Abstract
Objective
To investigate a targeted set of biochemical biomarkers as predictors of clinically relevant osteoarthritis (OA) progression.
Methods
Eighteen biomarkers were measured at baseline, 12 months (M) and 24 M in serum (s) and/or urine (u) of cases (n=194) from the OA initiative cohort with knee OA and radiographic and persistent pain worsening from 24 to 48 M and controls (n=406) not meeting both end point criteria. Primary analyses used multivariable regression models to evaluate the association between biomarkers (baseline and time-integrated concentrations (TICs) over 12 and 24 M, transposed to z values) and case status, adjusted for age, sex, body mass index, race, baseline radiographic joint space width, Kellgren-Lawrence grade, pain and pain medication use. For biomarkers with adjusted p<0.1, the c-statistic (area under the curve (AUC)), net reclassification index and the integrated discrimination improvement index were used to further select for hierarchical multivariable discriminative analysis and to determine the most predictive and parsimonious model.
Results
The 24 M TIC of eight biomarkers significantly predicted case status (ORs per 1 SD change in biomarker): sCTXI 1.28, sHA 1.22, sNTXI 1.25, uC2C-HUSA 1.27, uCTXII, 1.37, uNTXI 1.29, uCTXIα 1.32, uCTXIβ 1.27. 24 M TIC of uCTXII (1.47–1.72) and uC2C-Human Urine Sandwich Assay (HUSA) (1.36–1.50) both predicted individual group status (pain worsening, joint space loss and their combination). The most predictive and parsimonious combinatorial model for case status consisted of 24 M TIC uCTXII, sHA and sNTXI (AUC 0.667 adjusted). Baseline uCTXII and uCTXIα both significantly predicted case status (OR 1.29 and 1.20, respectively).
Conclusions
Several systemic candidate biomarkers hold promise as predictors of pain and structural worsening of OA.
INTRODUCTION
A cure for osteoarthritis (OA) remains elusive and the management of OA is largely palliative. The progression of knee OA is highly variable and difficult to predict by currently available clinical and imaging measures.1 At present, there are no disease-modifying OA drugs approved by the US Food and Drug Administration (FDA) or European Medicines Agency. The low responsiveness to change and at most, moderate correlation of radiographic data with clinical end points,2 represents one of the main obstacles to efficient development of structure-modifying therapies for OA. Although there are no FDA-qualified OA-related soluble biomarkers, there are many that have shown associations with some aspect of OA.1,3 The biomarkers selected for this project were agreed upon through consensus of the Osteoarthritis Research Society International (OARSI)/FDA Biomarkers Working Group.1 These biomarkers of bone and cartilage turnover, representing the best-qualified OA-related biomarkers to date, have all fulfilled one or more of the BIPEDS categories1 corresponding to Burden of disease, Investigational, Prognostic, Efficacy of Intervention, Diagnostic and Safety biomarkers, but few of them have been directly compared in the same sample set.
The objectives of this study were to investigate a targeted set of 18 biochemical biomarkers in serum (s) and/or urine (u) for their ability to predict knee OA-related radiographic and persistent pain progression over the 24–48 months (M) follow-up compared with baseline. The sample and data available through the osteoarthritis initiative (OAI) provided a unique opportunity to assess this strategy and to compare single and combinatorial uses of biomarker concentrations at baseline and time-integrated concentrations (TICs) over 12 and 24M. The ultimate goals of this work are to provide new drug development tools to effectively enrich OA clinical trials with faster progressors and to qualify biomarker candidates that might serve as surrogates of efficacy in OA trials.
PATIENTS AND METHODS
Study design
This nested case–control study (194 cases and 406 OA comparators) used data from the OAI (http://www.oai.ucsf.edu/) (see online supplementary text for further description). Details of the study design had been previously published.2,4
Definitions of radiographic and symptomatic progression
Two main outcome groups, with one study knee per subject, were defined: case knees (n=194) with clinically relevant (both radiographic and pain) progression and comparator OA knees lacking the combination of radiographic and pain progression (n=406). Comparator knees consisted of three subgroups: those with radiographic progression but not pain progression in either the study or contralateral knee (n=103); pain progression but not radiographic progression in either knee (n=103) and OA non-progressors (n=200) with no radiographic and no pain progression in either knee.
Biomarker assays
The general biological processes represented by the 18 chosen biomarkers (counting each analyte in each fluid as one biomarker) are listed in table 1 and include catabolic and anabolic processes of cartilage and bone. Several biomarkers were quantified in both serum and urine. Biomarkers analysed in serum alone were cartilage oligomeric matrix protein (COMP), hyaluronan (HA), C-propeptide of type II collagen, N-terminal propeptide of collagen IIA (PIIANP), chondroitin sulfate 846 epitope, the C-terminal crosslinked telopeptide of type I collagen (CTXI) and matrix metalloproteinase 3. Biomarkers analysed in serum and urine were Col2-3/4 C-terminal cleavage product of types I and II collagen (C1, 2C), Col2-3/4 C-terminal cleavage product of human type II collagen (C2C competitive assay in serum, C2C-HUSA sandwich assay in urine), nitrated epitope of the α-helical region of type II collagen (Coll2-1 NO2) and the crosslinked N-telopeptide of type I collagen (NTXI). Biomarkers analysed in urine alone were the C-terminal crosslinked telopeptide type II collagen (CTXII), and alpha and beta isomerised versions of the CTXI (CTXIα and CTXIβ).
Table 1.
Concentrations and performance of the panel of soluble biomarkers analysed for the FNIH OA Biomarkers Consortium project
| Biomarkers (units) | Comparators Baseline mean (SD) Median (range) (n=406) |
Cases Baseline mean (SD) Median (range) (n=194) |
CV (%) |
Number of samples below LLOQ (out of n=1785) |
Biological process indicated | Manufacturer (catalogue #) |
|---|---|---|---|---|---|---|
| Serum C1, 2C (µg/mL) | 0.38 (0.14) | 0.39 (0.15) | 23.3 | 3 | Types I and II collagen degradation | IBEX (60-1002-001) |
| 0.37 (0.04–1.34) | 0.38 (0.05–1.09) | |||||
|
| ||||||
| Serum-C2C (ng/ml) | 208.9 (46.8) | 212.0 (54.9) | 11.7 | 0 | Type II collagen degradation | IBEX (60-1001-001) |
| 202.0 (100.0–395.0) | 204.0 (102.0–423.0) | |||||
|
| ||||||
| Serum Coll2-1 NO2 (nM) | 8.93 (5.45) | 8.91 (4.99) | 13.6 | 19 | Type II collagen degradation and inflammation | Artialis (ACP022) |
| 7.97 (0.00–45.17) | 8.20 (0.00–37.98) | |||||
|
| ||||||
| Serum CPII (ng/mL) | 945.7 (391.0) | 944.2 (363.2) | 12.2 | 0 | Type II collagen synthesis | IBEX (60-1003-001) |
| 889.0 (252.0–4063.0) | 894.0 (204.0–3006.0) | |||||
|
| ||||||
| Serum CS846 (ng/mL) | 76.8 (52.8) | 79.2 (60.2) | 16.8 | 1354 | Cartilage aggrecan synthesis/turnover | IBEX (60-1004) |
| 66.0 (1.0–383.0) | 65.0 (0.0–412.0) | |||||
|
| ||||||
| Serum CTX1 (ng/mL) | 0.39 (0.22) | 0.42 (0.21) | 4.9 | 123 | Bone resorption | IDS (AC-02F1) |
| 0.34 (0.08–1.79) | 0.39 (0.07–1.23) | |||||
|
| ||||||
| Serum COMP (ng/mL) | 783.4 (302.5) | 761.1 (280.1) | 5.2 | 5 | Cartilage degradation | BioVendor (RD194080200) |
| 739.0 (156.0–2007.0) | 705.0 (168.0–1903.0) | |||||
|
| ||||||
| Serum HA (ng/mL) | 45.2 (40.2) | 49.5 (36.5) | 7.4 | 1170 | Osteophyte burden, synovitis | Corgenix (029-001) |
| 34.0 (3.0–297.0) | 38.0 (4.0–193.0) | |||||
|
| ||||||
| Serum MMP3 (ng/mL) | 16.9 (11.2) | 17.5 (11.3) | 9.6 | 27 | Total (active and inactive) metalloprotease involved with joint tissue degradation | Invitrogen (Life Technologies) (KAC1541) |
