Abstract
Streptococcus mutans (S. mutans) has been proved to be the main aetiological factor in dental caries. Curcumin, a natural product, has been shown to exhibit therapeutic antibacterial activity, suggesting that curcumin may be of clinical interest. The objective of this study is to evaluate the inhibitory effects of curcumin on metabolism and biofilm formation in S. mutans using a vitro biofilm model in an artificial oral environment. S. mutans biofilms were treated with varying concentrations of curcumin. The biofilm metabolism and biofilm biomass were assessed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide assay and the crystal violet assay. Confocal laser scanning microscopy was used to analyse the composition and extracellular polysaccharide content of S. mutans biofilm after curcumin treatment. The biofilm structure was evaluated using a scanning electron microscope. The gene expression of virulence-related factors was assessed by real-time PCR. The antibiofilm effect of curcumin was compared with that of chlorhexidine. The sessile minimum inhibitory concentration (SMIC50%) of curcumin against S. mutans biofilm was 500 μM. Curcumin reduced the biofilm metabolism from 5 min to 24 h. Curcumin inhibited the quantity of live bacteria and total bacteria in both the short term (5 min) and the long term. Moreover, curcumin decreased the production of extracellular polysaccharide in the short term. The expression of genes related to extracellular polysaccharide synthesis, carbohydrate metabolism, adherence, and the two-component transduction system decreased after curcumin treatment. The chlorhexidine-treated group showed similar results. We speculate that curcumin has the capacity to be developed as an alternative agent with the potential to reduce the pathogenic traits of S. mutans biofilm.
1. Introduction
Dental caries, one of the most prevalent infectious diseases worldwide, is a biofilm-mediated, sugar-driven, multifactorial, dynamic disease [1]. Although the pathobiology of dental caries is complicated, it is widely recognized that the formation of dental plaque biofilm is one of the important causes of dental caries [2, 3]. Biofilm formation creates an anaerobic and acidic environment that results in the formation and development of caries [4]. Therefore, the eradication of dental biofilm is an effective method of controlling caries.
Streptococcus mutans (S. mutans) has been proved to be the main aetiological factor in dental caries [5, 6]. By adhering to solid surfaces, S. mutans can colonize the oral cavity and form a bacterial biofilm [7]. Dental biofilms are microbial aggregates encased in a self-produced extracellular polymer matrix. The phenotype of the bacteria involved in biofilm formation is quite different from the phenotype in the planktonic state [8]. The bacteria in a dental biofilm are far more resistant to unfavourable growth conditions, such as biocides and hydrodynamic shear forces [9, 10]. The extracellular polysaccharide (EPS) produced by S. mutans through glycosyltransferases (Gtfs) has been shown to form the matrix of the biofilm [11]. The EPS mediates the irreversible adherence between bacteria to form a high-cell-density biofilm [12], which increases the bacterial resistance to antibiotics.
Currently, many natural products have been considered as alternative or adjunctive anticaries therapies, particularly botanically derived molecules, which offer advantages over synthetic derivatives due to their natural evolution and diminished likelihood of resistance [13]. Curcumin, the major constituent of Curcuma longa L. or turmeric, has been confirmed as a potential therapeutic antibacterial agent [14]. Curcumin has been reported to inhibit various bacteria: Staphylococcus aureus, Salmonella paratyphi, Trichophyton gypseum, and Mycobacterium tuberculosis [14]. Recent publications have also reported that curcumin is active against a plethora of drug-resistant bacterial strains [14].
Studies have shown the effects of curcumin on oral bacteria associated with dental disease. Song et al. [15] found that curcumin could significantly inhibit the adhesiveness of S. mutans by its effects on collagen and fibronectin. Hu et al. [16, 17] reported that curcumin is an S. mutans sortaseA inhibitor and has promising anticaries characteristics based on an antiadhesion-mediated mechanism. Manoil et al. [18] found that blue light-activated curcumin can photoinactivate planktonic S. mutans, but the effect on S. mutans biofilm was poor. These studies proposed the potential use of curcumin as an antibacterial agent and underlined the need to explore the mechanism by which curcumin acts on oral bacteria.
Our research aims to explore the effects of curcumin on the biofilm formed by S. mutans and compares the results with the effects of chlorhexidine. The study contributes to the possibility of a natural medicine with fewer side effects and stronger antibacterial effects, thereby promoting the wider clinical application of curcumin.
2. Materials and Methods
2.1. Bacterial Strain and Growth Conditions
S. mutans strain UA159 (ATCC700610) was provided by the Guangdong Microbial Culture Collection Center. The S. mutans growth in brain-heart infusion (BHI, Difco, Detroit, MI, USA) broth medium was measured. For biofilm formation, an overnight culture of bacteria was adjusted to OD = 1.0 (1∗108 CFU/ml). The adjusted bacteria were inoculated into 96-well flat-bottom plates. The medium was 1% BHIS, that is, BHI with the addition of 1% (wt/vol) sucrose. The bacteria were grown at 37°C under anaerobic conditions (5% CO2, 10% H2, 85% N2).
2.2. Minimum Inhibitory Concentration Assay
The minimum inhibitory concentration (MIC) of curcumin against planktonic S. mutans was determined by microdilution methods following the Clinical Laboratory Standards Institute (CLSI) procedure [19], with some modifications as described below. Curcumin from a 250 mM stock in dimethyl sulfoxide (DMSO) was diluted in BHI. The resulting solution was then serially diluted in sterile BHI broth in a 96-well round-bottom plate to obtain 2000, 1000, 500, 250, 125, 62.5, and 31.25 μM curcumin in BHI culture medium. The corresponded concentrations of DMSO were used as control. Each well contained 100 μl of serially diluted curcumin, to which 100 μl of an overnight culture of S. mutans was added before incubation at 37°C under anaerobic conditions (5% CO2, 10% H2, 85% N2) for 24 h. Three parallel samples were prepared at each concentration, and BHI without curcumin was used as a control. The MIC was defined as the lowest concentration at which no visible bacteria grew in the broth.
