Abstract
Tension generated in the circular mitochondrial genome during replication and transcription points to the need for mtDNA topoisomerase activity. Here we report a 601-aa polypeptide highly homologous to nuclear topoisomerase I. The N-terminal domain of this novel topoisomerase contains a mitochondrial localization sequence and lacks a nuclear localization signal. Therefore, we refer to this polypeptide as top1mt. The pattern of top1mt expression matches the requirement for high mitochondrial activity in specific tissues. top1mt is a type IB topoisomerase that requires divalent metal (Ca2+ or Mg2+) and alkaline pH for optimum activity. The TOP1mt gene is highly homologous to the nuclear TOP1 gene and consists of 14 exons. It is localized on human chromosome 8q24.3.
DNA topoisomerases are enzymes that modify DNA topology by introducing reversible breaks in the DNA phosphodiester backbone. Topoisomerases are ubiquitous and essential for DNA strand separation, chromatin compaction, and DNA decatenation during replication and recombination. The first topoisomerase was identified in Escherichia coli in 1971 (1), and the existence of additional topoisomerases led to their classification into two types based on their enzymatic intermediates. The catalytic intermediate of type I topoisomerases is a DNA single-strand break. Type I enzymes have been subdivided further into two groups: type IA and type IB. Type IA enzymes break the DNA by forming a covalent bond to the 5′ end of the broken DNA, whereas type IB topoisomerases bind covalently to the 3′ end of the break (2, 3). The catalytic intermediate of type II topoisomerases is a DNA double-strand break with one enzyme molecule bound to each of the 5′ ends of the double-strand break (2–4). Until the present report, to our knowledge, two topoisomerase II genes (TOP2α and TOP2β), two topoisomerase III genes (TOP3α and TOP3β), and only one topoisomerase I gene (TOP1) had been identified in human cells.
Human mitochondria contain ≈5–10 copies per mitochondrion of a covalently closed duplex DNA genome (mtDNA; refs. 5 and 6) consisting of 16,569 base pairs (7, 8). The mitochondrial genome encodes 22 transfer RNAs, 13 mRNAs, and 12S and 16S ribosomal RNAs (7). The biogenesis of a functional mitochondrion requires both mitochondrial and nuclear gene products. For instance, the mtDNA polymerase (POLγ; ref. 9) and the essential mitochondrial transcription factor (mtTAF; ref. 10) are nuclear genes.
Because mtDNA molecules are closed circular and therefore topologically constrained, there must be a mechanism to relieve the DNA strain that arises during replication. The presence of mitochondrial top1 activity has been reported in mammalian cells (human leukemia cells, human platelets, calf thymus, bovine and rat liver, and mouse leukemia L cells; refs. 11–19), Xenopus oocytes (20), plants (21, 22), neurospora crassa (23), and yeast Saccharomyces cerevisiae (YSC; refs. 24–26). However, to our knowledge, until the present report, the small amounts of mitochondrial protein associated with this top1 activity have precluded sequencing of the corresponding polypeptide and characterization of its gene.
We now report the existence of an mtDNA topoisomerase I gene TOP1mt, which is localized on human chromosome 8q24, and the characterization of its corresponding polypeptide (top1mt).
Materials and Methods
Cloning of top1mt.
DNA manipulation, PCR, and DNA sequencing were performed according to standard protocols. Clones of AI 872335 and AW182340 were ordered from Incyte Genomics (St. Louis). The missing 5′-end portion of TOP1mt from clone AI 872335 was amplified from HeLa cells by using a Generacer kit (Invitrogen). The 5′ end was joined to the sequence derived from clone A1872335 to generate full-length TOP1mt. All oligonucleotide sequences used for cDNA identification are available on request.
Constructs.
To facilitate its cloning, expression, and purification, the starting ATG codon of TOP1mt was changed to an NdeI site. The His-tagged top1mt expression vector was constructed by cloning TOP1mt into the NdeI and XhoI sites of pET15b vector (Novagen). The green fluorescent protein (GFP)-tagged TOP1mt vector pEGFP-TOP1mt was constructed by cloning TOP1mt cDNA into the NheI and EcoRI sites of the pEGFP-N2 vector (CLONTECH). Variants of this vector, containing the N-terminal domain (NTD) or top1mt without the NTD, were constructed similarly.
Fluorescence Microscopy.
The different constructs of pEGFP were transiently transfected into subconfluent malignant glioma M059J cell cultures by using the Fugene kit (Roche Molecular Biochemicals). The subcellular localization of GFP fusion proteins was monitored by fluorescence microscopy 24 h after transfection.
Northern Blots.