| 14.6 (0.0–99.9) | 14.4 (1.0–76.3) | |||||
|
| ||||||
| Serum NTXI (nm BCE) | 14.8 (4.8) | 15.5 (4.9) | 7.01 | 8 | Bone resorption | ALERE -Osteomark (Inverness Medical) (9021) |
| 14.0 (3.0–43.0) | 15.0 (4.0–28.0) | |||||
|
| ||||||
| Serum PIIANP (ng/mL) | 2677.2 (752.5) | 2581.1 (783.7) | 12.3 | 0 | Type II collagen synthesis | Merck Group/Millipore (EZPIIANP-53K) |
| 2626.5 (160.0–5656.0) | 2524.0 (781.0–5219.0) | |||||
|
| ||||||
| Urine Col2-1 NO2 (nM/mmol Cr) | 0.024 (0.015) | 0.025 (0.014) | 9.3 | 659 | Type II collagen degradation and inflammation | Artialis (ACP021) |
| 0.020 (0.003–0.108) | 0.021 (0.002–0.120) | |||||
|
| ||||||
| Urine C1, 2C (ng/mmol Cr) | 0.011 (0.011) | 0.010 (0.011) | 21.5 | 805 | Types I and II collagen degradation | IBEX (60-1002-001) |
| 0.009 (0.000–0.063) | 0.007 (0.000–0.045) | |||||
|
| ||||||
| Urine C2C-HUSA (ng/mmol Cr) | 152.7 (90.3) | 165.6 (103.0) | 6.24 | 104 | Type II collagen degradation | IBEX (60-1017) |
| 137.9 (0.0–763.3) | 149.1 (0.0–695.1) | |||||
|
| ||||||
| Urine CTXII (ng/mmol Cr) | 287.7 (190.7) | 333.1 (210.5) | 5.21 | 271 | Type II collagen degradation | IDS (AC-10F1) |
| 233.5 (0.0–1794.1) | 283.7 (59.4–1446.4) | |||||
|
| ||||||
| Urine NTXI (nM BCE/mmol Cr) | 32.6 (18.2) | 34.9 (16.6) | 3.03 | 16 | Bone resorption | ALERE -Osteomark (Inverness Medical) (9006) |
| 29.1 (6.2–152.0) | 32.1 (8.6–116.4) | |||||
|
| ||||||
| Urine CTXIα (µg/mmol Cr) | 0.41 (0.33) | 0.46 (0.33) | 3.91 | 961 | Turnover of newly formed bone | IDS (AC-04F1) |
| 0.34 (0.00–2.76) | 0.39 (0.00–2.56) | |||||
|
| ||||||
| Urine CTX1 β (µg/mmol Cr) | 2.19 (1.67) | 2.34 (1.61) | 7.67 | 465 | bone resorption | IDS (AC-05F1) |
| 1.79 (0.00–11.08) | 2.04 (0.00–7.98) | |||||
|
| ||||||
| Urine creatinine (mmol/L) | 7.32 (5.23) | 6.94 (5.20) | 3.02 | 0 | Normalization analyte for urinary biomarkers | Quidel MicroVue (8009) |
| 6.00 (0.30–33.90) | 5.50 (0.40–25.50) | |||||
Urine biomarkers are normalised to urinary creatinine.
BCE, bone collagen equivalents; COMP, cartilage oligomeric matrix protein; CPII, C-propeptide of type II collagen; Cr, creatinine; CS846, chondroitin sulfate 846 epitope; CTXI, C-terminal crosslinked telopeptide of type I collagen; CTXII, C-terminal crosslinked telopeptide type II collagen; CV, interassay coefficient of variation; FNIH, Foundations for National Institutes of Health; HA, hyaluronan; LLOQ, lower limit of quantification; NTXI, N-telopeptide of type I collagen; OA, osteoarthritis.
All biomarker assays were performed by LabCorp Clinical Trials, a Clinical Laboratory Improvement Amendments (CLIA) and College of American Pathologists (CAP) certified division within LabCorp, with the exception of urine Col2-1 NO2, which was measured by Artialis, a Good Laboratory Practice-certified facility. All assays were run blinded to the clinical information. The biochemical markers measured in this study, the kit manufacturers and catalogue numbers and reported lower limits of quantification are listed in table 1. All biomarker data are freely available on the OAI website (https://oai.epi-ucsf.org/datarelease/).
Statistical analysis
Due to the known dynamic nature of OA-related biochemical markers,5,6 their concentrations were expressed in terms of TICs over 12 and 24 M from baseline; these measures are equivalent to the area under the curve defined by the individual values for the specific time interval. TICs were chosen over change scores to provide a means of evaluating the longitudinal information content of the biomarkers that overcomes the difficulty inherent in analysing dynamic biomarker data and combines the information contained in the baseline value. Each measure was transposed to z values (created by subtracting the biomarker value for a subject from the total group mean and dividing by the SD) in order for 1 unit of change to be comparable across the biomarkers.
The prespecified primary analysis evaluated the ability of biomarker concentrations (baseline and TICs) to predict case status (n=194 cases with pain and radiographic joint space loss (JSL) progression vs n=406 comparator knees lacking the combination of pain and radiographic progression over 48 M). The prespecified secondary analysis compared each of the three progressor subgroups (combination pain and JSL progression, pain only or JSL only progression) with the non-progressor (neither radiographic nor pain progressor) reference group. Models were adjusted for the following covariates: age, sex, body mass index (BMI), race, baseline radiographic joint space width, Kellgren-Lawrence grade, pain and pain medication use. The discriminative ability was assessed using the c-statistic (area under the curve (AUC)), category-less net reclassification index (NRI) and the integrated discrimination improvement (IDI) index,7,8 comparing the biomarker plus covariates model to the covariates-only model. We created a 5-level categorical variable to evaluate the dose–response relationship between the biomarker and risk of case status; categories were based on z-score-based deviation from the mean. Finally, we built hierarchical multivariable models for the best urine biomarkers, serum biomarkers and combination of serum and urine biomarkers.
RESULTS
Primary analysis
Cases and comparators were well-matched by age, sex, BMI, Kellgren-Lawrence grade of baseline knee OA severity and baseline Western Ontario and McMaster Universities Arthritis Index (WOMAC) pain (table 2). Three configurations of biomarker concentrations were analysed for the primary analysis: baseline biomarkers and the TIC from baseline to 12 M and from baseline to 24 M (table 3). Several serum and urine biomarkers yielded statistically significant associations with case status. Baseline, 12 and 24 M TICs of uCTXII, indicative of type II collagen degradation, were all associated with case status; the highest OR for any single biomarker predictor of case status was provided by 24 M TIC of uCTXII (OR 1.37, 95% CIs 1.15 to 1.65, p=0.0006). Twelve and 24 M TICs of sCTXI, sNTXI and uNTXI, all indicative of type I collagen degradation and bone resorption, were positively associated with case status. We also evaluated two isomerised forms of CTXI, uCTXIα and uCTXIβ, indicative of turnover of newly synthesised type I collagen versus turnover of older bone, respectively.9 Concentrations of both urinary CTXI isoforms over 12 and 24 M yielded comparable results to sCTXI for prediction of case status. However, in contrast to sCTXI and uCTXIβ, uCTXIα at baseline also significantly predicted case status.
Table 2.
Characteristics of study participants and association of baseline biomarker concentrations with covariates for the whole cohort
| Covariates | Comparators (n=406) | Cases (n=194) | Biomarker associations with covariates* |
|---|---|---|---|
| Age, mean years (SD) | 61.3 (8.9) | 62.0 (8.8) | sCOMP, sHA, sMMP3, uC2C-HUSA, uC1, 2C, uCTXII |
| Sex, n (%) female | 243 (60%) | 110 (57%) | sC1, 2C, sCS846, sCTXI, s+uNTXI, sMMP3, uCol2-1 NO2, uC1, 2C, uC2C-HUSA, uCTXII, uCTXIα, uCTXIβ |
| BMI, mean kg/m2 (SD) | 30.7 (4.8) | 30.7 (4.8) | sCTXI, sMMP3, sPIIANP, uNTXI, uCTXIα |
| History of knee injury, n (%) yes | 145 (36%) | 68 (35%) | |
| Baseline knee Kellgren-Lawrence grade, n (%) | |||
| KL1 | 51 (13%) | 24 (12%) | |
| KL2 | 222 (55%) | 84 (43%) | |
| KL3 | 133 (33%) | 86 (44%) | |
| White race, n (%) | 320 (79%) | 1 55 (80%) | sC1, 2C, s+uCol2-1 NO2, sCTXI, sHA, uC2C-HUSA |
| Baseline use of pain medication, n (%) yes | 114 (28%) | 63 (32%) | sCPII, uCol2-1 NO2, uCTXII, uNTXI, uCTXIα |
| Baseline WOMAC pain score, mean (SD) | 13.0 (16.7) | 10.2 (13.0) | sPIIANP, uCTXII |
| Baseline joint space width, mean mm (SD) | 3.9 (1.1) | 3.8 (1.4) | sHA, uC2C-HUSA |
Detailed results of associations of biomarkers with covariates are provided in online supplementary table S2.
