Abstract
Metastasis significantly reduces the survival rate of osteosarcoma (OS) patients. Therefore, identification of novel targets remains extremely important to prevent metastasis and treat OS. In this report, we show that SPARCL1 is downregulated in OS by epigenetic methylation of promoter DNA. In vitro and in vivo experiments revealed that SPARCL1 inhibits OS metastasis. We further demonstrated that SPARCL1-activated WNT/β-catenin signaling by physical interaction with various frizzled receptors and lipoprotein receptor-related protein 5/6, leading to WNT–receptor complex stabilization. Activation of WNT/β-catenin signaling contributes to the SPARCL1-mediated inhibitory effects on OS metastasis. Furthermore, we uncovered a paracrine effect of SPARCL1 on macrophage recruitment through activated WNT/β-catenin signaling-mediated secretion of chemokine ligand5 from OS cells. These findings suggest that the targeting of SPARCL1 as a new anti-metastatic strategy for OS patients.
Introduction
Osteosarcoma (OS) is the most common primary bone malignant tumor and has a high propensity to metastasis.1, 2 The heterogeneous nature of OS is indicated by the presence of cells arrested at different stages of differentiation.3, 4 Although osteosarcoma is a rare type of tumor, it represents ~55% of childhood and adolescent malignant bone tumors.5
WNT/β-catenin signaling is activated when WNT ligands bind to frizzled (FZD) receptors and co-receptors (LRP5/6) on the cellular surface.6 This signal transduction is increasingly complex and can be regulated both extracellularly and intracellularly.7 WNT/β-catenin signaling is involved in osteoblast differentiation and has been linked to the development of epithelial carcinoma.8, 9 However, its role in mesenchymal tumors remains unclear.10, 11, 12, 13 Conflicting reports have been published concerning the role and effect of this pathway in OS.14, 15, 16, 17, 18, 19, 20
SPARC (secreted protein acidic and rich in cysteine) protein expression affects osteoblast differentiation, tumorigenesis and tumor metastasis.21 SPARCL1, a member of the SPARC family, plays important roles in many physiological and pathological conditions.22, 23 In addition, SPARCL1 mitigates fibroblast adhesion and inhibits migration.24 Abnormal expression of SPARCL1 is closely related to the metastasis and prognosis of a variety of tumors.25, 26, 27, 28 Moreover, Mintz et al.29 found that SPARCL1 showed significantly different expression patterns between Huvos I/II (n=15) and III/IV (n=15) OS tissues. However, the mechanisms underlying the regulation of cancer development by SPARCL1 and the membrane receptor involved remain elusive.
In the present study, we show that SPARCL1 is downregulated in OS via epigenetic methylation of its promoter DNA. SPARCL1 inhibited OS metastasis through activation of canonical WNT/β-catenin signaling. We further demonstrated that SPARCL1 interacted physically with FZDs and LRP5/6, and stabilized the interaction between canonical WNT ligands and their receptors. We also show that SPARCL1-mediated activation of WNT/β-catenin signaling promotes the recruitment of macrophages via an increase in chemokine ligand5 (CCL5) production in human OS cells.
Results
SPARCL1 is downregulated in OS via epigenetic promoter DNA methylation
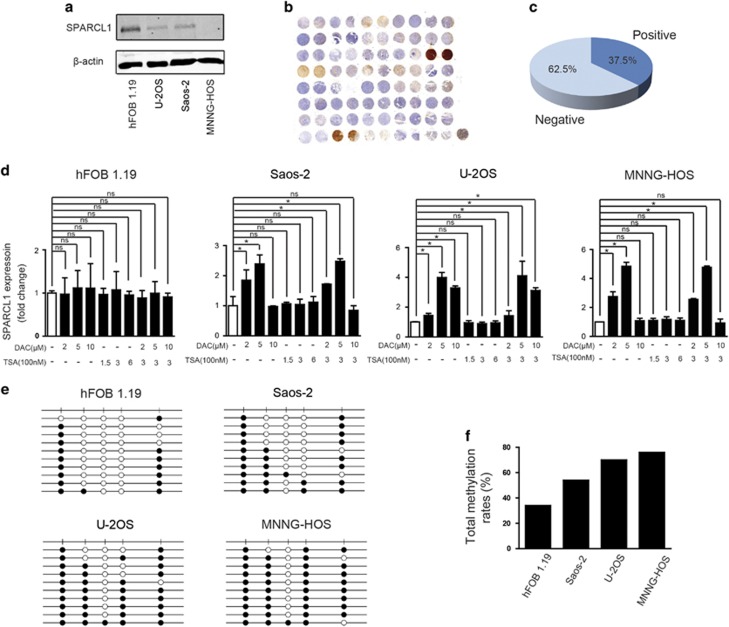
To investigate the expression pattern of SPARCL1 in OS, we first compared the level of the SPARCL1 protein in the normal human osteoblast cell line, hFOB1.19, with that in OS cell lines (Saos-2, U-2OS and MNNG-HOS). The SPARCL1 protein level was significantly lower in the OS cell lines (Figure 1a). We further examined SPARCL1 levels in a human osteosarcoma tissue microarray (TMA) (n=40) by immunohistochemistry (IHC). SPARCL1 was almost absent in most of the samples (25/40, 62.5%), but was detected in 37.5% (11/40) of the samples (Figures 1b and c). Genetic or epigenetic mechanisms or both might cause dysregulation of SPARCL1 expression in OS. SPARCL1 is located on chromosome 4q22.1 and according to published studies; there is no deletion in this region in OS.30, 31 We then explored the epigenetic regulation of SPARCL1 expression in OS. Three OS cell lines (Saos-2, U-2OS and MNNG-HOS) and an osteoblast cell line, hFOB1.19, were treated with a specific methyltransferase inhibitor, 5-aza-2′-deoxycytidine (DAC), and a histone deacetylase inhibitor, trichostatin A (TSA), both separately and in combination. The mRNA expression level of SPARCL1 in OS cells was increased significantly by DAC treatment in all tested OS cell lines, but not by TSA treatment (Figure 1d). Meanwhile, the expression of SPARCL1 increased only slightly in osteoblast cells treated with DAC (Figure 1d). To further validate the methylation-mediated downregulation of SPARCL1 in OS, we performed bisulfite sequencing PCR of DNA from Saos-2, U-2OS, MNNG-HOS and hFOB1.19 cells (Figure 1e). The methylation status of five CpG sites in the SPARCL1 promoter in these cell lines is shown in Figure 1f. The SPARCL1 promoter in OS cells showed higher levels of methylation compared to that in human osteoblast cells (hFOB1.19: 34% Saos-2: 54% U-2OS: 70% and MNNG-HOS: 76%). Taken together, these results demonstrated that SPARCL1 was downregulated in OS via epigenetic methylation of its promoter.
Figure 1.
SPARCL1 is downregulated in OS owing to epigenetic silencing by promoter DNA methylation. (a) SPARCL1 expression patterns in OS cell lines (Saos-2, U-2OS and MNNG-HOS) and normal osteoblast cell line (hFOB1.19) by western blotting. (b) The IHC staining of SPARCL1 in OS human osteosarcoma TMA (n=40, duplicate cores per case). (c) Statistical analysis of IHC based on the protein expression level in 40 OS cases. (d) The mRNA expression of SPARCL1 was evaluated by real-time qPCR in OS cell lines (Saos2, U-2OS and MNNG-HOS) and osteoblast cell line (hFOB1.19) treated with vehicle, DAC, TSA or DAC plus TSA (n=3). Values are means±s.d., *P<0.05; ns indicates no significance. (e) Bisulfite-sequencing results of human osteoblast cell line hFOB1.19 and three OS cell lines (Saos2, U-2OS and MNNG-HOS). Five CpG sites were sequenced. Open circles indicate unmethylated and solid circles represent methylated CpG dinucleotides. (f) Total methylation rates of SPARCL1 promoter in the human osteoblast cell line hFOB1.19 and three OS cell lines (Saos2, U-2OS and MNNG-HOS) (n=1) are shown.