Probes were prepared with the Rediprime random primer labeling system (Amersham Pharmacia Biotech) and then column-purified. Blots were prehybridized with 15 ml of Ultrahyb hybridization buffer (Ambion, Austin, TX) for 15 min at 42°C. Twenty microliters of the 50 μl probe reaction was added, and hybridization was allowed to proceed for 12 h at the same temperature. Blots were washed and exposed to PhosphorImager screens (Molecular Dynamics) overnight. The screens were then scanned.
Fluorescence in Situ Hybridization (FISH) Localization of the Human TOP1mt Gene.
A P1 clone carrying the whole TOP1mt gene was screened by using two sets of PCR pairs, one set from the first exon and another from the last exon. The P1 clone probe, labeled with biotin, was used for FISH of chromosomes derived from methotrexate-synchronized normal peripheral lymphocytes. The conditions of hybridization, detection of fluorescence signals, digital image acquisition, image processing and analysis, and direct localization of signals on banded chromosomes were carried out as described (27).
DNA Relaxation Assays.
Reactions contained 0.3 μg of supercoiled SV40 DNA in 10 μl final volume reaction buffer (10 mM Tris⋅HCl, pH 7.5/50 mM KCl/0.1 mM EDTA/15 μg/ml BSA) and purified topoisomerase. Reactions were performed at 30°C for 30 min and terminated by addition of 0.5% SDS. Loading buffer (1.1 μl of 10X) was added, and the reaction mixture was loaded onto a 1% agarose gel made in 1X 90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3 (TBE) buffer. After electrophoresis, DNA bands were stained with ethidium bromide and visualized by transillumination with UV light (300 nm).
DNA Cleavage Assays.
A 22-mer oligonucleotide with a single top1 cleavage site (refs. 28–31; MWG Biotech, High Point, NC) was labeled either at the 3′ end with [α-32P]cordycepin and terminal transferase or at the 5′ end with [γ-32P]ATP with polynucleotide kinase (31). Incubations with top1mt or top1 were at 25°C for 30 min in the presence of 10 μM camptothecin (CPT). Reactions were terminated by adding 0.5% SDS. Samples were either resolved on sequencing gel for visualizing cleavage products or by SDS/PAGE gel for visualizing DNA–protein complexes. Analyses were done by using a PhosphorImager and imagequant software (Molecular Dynamics).
Results
Cloning of Human Mitochondrial TOP1 cDNA.
A systematic search of the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) database, using the blast search engine, revealed the existence of a clone (AW182340) that encodes a novel human TOP1 sequence. Clone AW182340 contains a segment of 469 nucleotides that codes for a polypeptide homologous to the C-terminal region of the known top1. Full sequencing of this clone yielded a 626-bp segment coding for a polypeptide homologous to the C-terminal segment of top1 between residues 580 and 765 (Fig. 1a). Further analysis revealed the existence of a second clone (AI872335) with a 381-nt segment highly homologous to clone AW182340. Because of its relatively large insert size (≈2,000 base pairs), this clone was selected for full sequencing. It yielded a cDNA coding for a novel top1 polypeptide corresponding to residues 206 to 765 of the known top1 (Fig. 1a). By using Generacer, the 5′ segment of the novel top1 cDNA was obtained from HeLa cells by reverse transcription (RT)-PCR with primers derived from clone AI872335. Put together, the novel top1 cDNA consists of 1,923 nucleotides that encode a 601-aa polypeptide [Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org (GenBank accession no. AF349017)].
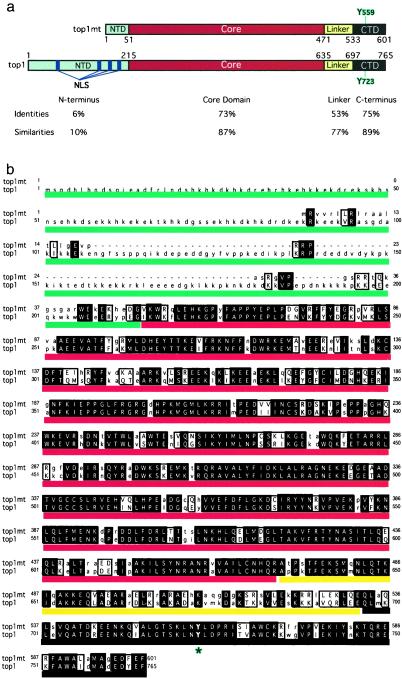
Figure 1.
