alteration of DHRS2 in ESCC cell lines and tumor tissues. The results revealed that copy number loss of DHRS2 existed in ESCC cell lines and ESCC tumor tissues (Supplementary Figures 1B and C).
DHRS2 suppressed tumor cell growth in vitro and in vivo
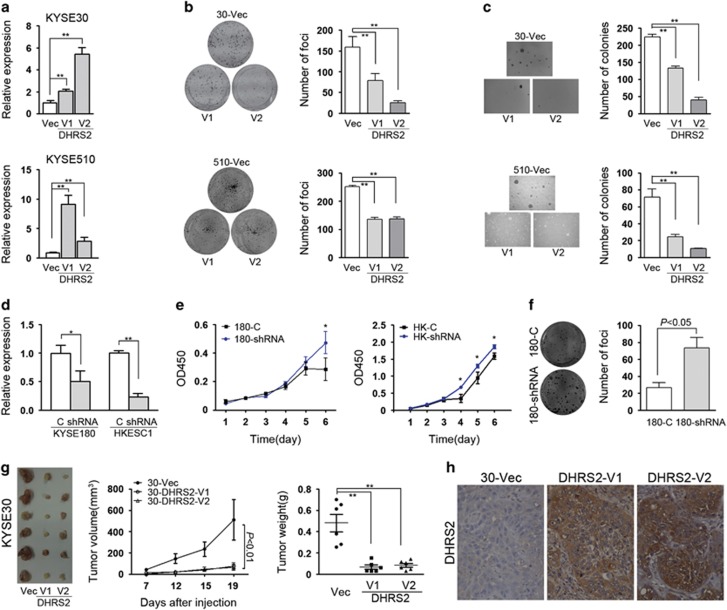
To further explore whether DHRS2 affects tumor cell growth, DHRS2-V1 or DHRS2-V2 was stabled overexpressed into KYSE30 and KYSE510 cells, respectively. Expression of DHRS2 was examined by quantitative PCR (Figure 2a). Both DHRS2-V1 and DHRS2-V2 reduced significantly foci formation rate (P<0.01, Figure 2b) and colony formation rate in soft agar (P<0.01, Figure 2c) in both KYSE30 and KYSE510 cells. Short hairpin RNA (shRNA) targeting DHRS2 was also stably transfected into KYSE180 and HKESC1, respectively (Figure 2d). Cell growth assay and foci formation results showed that DHRS2 knock-down could increase the tumorgenecity in KYSE180 and HKESC1 cells (Figures 2e and f).
Figure 2.
DHRS2 suppressed tumor cell growth in vitro and in vivo. (a) The relative quantification of DHRS2-V1 and DHRS2-V2 in transfected KYSE30 and KYSE510 cells compared with vector control cells (-Vec) respectively (**P<0.01). (b, c) Foci formation assay (b) and soft agar assay (c) demonstrated that DHRS2-V1 and DHRS2-V2 inhibited the anchorage-dependent and -independent cell growth ability. The results were summarized as mean±s.e.m. of three independent assays (**P<0.01). (d) The DHRS2 RNA level decreased in DHRS2 knock-down KYSE180 and HKESC1 (shRNA) cells compared with vector control (c) cells (*P<0.05; **P<0.01). (e) Cell proliferation increased in DHRS2 knock-down cells compared with control cells (*P<0.05). (f) Foci formation ability increased in DHRS2 knock-down cells compared with control cells. (g) The representative pictures of xenografts formed in nude nice (n=6). Tumor volume and tumor weight significantly decreased in DHRS2-V1 and DHRS2-V2-transfected KYSE30 cells (**P<0.01). (h) The representative pictures of DHRS2 staining of xenograft sections (original magnification: × 200).
Xenograft formation assay was also performed to investigate whether DHRS2 could suppress tumor growth in vivo. Vector control cells and DHRS2-V1 and DHRS2-V2 overexpressed KYSE30 cells were injected into the left, middle and right flanks of 4-week-old immunodeficient mice, respectively. Mice were killed 19 days after injection and xenografts were isolated for further analysis. The tumor volume and tumor weight of the xenografts induced by DHRS2-V1 or V2 significantly decreased compared with the xenografts induced by vector control cells (P<0.01, Figure 2g). The immunostaining results confirmed that expression of DHRS2 in xenografts induced by DHRS2-overexpressed cells was much stronger than that in the vector-transfected cells (Figure 2h).