The sessile minimum inhibitory concentration (SMIC) of curcumin against biofilm S. mutans was determined by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay. Overnight S. mutans UA159 cultures were adjusted with 1% BHIS to an initial optical density at 600 of 0.1 (approximately 107 CFU/ml), and equal volumes (200 μl) of diluted bacteria were added to 96-well plates and incubated for 24 h at 37°C in a humidified atmosphere containing 5% CO2. After 24 h, the planktonic bacteria were removed carefully, and the biofilms formed on the plates were washed with sterile phosphate buffered saline (PBS). The stock solution of curcumin was diluted in BHIS broth to concentrations ranging from 15.625 μM to 1000 μM and added to the test wells. The corresponded concentrations of DMSO were used as control. After 24 h of incubation, the BHIS broth with curcumin was removed carefully, and the biofilms formed on the plates were washed again with sterile PBS. To quantify the biofilms, 200 μl of 0.5 mg/ml MTT was added to each well and incubated at 37°C in the dark in a humidified atmosphere for 3 h. Then, the supernatants were removed, and 100 μl of 100% DMSO was added to each well. For each well, the absorbance at 570 nm was recorded using a microplate reader. The percentage of biofilm viability was calculated as follows: (absorbance of treated group/absorbance of control group) × 100% [22].
2.3. Assays of the Viability and Biomass of S. mutans Biofilm
The biofilm viability and biomass were tested at 5 min, 10 min, 15 min, 30 min, 60 min, and 24 h using the MTT assay and the crystal violet (CV) assay [20]. The SMIC50% concentration of curcumin was selected for the assay, and 0.12% (w/t) chlorhexidine was used in the experiment for comparison and the 0.12% (w/t) water was used as control. For the CV assay, the contents of the microplate were removed after incubation, and the wells were washed with PBS, fixed with 95% methanol, washed again, and stained with 0.1% (wt/vol) crystal violet solution for 15 min at room temperature. Subsequently, the microplates were vigorously tapped on napkins to remove any excess liquid and air-dried. The remaining CV was dissolved in 100 μl of 100% ethanol for 15 min at room temperature, and, finally, 75 μl from each sample was transferred to a new 96-well plate, and the extract was read at 600 nm in a spectrophotometer [20, 21].
2.4. Analysis of the Bacteria Composition and Extracellular Polysaccharide (EPS) of S. mutans Biofilm
2.4.1. Analysis of Live and Dead Bacteria by Confocal Laser Scanning Microscopy (CLSM)
As described previously, a 24 h biofilm of S. mutans was formed on a 15 mm confocal dish at 37°C in a humidified atmosphere containing 5% CO2. The planktonic bacteria were removed and the biofilm was washed twice with sterile PBS. The biofilm was treated with 500 μM curcumin, 0.12% chlorhexidine, or 1% BHIS as a control for 5 min or 24 h. After treatment, the biofilm was stained with L-7012 LIVE/DEAD® BacLight™ Bacterial Viability Kits (Molecular Probes, Eugene, OR, USA). The live/dead stain, stored at −20°C, was warmed to room temperature and centrifuged before use. The staining solution included two components, SYTO9 and propidium iodide, which were mixed in equal quantities and applied to the dish for 15 min. The excitation wavelengths for SYTO9 and propidium iodide are 488 and 543 nm. The biovolumes of live and dead cells were quantified from the entire stack using COMSTAT image-processing software. The biovolume is defined as the volume of the biomass (μm3) divided by the area of the substratum (confocal dish) (μm2) [22].
2.4.2. Analysis of EPS by CLSM
As described previously, a 24 h biofilm of S. mutans was grown in 15-mm confocal dishes, protected from light, with 1 μM Alexa Fluor 647® red fluorescent dye (Invitrogen Corp., Carlsbad, CA, USA) to label the EPS [23]. After incubation for 24 h at 37°C in a humidified atmosphere containing 5% CO2, treatments consisting of 500 μM curcumin, 0.12% chlorhexidine, and 1% BHIS as a control were added to separate dishes. Then, after incubation for 5 min or 24 h, the biofilms were gently washed with normal saline and incubated with 1 μM SYTO9 green fluorescent dye at room temperature for 15 min to label the live bacteria. The image collection gates were set to 655–690 nm for Alexa Fluor 647 and 495–515 nm for SYTO9. Images were obtained by CLSM and analysed using the image-processing software COMSTAT [24].
2.5. Scanning Electron Microscopy (SEM)
S. mutans 24 h biofilm was incubated in 1% BHIS medium with or without 500 μM curcumin or 0.12% chlorhexidine at 37°C for 5 min or 24 h. Next, the S. mutans biofilms were washed twice with PBS, fixed with 2.5% glutaraldehyde overnight at 4°C, and washed with PBS. After dehydration by an alcohol gradient (30%, 50%, 70%, 80%, 85%, 90%, 95%, and 100%), the biofilm was dried in a desiccator and sputter-coated with gold. Then, the sample were examined at 2000x, 5000x, and 10,000x magnification by SEM [23, 24].
2.6. Assay of Gene Expression by Real-Time PCR
Biofilms grown for 24 h were treated with drugs for 5 min or 24 h. The biofilms were harvested by centrifugation at 12,000 rpm for 5 min, and the total RNA was isolated by ultrasonic crushing and using the RNeasy Mini Kit (QIAGEN, Valencia, CA, USA). The total RNA concentration and purity were determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Pittsburgh, PA, USA). The reverse transcription of the total RNA and quantitative real-time PCR were performed similarl to the procedures described in previous studies [23]. The primer sequences were based on the previous literature and are listed in Table 1 [17, 24–27]. The 2−ΔΔCt method was used to calculate the gene expression fold changes in S. mutans.
Table 1.
Nucleotide sequence of primers used for qRT-PCR.