SPARCL1 inhibits OS metastasis in vitro and in vivo
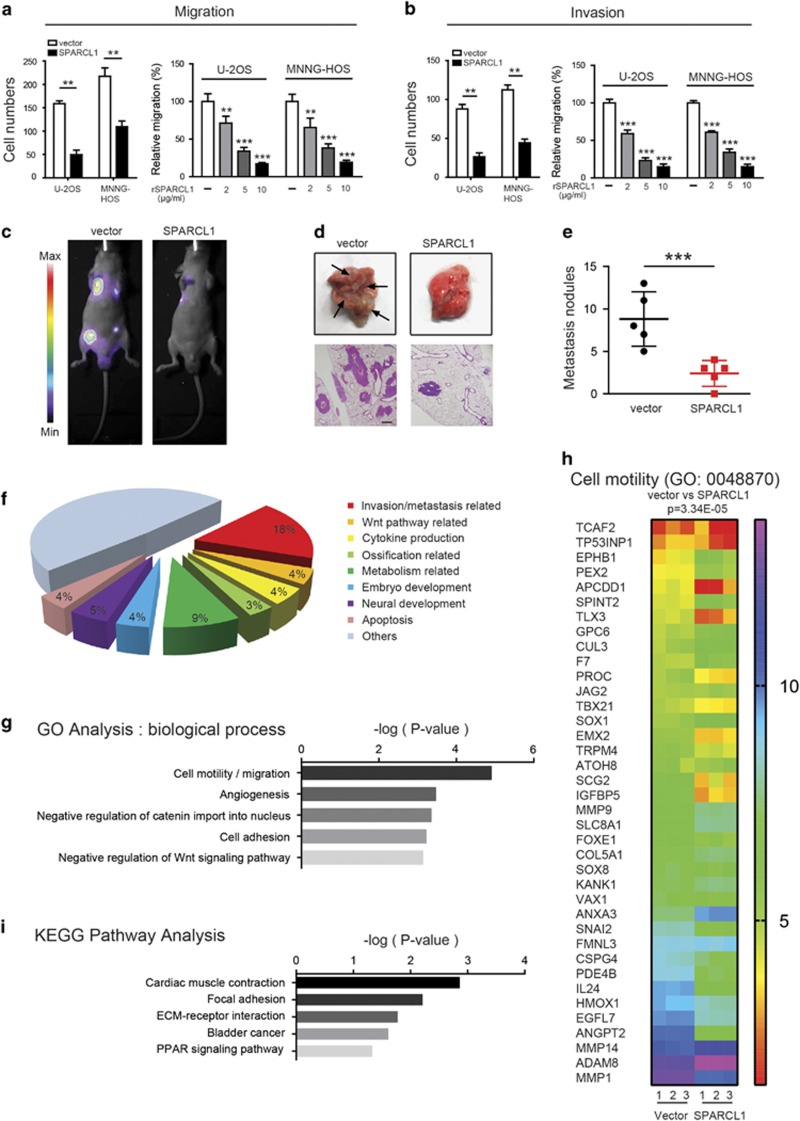
We next determined the biological functions of SPARCL1 in OS cells (U-2OS and MNNG-HOS) and in an osteoblast cell line (hFOB1.19). For U-2OS and MNNG-HOS cells, we established stable SPARCL1-overexpressing cells. Quantitative real-time polymerase chain reaction (qPCR) and western blotting (Supplementary Figure 1a) confirmed the efficiency of SPARCL1 overexpression in MNNG-HOS and U-2OS cells. We then investigated the effects of SPARCL1 on OS cell migration and invasion in vitro. Overexpression of SPARCL1 inhibited the migration and invasion of MNNG-HOS and U-2OS cells significantly (Figures 2a and b and Supplementary Figure 1b). SPARCL1 is an extracellular matrix protein; therefore, we further confirmed the suppressive role of SPARCL1 in OS cell migration and invasion, using recombinant human SPARCL1 (rSPARCL1); PBS was used as a control. The migration and invasion abilities of U-2OS and MNNG-HOS cells were inhibited significantly in a dose-dependent manner by rSPARCL1 (Figures 2a and b and Supplementary Figure 1f). While overexpression of SPARCL1 or adding rSPARCL1 did not significantly affect the migration and invasion abilities of the osteoblast cells (Supplementary Figures 1c–e).
Figure 2.
SPARCL1 inhibits osteosarcoma metastasis in vitro and in vivo. (a) SPARCL1 suppressed osteosarcoma cells (U-2OS and MNNG-HOS) migration in vitro. In the left panel, cellular migration ability in overexpression and control groups (U-2OS and MNNG-HOS cells) were detected by transwell migration assay. In the right panel, MNNG-HOS and U-2OS cells treated with different concentrations of rSPARCL1 or PBS were also used to perform transwell migration assay (n=3). Six randomly selected fields were photographed and the numbers were counted. Representative images of cellular migration conditions in each group are shown in Supplementary Figures 1c and f. Values are means±s.d., **P<0.01, ***P<0.001. (b) SPARCL1 suppressed osteosarcoma cells (U-2OS and MNNG-HOS) invasion in vitro. Similarly, cellular invasion ability was detected by transwell invasion assay in the left panel. In the right panel, MNNG-HOS and U-2OS cells treated with different concentrations of rSPARCL1 or PBS were used to perform transwell invasion assay (n=3). Representative images of invaded cells in each group are shown in Supplementary Figures 1c and f. Values are means±s.d., **P<0.01, ***P<0.001. (c) Control and SPARCL1-overexpression MNNG-HOS (Luc) cells were injected into the tail vein of BALB/C nude mice. After 30 days, the in vivo tumor metastasis was imaged. Representative images of mice in each treatment group are shown. (d) Representative images of collected lungs in control and SPARCL1-overexpression groups are shown in the upper panel. Black arrows indicate metastases. Representative photographs of hematoxylin and eosin staining of lung tissues are also shown in the lower panel. Scale bars, 50 μm. (e) Numbers of lung metastatic foci were counted in each group. Values are means±s.d., ***P<0.001. (f) Functional classifications of the 322 DEGs based on GO analysis. Shown are pie charts of gene categories (percentage indicated: invasion/metastasis-related genes: 18% WNT pathway-related genes: 4% cytokine production-related genes: 5% ossification-related genes: 3% metabolism-related genes: 9% embryo development-related genes: 4% neural development-related genes: 5% and apoptosis-related genes: 4%). (g) Representative GO (GO: biological process) categories affected by SPARCL1 expression in MNNG-HOS cells. (h) Heat map of cell motility molecules (GO: 0048870) from gene expression profiling results based on GO analysis (control groups vs SPARCL1-overexpressing groups, P=3.34E−5). (i) Representative KEGG pathway categories affected by SPARCL1 expression in MNNG-HOS cells.
We also examined the role of SPARCL1 in OS metastasis in vivo. Vector-Luc and SPARCL1-Luc transfected MNNG-HOS cells were injected into the tail veins of BALB/C nude mice. Tumor metastasis status was examined 30 days after the injection, using an imaging system that detected the luciferase signal. The representative bioluminescent images of the different groups are shown in Figure 2c. As shown in Figure 2d and Supplementary Figure 2a, representative pictures of resected lungs and hematoxylin and eosin staining of lung tissues confirmed that the incidence of metastasis in the lung decreased dramatically in the SPARCL1-overexpressing group compared with that in the control group (Supplementary Figure 2b). The number of metastatic lung nodules in the SPARCL1-overexpressing group was smaller than that in the control group (Figure 2e).
To explore the mechanism involved in the inhibition of OS metastasis by SPARCL1, we performed transcriptome analysis to compare the gene expression patterns in control MNNG-HOS cells and SPARCL1-overexpressing MNNG-HOS cells (GSE97572). To ensure the accuracy of the data analysis, differentially expressed genes (DEGs) were defined as those having both an average fold change >2.0 and P-values <0.05. As shown in Figure 2f, functional classifications of the 322 DEGs based on gene ontology (GO) analysis demonstrated distinct gene expression patterns. As expected, as many as 18% of them were involved in ‘invasion/metastasis’ (Figure 2f). The DEGs in different functional categories are listed in Supplementary Table 2. The lower expression of matrix metallopeptidase 1 (MMP1), matrix metallopeptidase 3 (MMP3), matrix metallopeptidase 7 (MMP7), matrix metallopeptidase 13 (MMP13) and snail family transcriptional repressor 2 (SNAI2) in SPARCL1-overexpressing MNNG-HOS cells compared with that in control MNNG-HOS cells was confirmed by qPCR (Supplementary Figure 3e). The results of GO analysis (biological process, molecular function and cellular component) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis also provided strong support to our earlier observations that SPARCL1 inhibits OS cell migration and invasion (Figures 2g and h and Supplementary Figures 3a and 3b). The analysis clearly identified ontology categories and pathways involved in cell motility, cell adhesion, focal adhesion and the extracellular matrix (Figures 2g and h and Supplementary Figures 3a–c). Meanwhile, the heat map shown in Figure 2h indicated that SPARCL1 target genes inhibit cell motility (GO0048870, vector compared with SPARCL1 overexpression, P=3.34E−5), which was also consistent with our previous biological studies. Collectively, these results indicated that SPARCL1 inhibits OS metastasis.