DHRS2 inhibited cell motility in vitro and in vivo
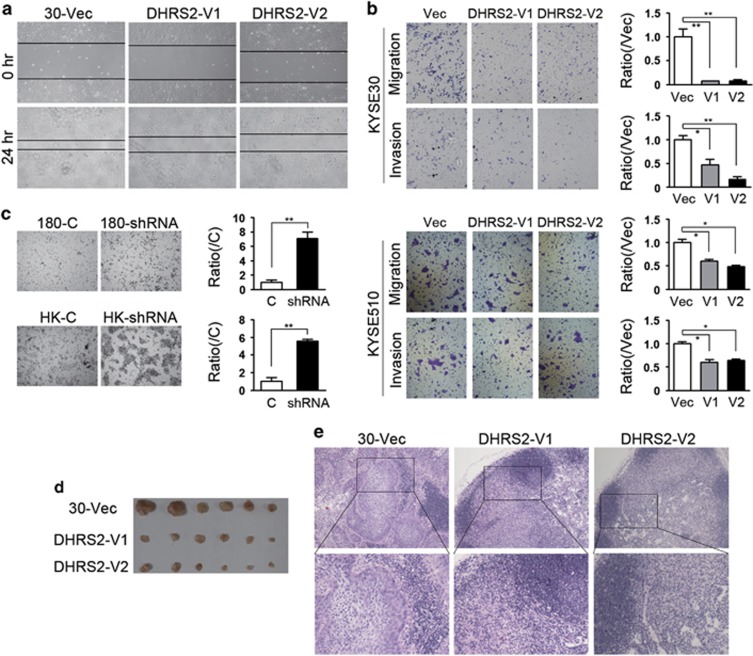
As downregulation of DHRS2 was correlated with tumor invasion and lymph node metastasis, we further explored whether DHRS2 could affect tumor cell motility. The scratch assay indicated that DHRS2-V1 and V2-transfected KYSE30 cells obtained slower closure of the scratched ‘wound’ compared with vector control (Figure 3a). Cell migration experiment demonstrated that the number of cells migrated decreased in the DHRS2-V1 and V2-transfected KYSE30 cells (Figure 3b). Cell invasion assay demonstrated that the number of cells invaded through Matrigel also significantly decreased in DHRS2-V1 and V2-transfected cells (Figure 3b). The results in DHRS2-V1 and V2 overexpressed KYSE510 cells were consistent with KYSE30 cells (Figure 3b). When DHRS2 was silenced in KYSE180 and HKESC1 cells, the number of cells migrated increased in DHRS2 knock-down cells (P<0.01, Figure 3c).
Figure 3.
DHRS2 inhibited cell motility in vitro and in vivo. (a) Wound-healing ability decreased in DHRS2-V1 and DHRS2-V2-transfected KYSE30 cells compared with vector control cells. (b) The cell migration and invasion ability decreased in DHRS2-V1 and DHRS2-V2-transfected KYSE30 and KYSE510 cells compared with vector control cells. The results were summarized of three independent assays (*P<0.05; **P<0.01). (c) The cell migration increased in DHRS2 knock-down KYSE180 and HKESC1 cells (shRNA) compared with control cells (c) (**P<0.01). (d) The size of popliteal lymph nodes decreased in DHRS2-V1 and DHRS2-V2 animal groups compared with vector control group. (e) The representative pictures of HE staining of popliteal lymph nodes of the DHRS2-V1, DHRS2-V2 and vector control groups (original magnification: × 200). The boxed regions were magnified as the bottom pictures.
To determine whether DHRS2 can affect tumor metastasis in vivo, lymph node metastasis animal models were performed in nude mice. DHRS2-V1 and V2-transfected cells were subcutaneously inoculated into the footpad of the left hind limb, respectively (n=6 for each group). Two months later, the mice were killed, and popliteal lymph nodes were isolated and fixed. No apparent nodule was observed in surface of lung and liver. The popliteal lymph nodes size significantly decreased in DHRS2-V1 and V2 groups, compared with that in control cells (Figure 3d). Tumor cells infiltration decreased in lymph nodes of DHRS2-V1 and V2 groups compared with that in control cells (Figure 3e).
DHRS2 affected cell cycle and induced apoptosis
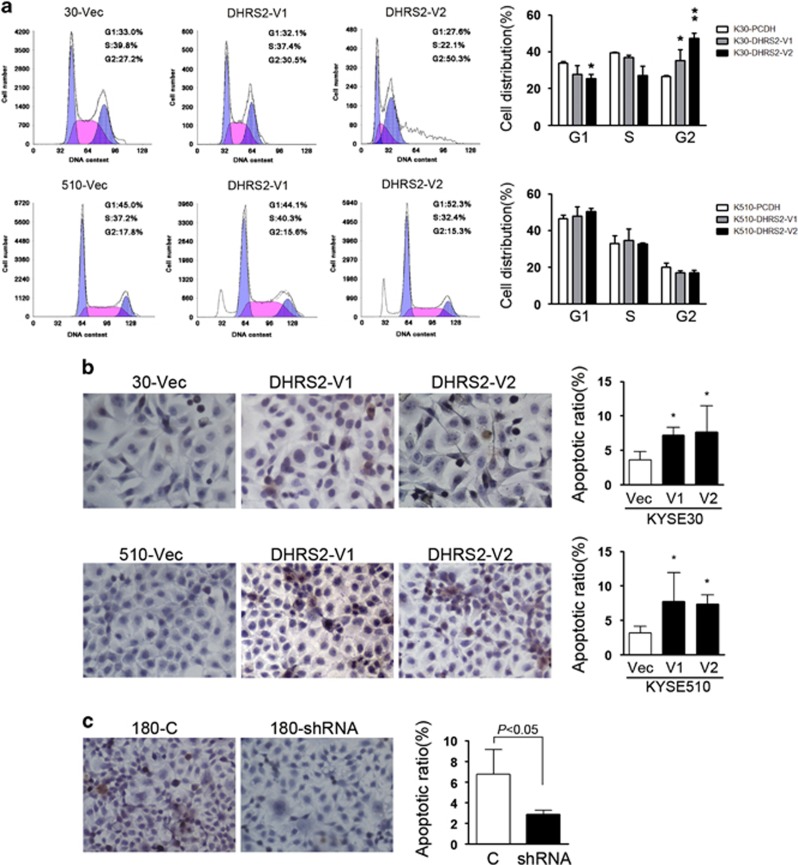
The cell cycle distribution was compared by flow cytometry among DHRS2-V1, V2 and vector-transfected cells. The results found that KYSE30-DHRS2-V1 and DHRS2-V2 cells were arrested at G2/M checkpoint, exhibited as cells accumulation in G2/M phase (Figure 4a). In KYSE510 cells, no significant change was observed in cells distribution except that pre-G1 was observed in DHRS2-V1 and V2-transfected cells (Figure 4a).