| Gene | Primer sequence (5′-3′) | |
|---|---|---|
| Forward | Reverse | |
| gtfB | ACACTTTCGGGTGGCTTG | GCTTAGATGTCACTTCGGTTG |
| gtfC | CCAAAATGGTATTATGGCTGTCG | GAGTCTCTATCAAAGTAACGCAGT |
| gtfD | TTGACGGTGTTCGTGTTGAT | AAAGCGATAGGCGCAGTTTA |
| fruA | TGTAGGTCTCGGTTTGTGGGAC | TCTTGAGCCAATGCTTCTGGTG |
| gbpB | AGCAACAGAAGCACAACCATCAG | CCACCATTACCCCAGTAGTTTCC |
| Ftf | CTGACATAACTACGCCAAAG | TGCTTAAATTAATACCAGCTTC |
| luxS | CCAGGGACATCTTTCCATGAGAT | ACGGGATGATTGACTGTTCCC |
| comC | GACTTTAAAGAAATTAAGACTG | AAGCTTGTGTAAAACTTCTGT |
| comD | CTCTGATTGACCATTCTTCTGG | CATTCTGAGTTTATGCCCCTC |
| comE | CCTGAAAAGGGCAATCACCAG | GGGGCATAAACTCAGAATGTGTCG |
| VicR | TGACACGATTACAGCCTTTGATG | CGTCTAGTTCTGGTAACATTAAGTCCAATA |
| atpH | ACCATACATTTCAGGCTG | TTTTAGCACTTGGGATTG |
| scrA | GATTGCCCTCAGCAGTTGACAT | GCTGGGAAACTTTGATGGAGAC |
| scrB | ACAGCCTGTCCTGATTTATAGTC | CTGGTAACCCAATCCATGAGAC |
| srtA | GAAGCTTCCTGTAATTGGCG | TTCATCGTTCCAGCACCATA |
| spaP | GACTTTGGTAATGGTTATGCATCAA | TTTGTATCAGCCGGATCAAGTG |
| 16sRNA | CTTACCAGGTCTTGACATCCCG | ACCCAACATCTCACGACACGAG |
2.7. Statistical Analysis
Each experiment was independently repeated at least three times. GraphPad Prism version 5.04 (GraphPad Software, San Diego, CA) was used to assess the data. The differences between the experimental group and the untreated control group were statistically analysed using the SPSS 17.0 software. The data were assessed to determine whether they were normally distributed. One-way analysis of variance (ANOVA) and Tukey's test were performed for multiple groups. The unpaired t-test was used for two groups. The significance level of the P value was <0.05.
3. Results
3.1. Determination of the MIC of Curcumin against Planktonic and Biofilm S. mutans
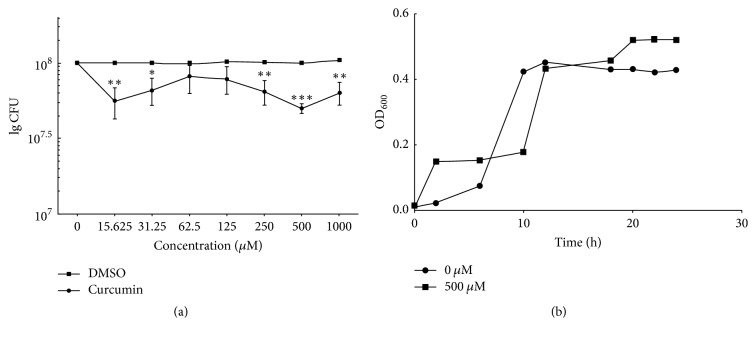
The results showed that curcumin inhibited the growth of planktonic S. mutans with an MIC of 125 μM. After treatment with concentrations higher than 125 μM, no visible S. mutans growth was observed. The MTT assay showed that, after treatment with serial dilutions of curcumin, the biofilm viability decreased significantly, by 50%, at 500 μM. The DMSO has no effect on viability of biofilm (Figure 1(a)). The growth curve at concentrations of 500 μM and for the control showed similar trends. The bacteria in both cases reached the stationary phase at 12 h (Figure 1(b)). The variation in biofilm biomass between 500 μM and 0 was independent of the growth kinetics. Therefore, a concentration of 500 μM was defined as causing 50% inhibition (SMIC50%) and selected for the next study.
Figure 1.
(a) Antibiofilm effects of different concentrations of curcumin and the correspondent concentrations of DMSO on S. mutans biofilm. The bacteria were inoculated in a 96-well microtiter plate containing 1% BHIS medium to form a 24 h biofilm, then washed with PBS, and incubated in 1% BHIS with different concentrations of curcumin (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). After 24 h of incubation, the Colony-Forming Units (CFU) of lived bacteria in biofilm were evaluated by the MTT assay. (b) Effect of SMIC50 curcumin on the growth of S. mutans.
3.2. Inhibition of the Metabolism of Biofilm Formation by Curcumin
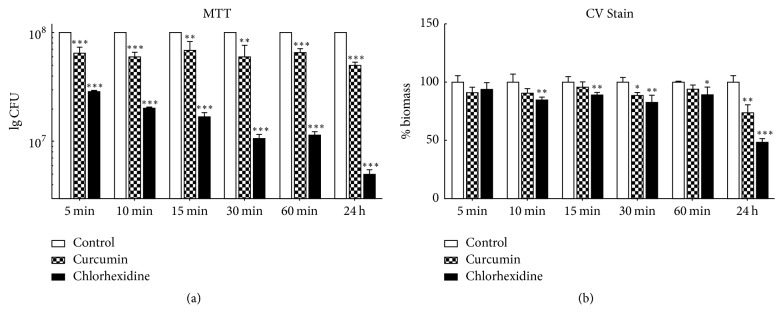
Both MTT and CV assays were conducted to evaluate the alteration in relevant biofilm characteristics of S. mutans under the influence of curcumin. In addition, 0.12% chlorhexidine treatment was used to provide a comparison group. The results of the MTT assay showed that curcumin decreased the biofilm metabolism at both 5 min and 24 h compared to that of untreated biofilm (P < 0.001, Figure 2(a)). Chlorhexidine treatment produced the same trend as curcumin, and the biofilm metabolism decreased significantly more than for curcumin (P < 0.001, Figure 2(a)). The CV assay showed that curcumin and chlorhexidine both resulted in reduced S. mutans biomass at 24 h. Chlorhexidine resulted in a significantly greater decrease than curcumin (P < 0.001, Figure 2(b)). As curcumin decreased the metabolism of the S. mutans biofilm in a short time (5 min), we next compared the short-term (5 min) and long-term (24 h) effects of curcumin on S. mutans biofilm.
Figure 2.
Antibiofilm effect of different curcumin exposure times on S. mutans biofilm. A 24 h biofilm was incubated in curcumin at the SMIC50 for different times. The Colony-Forming Units (CFU) of lived bacteria in biofilm were evaluated by the MTT assay (a), and the percentage of biofilm biomass was evaluated by the crystal violet (CV) assay (b). The data represent the mean ± SD of three independent tests. The asterisks (∗) indicate significant differences (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001).