Comparison between the novel human top1 (top1mt) and the previously known top1 (top1) polypeptides. (a) Domains are defined according to the nuclear top1 (33, 47). The positions of catalytic tyrosines are outlined in blue. The NTD, core domain, linker, and C-terminal domain are colored in blue, red, yellow, and black, respectively. Amino acid identities and homologies are indicated for each domain. There is no homology between the two NTDs, whereas the other domains are highly conserved. (b) Polypeptide sequence alignment for top1mt (upper lines) and the previously known human top1 (top1, lower lines). Identical matches are in uppercase, bold, and shaded; similar amino acids are in uppercase but not shaded. The unmatched amino acids are in lowercase. The catalytic tyrosines (Y723 for top1 and Y559 for top1mt) are indicated by an asterisk. Colored lines correspond to a: blue for the NTD, red for the core, yellow for the linker, and black for the C-terminal domain.
Fig. 1b shows the amino acid sequence comparison between the novel top1 (top1mt) and the previously known top1 (top1). The similarity between the two polypeptides is striking, except for the NTD of top1mt, which is much shorter than the NTD of top1. Overall, the top1mt shows a 52% identity and 64% similarity with top1. The segment of the C-terminal domain containing the top1 catalytic tyrosine (Y723) is highly conserved between the two top1 polypeptides, which suggests that Y559 is the catalytic tyrosine for top1mt.
Mitochondrial Localization of the Novel top1 Polypeptide.
In contrast to the previously identified top1, which contains several nuclear localization sequences (NLS) in its NTD (see Fig. 1a; refs. 32–34), the novel top1 lacks an amino acid sequence corresponding to an NLS. Examination of the NTD sequence from the novel top1 revealed the presence of positively charged residues that could fold in a positively charged amphiphilic helix suggestive of a mitochondrial targeting signal (35). Computer analysis, using mitoprot (http://www.mips.biochem.mpg.de/proj/medgen/mitop/) analysis, gave a 98% probability that the top1mt NTD contained a mitochondrial localization sequence. To test the cellular localization of the novel top1, an expression vector was constructed by fusing (in-frame) the GFP at the C terminus of the novel top1. The construct (top1mt-GFP) was transiently transfected into human glioma M059J cells, and the subcellular localization of the GFP fusion proteins was monitored by fluorescence microscopy. The fusion protein seemed to be concentrated in perinuclear structures but not in the nucleus, consistent with a mitochondrial distribution (Fig. 2b Left). Colocalization of the top1mt protein with the mitochondrial marker MitoTracker red CM-H2Xros (Molecular Probes; Fig. 2b Right) demonstrated the mitochondrial localization of the top1mt polypeptide.
Figure 2.
Mitochondrial localization of top1mt. a shows the constructs used: top1mt-GFP, NTD-GFP, and ΔNTD-top1mt-GFP. b and c show fluorescence microscopy images 24 h after transient transfection of human glioblastoma M059J cells with the constructs indicated above each. Mitochondria were localized by using MitoTracker red CM-H2Xros.
To better determine which part of the top1mt is responsible for its mitochondrial localization, we either fused the NTD of top1mt with GFP (NTD-GFP construct) or fused the core, linker, and C-terminal domains of top1mt with GFP (ΔNTD-top1mt-GFP construct; Fig. 2a). The NTD-GFP polypeptide, like top1mt-GFP, colocalized with the mitochondrial marker, whereas the ΔNTD-top1mt-GFP polypeptide showed a diffuse cytosolic cellular localization (Fig. 2c). These experiments demonstrate that the NTD of top1mt is necessary and sufficient for mitochondrial targeting.
top1mt Is a Type IB Topoisomerase Stimulated by Divalent Metals (Ca2+ or Mg2+) and Alkaline pH.
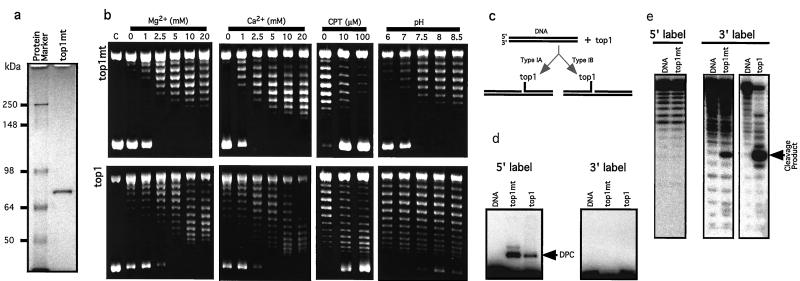
We next tested whether top1mt possesses topoisomerase activity. A His-tagged top1mt construct was made to express the protein. The recombinant protein was purified to apparent homogeneity after two-column purification (Fig. 3a).
Figure 3.