SPARCL1 activates the WNT/β-catenin pathway by promoting nuclear translocation of β-catenin
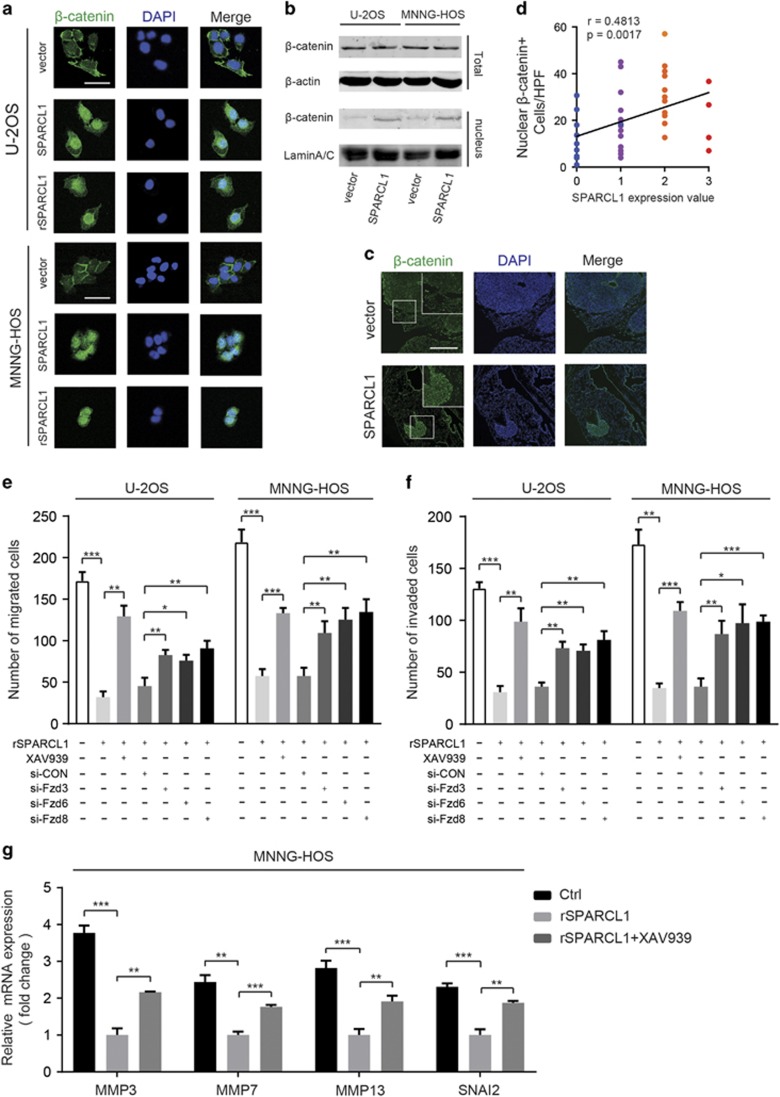
The aforementioned results of GO analysis (biological process) demonstrated that the control cells’ gene expression signature involves a drive toward negative regulation of catenin import into the nucleus and negative regulation of WNT signaling, compared with that in the SPARCL1-overexpressing cells (Figure 2g and Supplementary Figure 3d). It has also been reported that WNT/β-catenin plays an important role in the process of osteoblast differentiation and that this pathway is inactivated in OS.8, 14, 15, 16, 17, 18, 19 To further reveal the potential mechanism of SPARCL1-mediated inhibition of OS metastasis, we first examined whether SPARCL1 could activate the canonical WNT/β-catenin pathway, by a dual-luciferase reporter gene assay (Supplementary Figure 4a). MNNG-HOS and U-2OS cells were treated with 10 μg/ml rSPARCL1 or vehicle (PBS). The TOP Flash reporter activity was increased significantly in the rSPARCL1 group (P<0.001) (Supplementary Figure 4a). We obtained similar results using HEK293T cells (Supplementary Figure 4b). We then carried out immunofluorescence (IF) staining of OS cells overexpressing SPARCL1, cells treated with rSPARCL1 protein and control cells. Nuclear localization of β-catenin (green) was more pronounced in the SPARCL1-overexpressing cells and rSPARCL1-treated cells than in the control cells (Figure 3a). Western blotting was also performed to detect the distribution of β-catenin. As shown in Figure 3b, there was no significant difference in the total amount of β-catenin in the control and SPARCL1-overexpressing cells (U-2OS and MNNG-HOS). Nuclear β-catenin was increased obviously in SPARCL1-overexpressing cells compared to that in the control cells, as revealed by the stronger band corresponding to β-catenin (Figure 3b). Relative amounts of the nuclear β-catenin proteins were quantified and were shown in Supplementary Figure 4c. We next examined the distribution of β-catenin in lung tissues from a metastatic xenograft animal model of OS, by tissue IF. The results showed a much greater extent of nuclear localized β-catenin (green) in the SPARCL1-overexpressing group than in the control group (Figure 3c). Finally, we investigated the relationship between the expression of SPARCL1 and the nuclear localization of β-catenin in 40 osteosarcoma tissues by IHC staining. The results demonstrated a positive correlation between the expression of SPARCL1 and nuclear localization of β-catenin (r=0.4813, P=0.0017) (Figure 3d and Supplementary Figure 4d). To further confirm that activated WNT/β-catenin signaling is involved in SPARCL1-mediated biological functions in OS, a small molecule inhibitor of WNT/β-catenin signaling (XAV-939) and siRNAs targeting Fzd3, Fzd6 or Fzd8 were used. Inhibiting the WNT/β-catenin pathway with XAV-939 almost completely reversed the inhibitory effects of SPARCL1 on OS cell migration and invasion (Figures 3e and f and Supplementary Figure 5a). In addition, compared with the si-control, siRNA-mediated silencing of individual frizzled genes (Fzd3, Fzd6 or Fzd8) also reversed the inhibitory effect of SPARCL1 to some extent (Figures 3e and f and Supplementary Figures 4e, 4f and 5a). Meanwhile, qPCR analyses revealed significant downregulation of several genes associated with migration and invasion (MMP3, MMP7, MMP13 and SNAI2) in OS cells treated with rSPARCL1 compared with that in the control cells (U-2OS and MNNG-HOS) (Figure 3g and Supplementary Figure 5b). These observations were similar to our transcriptome analysis data. Moreover, inhibiting the WNT/β-catenin pathway using XAV-939 partly reversed the downregulation effect of these genes in U-2OS and MNNG-HOS cells (Figure 3g and Supplementary Figure 5b). These data indicated that SPARCL1 activates the WNT/β-catenin pathway by promoting nuclear translocation of β-catenin. Blocking the WNT/β-catenin signaling reversed the inhibitory effects of SPARCL1 on OS cell migration and invasion.
Figure 3.
SPARCL1 activates the WNT/β-catenin pathway by promoting nuclear translocation of β-catenin. (a) The distribution of β-catenin in OS cells (U-2OS and MNNG-HOS) transfected with Lenti-vector, Lenti-SPARCL1 or treated with 10 μg/ml rSPARCL1 were analyzed by IF staining. β-catenin is shown by green fluorescence, and the cell nuclei was stained with DAPI (blue fluorescence), Scale bars, 100 μm. (b) In the upper panel, the expressions of β-catenin from whole cell lysates in control and SPARCL1-overexpression cells (U-2OS and MNNG-HOS) were detected by western blotting. Nuclear proteins of control and SPARCL1-overexpression groups (U-2OS and MNNG-HOS cells) were extracted. In the lower panel, the nuclear β-catenin expressions in each group were also detected by western blotting. (c) The distribution of β-catenin in mice lung tissues was examined by IF. Representative images of β-catenin (green) in mice lung tissues are shown, the cell nuclei was stained with DAPI (blue). Scale bars, 500 μm. (d) Spearman correlation analysis between SPARCL1 expression and number of nuclear β-catenin+ OS cells based on the IHC staining (r=0.4813, P=0.0017) (HPF, high power field). (e and f) The cell migration and invasion abilities were determined by transwell migration and invasion assay in U-2OS and MNNG-HOS cells treated with 10 μg/ml rSPARCL1, 10 μM XAV-939, si-NC, si-Fzd3, si-Fzd6 or si-Fzd8 (n=3). Representative images of migrated or invaded cells are shown in Supplementary Figure 5a. Scale bars, 200 μm, values are mean±s.d., *P<0.05, **P<0.01, ***P<0.001. (g) Real-time qPCR analyses of relative expression of indicated genes in MNNG-HOS cells and MNNG-HOS cells treated with XAV939 combined 10 μg/ml rSPARCL1 normalized to MNNG-HOS cells treated with rSPARCL1 alone (n=3). Values are mean±s.d., *P<0.05, **P<0.01, ***P<0.001.
SPARCL1 stabilizes ligand–receptor interactions for the WNT/β-catenin pathway
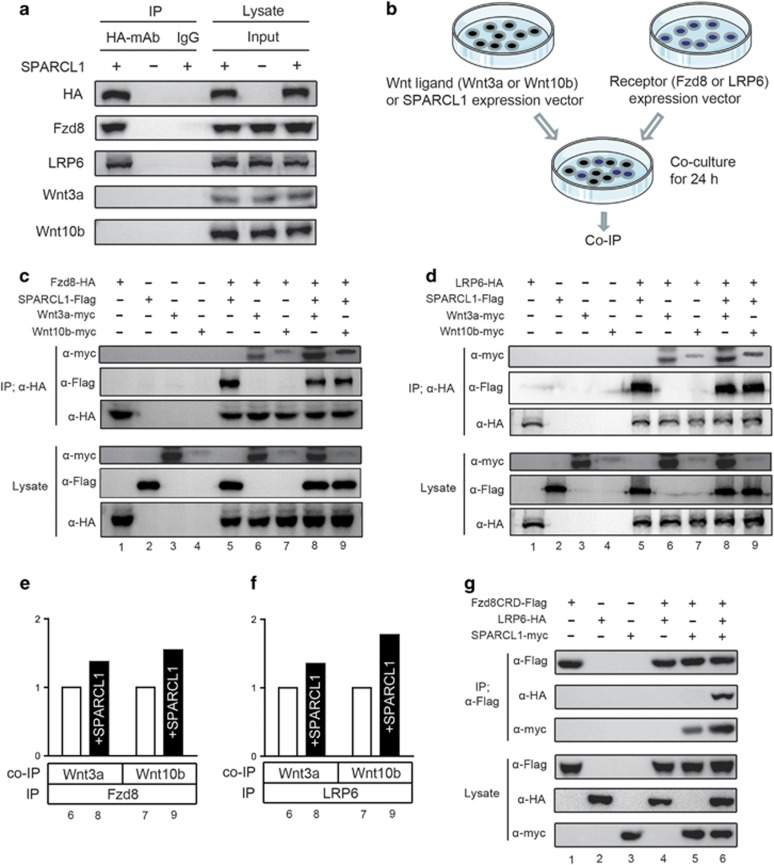
To investigate the mechanism by which SPARCL1 controls WNT/β-catenin signaling more deeply, we examined the interaction of SPARCL1 with various extracellular components of this signaling pathway. Using a co-immunoprecipitation (Co-IP) approach, we found that SPARCL1 physically interacted with endogenous LRP5/6 and FZDs (Fzd3, Fzd6 and Fzd8) in HEK293T cells (Figure 4a and Supplementary Figure 5c). However, no direct interaction was observed between SPARCL1 and canonical WNT proteins (Wnt3a and Wnt10b) (Figure 4a). This interaction between SPARCL1 and endogenous LRP5/6 and FZDs was also confirmed in OS cell lines (Supplementary Figure 5d). We further determined whether SPARCL1 modulated the WNT ligand–receptor interaction. For this purpose, we performed an interaction assay based upon the immunoprecipitation of tagged proteins from the lysates of co-cultured HEK293T cells that were transfected individually with expression plasmids for SPARCL1-Flag, Fzd8-HA, LRP6-HA, Wnt3a-myc or Wnt10b-myc (co-culture IP) (Figure 4b). Wnt3a and Wnt10b were used as canonical WNT ligands. We also used LRP6 and Fzd8 as representative pathway-specific receptors. We first examined whether SPARCL1 influenced the interaction of canonical WNT ligands (Wnt3a or Wnt10b) and Fzd8 proteins. As shown in Figure 4c, Wnt3a co-precipitated with Fzd8, and SPARCL1 stabilized the binding between Wnt3a and Fzd8, as indicated by the stronger band of precipitated Wnt3a (lanes 6 and 8). The relative amounts of the precipitated Wnt3a were quantified and are shown in Figure 4e. A similar enhancement of the WNT–FZD interaction by SPARCL1 was observed in combinations of Wnt10b and Fzd8 (Figures 4c and e) (lanes 7 and 9). We next examined the effect of SPARCL1 on the interaction between WNT proteins and the pathway-specific co-receptor, LRP6. As shown in Figures 4d and f, Wnt3a was co-precipitated with LRP6, and this interaction was enhanced in the presence of SPARCL1, as indicated by the stronger bands in lanes 6 and 8. Similarly, SPARCL1 also stabilized the interaction between Wnt10b and LRP6 (Figures 4d and f) (lanes 7 and 9). To further clarify whether SPARCL1 bound to both LRP6 and Fzd8 and formed a complex, another co-culture IP assay was designed (Figure 4g). Previous studies reported that LRP6 and Fzd8 interacted specifically with each other in a WNT-independent manner and this interaction was mediated by Fzd8 beyond the Fzd8 cysteine-rich domain (CRD).32 Therefore, in our co-culture IP assay, co-cultured HEK293T cells were transfected individually with expression plasmids for SPARCL1-myc, Fzd8CRD-Flag and LRP6-HA. As shown in Figure 4g, Fzd8CRD did not co-precipitate with LRP6, as previously reported (lane 4).32, 33 Meanwhile, in the presence of SPARCL1, Fzd8CRD co-precipitated with both LRP6 and SPARCL1 (lane 6). These results suggested that SPARCL1 formed a complex comprising SPARCL1-LRP6-Fzd8 and stabilized the interaction between canonical WNT ligands and their receptors (Figure 7).