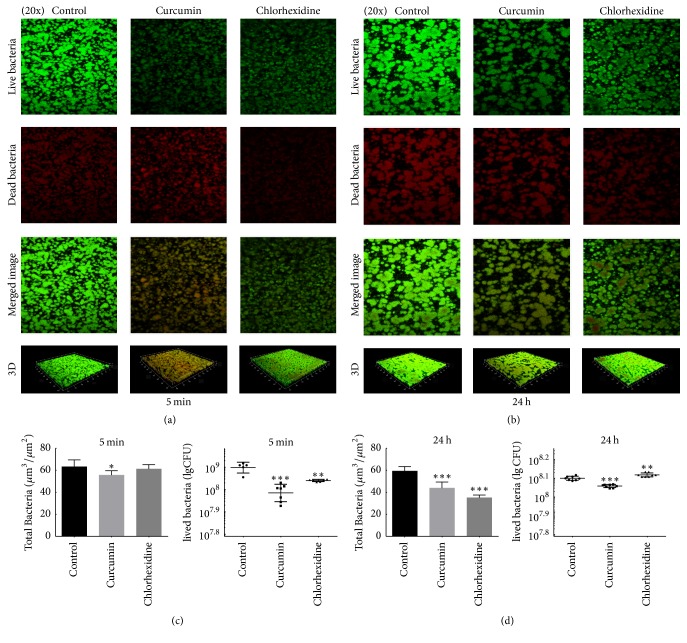
3.3. Evaluation of the Effect on the Live/Dead Bacteria Ratio in S. mutans Biofilm
The effects of curcumin and chlorhexidine on the live/dead bacteria ratio in S. mutans biofilm were assayed by CLSM (Figure 3). In the confocal micrographs, green florescence indicates lived cells, while red fluorescence indicates dead cells. The images reflect different green and red florescence intensities from those in the control group. Lower green-stained density than in the control group was observed in both treated groups at 5 min and 24 h (Figures 3(a) and 3(b)). At 5 min, the live bacteria in the curcumin group and chlorhexidine group were reduced, while the total bacteria in the curcumin group were reduced (Figure 3(c)). At 24 h, the live bacteria in the curcumin group were reduced, while the total bacteria were reduced in both groups (Figure 3(d)).
Figure 3.
CLSM imaging of S. mutans biofilm grown in 1% BHIS. After 24 h of growth, the biofilm was treated with 1% BHIS (control), 500 μM curcumin, or 0.12% chlorhexidine for 5 min (a) and 24 h (b). Each micrograph represents 4 optical sections: green representing live bacteria, red representing dead bacteria, combined green and red from two channel images, and three-dimensional reconstructions of the control biofilm without any treatment, the 500 μM curcumin-treated biofilm, and the 0.12% chlorhexidine-treated biofilm. The total bacteria biomass and the Colony-Forming Units (CFU) of lived bacteria are quantified in (c) and (d). The data represent the mean ± SD. The asterisks (∗) indicate significant differences (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001).
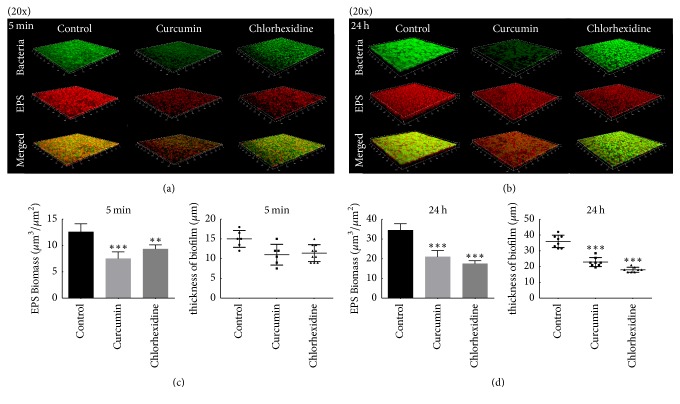
3.4. Evaluation of the Effect on the EPS of S. mutans Biofilm
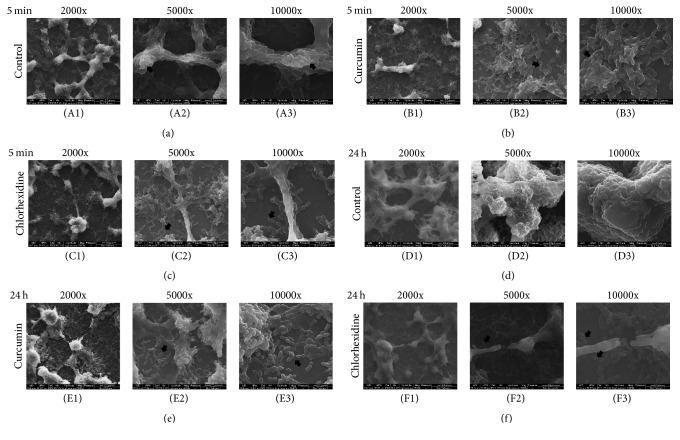
The three-dimensional reconstruction images obtained from CLSM of the S. mutans biofilm (Figures 4(a) and 4(c)) confirmed that the EPS decreased with both curcumin and chlorhexidine treatment after 5 min (P < 0.05), and this trend continued at 24 h. The thickness of the biofilm showed no difference among the three groups at 5 min but was visibly reduced at 24 h (Figures 4(b) and 4(d)). The SEM image confirmed the change in EPS (Figure 5). Treatment with curcumin or chlorhexidine was able to reduce the overall quantity of S. mutans EPS compared to the untreated control, as indicated by arrows. Furthermore, long-term treatment (24 h) resulted in a more obvious reduction than short-term treatment (5 min).
Figure 4.
CLSM images of S. mutans biofilm. (a, b) Three-dimensional reconstructions of the untreated (control) biofilm, the 500 μM curcumin-treated biofilm, and the 0.12% chlorhexidine-treated biofilm at 5 min (a) and 24 h (b). EPS was labelled in red (Alexa Fluor 647), bacterial cells were labelled in green (SYTO9), and red and green superimposed appear as yellow. Images were obtained at 20x magnification. (c) Image representing the volume of EPS, calculated according to 5 random sites of each sample, repeated three times. (d) Change in biofilm thickness at 5 min and 24 h, respectively, calculated from data obtained from fluorescence CLSM. Data are presented as the mean ± SD. The asterisks (∗) indicate significant differences (∗∗P < 0.01, ∗∗∗P < 0.001).
Figure 5.
Morphological characteristics of S. mutans biofilm treated with or without drugs. Representative SEM images of 24 h S. mutans biofilm grown in curcumin for 5 min (b) and 24 h (e) and grown in chlorhexidine for 5 min (c) and 24 h (f). The image of 24 h S. mutans biofilm grown in 1% BHIS for 5 min (a) and 24 h (d) as a control. Magnifications of 2000x, 5000x, and 10,000x are shown for each condition. The black arrows highlight the EPS of S. mutans, which was markedly reduced.