Biochemical characterization of top1mt. (a) Recombinant top1mt was expressed as a His-tagged protein, purified through nickel column and Q-Sepharose FPLC chromatography (Amersham Pharmacia Biotech). Coomassie blue staining shows that recombinant top1mt was homogeneous (≈72 kDa). (b) top1mt is a type I topoisomerase whose relaxation activity is inhibited by CPT, and stimulated by Ca2+ or Mg2+ in combination with a higher pH optimum than that of top1. Parallel experiments were performed with top1mt (Upper) and top1 (Lower). Experimental conditions are indicated above lanes. (c) Schematic representation of the covalent intermediates formed by types IA and IB topoisomerases. Type IA enzymes link to the 5′ terminus of the broken DNA, whereas type IB enzymes link to the 3′ terminus (3). (d) SDS/PAGE gel: both top1mt (72 kDa, His-tagged) and top1(68 kDa, His-tagged, N-terminal truncated) formed a DNA–protein complex (DPC) with 5′ end-labeled DNA (Left). No DPC was detected with 3′end-labeled DNA (Right). (e) Sequencing gel: both top1mt and top1 release a 3′ end-labeled oligonucleotide (Right); cleavage is not detectable when the DNA is 5′ end-labeled (Left).
Relaxation assays were performed by using native supercoiled SV40 DNA in the absence of added ATP and divalent metal. Among eukaryotic topoisomerases, only type I topoisomerases should be active under such conditions (4, 36). The top1mt had only weak DNA relaxation activity (lanes labeled 0 in Fig. 3b Upper Left). Addition of MgCl2 or CaCl2 markedly stimulated topoisomerase activity (Fig. 3b Upper Left). Similarly, we found that nuclear top1 was stimulated by MgCl2 (37) or CaCl2, although top1 was consistently more active than top1mt in the absence of divalent metal. top1mt was inhibited by the specific top1 inhibitor CPT (Fig. 3b Center Right) (38), a finding consistent with the high homology between top1mt and top1 (see Fig. 1). CPT also enhanced top1mt-mediated DNA cleavage (data not shown).
The two top1 enzymes differ in their pH requirement. The top1mt is most active at higher pH optimum (pH 8 or 8.5; Fig. 3b Upper Right) than nuclear top1, which is active over a wide pH range and most effective below pH 8, down to pH 6 (Fig. 3b Lower Right). This differential pH optimum for the two enzymes matches the physiological pH values measured in the nucleus and mitochondrion, respectively (39).
Type IA and IB topoisomerases are well known for their differences in DNA cleavage. Type IA enzymes cleave the DNA by forming a covalent bond to the 5′ end of the broken DNA, whereas type IB enzymes form a covalent bond to the 3′ end of the broken DNA (ref. 3; Fig. 3c). In human cells, topoisomerases IIIα and IIIβ belong to the type IA class, whereas top1 was until now the only type IB enzyme. To characterize top1mt, we used a duplex oligonucleotide containing a high-affinity top1 cleavage site (28–31). For both top1mt (72 kDa) and top1 (68 kDa, N terminus-truncated form), covalent DNA–protein complexes were detectable by SDS/PAGE only when the oligonucleotide was labeled at the 5′ end (Fig. 3d). Conversely, DNA cleavage in the absence of proteinase K was detectable only when the DNA was labeled at the 3′ end (Fig. 3e). These results indicate that top1mt cleaves DNA at the high-affinity top1 site and is a type IB topoisomerase.
Expression of top1mt RNA Is High in Tissues Rich in Mitochondria.
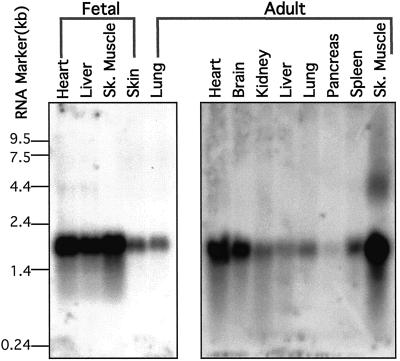
Northern blot analyses were carried out by using the full-length TOP1mt cDNA clone as a probe. As seen in Fig. 4, TOP1mt expression is highest in skeletal muscle, heart, brain, and fetal liver. Thus, the expression pattern of TOP1mt matches the requirement for high mitochondrial activity in these organs.
Figure 4.
Northern blot for TOP1mt expression. Blots were probed with our full-length TOP1mt cDNA. Two membranes are shown (Invitrogen). The four lanes on the Left are from human fetal tissues. The nine lanes on the Right are from human adult tissues. The size markers (in kb) are indicated Left.
The Human Mitochondrial Topoisomerase I Gene (TOP1mt) Is Located at 8q24.3 and Consists of 14 Exons That Are Conserved in Length with the Previously Known Nuclear TOP1.