Figure 4.
Stabilization of ligand–receptor interaction for the WNT/β-catenin pathway by SPARCL1. (a) Co-IP assay between SPARCL1 and canonical WNT components (receptor: Fzd8 or LRP6; ligand: Wnt3a or Wnt10b). HEK293T cells were transfected with SPARCL1-HA or vector control. The input on the right panel shows the levels of transfected HA-SPARCL1 and endogenous WNT components (Fzd8, LRP6, Wnt3a and Wnt10b) in HA-tagged SPARCL1 or vector control. (b) Brief scheme of the co-culture IP procedure. HEK293T cells were individually transfected with expression plasmids for SPARCL1 or various WNT components (receptor expression plasmid: LRP6 or Fzd8; WNT ligand expression plasmid: Wnt3a or Wnt10b). Eight hours after transfection, cells with receptors and ligands were mixed and co-culture for 24 h, followed by immunoprecipitation assay. (c) SPARCL1 stabilized the binding between canonical WNT protein (Wnt3a or Wnt10b) and Fzd8. HA-tagged Fzd8-expressing cells were co-cultured with myc-tagged WNT ligands-expressing (Wnt3a or Wnt10b) cells and Flag-tagged SPARCL1-expressing cells both separately and in combination. The HA-tagged Fzd8 was immunoprecipitated in this experiment. (d) SPARCL1 stabilized the binding between canonical WNT protein (Wnt3a or Wnt10b) and LRP6. Similarly, HA-tagged LRP6-expressing cells were co-cultured with WNT ligands-expressing cells and Flag-tagged SPARCL1-expressing cells both separately and in combination. (e and f) Densitometric analysis showed the relative amounts of precipitated WNT ligand (Wnt3a or Wnt10b) interacted with Fzd8 or LRP6 affected by SPARCL1. Values are normalized to intensities without SPARCL1 as 1. (g) Fzd8 CRD co-precipitated LRP6 in the presence of SPARCL1. Flag-tagged Fzd8CRD-expressing cells were co-cultured with myc-tagged SPARCL1-expressing cells and HA-tagged LRP6-expressing cells both separately and in combination. To further confirm the SPARCL1-LRP6-Fzd8 complex model, the Flag-tagged Fzd8CRD was immunoprecipitated in this experiment.
Correlation between SPARCL1 expression and M1 macrophages infiltration in human OS tissues or xenograft OS tissues
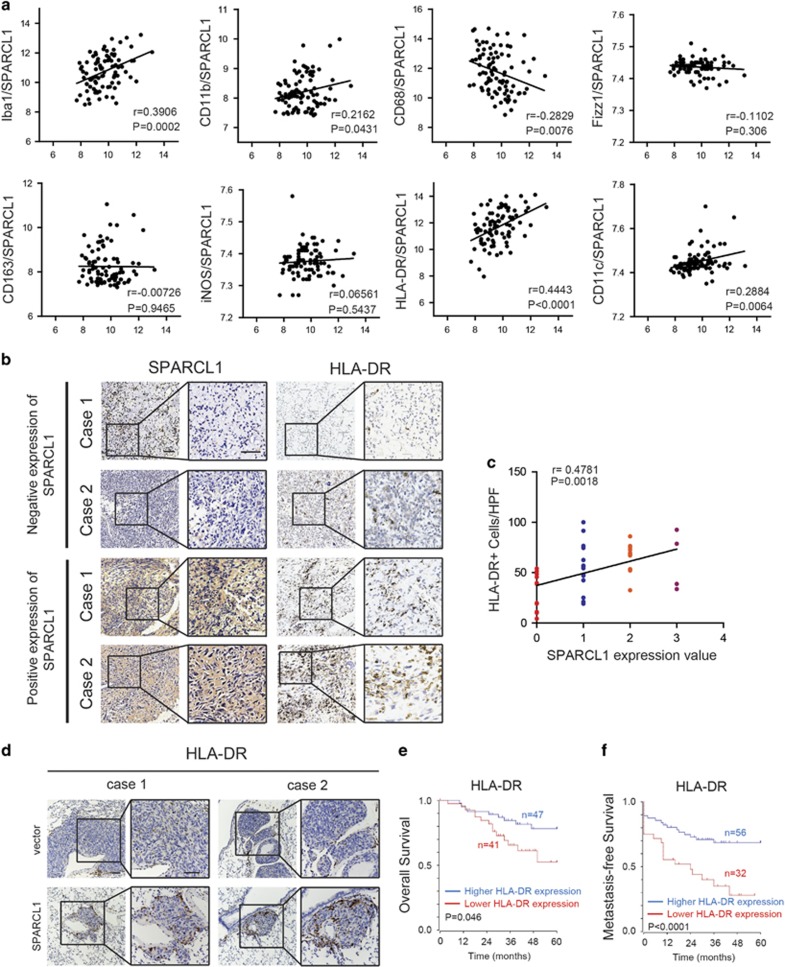
It is generally accepted that an appropriate innate immune response restricts the growth and spread of tumors.34 We first used a bioinformatics tool to explore the relationship between SPARCL1 and the immune process in an OS database (http://hgserver1.amc.nl). GO analysis and KEGG pathway analysis indicated that SPARCL1 was related closely to the immune process via its involvement in antigen presentation and the chemokine pathway in OS (Supplementary Figures 6a–c). We also noted a positive correlation between SPARCL1 and M1 HLA-DR+ macrophages (r=0.4443, P<0.0001) and M1 CD11c+ macrophages (r=0.2884, P=0.0064) (Figure 5a). However, there was no significant correlation between SPARCL1 and total tumor-associated macrophages (TAMs) (CD11b+, CD68+ and Iba-1+) or M2 macrophages (CD163+, Fizz1+ and iNOS+) (Figure 5a). Therefore, we hypothesized that M1 macrophages might be involved in SPARCL1-mediated cellular processes in OS. To further investigate this correlation between SPARCL1 and macrophages in OS, we performed immunostaining of HLA-DR in 40 primary OS tissues. IHC staining showed a positive correlation between SPARCL1 expression and the number of M1 HLA-DR+ macrophages (r=0.4781, P=0.0018) (Figures 5b and c). As expected, SPARCL1-overexpressing metastatic lung nodules exhibited increased numbers of M1 HLA-DR+ macrophages compared with those in the control group (Figure 5d). Kaplan–Meier analysis also showed that higher HLA-DR expression was associated with markedly increased overall and metastasis-free survival rates (Figures 5e and f). These data demonstrated that SPARCL1 levels correlated positively with the infiltration of M1 macrophages in osteosarcoma.
Figure 5.
Correlation between SPARCL1 expression and M1 macrophages infiltration in OS. (a) Pearson correlation analysis between SPARCL1 and TAMs by analyzing OS gene expression database (http://hgserver1.amc.nl). Ibal1, CD11b and CD68 are total TAM-associated makers; Fizz1, CD163 and iNOS are M2-associated makers; M1-associated makers are HLA-DR and CD11c. (b) Representative images of IHC staining of SPARCL1 and HLA-DR in 40 OS cases. Two cases of negative expression of SPARCL1 and HLA-DR were shown in upper panel and lower panel revealed two cases of positive expression of SPARCL1 and HLA-DR. Scale bars, 100 μm. (c) Spearman correlation analysis between SPARCL1 score and number of M1 HLA-DR+ macrophages based on the IHC staining of 40 osteosarcoma cases (r=0.4781, P=0.0018) (HPF, high power field). (d) The more infiltration of macrophages in SPARCL1-overexpressing metastatic lung nodules compared with vector group. Representative images of IHC staining against HLA-DR in mice lung tissues are shown. Scale bars, 100 μm. (e and f) Kaplan–Meier analysis of overall and metastasis-free survival rate related to the expression of HLA-DR expression in 88 OS cases based on a human osteosarcoma gene expression database (http://hgserver1.amc.nl).