3.5. Evaluation of the Effect on the mRNA Levels of S. mutans Biofilm Virulence Factors
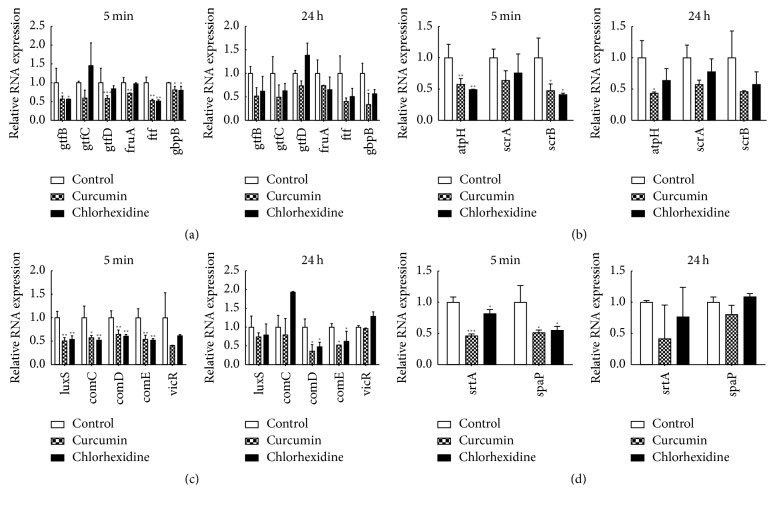
Compared with the levels observed in S. mutans biofilms grown in 1% BHIS, the expression levels of gtfB, gtfC, and gtfD in S. mutans biofilms exposed to 500 μM curcumin for 5 min were significantly decreased by 0.57-, 0.59-, and 0.59-fold, respectively. At 24 h, the expression levels of gtfB, gtfC, and gtfD in S. mutans were also decreased, and there was no significant difference from the levels at 5 min. Meanwhile, the expression of gtfB, gtfC, and gtfD was also downregulated under the influence of 0.12% chlorhexidine for 5 min and 24 h; there was no significant difference between two time points (Figure 6(a)).
Figure 6.
Results of qRT-PCR to examine the gene expression of different virulence systems in S. mutans UA159. (a) EPS synthesis system; (b) carbohydrate metabolism system; (c) quorum-sensing system; (d) two-component transduction system. All targets were amplified using primers. Different gene expression levels were normalized to the level of 16sRNA gene transcripts. Data are presented as the mean ± SD. The asterisks (∗) indicate significant differences (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001).
In Figure 6(b), we observed no significant difference in the expression of scrA at either 5 min or 24 h. However, the expression of atpH and scrB decreased in the presence of curcumin (P < 0.05). The expression of atpH decreased further over time. In contrast, the expression of scrB increased over time.
The entire set of virulence genes involved in quorum-sensing signalling mechanisms was found to be downregulated after treatment with curcumin. Treatment with curcumin for 5 min repressed the expression levels of luxS, comC, comD, and comE to 50.7%, 57.4%, 64.8%, and 54.1%, respectively, as compared to the control values. Likewise, chlorhexidine repressed the gene expression levels to 50.1%, 52.3%, 60.7%, and 52.3%, respectively. After treatment with the drugs for 24 h, only the expression of comDE showed a significant difference.
The expression levels of srtA and spaP in the biofilms treated with curcumin for 5 min were significantly downregulated by approximately onefold compared with the levels in untreated biofilm, and the same tendency was shown by chlorhexidine (Figure 6(d)). After 24 h of treatment with either drug, the expression levels of srtA and spaP showed no significant difference.
4. Discussion
Dental caries, a common infectious disease worldwide, is caused by biofilm known as dental plaque that result from the adhesion of bacteria to tooth surfaces. The effective eradication of cariogenic biofilm and the pathogenic microorganisms within is the key way to prevent tooth decay [28]. S. mutans is the main aetiological microorganism involved in biofilm [29]. In this study, we used curcumin, a natural food-grade product, to control the formation of S. mutans biofilm.
Previous studies have shown that curcumin could inhibit the viability of S. mutans with an MIC value of 125–175 μM [17]. Similar MIC results were obtained in this study, but the SMIC values here were much higher than the MIC. As is well known, it is more difficult to inhibit the viability of S. mutans biofilm than planktonic bacteria because the planktonic bacteria are more susceptible to the effects of commonly used antimicrobial drugs than the bacteria embedded in a biofilm [30, 31].
Accordingly, we evaluated the effect of curcumin on S. mutans biofilm over time. At the SMIC50, curcumin was shown to exert both a short-term effect and a long-term effect on the viability of S. mutant biofilm, which was also true for 0.12% chlorhexidine. Previous studies showed the effect of chlorhexidine on S. mutant biofilm. Yang et al. [32] reported that S. mutans biofilm was susceptible to 0.12% chlorhexidine, with a biomass reduction of over 80% observed after treatment with 0.12% chlorhexidine for 1 h. Tamura et al. [33] found that 8 μg/ml chlorhexidine was effective in removing biofilm after 24 h. All of these results illustrate that chlorhexidine has antibiofilm effects, which was consistent with our results. The short-term effect of curcumin on S. mutans biofilm also suggested that curcumin has the potential to be a new antibiofilm drug because it exhibited similar effects to chlorhexidine.
S. mutans has a regulatory network to integrate its cellular response to environmental change [6]. This study found that, after treatment with curcumin, the amounts of viable bacteria and total bacteria were reduced, as confirmed by the results of CLSM. Because curcumin exhibited no effect on the growth kinetics of S. mutans, the reduced quantity of live bacteria indicated a direct effect of curcumin on S. mutans.
A biofilm is a highly dynamic and structured community of microbial cells that are enmeshed in an extracellular matrix of polymeric substances such as exopolysaccharides, proteins, and nucleic acids [34]. As reported previously, the production of EPS by S. mutans on a surface enhances the local accumulation and clustering of microbes. In addition, as the biofilm develops, the spatial heterogeneities resulting from EPS synthesis form a complex 3D matrix architecture and create environmental and protective niches within the biofilm that can directly modulate caries pathogenesis [34, 35]. Curcumin was shown to destroy the structure of EPS in the short term, decreasing the biomass of EPS, the quantity of total bacteria, and the percentage of lived bacteria. With prolonged curcumin exposure, the structure of the EPS was badly damaged, and the thickness of the biofilm and the number of total bacteria were decreased demonstrably. Khan et al. reported that the extract of Trachyspermum ammi seeds could modulate the expression of specific virulence genes by S. mutans, which disrupted the accumulation and structural organization of EPS, ultimately affecting the cariogenicity of S. mutans [36]. Xu et al. found that the tea catechin epigallocatechin gallate could inhibit the formation of S. mutans biofilm by suppressing the gtfs gene [37]. These studies serve as a reminder that the effects of natural products on the EPS of S. mutans biofilm are usually exerted by regulating cariogenic genes.