Figure 4.
DHRS2 affected cell cycle and increased apoptosis. (a) The cell distribution was determined by FACS in DHRS2-V1 and DHRS2-V2-transfected KYSE30 and KYSE510 cells compared with vector control cells (*P<0.05). (b) Representative pictures of in situ terminal deoxyribonucleotidyl transferase-mediated dUTP-digoxigenin nick and labeling (TUNEL) assay showed that apoptotic cells increased in DHRS2-V1 and DHRS2-V2-transfected KYSE30 and KYSE510 cells compared with vector control cells. Data were summarized of three independent assays (right) (*P<0.05). (c) Apoptotic cells decreased in DHRS2 knock-down KYSE180 cells compared with control cells (c). Data was summarized of three independent assays (right).
We next elucidated whether DHRS2-V1 and V2 inhibited cell proliferation through inducing apoptosis. The in situ terminal deoxyribonucleotidyl transferase-mediated dUTP-digoxigenin nick and labeling assays showed that the number of apoptotic cells increased in DHRS2-V1 and DHRS2-V2-transfected KYSE30 and KYSE510 cells compared with 30-Vec and 510-Vec, respectively (P<0.05, Figure 4b). Conversely, silencing DHRS2 in KYSE180 (180-shRNA) decreased the number of apoptotic cells compared with 180-C (control) cells (P<0.05, Figure 4c).
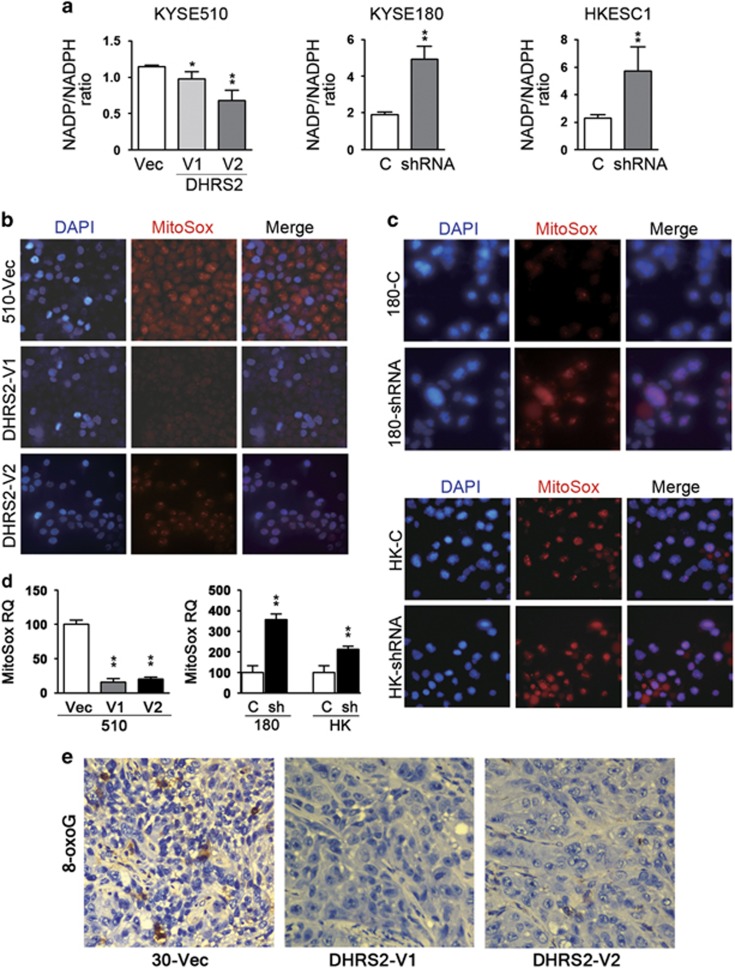
DHRS2 decreased NADP/NADPH ratio and reactive oxygen species (ROS)
As DHRS2 is a member of SDR family and an NADPH-dependent dicarbonylreductase,20 the nicotinamide adenine dinucleotide phosphate (oxidized/reduced) (NADP/NADPH) ratio was examined in DHRS2-overexpressed cells and knock-down cells. The results indicated that the NADP/NADPH ratio decreased in KYSE510 cells with DHRS2-V1 and V2 overexpression (Figure 5a), compared with 510-Vec cells. When DHRS2 was silenced, the ratio increased compared with control cells (Figure 5a).
Figure 5.
DHRS2 decreased ROS in vitro. (a) NADP/NADPH ratio decreased in DHRS2-V1 and DHRS2-V2-transfected KYSE510 cells compared with vector control cells; NADP/NADPH ratio increased in DHRS2 knock-down KYSE180 and HKESC1 cells (*P<0.05; **P<0.01). (b) MitoSox Red staining decreased in DHRS2-V1 and DHRS2-V2-transfected KYSE510 cells compared with vector control cells (original magnification: × 200). (c) MitoSox Red staining increased in DHRS2 knock-down KYSE180 and HKESC1 cells compared with control cells (c) (original magnification: × 200). (d) The mitoSox red staining was quantified by Image J software (http://rsb.info.nih.gov/ij/) and summarized (**P<0.01). (e) DNA lesions resulting from ROS were detected in the xenograft sections by immunostaining with 8-oxoG (original magnification: × 200).