On the basis of our TOP1mt cDNA clone (Fig. 1), two sets of primers were designed to screen a genomic library. The first set corresponded to the 5′ end of the TOP1mt cDNA, the other to the 3′ end. A P1 genomic clone (≈100 kb) containing the whole TOP1mt gene sequence was obtained. This clone was then used as a probe for FISH. In two FISH experiments with lymphocytes from different individuals, specific fluorescence signals were observed in virtually all 50 metaphases randomly selected in each sample. The signal was located in the terminal region of the long arm of chromosome 8 (on both chromatids; Fig. 5). Double fluorescence signals were not observed on any other chromosome, and nonspecific brighter fluorescent spots were sporadic. The location of the signals was determined directly in 20 metaphases with 4′,6-diamidino-2-phenylindole (DAPI)-enhanced G-like banding. The chromosome location of the TOP1mt gene is 8q24.3.
Figure 5.
FISH localization of TOP1mt. Metaphase chromosome spreads were derived from methotrexate-synchronized normal human peripheral leukocytes after hybridization with a genomic DNA probe and 4′,6-diamidino-2-phenylindole (DAPI) counterstaining. Both chromosomes with symmetrical FITC signals on sister chromatids (arrows) were identified as chromosome 8 on DAPI-inverted G-banded spreads, and fluorescent signals were localized at band 8q24.3.
Having obtained the full-length TOP1mt gene, we sequenced the flanking region of each exon to determine the exon/intron boundaries and sizes. Sequences for the genomic clone have been deposited in GenBank (accession nos. AF349018–AF349031). Table 1 shows a comparison of the overall gene structure for the TOPmt and TOP1 genes. The last 13 exons of both genes have the exact same size and intron frame. Also, the nucleotide homology is remarkably high (between 60 and 75%), and one stretch of 15 consecutive nucleotides in exon 9 of TOP1mt is common to TOP1mt and nuclear TOP1. By contrast, intron sizes are very different in the two genes.
Table 1.
Exon and intron sizes for the TOP1 (nuclear) and TOP1mt (mitochondrial) genes
| Number of exons
|
Exon sizes
|
Homology, % | Intron phase
|
Intron sizes
|
||||
|---|---|---|---|---|---|---|---|---|
| TOP1 | TOP1mt | TOP1 | TOP1mt | TOP1 | TOP1mt | TOP1 | TOP1mt | |
| 1 | 33 (280) | 0 | 330 | |||||
| 2 | 25 | 1 | 31,938 | |||||
| 3 | 97 | 2 | 14,680 | |||||
| 4 | 124 | 0 | 1,287 | |||||
| 5 | 56 | 2 | 2,447 | |||||
| 6 | 96 | 2 | 984 | |||||
| 7 | 76 | 0 | 3,221 | |||||
| 8 | 1 | 107 | 122 (141) | 27 | 2 | 2 | 7,903 | 3,400 |
| 9 | 2 | 116 | 116 | 60 | 1 | 1 | 4,632 | 1,752 |
| 10 | 3 | 122 | 122 | 65 | 0 | 0 | 873 | 3,005 |
| 11 | 4 | 123 | 123 | 62 | 0 | 0 | 1,718 | 688 |
| 12 | 5 | 188 | 188 | 75 | 2 | 2 | 965 | 716 |
| 13 | 6 | 145 | 145 | 64 | 0 | 0 | 11,428 | 342 |
| 14 | 7 | 144 | 144 | 70 | 0 | 0 | 1,044 | 2,612 |
| 15 | 8 | 186 | 186 | 75 | 0 | 0 | 1,215 | >2,500 |
| 16 | 9 | 69 | 69 | 72 | 0 | 0 | 838 | 180 |
| 17 | 10 | 115 | 115 | 72 | 1 | 1 | 1,776 | 1,596 |
| 18 | 11 | 128 | 128 | 71 | 0 | 0 | 3,399 | 177 |
| 19 | 12 | 95 | 95 | 64 | 2 | 2 | 215 | >5,000 |
| 20 | 13 | 150 | 150 | 67 | 2 | 2 | 1,039 | 524 |
| 21 | 14 | 103 (1298) | 103 (184) | 75 | ||||
Exon and intron sizes are in bp. For exons 1 and 14 of TOP1mt, and exons 1 and 21 of TOP1, sequence lengths given inside parentheses include noncoding regions. Sizes given without parentheses are for the coding regions, which were used for homology calculation. Intron sizes for TOP1 were derived from the National Center for Biotechnology Information (NCBI) database. Intron sizes of the TOP1mt gene are based on the human genome data (NCBI database) and sequencing data from our genomic clone (GenBank accession nos. AF349018–AF349031).