SPARCL1-mediated activation of the WNT/β-catenin pathway promotes macrophages recruitment by increasing CCL5 production in human OS cells
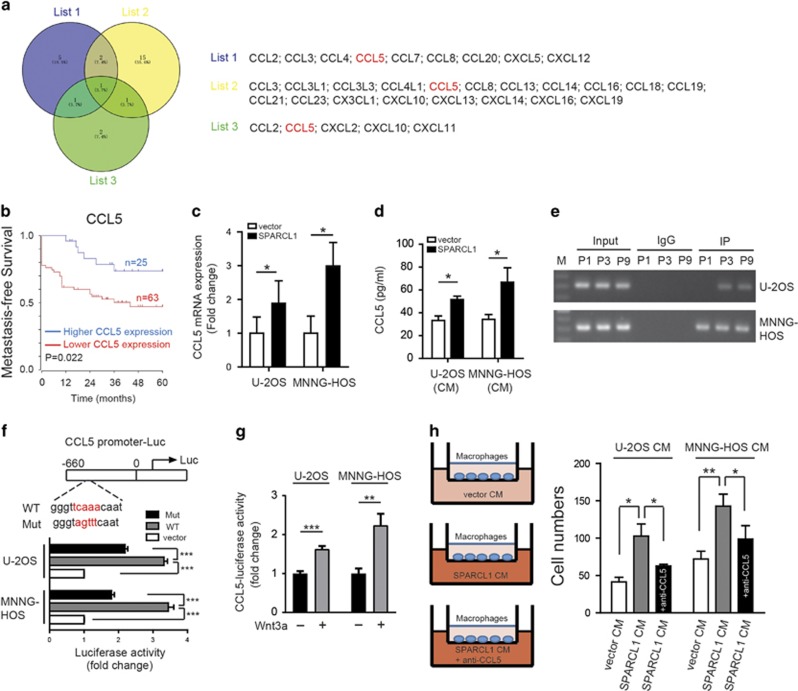
We next investigated the mechanism by which SPARCL1 promotes macrophages recruitment. Many chemokines play crucial roles in macrophages recruitment; therefore, we attempted to identify the chemokines involved in SPARCL1-mediated recruitment of macrophages. As shown in Figure 6a, our literature review revealed nine chemokines (list 1) that have the ability to recruit macrophages.35, 36, 37 We then analyzed the correlation between the expression of SPARCL1 and these chemokines by searching the human OS database, which identified 19 chemokines (list 2) that have a positive correlation with SPARCL1 (Supplementary Figure 6c). Finally, we identified five chemokines (list 3) that might be regulated by the WNT/β-catenin pathway, as reported in the literature.38, 39 CCL5 was the only common chemokine among the three lists (Figure 6a). Correlation analysis of the relationship between SPARCL1 expression and CCL5 in OS revealed a positive association (r=0.2478, P=0.0199) (Supplementary Figure 6d). As shown in Figure 6b and Supplementary Figure 6e, there was no significant difference in overall survival rate (P=0.154); however, patients with higher CCL5 levels had a significantly higher metastasis-free survival rate (P=0.022) in 88 cases of OS, based on a human osteosarcoma gene expression database. Furthermore, we found that induction of SPARCL1 overexpression increased CCL5 mRNA expression significantly in OS cells (U-2OS and MNNG-HOS) (Figure 6c). Enzyme-linked immunosorbent assay (ELISA) (Figure 6d) confirmed a significant increase in the CCL5 protein level resulting from SPARCL1 overexpression.
Figure 6.
SPARCL1-activated osteosarcoma WNT/β-catenin signaling promotes macrophages recruitment by increasing CCL5 production. (a) Venn diagram depicting overlapping chemokines in three lists. List 1 included nine chemokines that have the ability to recruit macrophages. List 2 revealed 19 chemokines that have a positive correlation with SPARCL1 after analyzing in the OS database (http://hgserver1.amc.nl). List 3 contained five chemokines that might be regulated by the WNT/β-catenin pathway, as reported in the literatures. (b) Kaplan–Meier analysis of metastasis-free survival rate was related to the expression of CCL5 expression in 88 OS cases based on a human osteosarcoma gene expression database (http://hgserver1.amc.nl). (c) Expression of CCL5 mRNA in established SPARCL1-overexpression cell lines (MNNG-HOS and U-2OS) and control cells (n=3). The results shown are mean±s.d., *P<0.05. (d) Amount of secreted CCL5 in 48-h serum-free SPARCL1-overexpression compared control OS cell supernatants, assessed by ELISA (CM, conditioned medium) (n=3). The results shown are mean±s.d., *P<0.05. (e) A ChIP assay in U-2OS and MNNG-HOS cell lines was performed to confirm the potential TCF4-binding site in the CCL5 promoter region. Integration maps of chip assay are shown. IgG and input fractions were used as controls (M, maker; P1, primer 1; P3, primer 3; P9, primer 9). (f) Luciferase activities of OS cells (U-2OS and MNNG-HOS) in luciferase reporter plasmid containing wild type and mutant CCL5 promoter (mutation site: red) (n=3). The data shown are mean±s.d., ***P<0.001. (g) Luciferase reporter gene assay of CCL5 in OS cells (U-2OS and MNNG-HOS) treated with 200 ng/ml rWnt3a or vehicle (PBS containing 0.1 % bovine serum albumin) (n=3). The results shown are the mean±s.d. of relative firefly/Renilla ratio, **P<0.01, ***P<0.001. (h) Chemotaxis assay was performed using the transwell system. As shown in left panel, primed THP-1 cells (macrophages) were seeded in the upper chamber and CM from SPARCL1-overexpressing or control cells (U-2OS and MNNG-HOS) in the lower chamber. In order to explore the chemotaxis effect of CCL5, its neutralizing antibodies were added to the CM from SPARCL1-overexpressing cells (n=3). As shown in right panel, the migrated cells were counted. Representative images of migrated cells are shown in Supplementary Figure 6h. The data shown are mean±s.d., *P<0.05, **P<0.01.
We next investigated how SPARCL1-mediated activation of WNT/β-catenin signaling regulates the expression of CCL5. Transcription factor 4 (TCF4) is a classical transcription factor of WNT/β-catenin signaling; therefore, a chromatin immunoprecipitation (ChIP) assay was performed to examine whether TCF4 was indeed involved directly in the regulation of CCL5 expression. The results showed that TCF4 had two binding sites in the CCL5 promoter (at −603/−301 and −2420/−2110 relative to the transcription start site) common to both U-2OS and MNNG-HOS cells (Figure 6e). We constructed luciferase reporter plasmids containing about 660 bp of the wild type and mutant CCL5 promoters. The dual-luciferase reporter assays indicated that the transcriptional activity of the CCL5 promoter was induced significantly by TCF4 in U-2OS and MNNG-HOS cells (P<0.001), and was decreased significantly in cells harboring the CCL5 promoter mutation construct (P<0.001) (Figure 6f). To further confirm the relationship between WNT/β-catenin pathway and CCL5, a CCL5 dual-luciferase reporter gene assay was performed. MNNG-HOS and U-2OS cells were treated with rWnt3a or vehicle (PBS containing 0.1% bovine serum albumin). The reporter activity was increased significantly in the rWnt3a group compared with that in the control group (for U-2OS cells, P<0.001; for MHHG-HOS cells, P<0.01) (Figure 6g). Finally, we performed chemotaxis assays with primed THP-1 cells (macrophages) in the upper chamber and conditioned medium (CM) from cultures of SPARCL1-overexpressing or control OS cells in the lower chamber (Figure 6h, left panel). We found that CM from SPARCL1-overexpressing cells induced increased migration of macrophages compared with that from the control cells (Figure 6h and Supplementary Figure 6h). The chemotactic effect of SPARCL1-overexpressing U-2OS CM and MNNG-HOS CM on macrophages was blocked partially using an anti-CCL5 monoclonal antibody (Figure 6h and Supplementary Figure 6h). These data suggested a regulatory role for SPARCL1 in the promotion of chemotaxis via the WNT/β-catenin pathway, leading to an increase in macrophage infiltration in OS.
Discussion
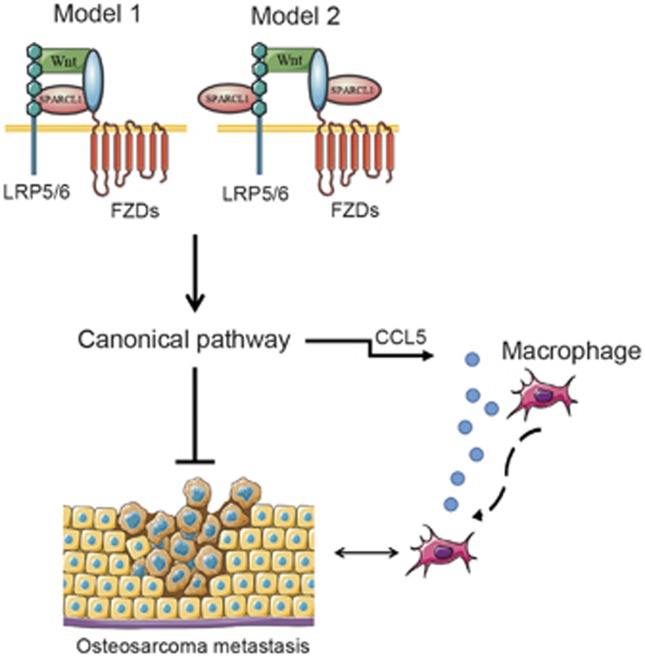
It is widely accepted that SPARC enhances osteogenesis by activating the WNT/β-catenin pathway.40 A relationship between SPARCL1, a member of the SPARC family, and WNT/β-catenin has not been reported in the literature. In the present study, we showed that SPARCL1, an extracellular component of the WNT pathway, contributes to the activation of the canonical pathway by promoting nuclear translocation of β-catenin. SPARCL1 might bridge the Fzd8 and LRP6 proteins through trimer formation, further stabilizing WNT ligand–receptor interactions (Figure 7).