As DHRS2 was reported to be associated with oxidative stress,21 we next examined the mitochondrial ROS by MitoSox Red staining assay. The results found that mitochondrial ROS decreased in DHRS2-overexpressed cells (Figures 5b and d). When DHRS2 was knocked-down in KYSE180 and HKESC1 cells, ROS increased compared with control cells (Figure 5c and d). ROS was also examined in xenograft sections by immunostaining with 8-oxoguanine antibody to detect DNA damage resulting from ROS.22 Our results indicated that DNA lesions resulting from ROS decreased in DHRS2-V1 and DHRS2-V2-transfected KYSE30 cells compared with vector control cells in in vivo assay (Figure 5e).
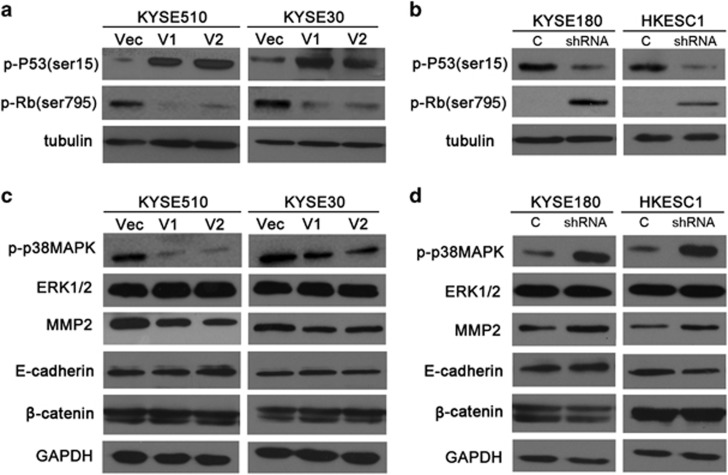
DHRS2 stablized P53 and decreased p38 mitogen activated protein kinase (MAPK) phosphorylation
As DHRS2 was reported to stabilize P53 in osteosarcoma cell line,15 we next examined whether P53 was stablized in DHRS2-overexpressed ESCC cells. Our results showed that both DHRS2-V1 and V2 could stabilize P53 through phosphorylation at Ser15 and decrease the phosphorylation of Rb at ser795 (Figure 6a and Supplementary Figure 2A). When DHRS2 was silenced, the P53 phosphorylation (Ser15) decreased and Rb phosphorylation (ser795) increased compared with control cells (Figure 6b and Supplementary Figure 2B). No significant change was observed in MDM2 protein level in DHRS2 knock-down cells compared with control cells (Supplementary Figure 2C).
Figure 6.
DHRS2 stabilized P53 and decreased p38MAPK phosphorylation. (a) The protein levels of p-P53(ser15) and p-Rb(ser795) were determined in DHRS2-V1 and DHRS2-V2-transfected KYSE510 and KYSE30 cells compared with vector control cells. (b) The protein levels of p-P53(ser15) and p-Rb(ser795) were determined in DHRS2 knock-down KYSE180 and HKESC1 cells. Tubulin was set as loading control. (c) The protein levels of p-p38MAPK, extracellular signal-regulated kinases 1/2 (ERK1/2), MMP2, E-cadherin and β-catenin were determined in DHRS2-V1 and DHRS2-V2-transfected KYSE510 and KYSE30 cells compared with vector control cells. (d) The protein levels of p-p38MAPK, ERK1/2, MMP2, E-cadherin and β-catenin were determined in DHRS2 knock-down KYSE180 and HKESC1 cells. GAPDH was set as loading control.
As DHRS2 could downregulate ROS and ROS is known to actively participate in the cell migration process,23 we examined the activation of MAPK family of proteins. As shown by Figure 6c, phosphorylation of p38MAPK at Thr180/Tyr182 decreased in DHRS2-V1 and DHRS2-V2-overexpressed cells (Figure 6c and Supplementary Figure 2D). The phosphorylation of p38MAPK increased when DHRS2 was knocked-down in KYSE180 and HKESC1 cells (Figure 6d and Supplementary Figure 2E). No significant change of extracellular signal-regulated kinases 1/2 was observed in DHRS2-overexpressed cells or knock-down cells. Matrix metalloproteinase 2 (MMP2) decreased in DHRS2-V1 and V2-transfected cells and increased when DHRS2 was knocked-down (Figures 6c and d and Supplementary Figures 2D and E). No significant change was observed on E-cadherin and β-catenin protein levels (Figures 6c and d and Supplementary Figures 2D and E).