WOMAC, Western Ontario and McMaster Universities Arthritis Index.
Table 3.
Prediction of case status (primary analysis) at 48 M by biomarker concentrations at baseline and by TICs of biomarkers over 12 and 24 M
| Biomarker | Baseline concentration | 12 M TIC | 24 M TIC | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| Mean (SD) median z score | OR (95% CI) p Value |
Mean (SD) median z score | OR (95% CI) p Value |
Mean (SD) median z score | OR (95% CI) p Value |
||||
|
|
|
|
|||||||
| Comparators | Cases | Comparators | Cases | Comparators | Cases | ||||
| Serum C1, 2C | −0.03 (0.98) | 0.06 (1.04) | 1.09 (0.91 to 1.30) | −0.01 (1.00) | 0.02 (1.00) | 1.02 (0.85 to 1.22) | −0.02 (1.01) | 0.05 (0.98) | 1.07 (0.89 to 1.27) |
| −0.09 | −0.02 | 0.3510 | −0.04 | −0.04 | 0.8354 | −0.15 | −0.01 | 0.4686 | |
|
| |||||||||
| Serum C2C | −0.02 (0.94) | 0.04 (1.11) | 1.05 (0.88 to 1.26) | −0.01 (0.98) | 0.02 (1.05) | 1.00 (0.83 to 1.20) | −0.03 (0.97) | 0.05 (1.05) | 1.07 (0.89 to 1.28) |
| −0.16 | −0.12 | 0.5772 | −0.12 | −0.09 | 0.9940 | −0.07 | −0.03 | 0.4779 | |
|
| |||||||||
| Serum Coll2-1 NO2 | 0.00 (1.03) | −0.00 (0.94) | 1.00 (0.83 to 1.21) | 0.01 (1.02) | −0.01 (0.96) | 0.98 (0.80 to 1.19) | −0.00 (1.02) | 0.00 (0.95) | 1.02 (0.85 to 1.23) |
| −0.18 | −0.13 | 0.9601 | −0.19 | −0.18 | 0.8195 | −0.23 | −0.15 | 0.8238 | |
|
| |||||||||
| Serum CPII | 0.00 (1.02) | −0.00 (0.95) | 0.98 (0.81 to 1.18) | 0.01 (1.00) | −0.02 (1.00) | 0.95 (0.78 to 1.16) | 0.01 (1.02) | −0.01 (0.95) | 0.98 (0.81 to 1.19) |
| −0.15 | −0.13 | 0.8017 | −0.11 | −0.18 | 0.6212 | −0.14 | −0.18 | 0.8179 | |
|
| |||||||||
| Serum CS846 | −0.01 (0.96) | 0.03 (1.09) | 1.06 (0.89 to 1.25) | −0.01 (0.95) | 0.03 (1.11) | 1.06 (0.89 to 1.26) | −0.02 (0.93) | 0.04 (1.13) | 1.07 (0.90 to 1.26) |
| −0.21 | −0.23 | 0.5286 | −0.19 | −0.26 | 0.5024 | −0.20 | −0.25 | 0.4560 | |
|
| |||||||||
| Serum CTXI | −0.05 (1.01) | 0.10 (0.96) | 1.18 (0.99 to 1.41) | −0.07 (0.98) | 0.16 (1.04) | 1.29 (1.08 to 1.54) | −0.07 (0.97) | 0.15 (1.05) | 1.28 (1.08 to 1.53) |
| −0.28 | −0.04 | 0.0583 | −0.24 | 0.02 | 0.0057 | −0.25 | 0.02 | 0.0051 | |
|
| |||||||||
| Serum COMP | 0.02 (1.02) | −0.05 (0.95) | 0.89 (0.74 to 1.07) | 0.02 (1.02) | −0.05 (0.95) | 0.90 (0.74 to 1.09) | 0.02 (1.03) | −0.04 (0.94) | 0.91 (0.76 to 1.09) |
| −0.13 | −0.24 | 0.2254 | −0.10 | −0.26 | 0.2687 | −0.07 | −0.25 | 0.3083 | |
|
| |||||||||
| Serum HA | −0.04 (1.03) | 0.07 (0.93) | 1.07 (0.89 to 1.29) | −0.06 (1.00) | 0.12 (1.00) | 1.18 (0.98 to 1.44) | −0.06 (0.98) | 0.13 (1.02) | 1.22 (1.01 to 1.48) |
| −0.32 | −0.22 | 0.4466 | −0.35 | −0.14 | 0.0877 | −0.36 | −0.12 | 0.0415 | |
|
| |||||||||
| Serum MMP3 | −0.02 (1.00) | 0.03 (1.00) | 0.99 (0.81 to 1.22) | −0.03 (0.99) | 0.07 (1.03) | 1.06 (0.86 to 1.31) | −0.04 (0.99) | 0.08 (1.02) | 1.09 (0.88 to 1.34) |
| −0.22 | −0.24 | 0.9416 | −0.22 | −0.15 | 0.5978 | −0.22 | −0.14 | 0.4345 | |
|
| |||||||||
| Serum NTXI | −0.05 (0.99) | 0.10 (1.01) | 1.18 (0.99 to 1.41) | −0.07 (0.97) | 0.16 (1.05) | 1.28 (1.07 to 1.53) | −0.07 (0.97) | 0.14 (1.05) | 1.25 (1.05 to 1.48) |
| −0.22 | −0.01 | 0.0591 | −0.21 | 0.00 | 0.0064 | −0.20 | −0.09 | 0.0131 | |
|
| |||||||||
| Serum PIIANP | 0.04 (0.99) | −0.09 (1.03) | 0.88 (0.74 to 1.06) | 0.06 (0.98) | −0.13 (1.04) | 0.83 (0.69 to 1.00) | 0.04 (0.96) | −0.08 (1.07) | 0.89 (0.75 to 1.07) |
| −0.03 | −0.16 | 0.1729 | 0.09 | −0.18 | 0.0490 | 0.08 | −0.10 | 0.2102 | |
|
| |||||||||
| Urine Coll2-1 NO2 creatinine adjusted | −0.02 (1.02) | 0.03 (0.96) | 1.05 (0.88 to 1.24) | −0.03 (1.01) | 0.06 (0.98) | 1.10 (0.92 to 1.31) | −0.03 (1.01) | 0.07 (0.97) | 1.11 (0.93 to 1.31) |
| −0.27 | −0.20 | 0.6075 | −0.29 | −0.12 | 0.2878 | −0.28 | −0.13 | 0.2384 | |
|
| |||||||||
| Urine C1, 2C creatinine adjusted | 0.03 (1.02) | −0.06 (0.96) | 0.91 (0.76 to 1.09) | 0.01 (1.02) | −0.03 (0.96) | 0.97 (0.80 to 1.16) | 0.00 (1.02) | −0.01 (0.97) | 0.99 (0.83 to 1.19) |
| −0.14 | −0.30 | 0.3166 | −0.11 | −0.09 | 0.7105 | −0.11 | −0.10 | 0.9339 | |
|
| |||||||||
| Urine C2C-HUSA creatinine adjusted | −0.04 (0.95) | 0.09 (1.09) | 1.12 (0.94 to 1.34) | −0.05 (0.98) | 0.11 (1.04) | 1.16 (0.96 to 1.39) | −0.08 (0.94) | 0.16 (1.11) | 1.27 (1.06 to 1.53) |
| −0.20 | −0.08 | 0.2030 | −0.25 | −0.05 | 0.1231 | −0.25 | −0.06 | 0.0108 | |
|
| |||||||||
| Urine CTXII creatinine adjusted | −0.07 (0.96) | 0.15 (1.06) | 1.29 (1.08 to 1.55) | −0.08 (0.98) | 0.19 (1.02) | 1.35 (1.12 to 1.62) | −0.09 (0.98) | 0.19 (1.01) | 1.37 (1.15 to 1.65) |
| −0.35 | −0.09 | 0.0049 | −0.30 | 0.03 | 0.0015 | −0.31 | 0.03 | 0.0006 | |
|
| |||||||||
| Urine NTXI creatinine adjusted | −0.04 (1.03) | 0.09 (0.94) | 1.17 (0.98 to 1.39) | −0.06 (1.00) | 0.12 (0.98) | 1.24 (1.03 to 1.49) | −0.07 (1.00) | 0.14 (0.98) | 1.29 (1.08 to 1.54) |
| −0.24 | −0.07 | 0.0842 | −0.18 | −0.11 | 0.0199 | −0.22 | −0.03 | 0.0057 | |
|
| |||||||||
| Urine CTXIa creatinine adjusted | −0.05 (0.99) | 0.11 (1.01) | 1.20 (1.01 to 1.43) | −0.07 (0.98) | 0.15 (1.03) | 1.28 (1.07 to 1.53) | −0.08 (0.97) | 0.17 (1.04) | 1.32 (1.11 to 1.58) |
| −0.26 | −0.10 | 0.0364 | −0.27 | −0.02 | 0.0065 | −0.30 | 0.04 | 0.0020 | |
|
| |||||||||
| Urine CTXIp creatinine adjusted | −0.03 (1.01) | 0.06 (0.97) | 1.14 (0.96 to 1.36) | −0.06 (0.97) | 0.13 (1.06) | 1.27 (1.06 to 1.52) | −0.06 (0.95) | 0.13 (1.08) | 1.27 (1.06 to 1.51) |
| −0.27 | −0.12 | 0.1408 | −0.27 | −0.11 | 0.0099 | −0.27 | −0.09 | 0.0086 | |
Odds shown per 1 SD increase in biomarker.