Figure 7.
Schematic of SPARCL1 suppresses OS metastasis and recruits macrophages by activation of canonical WNT/β-catenin signaling through stabilization of WNT–receptor complex. Once SPARCL1 is involved, it stabilizes canonical WNT ligand–receptor complex through physical interaction with FZDs and LRP5/6. Model 1, SPARCL1 bridges the FZDs and LRP5/6 proteins through a complex formation. Model 2, SPARCL1 may interact with both FZDs and LRP5/6, respectively, but not form a complex. In the presence of SPARCL1, enhanced WNT/β-catenin signaling directly inhibits OS metastasis. Additionally, SPARCL1-mediated activation of WNT/β-catenin signaling promotes macrophages recruitment to the osteosarcoma microenvironment by increasing CCL5 production in human OS cells.
SPARCL1 inhibits metastatic progression in many kinds of tumors and may act as a tumor suppressor, which is consistent with our results in OS.25, 26, 27, 28 The role of canonical WNT signaling in epithelial carcinoma development has been studied intensively.9 However, the functional role of this signaling pathway in mesenchymal tumors, especially OS, remains controversial, with some studies suggesting an oncogenic role, and others supporting an anti-tumorigenic effect.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20 Several studies have revealed a clear correlation between tumorigenesis and defective differentiation in OS,14, 15, 16, 17, 18, 19 indicating the involvement of WNT/β-catenin in OS development. Goldstein et al.19 showed that activating WNT/β-catenin signaling by blocking DKK1 using an antibody inhibited OS growth and metastasis of orthotopically implanted, patient-derived OS, in an animal model. In accordance with this, we also demonstrated that SPARCL1-activated WNT/β-catenin signaling suppressed OS metastasis.
Growing evidence suggests that immune cells, in particular TAMs, are important participants in tumor progression and metastasis.35, 36, 37 TAMs comprise distinct subsets that coexist in tumors and adapt to the changing milieu.41, 42 A high density of M2 macrophages has been associated with poor prognosis and tumor metastasis.43 In contrast, a high density of M1 macrophages exerted an anti-metastatic effect and correlated with increased patient survival.44 This has provoked interest in developing therapies aimed at skewing the population of macrophages from the M2 to the M1 type.41, 42, 43, 44 It is worth noting that a randomized clinical trial involving 662 patients with OS, which examined the effect of activating macrophages and monocytes on standard chemotherapy, revealed a significant improvement in overall survival at 6 years.45 The pro-tumor role of TAMs has been observed in most epithelial tumor types; however, in OS (mesenchymal origin), TAMs are associated with anti-metastatic functions,46 in which TAMs are presumably polarized to an M1 type. Certain extracellular matrix molecules have been shown to act as inflammatory stimuli for the recruitment of innate immune cells and the expression of pro-inflammatory genes.47 Our work identified one potential mechanism by which SPARCL1 can recruit macrophages via activation of the WNT/β-catenin signaling pathway in OS.
Our study revealed that SPARCL1 plays important roles in OS metastasis and recruitment into the tumor microenvironment, and revealed the underlying mechanism of the regulation of the canonical WNT/β-catenin signaling, in which SPARCL1 stabilizes the WNT–receptor complex. However, a key limitation of this study is that our mouse model is immune-deficient, which might have influenced the metastatic pattern and prevented a complete evaluation of the role of the immune system in metastasis. Furthermore, because of the low incidence of OS, it was difficult to obtain a larger sample size to carry out a more thorough evaluation of clinical pathology and prognosis. The specific mechanisms prompting macrophages to polarize to the M1 phenotype, and whether a balance between M1- and M2-type functions is responsible, is unclear and will be further explored in future studies. Advances in this field will help to further decipher the complex roles of the canonical WNT pathway in the occurrence and development of OS.
In conclusion, SPARCL1 inhibited OS metastasis in vitro and in vivo. This effect correlated with the activation of WNT/β-catenin signaling. Activated WNT/β-catenin signaling also recruited macrophages via increasing CCL5 secretion (Figure 7). These findings suggest that canonical WNT signaling is anti-metastatic in OS, and support the targeting of SPARCL1 as a new anti-metastatic strategy for patients with OS.
Methods
Cell culture and reagents
Human U-2OS, Saos-2, MNNG-HOS, hFOB1.19, THP-1 and HEK293T cell lines were all purchased from Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All these cells were cultured following the instructions of the American Type Culture Collection (ATCC, Manassas, VA, USA). The antibodies used in this study were against SPARCL1 (ab107533, Abcam, Cambridge, MA, USA), β-actin (M1210-2, Huabio, Hangzhou, China), and lamin A/C (2032s, Cell Signaling, Danvers, MA, USA) for western blotting (WB); β-catenin (ab32572, Abcam) for WB, cell IF, and tissue IHC; β-catenin (GB13051, Goodbio, Shanghai, China) for tissue IF; HLA-DR (ab20181 and ab134038) for IHC; Wnt3a (ab28472, Abcam), Wnt10b (ab70816, Abcam), Frizzled3 (sc-68334, Santa Cruz, Dallas, TX, USA), Frizzled6 (5158, Cell Signaling), Frizzled8 (sc-33503, Santa Cruz), LRP5 (ab36121, Abcam), LRP6 (ab134146, Abcam) for co-IP, TCF4 (2565, Cell Signaling) for ChIP, CCL5 (21418, R&D Systems, Minneapolis, MN, USA) for chemotaxis assay; and HA-tag (3724, Cell Signaling), Flag-tag (8146, Cell Signaling), and myc-tag (2278, Cell Signaling) for co-culture IP. Secondary antibodies were purchased from Jackson ImmunoResearch (West Grove, PA, USA). Chemicals and biochemical used were: DAC (A3656, Sigma-Aldrich, St Louis, MO, USA), TSA (S1045, Selleck, Shanghai, China), XAV-939 (S1180, Selleck), recombinant human Wnt3a (5036-WN, R&D, Minneapolis, MN, USA) and recombinant human SPARCL1 (2728-SL, R&D).
R2 database analysis
The R2 database (http://hgserver1.amc.nl) was used to generate the Kaplan–Meier survival curves for patients with osteosarcoma. The tumor type was defined as osteosarcoma and mixed osteosarcoma - Kuijjer - 127 - vst - ilmnhwg6v2 data set was selected. The GO and KEGG pathway analysis generated by this database were used to comprehensively investigate the relationship between SPARCL1 and other genes. We also selected ‘correlate 2 genes’ in ‘Select type of analysis’ section and a Pearson rank correlation test was used to analyze the correlation between SPARCL1 and the markers of macrophages in 88 OS samples.
IHC and hematoxylin and eosin staining
A human osteosarcoma TMA containing 40 specimens (with duplicate cores for each sample) was purchased from Xi'an Alena Biotechnology Co., Ltd (Xi'an, China). Detailed information on the TMA can be found in Supplementary Table 1. IHC and hematoxylin and eosin staining were performed as previously reported.48 The following primary antibodies were used: SPARCL1 (1:100), β-catenin (1:100) and HLA-DR (1:150). The scoring was based on the intensity of positive staining using a 3-point scale: 0–5%, 0; 6–35%, 1; 36–70%, 2; and >70%, 3. Negative and positive expression levels were defined as a total score <2 and ⩾2, respectively. The scoring was performed in a blinded manner and determined independently by two senior pathologists.
IF
The IF cell assay was performed as previously described.49 For tissue IF staining, the samples were first deparaffinized, rehydrated and then subjected to heat-mediated antigen retrieval in citric acid (pH 6.0). All the cells and slides were incubated with specific antibodies against β-catenin and labeled with Alexa 488-conjugated secondary antibody (1:200). The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich). A confocal microscope (LSM 510, Zeiss, Jena, Germany) was used to photograph the images.
RNA isolation and qPCR
In accordance with the manufacturer’s instructions, TRIzol reagent (Takara, Dalian, China) was used to extract the total RNA, and the PrimeScript reverse transcription-polymerase chain reaction (RT-PCR) kit (Takara) was used to perform the RT. The qPCR was performed using a 7500 real-time PCR system (Applied Biosystems, Inc., USA) using the SYBR Premix Ex Taq (Takara) using the following recommended cycling settings: one initial cycle for 2 min at 95 °C followed by 40 cycles of 5 s at 95 °C and 31 s at 60 °C. The data were analyzed using the 2−△CT method and normalized to the glyceraldehyde 3-phosphate dehydrogenase expression level. The primer sequences used are all listed in Supplementary Table 3.
Short interfering RNA (siRNA) transfection
Cells were replated in six-well plates at 60–65% confluence and transfected with specific siRNA (si-Frizzled3, si-Frizzled6 and si-Frizzled8) using the Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific, Waltham, MA, USA). A non-targeted siRNA (si-NC) was used as the control. Transfection steps were performed following the manufacturer’s protocols. The siRNA oligonucleotides were all produced by GenePharma (Shanghai, China), and their sequences are listed in Supplementary Table 3.
DAC and TSA treatment
The different treatment concentrations and times used for the DAC or TSA were as previously described.48 Cells incubated with dimethyl sulfoxide were used as a control.
Bisulfite sequencing
The bisulfite sequencing assay was performed as previously described.48 The primer sequences of CpG-rich regions of the SPARCL1 promoter were: forward, 5′-GAGGTTTTGAGAATAAGAGTTTGAG-3′ and reverse, 5′-ACAAAAAATAAAAATTTAACAAACACC-3′.