Discussion
Deletion of 14q11.2 region is frequently detected in many tumors, including ESCC.2 Currently, we report the identification of DHRS2, one candidate tumor-suppressor gene at 14q11.2, in ESCC. DHRS2 is an enzyme that belongs to the short-chain dehydrogenase/reductase family. It is a superfamily of NAD/NADP-dependent oxidoreductases implicated in many metabolic processes that function in breaking-down steroids, retinoids, prostaglandins and xenobiotics.20 DHRS2 codes for a NADPH-dependent dicarbonylreductase enzyme active on three toxic a-dicarbonyl-compounds (3,4-hexanedione, 2,3-heptanedione and 1-phenyl-1,2-propanedione).20
Recently, it has been found that some unique SDR family members have been involved in tumors.13, 14 Presently, we compared the expression of DHRS2 in primary ESCC tumor vs paired non-tumor samples by immunostaining, and found that downregulation of DHRS2 was correlated significantly with tumor invasion, lymph node metastasis and staging. DHRS2 downregulation was significantly associated with worse prognosis of ESCC patients (P<0.001). As DHRS2 has different variants: DHRS2-V1 (300 aa) and DHRS2-V2 (280 aa), and V2 uses an alternate splicing site in the 3′ coding region compared with V1, resulting a shorter protein, functional studies were used to evaluate DHRS2-V1 and DHRS2-V2. The results found that both V1 and V2 overexpression could inhibit ESCC tumor growth and motility. When DHRS2 was silenced, the cell proliferation and motility decreased.
DHRS2 was reported to bind MDM2 and lead to the attenuation of MDM2-intermediated p53 degradation.15 In consistent with this report, the phosphorylation of p53 (ser15) increased in both DHRS2-V1 and V2-overexpressed cells. This phosphorylation impairs the ability of MDM2 to bind p53, indicating p53 accumulation and activation. The introduction of DHRS2-V1 and V2 leads to significant p53 stabilization and apoptotic cells increased. Inactive form of Rb (phosphorylated at ser795) also reduced in both DHRS2-V1 and V2-overexpressed cells. Rb could bind to repress the transcription factor E2F.24 The phosphorylation of Rb induces Rb to dissociate from E2F, permitting the transcription of cell proliferation promoting genes. These results indicate that the tumor inhibition function of DHRS2 might be via stabilizing p53 and downregulating of Rb phosphorylation.
DHRS2 belongs to SDR family and it has amino-acid domains participating in coenzyme NADP binding and the G/AxxxGxG consensus sequence, a NADP/NAD-binding region that was named Rossman fold.9 In addition, it also has a NADPH-dependent dicarbonylreductase enzyme activity. Results showed that the NADP/NADPH ratio decreased in DHRS2-overexpressed cells and increased in DHRS2 knocked-down cells. The increase of NADPH is beneficial for ROS quenching.25 Consistent with this, the mitochondrial ROS decreased in DHRS2-overexpressed cells.
In cancer cells, the antioxidant systems are often overwhelmed by increased ROS, resulting in oxidative stress. The variant degrees of oxidative stress exhibit to bring diverse outcomes in tumor cells: while high oxidative stress can lead to cell death, mild oxidative stress initiates cell signaling activation, including increased cell growth, migration and invasion.23, 26, 27, 28 It has been well established that ROS promotes signaling pathways for cell motility.29 During cell migration, MAPK family members, including p38MAPK, the c-jun NH-2 terminal kinase and extracellular signal-regulated kinase are activated in the process.23 The phosphorylation of p38MAPK decreased in DHRS2-overexpressed cells and increased in DHRS2 knock-down cells, whereas the extracellular signal-regulated kinase did not change significantly, indicating that DHRS2 inhibited cell migration and invasion through inhibiting p38MAPK activation. MMPs are reported to be regulated by ROS in several cancers, including pancreatic cancer,30 breast cancer31 and glioblastoma.32 MMPs could degrade extracellular matrix components, including Laminin, fiber adhesion protein, gelatin and proteoglycan core protein, promoting tumor cell invasion into surrounding tissues. In this research, MMP2 decrease also had a part in DHRS2-induced cell motility inhibition.
Taken together, our data show that DHRS2 downregulation is associated with worse prognosis in ESCC. Both DHRS2 variants (V1 and V2) could suppress tumor cell growth and cell motility. The underlying molecular mechanism regarding DHRS2’s tumor-suppressive function further extending the understanding of ESCC development and progression.
Materials and methods
Cells and tumor specimens
KYSE30, KYSE140, KYSE180, KYSE410, KYSE510 and KYSE520 were acquired from the German Resource Center for Biological Material (DSMZ) (Braunschweig, Germany).33 HKESC1, EC109 and EC9706, were supplied by Dr G Srivastava (Department of Pathology, The University of Hong Kong, Hong Kong, China) and Dr G Tsao (Department of Anatomy, The University of Hong Kong), respectively.34 NE1 (an immortalized esophageal epithelial cell line) was established in Dr G Tsao’s lab. ESCC cells were authenticated by cytogenetic methods as human origin in 2009.35 The cells were tested for mycoplasma contamination recently.
Fluorescence in situ hybridization
Nick translation method was adopted to label the bacterial artificial chromosome clone containing DHRS2 with Spectrum-orange-deoxyuridine triphosphate (Vysis, Abbott Laboratories, Abbott Park, IL, USA). FISH assay was conducted as described previously.36
Tissue microarray and immunohistochemistry
In previous report, we constructed a tissue microarray consisting of 300 pairs of ESCC primary tumor tissues and corresponding non-tumorous tissues.37 Seventy-three pairs of primary tumor and non-tumor samples were collected at Sun Yat-sen University Cancer Center, Guangzhou, China. Patients enrolled in the research had not received follow-up radiation or chemotherapeutic treatment. Patients’ age ranged from 40 to 80 years at the point of surgery. The research was approved by the Committees for Ethical Review of Research Involving Human Subjects in Sun Yat-sen University Cancer Center. Signed informed consents for collecting the patients’ specimens were acquired.