Values in bold represent associations of p≤0.05.
OR adjusted for age, sex, body mass index, race, baseline joint space width, Kellgren-Lawrence grade, baseline pain and pain medication use.
COMP, cartilage oligomeric matrix protein; CPII, C-propeptide of type II collagen; CS846, chondroitin sulfate 846 epitope; CTXI, C-terminal crosslinked telopeptide of type I collagen; CTXII, C-terminal crosslinked telopeptide type II collagen; HA, hyaluronan; NTXI, N-telopeptide of type I collagen; TIC, time-integrated concentration.
Secondary analyses
The biomarkers were further evaluated for their ability to predict individual group status at 48 M. Ten of the 18 biomarkers were significantly associated with the status of one or more of these groups (table 4; non-transformed biomarker concentrations for each group in online supplementary table S2). Baseline, 12 and 24 M TICs of uCTXII significantly predicted all groups. The highest odds were achieved by uCTXII 24M TIC predicting the combination of pain and JSL progression (OR 1.72, 95% CIs 1.36 to 2.18). Baseline, 12 MTIC and 24 MTIC uC2C-HUSA, a neoepitope resulting from type II collagen degradation, predicted the combination of pain and JSL progression, and the 12 and 24 M TIC predicted all groups. The 12 and 24M TICs of sPIIANP, the type II collagen synthesis marker, significantly predicted JSL and the combination of pain and JSL progression. The type I collagen markers indicative of bone resorption (sCTXI, uCTXIα, uCTXIβ, sNTXI and uNTXI) all significantly predicted the combination of pain and JSL progression, but not pain-only progression or JSL-only progression. The 12 and 24 M TIC of uC1, 2C, indicative of type I and II collagen degradation, predicted pain-only progression. Finally, the 12 and 24M TIC of sHA, indicative of joint inflammation, significantly predicted the combination of pain and JSL progression.
Table 4.
Odds of being a pain, radiographic JSL or combination (pain and JSL) progressor compared with neither pain nor JSL progression (secondary analysis) based on a 1 SD increase in biomarker concentrations
| Biomarker | Biomarker prediction of odds (95% CI) of progression compared with reference subjects without any progression | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Baseline concentration | 12 M TIC | 24 M TIC | |||||||
| JSL-only progressor (n=103) |
Pain-only progressor (n=103) |
Progressors (JSL and pain) (n=194) | JSL-only progressor (n=103) |
Pain-only progressor (n=103) |
Progressors (JSL and -pain) (n=194) |
JSL-only progressor (n=103) |
Pain-only progressor (n=103) |
Progressors (JSL and pain) (n=194) |
|
| Serum C1, 2C | 1.11 (0.86 to 1.43) | 0.98 (0.76 to 1.26) | 1.12 (0.91 to 1.37) | 1.12 (0.86 to 1.44) | 1.00 (0.78 to 1.28) | 1.05 (0.85 to 1.30) | 1.12 (0.87 to 1.45) | 1.03 (0.80 to 1.32) | 1.11 (0.90 to 1.37) |
| Serum-C2C (ng/ml) | 1.04 (0.81 to 1.34) | 0.92 (0.71 to 1.18) | 1.03 (0.84 to 1.27) | 1.04 (0.81 to 1.35) | 0.92 (0.71 to 1.19) | 0.98 (0.79 to 1.21) | 1.05 (0.81 to 1.35) | 0.97 (0.75 to 1.25) | 1.06 (0.86 to 1.31) |
| Serum Coll2-1 NO2 | 1.00 (0.75 to 1.32) | 1.05 (0.82 to 1.34) | 1.01 (0.82 to 1.26) | 0.94 (0.70 to 1.25) | 0.98 (0.76 to 1.26) | 0.95 (0.76 to 1.18) | 0.91 (0.68 to 1.22) | 0.97 (0.75 to 1.25) | 0.99 (0.80 to 1.22) |
| Serum CPII | 0.89 (0.65 to 1.21) | 1.16 (0.91 to 1.49) | 0.99 (0.79 to 1.24) | 1.00 (0.75 to 1.33) | 1.10 (0.85 to 1.43) | 0.96 (0.77 to 1.21) | 0.99 (0.73 to 1.33) | 1.15 (0.90 to 1.47) | 1.01 (0.80 to 1.27) |
| Serum CS846 | 0.76 (0.57 to 1.01) | 0.99 (0.78 to 1.25) | 1.00 (0.82 to 1.21) | 0.77 (0.58 to 1.03) | 1.01 (0.80 to 1.29) | 1.02 (0.83 to 1.24) | 0.80 (0.60 to 1.06) | 1.04 (0.82 to 1.31) | 1.05 (0.86 to 1.28) |
| Serum CTXI | 1.13 (0.87 to 1.46) | 1.14 (0.89 to 1.47) | 1.24 (1.01 to 1.53) | 1.10 (0.84 to 1.43) | 1.15 (0.89 to 1.47) | 1.36 (1.10 to 1.68) | 1.10 (0.85 to 1.42) | 1.09 (0.85 to 1.41) | 1.34 (1.09 to 1.65) |
| Serum COMP | 0.88 (0.69 to 1.14) | 0.96 (0.74 to 1.23) | 0.86 (0.70 to 1.06) | 0.94 (0.73 to 1.21) | 1.04 (0.80 to 1.33) | 0.90 (0.72 to 1.11) | 0.94 (0.73 to 1.21) | 1.07 (0.83 to 1.37) | 0.92 (0.74 to 1.13) |
| Serum HA | 1.19 (0.89 to 1.58) | 1.29 (0.98 to 1.69) | 1.21 (0.96 to 1.54) | 1.18 (0.89 to 1.57) | 1.14 (0.85 to 1.53) | 1.29 (1.02 to 1.64) | 1.17 (0.88 to 1.56) | 1.19 (0.88 to 1.60) | 1.34 (1.05 to 1.70) |