Discussion
In eukaryotic cells, DNA is localized in the nucleus and in mitochondria. mtDNA represents ≈3% of the DNA in human cells. It consists of 1,000–5,000 mitochondrial genome molecules per cell distributed in several hundred mitochondria (5, 6, 40). That each mtDNA is a circular duplex molecule of 16,659 base pairs, which needs to be transcribed, replicated, and repaired, points to the need for mitochondrial topoisomerase activities. The present study defines the cDNA and corresponding amino acid sequence of human mitochondrial type I topoisomerase (top1mt) and characterizes the biochemical properties, gene structure, and chromosomal localization of the enzyme.
The assignment of this novel top1 to mitochondria was initially suggested because the NTD of top1mt contains none of the nuclear localization signals previously identified in nuclear top1 (see Fig. 1), and because the amino acid sequence of the top1mt NTD is suggestive of a mitochondrial targeting sequence (35). More direct evidence was obtained by examining the cellular distribution of expression constructs fused to a GFP tracer. As shown in Fig. 2, the NTD of top1mt is necessary and sufficient to localize top1mt to mitochondria.
Previous reports on the biochemical characteristics of the mitochondrial topoisomerases have often diverged in their conclusions, probably because the mitochondrial topoisomerase I is in low abundance and difficult to purify in large quantities. The variety of organisms for which the enzymes have been identified could also contribute to some of the reported differences. Nevertheless, the size of top1mt is generally consistent with those of the previously reported mitochondrial top1 enzymes [i.e., between 60 and 70 kDa (11–15, 20, 23, 41, 42)]. The calcium dependence of the purified mitochondrial top1 is also consistent with previous biochemical reports (14, 15, 25, 41, 42). The optimum activity of top1mt at alkaline pH, whereas the pH optimum for nuclear top1 is at neutral or acidic pH, fits the known pH values for the mitochondrion and the nucleus, respectively (39). Activity of purified mitochondrial top1 at alkaline pH has also been reported (14, 15, 18, 19, 23, 25). Thus, it is likely that the top1mt cloned in this study corresponds to the previously isolated enzyme activities.
The top1mt is sensitive to CPT (25, 41, 42), a specific inhibitor of eukaryotic top1 that generates DNA strand-breaks by blocking the religation of top1-linked DNA breaks (38, 43). The CPT sensitivity of top1mt is not surprising considering the high degree of homology between top1mt and top1 (see Fig. 1), especially in the polypeptide domains that are known to be critical for CPT activity and which are probably involved in CPT binding to top1-DNA complexes (38). Whether top1mt contributes to the cytotoxicity and anticancer activity of CPT remains to be determined. However, small molecules [such as ethidium bromide (6) and ditercalinium (44)] that enter mitochondria are generally positively charged, whereas CPT and its clinically used derivatives are not (38).
The high degree of homology between top1mt and nuclear top1 is striking, at both the amino acid (Fig. 1) and nucleotide levels (Table 1). Biochemically, both top1mt and top1 are type IB enzymes (Fig. 3). Based on sequence homology with top1, we propose that the top1mt catalytic residue is Tyr-559 (Fig. 1). The exon structure of the two genes is completely conserved (Table 1), suggesting that the TOP1mt gene is not derived from any of the prokaryotic top1 genes, which are very different from their eukaryotic counterparts (3). The TOP1mt gene presumably arose by duplication and modification of an early nuclear TOP1 gene. The antiquity of this duplication event is reflected by the great difference between the nuclear and mitochondrial genes in the size and sequence of their introns (Table 1).
Recently, mitochondrial disorders have emerged as important causes of hereditary diseases such as late-onset neurodegeneration (including ataxia, dementia, blindness, and deafness), myopathies (including myocardiopathies), and metabolic abnormalities (including diabetes mellitus) (6, 40, 45). Mitochondrial diseases can result from mtDNA alterations, which are maternally inherited and accumulate with age. They can also result from recessive genetic alterations of nuclear genes. The importance of nuclear genome alterations in the genesis of mitochondrial diseases is not surprising, because only 13 of the estimated 1,000 mitochondrial proteins are encoded by the mitochondrial genome. These 13 mitochondrial genes together constitute a small fraction of the OXPHOS gene set involved in oxidative phosphorylation and electron transport (46). Thus far all of the characterized gene products involved in mtDNA replication and transcription are encoded in the nucleus. Included are the essential transcription factor (mtTAF) and the mitochondrial RNA and DNA polymerases (6). Thus, identification of the TOP1mt gene in the nuclear genome is consistent with this pattern for mitochondrial biogenesis. Whether human genetic disorders are associated with TOP1mt gene alterations is an open question. It is conceivable that defects in TOP1mt gene expression might regulate mtDNA copy number, and that loss of heterozygosity in the 8q24.3 region might predispose to mitochondrial diseases with neurodegenerative and myopathy components or to late-onset degenerative diseases such as Alzheimer's disease, Parkinson's syndrome, and diabetes.