In the immunohistochemistry assay, a 1:70 diluted anti-DHRS2 antibody (PA5-25258, Thermo Fisher Scientific, Rockford, IL, USA) was applied for DHRS2 immunostaining. DHRS2 staining score was compared between primary tumor and corresponding non-tumor specimens. Immunohistochemistry score was determined independently by two pathologists without knowing the clinicopathological information. A staining index (0–12) was obtained as the staining intensity (negative (0); weak (1); moderate (2); strong (3)) multiplying the proportion of positive staining (0–25% (1); 25–50% (2); 50–75% (3); 75–100% (4)). DHRS2 downregulation was defined as the score of tumor tissues was less than the score of corresponding non-tumor tissues.
Quantitative PCR
RNAprep Pure Cell/Bacteria Kit (TianGen Biotech, Beijing, China) was used to extract RNA from tissues and cells. Reverse transcription was accomplished with FastQuant RT Kit (TianGen Biotech). Quantitative PCR was performed on Roche 480 Fast Real-Time PCR system using SYBR Green Supermix. The quantitative data were normalized by internal control (18S or β-actin) and triplicate assays were performed. Dissociation curves were analyzed to exclude the possibility of nonspecific amplification products. dCT method was adopted to analyze the quantitative PCR data as described previously.38
Cell growth assays
The cell proliferation rate was assessed by plating cells into 96-well plate and OD450 was measured by XTT with CCK-8 (Dojindo Co., Tabaru, Japan). The anchorage-dependent assay (foci formation) and anchorage-independent assay (soft agar colony formation) were performed as described.35 The assays were repeated three times independently.
Cell migration and invasion experiments
For the scratch assay, DHRS2-V1, DHRS2-V2 or empty vector-transfected cells were cultured in a 100 mm dish until 95% confluence and then wounded with a pipette tip. Transwell Permeable Support (Corning, NY, USA) was used to evaluate cell migration ability. In all, 8 × 104 cells were re-suspended in medium (no serum) in the top chamber of the insert. The cells were attracted by the medium with 10% serum that was added into the bottom well. Twenty-four hours later, penetrated cells through the filter were fixed, stained, and the number of penetrated cells was counted under microscope. Three independent assays were repeated. In the invasion experiment, BioCoat Matrigel Invasion Chamber (BD Biosciences, Bedford, MA, USA) was used in compliance with the manufacturer’s protocol.
Animal experiments
The research was approved by Institutional Animal Care and Use Committee of Sun Yat-sen University Cancer Center. Animal tests were carried out according to the principles for the Welfare of Experimental Animals in Sun Yat-sen University Cancer Center. The tumorigenicity of DHRS2-transfected cells was assessed by xenograft formation assay. Randomly selected 4-week-old female BALB/c nu/nu mice (n=5) were used in the study. DHRS2-V1, DHRS2-V2-overexpressed KYSE30, as well as vector control cells (2 × 106) were inoculated subcutaneously into the mice. The growth of xenografts was examined two times per week. After killing, xenografts were isolated, processed and stained for hematoxylin–eosin and immunohistochemistry study.
The metastasis ability of cells was evaluated by injecting tumor cells into footpad of the mice. DHRS2-V1, DHRS2-V2 and vector-transfected KYSE30 cells (8 × 105) were injected into the left footpad of 4-week-old immunodeficient mice (female, n=6) (randomly selected), respectively. After 2 months, animals were killed and the lungs and livers were examined. The popliteal lymph nodes were isolated, fixed and subjected to hematoxylin–eosin staining.
Cell cycle analysis
KYSE30 and KYSE510 derivative cells (3 × 106) were fixed in pre-cooled 75% ethanol, incubated with propidium iodide (Sigma-Aldrich, Saint Louis, MO, USA). Cytomics FC 500 (BECKMAN COULTER, Fullerton, CA, USA) was used to analyze DNA content. Cell cycle analysis was performed by Modfit LT 2.0 (BECKMAN COULTER). Triplicate assays were repeated independently.
In situ cell death detection experiment
In situ Cell Death Detection Kit (Roche, Mannheim, Germany) was used to detect the apoptotic cells. Cells plated on the cover slides were rinsed with phosphate-buffered saline and fixed. After blocking and permeabilization, apoptosis was detected by the terminal deoxyribonucleotidyl transferase-mediated dUTP-digoxigenin nick and labeling method. The staining signal was transformed by Converter-POD and apoptotic cells were counted under microscope.
Mitochondrial ROS detection
Cells were plated on cover slides, stained with 5 μM MitoSOX Red (Molecular Probes, Eugene, OR, USA) for 10 min at 37 °C, rinsed, then counterstained with 4′, 6′-diamidino-2-phenylindole. Mitochondrial ROS was observed under fluorescence microscope (BX61, Olympus, Tokyo, Japan).
NADP/NADPH quantification
NADP/NADPH quantification was detected in DHRS2-V1 and DHRS2-V2-overexpressed and knock-down cells as the manufacturer’s protocol (Sigma-Aldrich).