| Serum MMP3 | 1.15 (0.87 to 1.52) | 1.03 (0.75 to 1.40) | 1.04 (0.81 to 1.34) | 1.07 (0.80 to 1.45) | 1.02 (0.74 to 1.41) | 1.09 (0.84 to 1.40) | 1.07 (0.79 to 1.45) | 0.99 (0.72 to 1.36) | 1.11 (0.86 to 1.43) |
| Serum NTXI | 1.03 (0.80 to 1.33) | 1.07 (0.83 to 1.38) | 1.18 (0.96 to 1.45) | 1.04 (0.80 to 1.35) | 1.08 (0.84 to 1.40) | 1.29 (1.05 to 1.60) | 1.08 (0.84 to 1.40) | 1.07 (0.83 to 1.38) | 1.27 (1.03 to 1.56) |
| Serum PIIANP | 0.74 (0.57 to 0.96) | 0.85 (0.66 to 1.09) | 0.79 (0.64 to 0.98) | 0.71 (0.55 to 0.92) | 0.88 (0.68 to 1.13) | 0.74 (0.60 to 0.92) | 0.70 (0.54 to 0.91) | 0.85 (0.66 to 1.09) | 0.79 (0.64 to 0.97) |
| Urine Coll2-1 NO2 creatinine adjusted | 1.04 (0.80 to 1.36) | 1.11 (0.87 to 1.42) | 1.10 (0.89 to 1.35) | 0.93 (0.71 to 1.22) | 1.04 (0.82 to 1.33) | 1.10 (0.90 to 1.36) | 0.92 (0.69 to 1.21) | 1.17 (0.92 to 1.49) | 1.16 (0.95 to 1.43) |
| Urine C1, 2C creatinine adjusted | 1.12 (0.87 to 1.44) | 1.25 (0.99 to 1.59) | 1.01 (0.82 to 1.25) | 1.07 (0.83 to 1.39) | 1.35 (1.06 to 1.72) | 1.10 (0.88 to 1.36) | 1.09 (0.84 to 1.41) | 1.39 (1.09 to 1.78) | 1.14 (0.92 to 1.41) |
| Urine C2C-HUSA creatinine adjusted | 1.23 (0.94 to 1.62) | 1.35 (1.04 to 1.74) | 1.27 (1.02 to 1.58) | 1.32 (1.01 to 1.74) | 1.38 (1.06 to 1.80) | 1.35 (1.07 to 1.69) | 1.40 (1.05 to 1.85) | 1.36 (1.03 to 1.80) | 1.50 (1.19 to 1.90) |
| Urine CTXII creatinine adjusted | 1.49 (1.12 to 1.98) | 1.55 (1.17 to 2.06) | 1.62 (1.27 to 2.07) | 1.47 (1.10 to 1.95) | 1.53 (1.16 to 2.01) | 1.67 (1.31 to 2.12) | 1.54 (1.16 to 2.04) | 1.53 (1.16 to 2.02) | 1.72 (1.36 to 2.18) |
| Urine NTXI creatinine adjusted | 1.01 (0.76 to 1.34) | 1.23 (0.96 to 1.58) | 1.22 (0.99 to 1.51) | 0.99 (0.74 to 1.31) | 1.26 (0.98 to 1.62) | 1.31 (1.06 to 1.64) | 1.03 (0.77 to 1.36) | 1.24 (0.96 to 1.60) | 1.37 (1.10 to 1.71) |
| Urine CTXIα creatinine adjusted | 1.02 (0.77 to 1.36) | 1.12 (0.87 to 1.45) | 1.22 (0.99 to 1.51) | 1.04 (0.78 to 1.38) | 1.18 (0.91 to 1.53) | 1.34 (1.08 to 1.67) | 1.08 (0.81 to 1.43) | 1.16 (0.89 to 1.51) | 1.39 (1.12 to 1.73) |
| Urine CTXIβ creatinine adjusted | 1.02 (0.78 to 1.33) | 1.08 (0.84 to 1.37) | 1.15 (0.94 to 1.41) | 1.03 (0.79 to 1.35) | 1.15 (0.90 to 1.48) | 1.32 (1.07 to 1.63) | 1.11 (0.85 to 1.45) | 1.16 (0.90 to 1.49) | 1.35 (1.09 to 1.66) |
Values in bold represent associations of p≤0.05.
ORs are for comparison with knees with neither radiographic JSL nor pain progression and were adjusted for all covariates (age, sex, body mass index, race, baseline joint space width, Kellgren-Lawrence grade, baseline pain and pain medication use).
COMP, cartilage oligomeric matrix protein; CPII, C-propeptide of type II collagen; CS846, chondroitin sulfate 846 epitope; CTXI, C-terminal crosslinked telopeptide of type I collagen; CTXII, C-terminal crosslinked telopeptide type II collagen; HA, hyaluronan; JSL, joint space loss; NTXI, N-telopeptide of type I collagen; TIC, time-integrated concentration.
Multivariable analyses: 24 M TIC
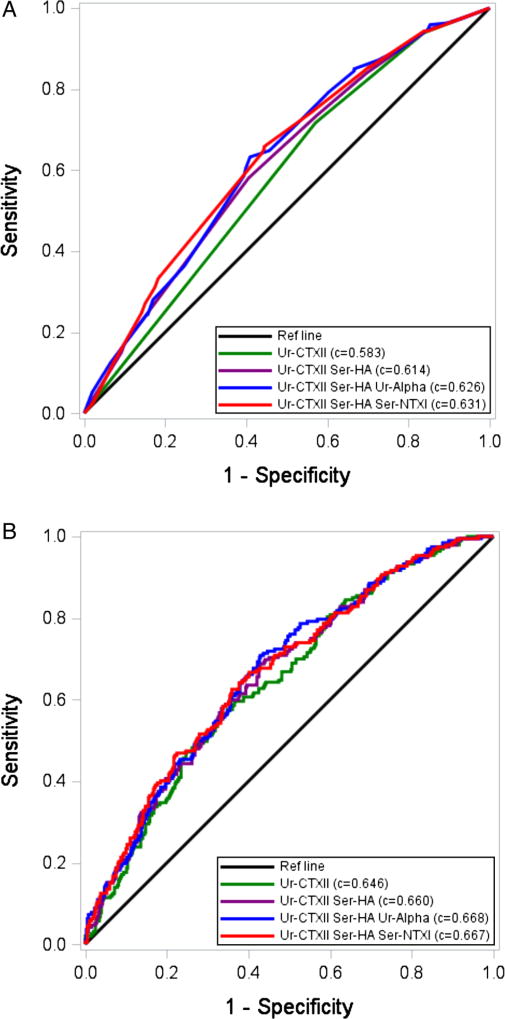
Based on univariable models, eight biomarkers met the p<0.10 threshold and were advanced for multivariable analyses: sCTXI, sHA, sNTXI, uC2C-HUSA, uCTXII, uNTXI, uCTXIα and uCTXIβ. Based on OR, the C-statistic AUC, IDI and NRI (see online supplementary table S5), four biomarkers were advanced for hierarchical modelling: uCTXII (collagen type II degradation: higher IDI, NRI and AUC compared with uC2C-HUSA); urinary CTXIα (collagen type I degradation: higher IDI, NRI and AUC compared with both sCTXI and uCTXIβ with sCTXI evaluated in sensitivity analysis); urinary NTXI (collagen type I degradation: higher IDI, NRI and AUC compared with sNTXI, with sNTXI evaluated in sensitivity analysis) and sHA. Table 5 shows the model building process. The final model consisted of uCTXII, sHA and sNTXI yielding an AUC of 0.631 (unadjusted for covariates). uNTXI did not perform well in multivariable models (p>0.99) and was replaced by sNTXI, which was statistically significant (p=0.041) when added to the model with uCTXII and sHA. The receiver operating characteristic curve for this combination is shown in figure 1. The cut-offs used to attain the final combinatorial prediction models are provided in online supplementary table S6.
Table 5.