As previously reported, the regulatory systems in S. mutans, such as the EPS synthesis system, carbohydrate metabolism system, quorum-sensing system, and two-component transduction system, are connected to the ecological balance and to bacterial carcinogenesis [6, 38]. The results showed that curcumin decreased the expression of cariogenic genes in the plaque of S. mutans. Although the majority of the 24 h qPCR results were not statistically significant, the P values between the control group and curcumin group were close to 0.05, and the decrease was pronounced. Thus, curcumin has an inhibitory effect on certain genes in S. mutans at 24 h.
Gtfs, encoded by the gtfBCD genes, are indispensable for the utilization of glucose and for EPS synthesis in S. mutans. They are important factors in biofilm formation and the development of caries [24]. GtfB synthesizes water-insoluble polysaccharide [39], which is the main component of the EPS. Downregulation of the expression of gtfB in curcumin might explain the decreased biomass of EPS in the short term. Oral bacterial aggregation is mediated by interactions between surface-associated glucan-binding proteins (GbpBs) that adhere to glucans, thereby promoting plaque formation [40]. The expression of gbpB was decreased in both treatment groups, which may relate to the reduced thickness of the biofilm. Fructosyltransferase (FTF) is an enzyme that converts sucrose to extracellular homopolymers of fructose, the fructans. FTF is the product of the fruA gene and is an exo-β-d-fructosidase that releases fructose from β(2.6)- and β(2.1)-linked fructans and cleaves fructose from sucrose and raffinose [41–43]. Both ftf and fruA expressions were dysregulated after short-term exposure to curcumin.
In S. mutans, the scrA gene in the phosphotransferase system (PTS) encodes a high-affinity permease, which internalizes sucrose. Then, intracellular sucrose-6-PO4 is first hydrolyzed by ScrB, a sucrose-6-PO4 hydrolase, to produce fructose and glucose-6-PO4. In addition, the downstream of scrA and scrB could encode a regulator to change the expression level of scrAB. Both scrAB genes were downregulated in our study, which might contribute to the reduction in the S. mutans biofilm. In the oral environment, S. mutans is subjected to rapid pH fluctuations arising from the availability of carbohydrates. The atpH gene in S. mutans encodes subunit C of a multisubunit enzyme (F1F0-ATPase) involved in intracellular pH regulation and acid tolerance [44, 45]. Gene expression analysis found that atpH was downregulated in S. mutans biofilm after treatment with curcumin for 5 min and 24 h. Thus, curcumin inhibits the acid tolerance ability of S. mutans and probably decreases the cariogenic traits of S. mutans.
The quorum-sensing system is an essential component of entire gene regulation networks responsible for the adaptation of bacteria in biofilms [46]. Two well-studied quorum-sensing systems exist in S. mutans: the ComCDE system, which can enable intraspecies cell-cell communication and has been proved to have a positive regulatory effect on the expression of biofilm-related genes such as gtfB, gtfC, and gbpB in S. mutans, and the LuxS system, which can catalyse the formation of the signal peptide AI-2 to mediate interspecies and intraspecies interaction in the multispecies plaque community [6, 47]. The findings of the current work suggest that curcumin can potentially contribute to reducing the quantity of lived bacteria in the biofilm and decrease the biomass of EPS, by decreasing the cariogenic potential of S. mutans biofilm, via inhibiting the expression of ComCDE system and LuxS system. The short-term inhibitory effect is more significant than the long-term effect. There is a decreasing tendency in the VicR expression of S. mutans after curcumin treatment, suggesting that curcumin may act by inhibiting the expression of VicR. The regulatory network of cariogenic genes in S. mutans is complex. It was reported that the ComR/ComS is the second quorum-sensing system in S. mutans [48]. The cell-to-cell communication system in S. mutans is not clear. Further research is required in this field.
S. mutans possesses a series of cell-surface proteins such as SpaP (also known as antigen I/II, Pac, P1 and antigen B), which are anchored to the cell envelope by sortase A (SrtA). Sortase A is encoded by the gene srtA and recognizes a motif consisting of leucine, proline, X, threonine, and glycine (LPXTG, where X is any amino acid) [33, 49]. Natural products attract substantial attention because they can inhibit srtA in vitro and in vivo but do not significantly decrease the viability of bacteria. The current study demonstrates that curcumin can inhibit the expression of spaP and srtA with an SMIC50 of 500 μM. The present study explored the effect of curcumin on S. mutans UA159. However, S. mutans has heterogeneity in different strains. Much more strains of S. mutans are needed to be test in future.
In conclusion, curcumin has both a short-term and a long-term antibacterial effect. Additionally, it is a food-grade natural product with a similar effect to that of chlorhexidine and it takes effect faster than 0.12% chlorhexidine. Curcumin could be an alternative strategy to treat oral disease. Curcumin is a promising antibacterial agent with potential for wider clinical application in the future.
Acknowledgments
This work was supported by Science and Technology Planning Project of Guangdong Province, China (2014A020212621 and 2017A020215064).
Contributor Information
Huancai Lin, Email: linhcgz@126.com.