Plasmid construction and transfection
The HA-tagged human SPARCL1 ORF (NM_004684.5) was subcloned into the CD510B-1 (pCDH-CMV-MCS-EF1-Puro) vector. SPARCL1-HA and mock vector were packaged into the virus and titers were determined. Target cells were infected with 1 × 108 lentivirus-transducing units and 6 μg/ml polybrene (Sigma-Aldrich, Shanghai, China). After 72 h, the infected cells were screened in the presence of 2.5 μg/ml puromycin. The qPCR and WB also verified the overexpression efficacy of SPARCL1. CD510B-1 vector was similarly transfected to serve as control.
Cell migration and invasion assay
In the migration assay, 2.5 × 104 cells were seeded into the upper chamber of the transwell plate (Millipore, La Jolla, CA, USA). The cell invasion assay was performed using Matrigel-coated filters (BD, La Jolla, CA, USA). Specific culture medium and recombinant SPARCL1 was added to the bottom chamber. Cells were allowed to migrate for 24 h or invade through the Matrigel for 48 h at 37 °C. The migrated or invaded cells were then fixed and stained with 0.1% crystal violet, six randomly selected fields were photographed, and the cell numbers were counted.
Chemotaxis assay
An in vitro chemotaxis assay was performed using the transwell system. In brief, THP-1 cells (5 × 104) primed using 100 ng/ml PMA (Sigma-Aldrich) were seeded into the upper chamber, and CM (serum-free) from SPARCL1-overexpressing or control cell cultures was placed in the lower chamber. The subsequent steps were identical to those used in the cell migration assay. Specifically, a neutralizing antibody against CCL5 was added to the CM from the SPARCL1-overexpressing cells to elucidate the chemotactic effect.
In vivo metastatic model and bioluminescent imaging
Ten male BALB/C nude mice (4-week-old) were reared and handled in compliance with the animal experimental protocols approved by the East China Normal University Animal Care Commission. For the in vivo metastasis model, the mice were randomly divided into two groups (vector and SPARCL1 groups) and injected with 1.5 × 106 cells via the tail veins. After 30 days, the in vivo tumor metastasis was imaged using bioluminescence. D-Luciferin (Xenogen, Hopkinton, MA, USA) was injected, and the bioluminescence was detected using a lumazone imaging system (MAG Biosystems, Tucson, AZ, USA). All the mice were subsequently killed, the lungs were isolated and the metastatic nodules were counted. The collected lungs were fixed and prepared for histological assessment.
Luciferase reporter gene assay
TOP-flash (TCF reporter plasmid) expression plasmids were purchased from Millipore (Cat. # 21-170). To verify the binding sites of TCF4 and CCL5, luciferase reporter plasmids containing wild type and mutant CCL5 promoters were constructed in the pGL4 plasmid (Supplementary Table 3) for the dual luciferase reporter assay. To further determine whether the WNT/β-catenin pathway stimulates CCL5 expression, a CCL5 dual-luciferase reporter gene plasmid (containing CCL5 promoter of ~1500 bp) was purchased from Asia-Vector Biotechnology Co., Ltd. (Shanghai, China). The cells were cultured in white-bottom 96-well plates and transfected with a mixture of reporter plasmid and Renilla, using the Lipofectamine 3000 transfection reagent (Thermo, USA) according to the manufacturer’s instructions. After 24 h, the cells were lysed in 1 × passive lysis buffer. Firefly and Renilla luciferase activities were measured using the Dual-Glo Luciferase reporter assay system (Promega, Madison, WI, USA), following the recommended protocol.
ELISA
ELISA was performed to detect human CCL5 using CCL5-specific ELISA kits (R&D Systems) following the manufacturer’s protocol.
ChIP assay
The ChIP assay was conducted using the Pierce Agarose ChIP kit (Thermo) following the manufacturer’s instructions. In brief, MNNG-HOS and U-2OS cells were fixed with 1% formaldehyde solution for 10 min at 37 °C, and the fixation reaction was quenched with glycine solution for 5 min. After crosslinking and cell pellet isolation, the lysis buffer and micrococcal nuclease were used to obtain the digested chromatin. Then, 5 μl of the supernatant containing the digested chromatin was transferred into a 1.5-ml tube, and this was the input sample. The corresponding antibody (TCF4-specific antibody or rabbit IgG) was added to the remaining supernatant and incubated for the IP reactions overnight at 4 °C. The IP elution and DNA recovery were performed following the manufacturer’s protocol. The DNA was analyzed by PCR, using the SYBR Green master mix and primers, which are listed in Supplementary Table 3.
Western blotting
Total or nuclear cellular protein was extracted using a protein extraction buffer (Beyotime, Shanghai, China) or nucleoprotein extraction kit (Sangon Biotech, C500009). Proteins were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto a nitrocellulose (NC) membrane. After blocking with 5% skimmed milk, the membranes were incubated with the following specific antibodies: anti-SPARCL1 (1:1000), anti-β-catenin (1:1000), anti-β-actin (1:2000), anti-lamin A/C (1:1000), anti-Wnt3a (1:800), anti-Wnt10b (1:800), anti-Fzd3 (1:100), anti-Fzd6 (1:1000), anti-Fzd8 (1:200), anti-LRP5 (1:200) and anti-LRP6 (1:2000), followed by species-specific secondary antibodies (1:10000). After incubating with the secondary antibodies for 50–60 min, the bands were detected using the Odyssey imaging system (LI-COR, Lincoln, NE, USA).
Co-IP and co-culture IP assays
For the co-IP assay, cell lysates of SPARCL1-HA or vector-transfected HEK293T cells were prepared as mentioned above. Protein A/G Sepharose (Santa Cruz Biotechnology) was preincubated with the anti-HA antibody for 30–60 min on a spinning wheel at 4 °C, followed by two washes. Then, the beads–antibody complex and protein lysate were suspended. All IPs were performed on the spinning wheel at 4 °C overnight. Then, the beads were collected by centrifugation at 3000 g, followed by three washes with lysis buffer. Then, the immunoprecipitates were analyzed using WB, following the steps described above.
For the co-culture IP assay, HEK293T cells (2 × 106) were first cultured in 10-cm dishes and individually transfected with 5 μg expression plasmids for SPARCL1 (SPARCL1-Flag or SPARCL1-myc) or various WNT components (receptor expression plasmids: LRP6-HA or Fzd8-HA or Fzd8CRD-Flag; ligand expression plasmids: Wnt3a-myc or Wnt10b-myc) using Lipofectamine 3000. All the plasmids were purchased from Shanghai Generay Biotech Co., Ltd. (Shanghai, China). After 8 h, the cells were mixed and seeded in six-well plates for the co-culture. To detect the interaction of one WNT receptor (Fzd8 or LRP6) and two ligands (Wnt3a or Wnt10b, or SPARCL1), cells with receptors and those with ligands were mixed in a 1:2:2 ratio. To examine the interaction between two WNT receptors (LRP6 and Fzd8) and SPARCL1, a 1:1:2 ratio was used. After co-culturing for 24 h, cell lysates were prepared as described above. Protein A/G Sepharose preincubated with an anti-HA or anti-Flag antibody was used for the different designed groups. The subsequent steps were identical to those of the IP assay mentioned above. Finally, the NC membranes were probed using anti-HA, anti-myc or anti-Flag monoclonal antibodies.
Gene expression profiling and data analysis
Control MNNG-HOS cells and SPARCL1-overexpressing MNNG-HOS cells (each n=3) were used to compare the gene expression patterns using transcriptomic analysis. The microarray data were generated by Shanghai Biotechnology Corp. In brief, according to the Agilent One-Color microarray-based gene expression analysis protocol (Agilent Technology), the labeled complementary RNA (cRNA) was purified using the RNeasy mini kit (Qiagen, Germantown, MD, USA). Hybridization was performed on the human whole genome 8 × 60 K oligonucleotide microarrays (GeneChip, Agilent), using reagents and protocols provided by the manufacturer. The array data were uploaded to the Gene Expression Omnibus database (GSE97572). The data analysis were performed using the statistical software R. DEGs were defined as genes presenting both an average fold change >2.0 and a P<0.05. Moreover, all the DEGs were subjected to GO (geneontology.org) and KEGG (www.genome.jp/kegg) analyses.
Statistical analysis
Data are shown as means±s.d. GraphPad Prism 5 was used to manipulate statistical analyses. Correlation of SPARCL1 expression with categorical clinical variables in patients with OS was done using χ2-test (SPSS 19.0 statistical software). After testing the homogeneity of variance, two-tailed Student’s t-test was used to compare the results from different groups. Spearman rank correlation test was used to analyze the correlation between SPARCL1 and HLA-DR+ macrophages. Values of P<0.05 were considered statistically significant.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its Supplementary Information files.
Acknowledgments
We are grateful to Dr Xiao-Li Zhou, Dr Xiao-Mei Yang, Dr Qin Yang, Dr Lei Zhu, Dr Li-Peng Hu and and Dr Xiao-Yan Cao for assistance with our experiments. This study was supported by grants from the Major Program of Changzhou Health Bureau (Grant number CJ20160047), the Nineteenth Batch of Science Technology Program of Changzhou (Grant number 2014YLC011) and the H-level Medical Talents Training Project (Grant number 2016CZBJ023).