Hierarchical models for 24 M TIC—serum and urine markers
| Model 1: uCTXII only |
Model 2: uCTXII, sHA |
Model 3: uCTXII, sHA, uCTXI ALPHA |
Model 4: uCTXII, sHA, uCTXI ALPHA, uNTXI |
Model 5: uCTXII, sHA, uCTXI ALPHA, sNTXI |
Model 6: uCTXII, sHA, sNTXI |
|
|---|---|---|---|---|---|---|
| AUC (biomarkers only, no cross-validation) | 0.583 | 0.614 | 0.626 | 0.625 | 0.635 | 0.631 |
| AUC (including covariates*, no cross-validation) | 0.646 | 0.660 | 0.668 | 0.669 | 0.672 | 0.667 |
| AUC (including covariates*, 10-fold cross-validation) | 0.602 | 0.613 | 0.617 | 0.610 | 0.617 | 0.618 |
| uCTXII | p=0.0006 | p=0.0057 | p=0.0625 | p=0.0662 | p=0.0636 | p=0.0129 |
| z≤−1 vs −1<z<−0.5 | 0.5 (0.2, 0.9) | 0.5 (0.2, 1.0) | 0.5 (0.3, 1.1) | 0.5 (0.3, 1.1) | 0.5 (0.2, 1.0) | 0.5 (0.2, 1.0) |
| z>−0.5 vs −1<z≤−0.5 | 1.5 (1.0, 2.3) | 1.4 (0.9, 2.1) | 1.2 (0.8, 1.9) | 1.2 (0.8, 1.9) | 1.2 (0.7, 1.8) | 1.3 (0.8, 2.0) |
| sHA | p=0.0755 | p=0.0503 | p=0.0507 | p=0.0623 | p=0.0893 | |
| z≤−0.5 vs −0.5<z≤1 | 0.7 (0.5, 1.1) | 0.7 (0.5, 1.1) | 0.7 (0.5, 1.1) | 0.7 (0.5, 1.1) | 0.8 (0.5, 1.1) | |
| z>1 vs −0.5<z≤1 | 1.4 (0.8, 2.4) | 1.5 (0.9, 2.5) | 1.5 (0.9, 2.5) | 1.5 (0.9, 2.5) | 1.4 (0.8, 2.4) | |
| uCTXI ALPHA | p=0.0853 | p=0.3263 | p=0.2818 | |||
| z≤−0.5 vs −0.5<z≤0.5 | 0.8 (0.5, 1.2) | 0.8 (0.5, 1.3) | 0.8 (0.5, 1.2) | |||
| z>0.5 vs −0.5<z≤0.5 | 1.4 (0.9, 2.1) | 1.4 (0.8, 2.4) | 1.2 (0.8, 1.9) | |||
| uNTXI | p=0.9975 | |||||
| z≤−0.5 vs −0.5<z≤1 | 1.0 (0.6, 1.7) | |||||
| z>1 vs −0.5<z≤1 | 1.0 (0.5, 1.9) | |||||
| sNTXI | p=0.2091 | p=0.0414 | ||||
| z>1 vs z≤1 | 1.4 (0.8, 2.4) | 1.6 (1.0, 2.6) |
ORs and p values for ORs are unadjusted for covariates: AUC *covariate models adjusted for all covariates (age, sex, body mass index, race, baseline joint space width, Kellgren-Lawrence grade, baseline pain and pain medication use).
AUC, area under the curve; TIC, time-integrated-concentration.
Figure 1.
Receiver operating characteristic curve and C-statistics (area under the curves (AUCs)) for models with up to four biomarkers. (A) Does not include covariates. (B) Includes covariates (sex, race, baseline body mass index, age, baseline Western Ontario and McMaster Universities Arthritis Index (WOMAC) pain and baseline medial minimum joint space width, baseline Kellgren-Lawrence grade).
We also examined the discriminative ability of the urine biomarkers only and serum biomarkers only. CTXII alone was the most predictive of the urine biomarkers, yielding an AUC of 0.583 (see online supplementary table S7). No additional urine biomarkers were statistically significant when added to the model with uCTXII alone. The final model for the serum-only model included sHA and sNTXI; the AUC was 0.601.
Multivariable analysis: baseline
We also explored the predictive capability of the baseline biomarkers in combination. Based on univariable models, a total of five biomarkers met the p<0.10 threshold and were advanced to multivariable modelling: uCTXII, sNTXI and uNTXI, sCTXI and uCTXIα (see online supplementary table S8). The final baseline model included uCTXII and sNTXI and had an AUC of 0.586.
DISCUSSION
Biomarkers have the potential to greatly impact the quality of life of patients with OA through improved individualised care and the identification of new treatment targets and mechanisms for more efficient trials of disease-modifying agents. The qualification of OA prognostic markers is of fundamental importance for achieving this potential.1,10 Although the qualification of a prognostic biomarker requires a large and long prospective trial, samples and data acquired through the OAI made it possible to evaluate a set of soluble biomarkers for their ability to predict OA progression based on clinical and imaging end points. Nine biomarkers were individually able to predict case status reflecting clinically relevant progression consisting of the combination of pain and JSL progression; these included sCTXI, uCTXIα and uCTXIβ, sNTXI, uNTXI, sPIIANP, sHA, uC2C-HUSA and uCTXII. Underscoring the face validity of these associations, the collagen catabolic biomarkers (uCTXII, uC2C-HUSA, sCTXI, uCTXIα, uCTXIβ, sNTXI and uNTXI) were all positively associated with OA progression, while the collagen anabolic marker (sPIIANP) was inversely associated with OA progression. The strongest predictions of case status were provided by the two catabolic markers uC2C-HUSA and uCTXII, indicative of articular cartilage degradation.
CTXII has been the only OA-related biomarker to achieve a ‘characterisation’ level of surrogacy (nomenclature of Wagner et al11 based on changing significantly in three pharmacological trials that met primary clinical end points12–14). In this study, all uCTXII time points (baseline, 12 M TIC and 24 M TIC) were associated with case status. Used alone, uCTXII yielded the highest OR for predicting case status (both radiographic and pain progression) from among the 18 biomarkers: OR 1.37 (24 M) for being a case compared with all other groups and OR 1.72 (24 M) for being a case compared with ‘pure’ non-progressors (neither radiographic nor pain progression). These ORs indicate that for every 1 SD increase in uCTXII, the odds of progression increased 37% and 72%, respectively. These results suggest that differences on the order of 1 SD appear clinically meaningful for this biomarker. Studies to date suggest that uCTXII likely reflects both articular cartilage and calcified cartilage turnover.9 Interestingly, results of uCTXII differed from the bone biomarkers in secondary analyses; in contrast to the bone biomarkers, uCTXII was associated with pain progression separate from radiographic JSL progression, whereas the traditional collagen type I bone resorption markers were not significantly associated with pain worsening independent of JSL worsening. Similarly, the other type II collagen biomarker, uC2C-HUSA, was also significantly associated with pain progression as well as JSL progression.
As shown here, individual biomarkers explained only a small proportion of the interindividual variation in disease progression.15,16 The use of their 24 M TICs in combination was examined and found to yield a slightly better predictive capability than any single biomarker: AUC 0.631 for the most parsimonious combination of three biomarkers (sHA, sNTXI and uCTXII) compared with AUC 0.583 for the 24 M TIC of the best single biomarker (uCTXII). Several studies have shown that the combination of two biomarkers, reflecting different pathways, were more effective for predicting disease progression than one marker alone in knee and hip OA.6,15,17–20 Taken together, these data suggested that a combination of several biomarkers, reflecting different pathophysiological pathways, would better predict disease progression than any single biomarker or clinical factor. In future analyses of the OA Consortium Biomarker data, we plan to evaluate soluble and imaging biomarkers in combination.
As recognised for cancer biomarkers,21,22 due to their dynamic nature, the use of TICs is well suited for evaluating their information content over time. Although the concentrations of OA biomarkers are recognised to vary dynamically,5,6,23 the practical challenges posed by longitudinal biomarker data for OA clinical use or clinical trial studies have not previously been widely appreciated or discussed in the OA biomarker literature. Nevertheless, the use of time-integrated biochemical biomarker levels (AUC) has been recommended in a primary textbook of Rheumatology as the preferred approach for assessing the progression of radiographic damage.24 This is because radiological and MRI measures provide a cumulative historical view of joint destruction, whereas biochemical biomarkers provide dynamic information on the rate of joint destruction or disease activity. Failure to use such a strategy for dynamic markers may be responsible for some lack of success in prior biomarker analyses in natural history studies, such as this one. Our preliminary analyses comparing change scores with TICs confirmed the utility and superiority of the TIC approach (data not shown). However, in a clinical trial with anticipated treatment effects, both TICs and change in biochemical markers over time would likely be appropriate.
Although two of the markers (CTXI and NTXI) are in vitro diagnostics approved for osteoporosis, none of these biochemical markers is yet approved for clinical use for OA. OA has long been known to be associated with bone abnormalities and a robust intercellular communication between cartilage and subchondral bone.25,26 From a bone perspective, early stage OA is associated with increased remodelling and bone loss, whereas late-stage OA is characterised by slow remodelling and subchondral densification.27 To date, a number of bone-acting agents have shown promise for treating OA.28 Metabolites of type I collagen (CTXI and NTXI) previously have been positively associated with knee OA progression.29 Non-isomerised CTXIα is localised to high turnover areas of subchondral bone and has recently been shown to be associated with high turnover in subchondral bone of osteoarthritic knees by scintigraphy and to be associated with progression of both osteophyte and joint space narrowing in knee OA.9 In this study, these bone biomarkers consistently predicted OA case status. Although they reflect bone turnover, secondary analyses revealed that these bone biomarkers were associated with radiographic JSL, which is known to be caused by both articular and meniscal cartilage degeneration and meniscal extrusion.30 Although they predicted the combination of JSL and pain progression, secondary analyses revealed that they did not independently predict pain progression in the absence of radiographic progression.