Yan Zhou, Email: zhouy10.3@163.com.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Pitts N. B., Zero D. T., Marsh P. D., et al. Dental caries. Nature Reviews Disease Primers. 2017;3 doi: 10.1038/nrdp.2017.30.17030 [DOI] [PubMed] [Google Scholar]
- 2.Pereira D. F. A., Seneviratne C. J., Koga-Ito C. Y., Samaranayake L. P. Is the oral fungal pathogen Candida albicans a cariogen? Oral Diseases. 2017 doi: 10.1111/odi.12691. [DOI] [PubMed] [Google Scholar]
- 3.Nyvad B., Crielaard W., Mira A., Takahashi N., Beighton D. Dental caries from a molecular microbiological perspective. Caries Research. 2013;47(2):89–102. doi: 10.1159/000345367. [DOI] [PubMed] [Google Scholar]
- 4.Marquis R. E. Oxygen metabolism, oxidative stress and acid-base physiology of dental plaque biofilms. Journal of Industrial Microbiology and Biotechnology. 1995;15(3):198–207. doi: 10.1007/BF01569826. [DOI] [PubMed] [Google Scholar]
- 5.Bowen W. H., Koo H. Biology of streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Research. 2011;45(1):69–86. doi: 10.1159/000324598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith E. G., Spatafora G. A. Gene regulation in S. mutans: complex control in a complex environment. Journal of Dental Research. 2012;91(2):133–141. doi: 10.1177/0022034511415415. [DOI] [PubMed] [Google Scholar]
- 7.Krzysciak W., Jurczak A., Koscielniak D., Bystrowska B., Skalniak A. The virulence of Streptococcus mutans and the ability to form biofilms. European Journal of Clinical Microbiology & Infectious Diseases. 2014;33(4):499–515. doi: 10.1007/s10096-013-1993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merritt J., Kreth J., Qi F., Sullivan R., Shi W. Non-disruptive, real-time analyses of the metabolic status and viability of Streptococcus mutans cells in response to antimicrobial treatments. Journal of Microbiological Methods. 2005;61(2):161–170. doi: 10.1016/j.mimet.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 9.Fricks-Lima J., Hendrickson C. M., Allgaier M., et al. Differences in biofilm formation and antimicrobial resistance of Pseudomonas aeruginosa isolated from airways of mechanically ventilated patients and cystic fibrosis patients. International Journal of Antimicrobial Agents. 2011;37(4):309–315. doi: 10.1016/j.ijantimicag.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Utsumi R. Bacterial Signal Transduction: Networks and Drug Targets. Vol. 631. New York, NY, USA: Springer; 2008. [DOI] [PubMed] [Google Scholar]
- 11.Koo H., Bowen W. H. Candida albicans and Streptococcus mutans: a potential synergistic alliance to cause virulent tooth decay in children. Future Microbiology. 2014;9(12):1295–1297. doi: 10.2217/fmb.14.92. [DOI] [PubMed] [Google Scholar]
- 12.Hamada S., Koga T., Ooshima T. Virulence factors of Streptococcus mutans and dental caries prevention. Journal of Dental Research. 1984;63(3):407–411. doi: 10.1177/00220345840630031001. [DOI] [PubMed] [Google Scholar]
- 13.Jeon J.-G., Rosalen P. L., Falsetta M. L., Koo H. Natural products in caries research: current (limited) knowledge, challenges and future perspective. Caries Research. 2011;45(3):243–263. doi: 10.1159/000327250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teow S.-Y., Liew K., Ali S. A., Khoo A. S.-B., Peh S.-C. Antibacterial Action of Curcumin against Staphylococcus aureus: A Brief Review. Journal of Tropical Medicine. 2016;2016:10. doi: 10.1155/2016/2853045.2853045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song J., Choi B., Jin E.-J., Yoon Y., Choi K.-H. Curcumin suppresses Streptococcus mutans adherence to human tooth surfaces and extracellular matrix proteins. European Journal of Clinical Microbiology & Infectious Diseases. 2012;31(7):1347–1352. doi: 10.1007/s10096-011-1448-y. [DOI] [PubMed] [Google Scholar]
- 16.Hu P., Huang P., Chen M. W. Curcumin reduces Streptococcus mutans biofilm formation by inhibiting sortase A activity. Archives of Oral Biolog. 2013;58(10):1343–1348. doi: 10.1016/j.archoralbio.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Hu P., Huang P., Chen W. M. Curcumin inhibits the sortase a activity of the streptococcus mutans UA159. Applied Biochemistry and Biotechnology. 2013;171(2):396–402. doi: 10.1007/s12010-013-0378-9. [DOI] [PubMed] [Google Scholar]
- 18.Manoil D., Filieri A., Gameiro C., et al. Flow cytometric assessment of Streptococcus mutans viability after exposure to blue light-activated curcumin. Photodiagnosis and Photodynamic Therapy. 2014;11(3):372–379. doi: 10.1016/j.pdpdt.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Pfaller M. A., Diekema D. J. Progress in antifungal susceptibility testing of Candida spp. by use of Clinical and Laboratory Standards Institute broth microdilution methods, 2010 to 2012. Journal of Clinical Microbiology. 2012;50(9):2846–2856. doi: 10.1128/JCM.00937-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weerasekera M. M., Wijesinghe G. K., Jayarathna T. A., et al. Culture media profoundly affect Candida Albicans and Candida tropicalis growth, adhesion and biofilm development. Memorias do Instituto Oswaldo Cruz. 2016;111(11):697–702. doi: 10.1590/0074-02760160294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J., Li T., Zhou X., et al. Characterization of the clustered regularly interspaced short palindromic repeats sites in Streptococcus mutans isolated from early childhood caries patients. Archives of Oral Biolog. 2017;83:174–180. doi: 10.1016/j.archoralbio.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 22.Sun M., Zhou Z., Dong J., Zhang J., Xia Y., Shu R. Antibacterial and antibiofilm activities of docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA) against periodontopathic bacteria. Microbial Pathogenesis. 2016;99:196–203. doi: 10.1016/j.micpath.2016.08.025. [DOI] [PubMed] [Google Scholar]
- 23.Huang X., Zhang K., Deng M., et al. Effect of arginine on the growth and biofilm formation of oral bacteria. Archives of Oral Biolog. 2017;82:256–262. doi: 10.1016/j.archoralbio.2017.06.026. [DOI] [PubMed] [Google Scholar]