Supplementary Material
Acknowledgments
We thank Dr. Kurt W. Kohn, for valuable suggestions during the course of this study, and Dr. John N. Weinstein, for critical reading of the manuscript and suggestions.
Abbreviations
- GFP
green fluorescent protein
- FISH
fluorescence in situ hybridization
- CPT
camptothecin
- NTD
N-terminal domain
Footnotes
References
- 1.Wang J C. J Mol Biol. 1971;55:523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]
- 2.Wang J C. J Biol Chem. 1991;266:6659–6662. [PubMed] [Google Scholar]
- 3.Wang J C. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 4.Gellert M. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- 5.Clayton D A. Int Rev Cytol. 1992;141:217–232. doi: 10.1016/s0074-7696(08)62067-7. [DOI] [PubMed] [Google Scholar]
- 6.Moraes C T. Trends Genet. 2001;17:199–205. doi: 10.1016/s0168-9525(01)02238-7. [DOI] [PubMed] [Google Scholar]
- 7.Anderson L. Proc Natl Acad Sci USA. 1981;78:2407–2411. doi: 10.1073/pnas.78.4.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bibb M J, Van Etten R A, Wright C T, Walberg M W, Clayton D A. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- 9.Zullo S J, Butler L, Zahorchak R J, Macville M, Wilkes C, Merril C R. Cytogenet Cell Genet. 1997;78:281–284. doi: 10.1159/000134672. [DOI] [PubMed] [Google Scholar]
- 10.Larsson N G, Barsh G S, Clayton D A. Mamm Genome. 1997;8:139–140. doi: 10.1007/s003359900373. [DOI] [PubMed] [Google Scholar]
- 11.Castora F J, Lazarus G M. Biochem Biophys Res Commun. 1984;121:77–86. doi: 10.1016/0006-291x(84)90690-9. [DOI] [PubMed] [Google Scholar]
- 12.Castora F J, Lazarus G M, Kunes D. Biochem Biophys Res Commun. 1985;130:854–866. doi: 10.1016/0006-291x(85)90495-4. [DOI] [PubMed] [Google Scholar]
- 13.Castora F J, Kelly W G. Proc Natl Acad Sci USA. 1986;83:1680–1684. doi: 10.1073/pnas.83.6.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazarus G M, Henrich J P, Kelly W G, Schmitz S A, Castora F J. Biochemistry. 1987;26:6195–6203. doi: 10.1021/bi00393a036. [DOI] [PubMed] [Google Scholar]
- 15.Lin J H, Lazarus G M, Castora F J. Arch Biochem Biophys. 1992;293:201–207. doi: 10.1016/0003-9861(92)90385-a. [DOI] [PubMed] [Google Scholar]
- 16.Lin J H, Castora F J. Arch Biochem Biophys. 1995;324:293–299. doi: 10.1006/abbi.1995.0042. [DOI] [PubMed] [Google Scholar]
- 17.Topcu Z, Castora F J. Biochim Biophys Acta. 1995;1264:377–387. doi: 10.1016/0167-4781(95)00180-8. [DOI] [PubMed] [Google Scholar]
- 18.Fairfield F R, Bauer W R, Simpson M V. J Biol Chem. 1979;254:9352–9354. [PubMed] [Google Scholar]
- 19.Fairfield F R, Bauer W R, Simpson M V. Biochim Biophys Acta. 1985;824:45–57. doi: 10.1016/0167-4781(85)90028-4. [DOI] [PubMed] [Google Scholar]
- 20.Brun G, Vannier P, Scovassi I, Callen J C. Eur J Biochem. 1981;118:407–415. doi: 10.1111/j.1432-1033.1981.tb06417.x. [DOI] [PubMed] [Google Scholar]
- 21.Echeverria M, Robert D, Carde J P, Litvak S. Plant Mol Biol. 1991;16:301–335. doi: 10.1007/BF00020561. [DOI] [PubMed] [Google Scholar]
- 22.Meissner K, Dorfel P, Borner T. Biochem Int. 1992;27:1119–1125. [PubMed] [Google Scholar]
- 23.Turna J, Pudzisova A, Osusky M, Supekova L, Kuchta T. Folia Microbiol. 1994;39:105–111. doi: 10.1007/BF02906803. [DOI] [PubMed] [Google Scholar]
- 24.Murthy V, Pasupathy K. Biochem Biophys Res Commun. 1994;198:387–392. doi: 10.1006/bbrc.1994.1054. [DOI] [PubMed] [Google Scholar]