Western blotting and antibodies, plasmids
Western blotting was carried out in compliance with the standard protocol. Antibodies used were: p-P53(ser15) (#9286), p-Rb(ser795) (#9301), p-p38MAPK(Thr180/tyr182) (#9212), extracellular signal-regulated kinases 1/2 (#4695), E-cadherin (#3195), β-catenin (#8481), tubulin (#2128) (Cell Signaling Technology, Danvers, MA, USA), GAPDH (ap7873a) and MMP2 (am1844a) (ABGENT, San Diego, CA, USA), DHRS2 (PA5-25258) (Thermo Fisher Scientific), MDM2 (M4308) (Sigma-Aldrich) and 8-oxoguanine (AB20646) (Abcam, Cambridge, UK). pCDH was bought from System Biosciences (Palo Alto, CA, USA). pCDH-DHRS2-V1 and DHRS2-V2 were constructed and sequenced. pLKO.1-DHRS2-shRNA targeting both variants was bought from Sigma-Aldrich.
Statistical analyses
SPSS software package (Version 13.0; SPSS, Inc., Chicago, IL, USA) was used in the study. We used Pearson chi-square test to examine the clinical correlation between DHRS2 downregulation and clinicopathological characters. We used Kaplan–Meier analysis to generate survival curves and log-rank test to calculate the significance. The Cox regression model was applied to identify independent prognostic factors. Results were reported as mean±s.e.m. P<0.05 was considered statistically significant.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (81472255 and 81472250); Guangdong Esophageal Cancer Institute Funding (M201511); Guangdong Science and Technology Foundation (2016A020214008).
Glossary
- DHRS2
short-chain dehydrogenase/reductase family, member 2
- ESCC
esophageal squamous cell carcinoma
- FISH
fluorescence in situ hybridization
- MAPK
mitogen activated protein kinase
- MDM2
mouse double minute 2 homolog.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015; 136: E359–386. [DOI] [PubMed] [Google Scholar]
- Shi ZZ, Jiang YY, Hao JJ, Zhang Y, Zhang TT, Shang L et al. Identification of putative target genes for amplification within 11q13.2 and 3q27.1 in esophageal squamous cell carcinoma. Clin Transl Oncol 2014; 16: 606–615. [DOI] [PubMed] [Google Scholar]
- Debiec-Rychter M, Sciot R, Pauwels P, Schoenmakers E, Dal Cin P, Hagemeijer A. Molecular cytogenetic definition of three distinct chromosome arm 14q deletion intervals in gastrointestinal stromal tumors. Genes Chromosomes Cancer 2001; 32: 26–32. [DOI] [PubMed] [Google Scholar]
- El-Rifai W, Sarlomo-Rikala M, Andersson LC, Miettinen M, Knuutila S. High-resolution deletion mapping of chromosome 14 in stromal tumors of the gastrointestinal tract suggests two distinct tumor suppressor loci. Genes Chromosomes Cancer 2000; 27: 387–391. [PubMed] [Google Scholar]
- Shao JY, Huang XM, Yu XJ, Huang LX, Wu QL, Xia JC et al. Loss of heterozygosity and its correlation with clinical outcome and Epstein-Barr virus infection in nasopharyngeal carcinoma. Anticancer Res 2001; 21: 3021–3029. [PubMed] [Google Scholar]
- Bjorkqvist AM, Wolf M, Nordling S, Tammilehto L, Knuuttila A, Kere J et al. Deletions at 14q in malignant mesothelioma detected by microsatellite marker analysis. Br J Cancer 1999; 81: 1111–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini S, Censini S, Guidotti S, Iacopetti P, Rocchi M, Bianchi M et al. A human short-chain dehydrogenase/reductase gene: structure, chromosomal localization, tissue expression and subcellular localization of its product. Biochim Biophys Acta 2002; 1574: 215–222. [DOI] [PubMed] [Google Scholar]
- Gabrielli F, Donadel G, Bensi G, Heguy A, Melli M. A nuclear protein, synthesized in growth-arrested human hepatoblastoma cells, is a novel member of the short-chain alcohol dehydrogenase family. Eur J Biochem 1995; 232: 473–477. [DOI] [PubMed] [Google Scholar]
- Gabrielli F, Tofanelli S. Molecular and functional evolution of human DHRS2 and DHRS4 duplicated genes. Gene 2012; 511: 461–469. [DOI] [PubMed] [Google Scholar]
- Bray JE, Marsden BD, Oppermann U. The human short-chain dehydrogenase/reductase (SDR) superfamily: a bioinformatics summary. Chemico-Biol Interact 09; 178: 99–109. [DOI] [PubMed] [Google Scholar]
- Oppermann U. Carbonyl reductases: the complex relationships of mammalian carbonyl- and quinone-reducing enzymes and their role in physiology. Annu Rev Pharmacol Toxicol 2007; 47: 293–322. [DOI] [PubMed] [Google Scholar]
- Wu X, Lukacik P, Kavanagh KL, Oppermann U. SDR-type human hydroxysteroid dehydrogenases involved in steroid hormone activation. Mol Cell Endocrinol 2007; 265-266: 71–76. [DOI] [PubMed] [Google Scholar]