In secondary analyses, sPIIANP 12 and 24 M TICs were associated with primary case status and JSL progression without pain progression compared with non-progressors. PIIANP is the NH2-propeptide of type IIA collagen. It is synthesised by chondroprogenitor cells and other non-chondrocytic cells during embryogenesis.31 Its synthesis is recapitulated in OA chondrocytes, possibly as an attempt to repair damaged cartilage.32 A switch in the splice form to type IIB collagen, characteristic of chondrocytes, occurs upon the commitment to chondrogenesis, eliminating the expression of exon-2 (encoding the antigen for PIIANP). Here, and as previously reported, a decrease in sPIIANP was correlated with progression of OA similar to that observed in the ‘uncoupling index’ of the degradative marker CTXII and PIIANP.17 While the percentage of type II collagen produced by OA chondrocytes is small, the PIIANP protein fragment, a von Willebrand Factor C domain containing 10 cysteines, may be very stable in the serum and thus easy to detect.
There are several limitations of this study. Systemic biomarkers such as these, measured in serum and urine, presumably report on all joints in the body albeit in different amounts according to the relative amounts of the epitope, level of turnover and disease status of each joint. Clear proof of this principle was provided in a study showing that uCTXII, sHA and sCOMP are all strongly associated with total body burden of osteophytes (accounting for hands, hips, knees and spine) with correlations ranging from R2 0.47–0.60 (p<10−6).33 Our current study only took into account the knee status and did not take into account OA status at other joint sites. Nevertheless, our approach, based on systemic biomarker analyses, is likely to be used in treatment trials focusing on an index joint. There was no clear reason that certain collagen biomarkers were associated with case status while others not. In general, the markers associated with case status had the best coefficient of variations, although this does not fully explain the differences. Possible explanations include intrinsic differences in the pathways generating the fragments, differences related to the presence or clearance of the fragment in serum or urine or methodological differences such as efficiency or specificity of fragment detection. The comparator group in the primary analyses (combined JSL progression-only group, pain progression-only group and ‘pure’ non-progressor group) could potentially lead to an underestimation of the value of these systemic markers in predicting radiographic progression and pain progression separately. Secondary analyses indicated that both uCTXII and uC2C-HUSA were associated with all three progressor groups, pain-only worsening, JSL-only and their combination. For these markers, the odds of predicting case status were stronger comparing cases with ‘pure’ non-progressors (uCTXII OR 1.72, uC2C-HUSA OR 1.50 based on 24M TIC) versus comparing cases with all comparators (uCTXII OR 1.37, uC2C-HUSA OR 1.27 based on 24 M TIC). Six other markers (sCTXI, sHA, sPIIANP, uNTXI, uCTX1α and uCTXIβ) also yielded stronger odds of predicting case status when cases were compared with ‘pure’ non-progressors; each of these markers also showed some association with the individual pain progression-only and JSL progression-only groups (although most did not surpass the p<0.05 level of significance). This discovery phase of the Foundations for National Institutes of Health (FNIH) project study involved analysis of a number of biomarkers; the primary analysis and biomarkers were clearly identified by an a priori analysis plan. Due to the limitation of sample size (we selected all possible cases from OAI), instead of formal adjustment for multiple comparisons we used a number of complementary statistical approaches to evaluate discriminatory power of each biomarkers including c-statistics, NRI and performed 10-fold cross-validation. It should be noted that while baseline and 12 M TICs evaluated the predictive ability of the biomarkers (defined outcomes were achieved by 24–48 M), the 24 M TICs potentially evaluated both predictive and concurrent validity. Finally, other approaches to data reduction, selection of variables for multivariate analysis and multivariate modelling may give results different from ours for the most predictive and parsimonious multivariable models.
In summary, this study is important for establishing a paradigm by which OA-related biomarker qualification can proceed. Several systemic candidate biomarkers hold promise as predictors of pain and radiographic structural worsening of OA over 48 M. The 12 and 24 M TICs of eight catabolic biomarkers, uCTXII, uC2C-HUSA, sHA, sNTXI, uNTXI, uCTXIα, uCTXIβ, sCTXI (OR 1.22–1.72 depending upon comparator group), and one anabolic biomarker, sPIIANP (OR 0.79–0.83 depending upon comparator group) significantly predicted 48 M case status. Although only modestly predictive of single knee progression, given their cost-effectiveness, they might have some utility for enriching OA trials for progressors or providing early proof of effectiveness of a drug to prevent OA progression. These results will require additional verification, ideally in extant clinical trials, to be formally qualified by the FDA as Drug Development Tools as described in FDA guidance.34,35 Additional biomarkers in development will no doubt add to what appears to be a very promising future.
Supplementary Material
Acknowledgments
The authors thank the Laboratory Analysts at LabCorp Clinical Trials, Li Cao and Des Delute who performed all biochemical analyses for this work. We also wish to thank Susan Rubin at University of California, San Francisco for expert assistance with sample management.
Funding In-kind donations to support biochemical testing was provided by Alere, ARTIALIS S.A., BioVendor—Laboratorni medicina a.s., IBEX Pharmaceuticals, Immunodiagnostic Systems and Quidel Corporation. Scientific and financial support for the Foundations for National Institutes of Health (FNIH) OA Biomarkers Consortium and the study are made possible through grants, direct and in-kind contributions provided by: AbbVie, Amgen, Arthritis Foundation, Bioiberica S.A., DePuy Mitek, Flexion Therapeutics, GlaxoSmithKline, Merck Serono, Rottapharm | Madaus, Sanofi, Stryker, The Pivotal OAI MRI Analyses study, NIH NHLBI HHSN2682010000. We thank the Osteoarthritis Research Society International for their leadership and expertise on the FNIH OA Biomarker Consortium project. The osteoarthritis initiative (OAI) is a public-private partnership comprising five contracts (N01-AR-2-2258, N01-AR-2-2259, N01-AR-2-2260, N01-AR-2-2261 and N01-AR-2-2262) funded by the National Institutes of Health. Funding partners include Merck Research Laboratories, Novartis Pharmaceuticals Corporation, GlaxoSmithKline and Pfizer. Private sector funding for the Consortium and OAI is managed by the FNIH. The statistical analysis and writing of this article was independent from and not contingent upon approval from the study sponsors or kit suppliers.
Footnotes
Correction notice This article has been corrected since it was published Online First. Text changes have been made to the first paragraph of the section ‘Biomarker assays.’ In tables 1,3 and 4 ‘serum C2C-HUSA’ has been corrected to ‘serum-C2C (ng/ml)’ and ‘Urine C2C-HUSA HUSA (ng/mmol Cr)’ has been corrected to ‘Urine C2C-HUSA (ng/mmol Cr).’
Contributors Study conception and design: VBK, DJH, MN, JEC, EL, JNK, EL and LJS. Acquisition of data: DH and MN. Analysis and interpretation of data: All authors. Writing of first manuscript draft: VBK. Critical manuscript revision and approval of final manuscript: All authors. JC had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Competing interests None declared.
Ethics approval University of California, San Francisco and all sites participating in the osteoarthritis initiative study.
Provenance and peer review Not commissioned; externally peer reviewed.
Data sharing statement All the data are made publically available for additional analyses and for comparison to other biomarkers that any researcher might do upon requesting the same sample set from the Biochemical Research Committee of the Osteoarthritis Initiative.