- 25.Tua A, Wang J, Kulpa V, Wernette C M. Biochimie. 1997;79:341–350. doi: 10.1016/s0300-9084(97)80028-4. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Kearney K, Derby M, Wernette C M. Biochem Biophys Res Commun. 1995;214:723–729. doi: 10.1006/bbrc.1995.2345. [DOI] [PubMed] [Google Scholar]
- 27.Zimonjic D B, Rezanka L, DiPaolo J A, Popescu N C. Cancer Genet Cytogenet. 1995;80:100–102. doi: 10.1016/0165-4608(94)00161-4. [DOI] [PubMed] [Google Scholar]
- 28.Bonven B J, Gocke E, Westergaard O. Cell. 1985;41:541–551. doi: 10.1016/s0092-8674(85)80027-1. [DOI] [PubMed] [Google Scholar]
- 29.Pourquier P, Ueng L-M, Fertala J, Wang D, Park H-J, Essigman J M, Bjornsti M-A, Pommier Y. J Biol Chem. 1999;274:8516–8523. doi: 10.1074/jbc.274.13.8516. [DOI] [PubMed] [Google Scholar]
- 30.Pourquier P, Ueng L-M, Kohlhagen G, Mazumder A, Gupta M, Kohn K W, Pommier Y. J Biol Chem. 1997;272:7792–7796. doi: 10.1074/jbc.272.12.7792. [DOI] [PubMed] [Google Scholar]
- 31.Pommier Y, Kohlhagen G, Laco G S, Sayer J M, Kroth H, Jerina D M. Proc Natl Acad Sci USA. 2000;97:10739–10744. doi: 10.1073/pnas.190312697. . (First Published September 19, 2000; 10.1073/pnas.190312697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alsner J, Svejstrup J Q, Kjeldsen E, Sørensen B S, Westergaard O. J Biol Chem. 1992;267:12408–12411. [PubMed] [Google Scholar]
- 33.Champoux J J. Prog Nucleic Acid Res Mol Biol. 1998;60:111–132. doi: 10.1016/s0079-6603(08)60891-0. [DOI] [PubMed] [Google Scholar]
- 34.Mo Y Y, Wang C, Beck W T. J Biol Chem. 2000;275:41107–41113. doi: 10.1074/jbc.M003135200. [DOI] [PubMed] [Google Scholar]
- 35.Hammen P K, Weiner H. J Exp Zool. 1998;282:280–328. [PubMed] [Google Scholar]
- 36.Wang J C. Annu Rev Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]
- 37.Liu L F, Miller K G. Proc Natl Acad Sci USA. 1981;78:3487–3491. doi: 10.1073/pnas.78.6.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pommier Y, Pourquier P, Urasaki Y, Wu J, Laco G. Drug Resistance Update. 1999;2:307–318. doi: 10.1054/drup.1999.0102. [DOI] [PubMed] [Google Scholar]
- 39.Lemasters J J, Chacon E, Ohata H, Harper I S, Nieminen A L, Tesfai S A, Herman B. Methods Enzymol. 1995;260:428–444. doi: 10.1016/0076-6879(95)60156-2. [DOI] [PubMed] [Google Scholar]
- 40.Wallace D C. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 41.Kosovsky M J, Soslau G. Biochim Biophys Acta. 1991;1078:56–62. doi: 10.1016/0167-4838(91)90092-e. [DOI] [PubMed] [Google Scholar]
- 42.Kosovsky M J, Soslau G. Biochim Biophys Acta. 1993;1164:101–107. doi: 10.1016/0167-4838(93)90117-a. [DOI] [PubMed] [Google Scholar]
- 43.Chen A Y, Liu L F. Annu Rev Pharmacol Toxicol. 1994;94:194–218. doi: 10.1146/annurev.pa.34.040194.001203. [DOI] [PubMed] [Google Scholar]
- 44.Esnault C, Brown S C, Segal-Bendirdjian E, Coulaud D, Mishal Z, Roques B P, Le Pecq J B. Biochem Pharmacol. 1990;39:109–122. doi: 10.1016/0006-2952(90)90654-4. [DOI] [PubMed] [Google Scholar]
- 45.Schapira A H. Biochim Biophys Acta. 1999;1410:99–102. doi: 10.1016/s0005-2728(98)00160-1. [DOI] [PubMed] [Google Scholar]
- 46.DiMauro S, Schon E A. Nat Genet. 1998;19:214–215. doi: 10.1038/883. [DOI] [PubMed] [Google Scholar]
- 47.Redinbo M R, Stewart L, Kuhn P, Champoux J J, Hol W G J. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.