- 24.Liu S., Qiu W., Zhang K., et al. Nicotine enhances interspecies relationship between streptococcus mutans and candida albicans. BioMed Research International. 2017;2017:9. doi: 10.1155/2017/7953920.7953920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bedran T. B. L., Grignon L., Spolidorio D. P., Grenier D. Subinhibitory concentrations of triclosan promote Streptococcus mutans biofilm formation and adherence to oral epithelial cells. PLoS ONE. 2014;9(2) doi: 10.1371/journal.pone.0089059.e89059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levesque C. M., Voronejskaia E., Huang Y.-C. C., Mair R. W., Ellen R. P., Cvitkovitch D. G. Involvement of sortase anchoring of cell wall proteins in biofilm formation by Streptococcus mutans. Infection and Immunity. 2005;73(6):3773–3777. doi: 10.1128/IAI.73.6.3773-3777.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nascimento M. M., Lemos J. A., Abranches J., Lin V. K., Burne R. A. Role of RelA of Streptococcus mutans in global control of gene expression. Journal of Bacteriology. 2008;190(1):28–36. doi: 10.1128/JB.01395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunze B., Reck M., Dötsch A., et al. Damage of Streptococcus mutans biofilms by carolacton, a secondary metabolite from the myxobacterium Sorangium cellulosum. BMC Microbiology. 2010;10, article no. 199 doi: 10.1186/1471-2180-10-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leung V., Dufour D., Lévesque C. M. Death and survival in Streptococcus mutans: Differing outcomes of a quorum-sensing signaling peptide. Frontiers in Microbiology. 2015;6 doi: 10.3389/fmicb.2015.01176.01176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tofiño-Rivera A., Ortega-Cuadros M., Galvis-Pareja D., Jiménez-Rios H., Merini L. J., Martínez-Pabón M. C. Effect of Lippia alba and Cymbopogon citratus essential oils on biofilms of Streptococcus mutans and cytotoxicity in CHO cells. Journal of Ethnopharmacology. 2016;194:749–754. doi: 10.1016/j.jep.2016.10.044. [DOI] [PubMed] [Google Scholar]
- 31.Jhajharia K., Parolia A., Shetty K. V., Mehta L. K. Biofilm in endodontics: a review. Journal of International Society of Preventive & Community Dentistry. 2015;5(1):1–12. doi: 10.4103/2231-0762.151956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang H., Bi Y., Shang X., et al. Antibiofilm activities of a novel chimeolysin against Streptococcus mutans under physiological and cariogenic conditions. Antimicrobial Agents and Chemotherapy. 2016;60(12):7436–7443. doi: 10.1128/AAC.01872-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamura H., Yamada A., Kato H. Molecular characterization of the dextran-binding lectin B gene dblB of Streptococcus criceti in Streptococcus mutans strain GS-5 with mutations in both gbpC and spaP genes. Genes & Genetic Systems. 2014;89(2):41–50. doi: 10.1266/ggs.89.41. [DOI] [PubMed] [Google Scholar]
- 34.Klein M. I., Hwang G., Santos P. H. S., Campanella O. H., Koo H. Streptococcus mutans-derived extracellular matrix in cariogenic oral biofilms. Frontiers in Cellular and Infection Microbiology. 2015;5, article no. 10 doi: 10.3389/fcimb.2015.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koo H., Falsetta M. L., Klein M. I. The exopolysaccharide matrix: a virulence determinant of cariogenic biofilm. Journal of Dental Research. 2013;92(12):1065–1073. doi: 10.1177/0022034513504218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan R., Adil M., Danishuddin M., Verma P. K., Khan A. U. In vitro and in vivo inhibition of Streptococcus mutans biofilm by Trachyspermum ammi seeds: An approach of alternative medicine. Phytomedicine. 2012;19(8-9):747–755. doi: 10.1016/j.phymed.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 37.Xu X., Zhou X. D., Wu C. D. Tea catechin epigallocatechin gallate inhibits Streptococcus mutans biofilm formation by suppressing gtf genes. Archives of Oral Biolog. 2012;57(6):678–683. doi: 10.1016/j.archoralbio.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 38.Stephenson K. Two-component and phosphorelay signal-transduction systems as therapeutic targets. Current Opinion in Pharmacology. 2002;2(5):507–512. doi: 10.1016/S1471-4892(02)00194-7. [DOI] [PubMed] [Google Scholar]
- 39.Aoki H., Shiroza T., Hayakawa H., Sato S., Kuramitsu H. K. Cloning of a Streptococcus mutans glycosyltransferase gene coding for insoluble glucan synthesis. Infection and Immunity. 1986;53(3):587–594. doi: 10.1128/iai.53.3.587-594.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He Z., Wang Q., Hu Y., et al. Use of the quorum sensing inhibitor furanone C-30 to interfere with biofilm formation by Streptococcus mutans and its luxS mutant strain. International Journal of Antimicrobial Agents. 2012;40(1):30–35. doi: 10.1016/j.ijantimicag.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 41.Ogawa A., Furukawa S., Fujita S., et al. Inhibition of Streptococcus mutans biofilm formation by Streptococcus salivarius FruA. Applied and Environmental Microbiology. 2011;77(5):1572–1580. doi: 10.1128/AEM.02066-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burne R. A., Penders J. E. C. Differential localization of the Streptococcus mutans GS-5 fructan hydrolase enzyme, FruA. FEMS Microbiology Letters. 1994;121(2):243–249. doi: 10.1016/0378-1097(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 43.Burne R. A., Schilling K., Bowen W. H., Yasbin R. E. Expression, purification, and characterization of an Exo-β-D-fructosidase of Streptococcus mutans. Journal of Bacteriology. 1987;169(10):4507–4517. doi: 10.1128/jb.169.10.4507-4517.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sadeghinejad L., Cvitkovitch D. G., Siqueira W. L., Merritt J., Santerre J. P., Finer Y. Mechanistic, genomic and proteomic study on the effects of BisGMA-derived biodegradation product on cariogenic bacteria. Dental Materials. 2017;33(2):175–190. doi: 10.1016/j.dental.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bender G. R., Sutton S. V. W., Marquis R. E. Acid tolerance, proton permeabilities, and membrane ATPases of oral streptococci. Infection and Immunity. 1986;53(2):331–338. doi: 10.1128/iai.53.2.331-338.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y.-H., Tian X.-L., Layton G., Norgaard C., Sisson G. Additive attenuation of virulence and cariogenic potential of Streptococcus mutans by simultaneous inactivation of the ComCDE quorum-sensing system and HK/RR11 two-component regulatory system. Microbiology. 2008;154(11):3256–3265. doi: 10.1099/mic.0.2008/019455-0. [DOI] [PubMed] [Google Scholar]
- 47.Cvitkovitch D. G., Li Y.-H., Ellen R. P. Quorum sensing and biofilm formation in Streptococcal infections. The Journal of Clinical Investigation. 2003;112(11):1626–1632. doi: 10.1172/JCI200320430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khan R., Rukke H. V., Filho A. P. R., et al. Extracellular identification of a processed type II ComR/ComS pheromone of Streptococcus mutans. Journal of Bacteriology. 2012;194(15):3781–3788. doi: 10.1128/JB.00624-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang P., Hu P., Zhou S. Y., Li Q., Chen W. M. Morin inhibits sortase A and subsequent biofilm formation in streptococcus mutans. Current Microbiology. 2014;68(1):47–52. doi: 10.1007/s00284-013-0439-x. [DOI] [PubMed] [Google Scholar]