References
- 1.Kraus VB, Burnett B, Coindreau J, et al. Application of biomarkers in the development of drugs intended for the treatment of osteoarthritis. Osteoarthr Cartil. 2011;19:515–42. doi: 10.1016/j.joca.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunter DJ, Nevitt M, Losina E, et al. Biomarkers for osteoarthritis: current position and steps towards further validation. Best Pract Res Clin Rheumatol. 2014;28:61–71. doi: 10.1016/j.berh.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Spil WE, DeGroot J, Lems WF, et al. Serum and urinary biochemical markers for knee and hip-osteoarthritis: a systematic review applying the consensus BIPED criteria. Osteoarthr Cartil. 2010;18:605–12. doi: 10.1016/j.joca.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 4.Hunter D, Nevitt M, Lynch J, et al. Longitudinal validation of periarticular bone area and 3D shape as biomarkers for knee OA progression? Data from the FNIH osteoarthritis biomarkers consortium. Ann Rheum Dis. 2016;75:1607–14. doi: 10.1136/annrheumdis-2015-207602. [DOI] [PubMed] [Google Scholar]
- 5.Sharif M, Kirwan JR, Elson CJ, et al. Suggestion of nonlinear or phasic progression of knee osteoarthritis based on measurements of serum cartilage oligomeric matrix protein levels over five years. Arthritis Rheum. 2004;50:2479–88. doi: 10.1002/art.20365. [DOI] [PubMed] [Google Scholar]
- 6.Sharif M, Kirwan J, Charni N, et al. A 5-yr longitudinal study of type IIA collagen synthesis and total type II collagen degradation in patients with knee osteoarthritis—association with disease progression. Rheumatology (Oxford) 2007;46:938–43. doi: 10.1093/rheumatology/kel409. [DOI] [PubMed] [Google Scholar]
- 7.Gu W, Pepe M. Measures to summarize and compare the predictive capacity of markers. Int J Biostat. 2009;5 doi: 10.2202/1557-4679.1188. Article 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pencina MJ, D’Agostino RB, Sr, D’Agostino RB, Jr, et al. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med. 2008;27:157–72. doi: 10.1002/sim.2929. discussion 207–12. [DOI] [PubMed] [Google Scholar]
- 9.Huebner JL, Bay-Jensen AC, Huffman KM, et al. Alpha C-telopeptide of type I collagen is associated with subchondral bone turnover and predicts progression of joint space narrowing and osteophytes in osteoarthritis. Arthritis Rheumatol. 2014;66:2440–9. doi: 10.1002/art.38739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kraus V, Blanco F, Englund M, et al. Call for standardized definitions of osteoarthritis and risk stratification for clinical trials and clinical use. Osteoarthr Cartil. 2015;23:1233–41. doi: 10.1016/j.joca.2015.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagner JA, Williams SA, Webster CJ. Biomarkers and surrogate end points for fit-for-purpose development and regulatory evaluation of new drugs. Clin Pharmacol Ther. 2007;81:104–7. doi: 10.1038/sj.clpt.6100017. [DOI] [PubMed] [Google Scholar]
- 12.Christgau S, Henrotin Y, Tankó LB, et al. Osteoarthritic patients with high cartilage turnover show increased responsiveness to the cartilage protecting effects of glucosamine sulphate. Clin Exp Rheumatol. 2004;22:36–42. [PubMed] [Google Scholar]
- 13.Gineyts E, Mo JA, Ko A, et al. Effects of ibuprofen on molecular markers of cartilage and synovium turnover in patients with knee osteoarthritis. Ann Rheum Dis. 2004;63:857–61. doi: 10.1136/ard.2003.007302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manicourt DH, Azria M, Mindeholm L, et al. Oral salmon calcitonin reduces Lequesne’s algofunctional index scores and decreases urinary and serum levels of biomarkers of joint metabolism in knee osteoarthritis. Arthritis Rheum. 2006;54:3205–11. doi: 10.1002/art.22075. [DOI] [PubMed] [Google Scholar]
- 15.Mazieres B, Garnero P, Gueguen A, et al. Molecular markers of cartilage breakdown and synovitis at baseline as predictors of structural progression of hip osteoarthritis. The ECHODIAH Cohort. Ann Rheum Dis. 2006;65:354–9. doi: 10.1136/ard.2005.037275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter DJ, Li J, LaValley M, et al. Cartilage markers and their association with cartilage loss on magnetic resonance imaging in knee osteoarthritis: the Boston Osteoarthritis Knee Study. Arthritis Res Ther. 2007;9:R108. doi: 10.1186/ar2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garnero P, Ayral X, Rousseau JC, et al. Uncoupling of type II collagen synthesis and degradation predicts progression of joint damage in patients with knee osteoarthritis. Arthritis Rheum. 2002;46:2613–24. doi: 10.1002/art.10576. [DOI] [PubMed] [Google Scholar]
- 18.Garnero P, Charni N, Juillet F, et al. Increased urinary type II collagen helical and C telopeptide levels are independently associated with a rapidly destructive hip osteoarthritis. Ann Rheum Dis. 2006;65:1639–44. doi: 10.1136/ard.2006.052621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cahue S, Sharma L, Dunlop D, et al. The ratio of type II collagen breakdown to synthesis and its relationship with the progression of knee osteoarthritis. Osteoarthr Cartil. 2007;15:819–23. doi: 10.1016/j.joca.2007.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dam EB, Loog M, Christiansen C, et al. Identification of progressors in osteoarthritis by combining biochemical and MRI-based markers. Arthritis Res Ther. 2009;11:R115. doi: 10.1186/ar2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhatt AN, Mathur R, Farooque A, et al. Cancer biomarkers—current perspectives. Indian J Med Res. 2010;132:129–49. [PubMed] [Google Scholar]
- 22.Sharma N, Shrivastav A, Shrivastav B. Telomerase as a biomarker for cancer diagnosis. Int J Biomed Adv Res. 2013;4:494–500. [Google Scholar]
- 23.Kumm J, Tamm A, Lintrop M, et al. The value of cartilage biomarkers in progressive knee osteoarthritis: cross-sectional and 6-year follow-up study in middle-aged subjects. Rheumatol Int. 2013;33:903–11. doi: 10.1007/s00296-012-2463-8. [DOI] [PubMed] [Google Scholar]
- 24.DeGroot J, Zuurmond A-M, Tak P-P. Biological markers. In: Firestein G, Budd R, Gabriel S, et al., editors. Kelley’s Textbook of Rheumatology. 9. Philadelphia: Elsevier; 2013. pp. 476–92. [Google Scholar]
- 25.Sharma AR, Jagga S, Lee SS, et al. Interplay between cartilage and subchondral bone contributing to pathogenesis of osteoarthritis. Int J Mol Sci. 2013;14:19805–30. doi: 10.3390/ijms141019805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chou CH, Lee CH, Lu LS, et al. Direct assessment of articular cartilage and underlying subchondral bone reveals a progressive gene expression change in human osteoarthritic knees. Osteoarthr Cartil. 2013;21:450–61. doi: 10.1016/j.joca.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burr DB, Gallant MA. Bone remodelling in osteoarthritis. Nat Rev Rheumatol. 2012;8:665–73. doi: 10.1038/nrrheum.2012.130. [DOI] [PubMed] [Google Scholar]
- 28.Castañeda S, Roman-Blas JA, Largo R, et al. Subchondral bone as a key target for osteoarthritis treatment. Biochem Pharmacol. 2012;83:315–23. doi: 10.1016/j.bcp.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 29.Bettica P, Cline G, Hart DJ, et al. Evidence for increased bone resorption in patients with progressive knee osteoarthritis: longitudinal results from the Chingford study. Arthritis Rheum. 2002;46:3178–84. doi: 10.1002/art.10630. [DOI] [PubMed] [Google Scholar]
- 30.Hunter DJ, Zhang YQ, Tu X, et al. Change in joint space width: hyaline articular cartilage loss or alteration in meniscus? Arthritis Rheum. 2006;54:2488–95. doi: 10.1002/art.22016. [DOI] [PubMed] [Google Scholar]
- 31.Oganesian A, Zhu Y, Sandell LJ. Type IIA procollagen amino propeptide is localized in human embryonic tissues. J Histochem Cytochem. 1997;45:1469–80. doi: 10.1177/002215549704501104. [DOI] [PubMed] [Google Scholar]
- 32.Aigner T, Zhu Y, Chansky HH, et al. Reexpression of type IIA procollagen by adult articular chondrocytes in osteoarthritic cartilage. Arthritis Rheum. 1999;42:1443–50. doi: 10.1002/1529-0131(199907)42:7<1443::AID-ANR18>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 33.Kraus VB, Kepler TB, Stabler T, et al. First qualification study of serum biomarkers as indicators of total body burden of osteoarthritis. PLoS ONE. 2010;5:e9739. doi: 10.1371/journal.pone.0009739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.FDA. Guidance for industry: qualification process for drug development tools. Maryland: Silver Spring; 2010. [Google Scholar]
- 35.FDA. Guidance for industry and FDA staff: qualification process for drug development tools. Maryland: Silver Spring; 2014. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.