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Hingorani P, Janeway K, Crompton BD, Kadoch C, Mackall CL, Khan J et al. Current state of pediatric sarcoma biology and opportunities for future discovery: a report from the sarcoma translational research workshop. Cancer Genet 2016; 209: 182–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nat Rev Cancer 2014; 14: 722–735. [DOI] [PubMed] [Google Scholar]
- Adamopoulos C, Gargalionis AN, Basdra EK, Papavassiliou AG. Deciphering signaling networks in osteosarcoma pathobiology. Exp Biol Med 2016; 241: 1296–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N, Song WX, Luo J, Haydon RC, He TC. Osteosarcoma development and stem cell differentiation. Clin Orthop Relat Res 2008; 466: 2114–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer 2009; 115: 1531–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell 2012; 149: 1192–1205. [DOI] [PubMed] [Google Scholar]
- Niehrs C. The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol 2012; 13: 767–779. [DOI] [PubMed] [Google Scholar]
- Canalis E. Wnt signalling in osteoporosis: mechanisms and novel therapeutic approaches. Nat Rev Endocrinol 2013; 9: 575–583. [DOI] [PubMed] [Google Scholar]
- Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene 2017; 36: 1461–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matushansky I, Hernando E, Socci ND, Mills JE, Matos TA, Edgar MA et al. Derivation of sarcomas from mesenchymal stem cells via inactivation of the Wnt pathway. J Clin Invest 2007; 117: 3248–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibault L, Perot G, Chibon F, Bonnin S, Lagarde P, Terrier P et al. New insights in sarcoma oncogenesis: a comprehensive analysis of a large series of 160 soft tissue sarcomas with complex genomics. J Pathol 2011; 223: 64–71. [DOI] [PubMed] [Google Scholar]
- Chen EY, DeRan MT, Ignatius MS, Grandinetti KB, Clagg R, McCarthy KM et al. Glycogen synthase kinase 3 inhibitors induce the canonical WNT/beta-catenin pathway to suppress growth and self-renewal in embryonal rhabdomyosarcoma. Proc Natl Acad Sci USA 2014; 111: 5349–5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancsok AR, Asleh-Aburaya K, Nielsen TO. Advances in sarcoma diagnostics and treatment. Oncotarget 2017; 8: 7068–7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Mohseny AB, Karperien M, Hogendoorn PC, Zhou G, Cleton-Jansen AM. Inactive Wnt/beta-catenin pathway in conventional high-grade osteosarcoma. J Pathol 2010; 220: 24–33. [DOI] [PubMed] [Google Scholar]
- Basu-Roy U, Seo E, Ramanathapuram L, Rapp TB, Perry JA, Orkin SH et al. Sox2 maintains self renewal of tumor-initiating cells in osteosarcomas. Oncogene 2012; 31: 2270–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu-Roy U, Basilico C, Mansukhani A. Perspectives on cancer stem cells in osteosarcoma. Cancer Lett 2013; 338: 158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Zhao W, Jiang Y, Liu D, Meng G, Cai Y. beta-catenin is a valuable marker for differential diagnosis of osteoblastoma and osteosarcoma. Hum Pathol 2014; 45: 1459–1465. [DOI] [PubMed] [Google Scholar]
- Du X, Yang J, Yang D, Tian W, Zhu Z. The genetic basis for inactivation of Wnt pathway in human osteosarcoma. BMC Cancer 2014; 14: 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein SD, Trucco M, Bautista Guzman W, Hayashi M, Loeb DM. A monoclonal antibody against the Wnt signaling inhibitor dickkopf-1 inhibits osteosarcoma metastasis in a preclinical model. Oncotarget 2016; 7: 21114–21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Kurenbekova L, Gao Y, Roos A, Creighton CJ, Rao P et al. NKD2, a negative regulator of Wnt signaling, suppresses tumor growth and metastasis in osteosarcoma. Oncogene 2015; 34: 5069–5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro N, Sousa SR, Brekken RA, Monteiro FJ. Role of SPARC in bone remodeling and cancer-related bone metastasis. J Cell Biochem 2014; 115: 17–26. [DOI] [PubMed] [Google Scholar]
- Sullivan MM, Sage EH. Hevin/SC1, a matricellular glycoprotein and potential tumor-suppressor of the SPARC/BM-40/Osteonectin family. Int J Biochem Cell Biol 2004; 36: 991–996. [DOI] [PubMed] [Google Scholar]
- Bradshaw AD. Diverse biological functions of the SPARC family of proteins. Int J Biochem Cell Biol 2012; 44: 480–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MM, Puolakkainen PA, Barker TH, Funk SE, Sage EH. Altered tissue repair in hevin-null mice: inhibition of fibroblast migration by a matricellular SPARC homolog. Wound Repair Regen 2008; 16: 310–319. [DOI] [PubMed] [Google Scholar]
- Hurley PJ, Hughes RM, Simons BW, Huang J, Miller RM, Shinder B et al. Androgen-regulated SPARCL1 in the tumor microenvironment inhibits metastatic progression. Cancer Res 2015; 75: 4322–4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naschberger E, Liebl A, Schellerer VS, Schutz M, Britzen-Laurent N, Kolbel P et al. Matricellular protein SPARCL1 regulates tumor microenvironment-dependent endothelial cell heterogeneity in colorectal carcinoma. J Clin Invest 2016; 126: 4187–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Zhang H, Ge W, Liu X, Loera S, Chu P et al. Secreted protein acidic and rich in cysteines-like 1 suppresses aggressiveness and predicts better survival in colorectal cancers. Clin Cancer Res 2012; 18: 5438–5448. [DOI] [PubMed] [Google Scholar]
- Claeskens A, Ongenae N, Neefs JM, Cheyns P, Kaijen P, Cools M et al. Hevin is down-regulated in many cancers and is a negative regulator of cell growth and proliferation. Br J Cancer 2000; 82: 1123–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz MB, Sowers R, Brown KM, Hilmer SC, Mazza B, Huvos AG et al. An expression signature classifies chemotherapy-resistant pediatric osteosarcoma. Cancer Res 2005; 65: 1748–1754. [DOI] [PubMed] [Google Scholar]
- Perry JA, Kiezun A, Tonzi P, Van Allen EM, Carter SL, Baca SC et al. Complementary genomic approaches highlight the PI3K/mTOR pathway as a common vulnerability in osteosarcoma. Proc Natl Acad Sci USA 2014; 111: E5564–E5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovac M, Blattmann C, Ribi S, Smida J, Mueller NS, Engert F et al. Exome sequencing of osteosarcoma reveals mutation signatures reminiscent of BRCA deficiency. Nat Commun 2015; 6: 8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren DN, Chen J, Li Z, Yan H, Yin Y, Wo D et al. LRP5/6 directly bind to Frizzled and prevent Frizzled-regulated tumour metastasis. Nat Commun 2015; 6: 6906. [DOI] [PubMed] [Google Scholar]
- Tamai K, Semenov M, Kato Y, Spokony R, Liu C, Katsuyama Y et al. LDL-receptor-related proteins in Wnt signal transduction. Nature 2000; 407: 530–535. [DOI] [PubMed] [Google Scholar]
- Berraondo P, Minute L, Ajona D, Corrales L, Melero I, Pio R. Innate immune mediators in cancer: between defense and resistance. Immunol Rev 2016; 274: 290–306. [DOI] [PubMed] [Google Scholar]
- Chanmee T, Ontong P, Konno K, Itano N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers 2014; 6: 1670–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Bae JS. Tumor-associated macrophages and neutrophils in tumor microenvironment. Mediators Inflamm 2016; 2016: 6058147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao NB, Lu MH, Fan YH, Cao YL, Zhang ZR, Yang SM. Macrophages in tumor microenvironments and the progression of tumors. Clin Dev Immunol 2012; 2012: 948098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halleskog C, Mulder J, Dahlstrom J, Mackie K, Hortobagyi T, Tanila H et al. WNT signaling in activated microglia is proinflammatory. Glia 2011; 59: 119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj D, Nager M, Camats J, David M, Benguria A, Dopazo A et al. Chemokines induce axon outgrowth downstream of hepatocyte growth factor and TCF/beta-catenin signaling. Front Cell Neurosci 2013; 7: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J, Sage EH. SPARC inhibits adipogenesis by its enhancement of beta-catenin signaling. J Biol Chem 2009; 284: 1279–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palma M, Lewis CE. Macrophage regulation of tumor responses to anticancer therapies. Cancer Cell 2013; 23: 277–286. [DOI] [PubMed] [Google Scholar]
- Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol 2011; 11: 723–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanganeh S, Hutter G, Spitler R, Lenkov O, Mahmoudi M, Shaw A et al. Iron oxide nanoparticles inhibit tumour growth by inducing pro-inflammatory macrophage polarization in tumour tissues. Nat Nanotechnol 2016; 11: 986–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity 2014; 41: 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers PA, Schwartz CL, Krailo MD, Healey JH, Bernstein ML, Betcher D et al. Osteosarcoma: the addition of muramyl tripeptide to chemotherapy improves overall survival—a report from the Children's Oncology Group. J Clin Oncol 2008; 26: 633–638. [DOI] [PubMed] [Google Scholar]
- Buddingh EP, Kuijjer ML, Duim RA, Burger H, Agelopoulos K, Myklebost O et al. Tumor-infiltrating macrophages are associated with metastasis suppression in high-grade osteosarcoma: a rationale for treatment with macrophage activating agents. Clin Cancer Res 2011; 17: 2110–2119. [DOI] [PubMed] [Google Scholar]
- Chiodoni C, Colombo MP, Sangaletti S. Matricellular proteins: from homeostasis to inflammation, cancer, and metastasis. Cancer Metastasis Rev 2010; 29: 295–307. [DOI] [PubMed] [Google Scholar]
- Fu Y, Feng MX, Yu J, Ma MZ, Liu XJ, Li J et al. DNA methylation-mediated silencing of matricellular protein dermatopontin promotes hepatocellular carcinoma metastasis by alpha3beta1 integrin-Rho GTPase signaling. Oncotarget 2014; 5: 6701–6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZG, Lambert CA, Servotte S, Chometon G, Eckes B, Krieg T et al. Effects of constitutively active GTPases on fibroblast behavior. Cell Mol Life Sci 2006; 63: 82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper and its Supplementary Information files.