- Li J, Liu J, Ren Y, Yang J, Liu P. Common chromosomal fragile site gene WWOX in metabolic disorders and tumors. Int J Biol Sci 2014; 10: 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Chen HY, Han T, Yang GZ, Feng D, Qi CY et al. Downregulation of DHRS9 expression in colorectal cancer tissues and its prognostic significance. Tumour Biol 2016; 37: 837–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisenroth C, Thorner AR, Enomoto T, Perou CM, Zhang Y. Mitochondrial Hep27 is a c-Myb target gene that inhibits Mdm2 and stabilizes p53. Mol Cell Biol 2010; 30: 3981–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton JJ, Davis LM, Lei W, Mo X, Leutz A, Ness SA. Distinct changes in gene expression induced by A-Myb, B-Myb and c-Myb proteins. Oncogene 2003; 22: 308–313. [DOI] [PubMed] [Google Scholar]
- Monge M, Colas E, Doll A, Gil-Moreno A, Castellvi J, Diaz B et al. Proteomic approach to ETV5 during endometrial carcinoma invasion reveals a link to oxidative stress. Carcinogenesis 2009; 30: 1288–1297. [DOI] [PubMed] [Google Scholar]
- Wang J, Shidfar A, Ivancic D, Ranjan M, Liu L, Choi MR et al. Overexpression of lipid metabolism genes and PBX1 in the contralateral breasts of women with estrogen receptor-negative breast cancer. Int J Cancer 2017; 140: 2484–2497. [DOI] [PubMed] [Google Scholar]
- Fang L, Cheng Q, Liu W, Zhang J, Ge Y, Zhang Q et al. Selective effects of a fiber chimeric conditionally replicative adenovirus armed with hep27 gene on renal cancer cell. Cancer Biol Ther 2016; 17: 664–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafqat N, Shafqat J, Eissner G, Marschall HU, Tryggvason K, Eriksson U et al. Hep27, a member of the short-chain dehydrogenase/reductase family, is an NADPH-dependent dicarbonyl reductase expressed in vascular endothelial tissue. Cell Mol Life Sci 2006; 63: 1205–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crean D, Felice L, Taylor CT, Rabb H, Jennings P, Leonard MO. Glucose reintroduction triggers the activation of Nrf2 during experimental ischemia reperfusion. Mol Cell Biochem 2012; 366: 231–238. [DOI] [PubMed] [Google Scholar]
- Wang P, Sun YC, Lu WH, Huang P, Hu Y. Selective killing of K-ras-transformed pancreatic cancer cells by targeting NAD(P)H oxidase. Chin J Cancer 2015; 34: 166–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochhawng L, Deng S, Pervaiz S, Yap CT. Redox regulation of cancer cell migration and invasion. Mitochondrion 2013; 13: 246–253. [DOI] [PubMed] [Google Scholar]
- Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev 1998; 12: 2245–2262. [DOI] [PubMed] [Google Scholar]
- Alberghina L, Gaglio D. Redox control of glutamine utilization in cancer. Cell Death Dis 2014; 5: e1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhar M, Engelberg D, Levitzki A. ROS, stress-activated kinases and stress signaling in cancer. EMBO Rep 2002; 3: 420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloire G, Legrand-Poels S, Piette J. NF-kappaB activation by reactive oxygen species: fifteen years later. Biochem Pharmacol 2006; 72: 1493–1505. [DOI] [PubMed] [Google Scholar]
- Nishikawa M. Reactive oxygen species in tumor metastasis. Cancer Lett 2008; 266: 53–59. [DOI] [PubMed] [Google Scholar]
- Hurd TR, DeGennaro M, Lehmann R. Redox regulation of cell migration and adhesion. Trends Cell Biol 2012; 22: 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binker MG, Binker-Cosen AA, Richards D, Oliver B, Cosen-Binker LI. EGF promotes invasion by PANC-1 cells through Rac1/ROS-dependent secretion and activation of MMP-2. Biochem Biophys Res Commun 2009; 379: 445–450. [DOI] [PubMed] [Google Scholar]
- Pelicano H, Lu W, Zhou Y, Zhang W, Chen Z, Hu Y et al. Mitochondrial dysfunction and reactive oxygen species imbalance promote breast cancer cell motility through a CXCL14-mediated mechanism. Cancer Res 2009; 69: 2375–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WT, Shen SC, Chow JM, Lin CW, Shia LT, Chen YC. Contribution of reactive oxygen species to migration/invasion of human glioblastoma cells U87 via ERK-dependent COX-2/PGE(2) activation. Neurobiol Dis 2010; 37: 118–129. [DOI] [PubMed] [Google Scholar]
- Shimada Y, Imamura M, Wagata T, Yamaguchi N, Tobe T. Characterization of 21 newly established esophageal cancer cell lines. Cancer 1992; 69: 277–284. [DOI] [PubMed] [Google Scholar]
- Wong ML, Tao Q, Fu L, Wong KY, Qiu GH, Law FB et al. Aberrant promoter hypermethylation and silencing of the critical 3p21 tumour suppressor gene, RASSF1A, in Chinese oesophageal squamous cell carcinoma. Int J Oncol 2006; 28: 767–773. [PubMed] [Google Scholar]
- Li Y, Chen L, Nie CJ, Zeng TT, Liu H, Mao X et al. Downregulation of RBMS3 is associated with poor prognosis in esophageal squamous cell carcinoma. Cancer Res 2011; 71: 6106–6115. [DOI] [PubMed] [Google Scholar]
- Guan XY, Sham JS, Tang TC, Fang Y, Huo KK, Yang JM. Isolation of a novel candidate oncogene within a frequently amplified region at 3q26 in ovarian cancer. Cancer Res 2001; 61: 3806–3809. [PubMed] [Google Scholar]
- Li Y, Zhu CL, Nie CJ, Li JC, Zeng TT, Zhou J et al. Investigation of tumor suppressing function of CACNA2D3 in esophageal squamous cell carcinoma. PLoS One 2013; 8: e60027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.