Abstract
Several members of the Rubiaceae and Violaceae families produce a series of cyclotides or macrocyclic peptides of 29–31 amino acids with an embedded cystine knot. We aim to understand the mechanism of synthesis of cyclic peptides in plants and have isolated a cDNA clone that encodes the cyclotide kalata B1 as well as three other clones for related cyclotides from the African plant Oldenlandia affinis. The cDNA clones encode prepropeptides with a 20-aa signal sequence, an N-terminal prosequence of 46–68 amino acids and one, two, or three cyclotide domains separated by regions of about 25 aa. The corresponding cyclotides have been isolated from plant material, indicating that the cyclotide domains are excised and cyclized from all four predicted precursor proteins. The exact processing site is likely to lie on the N-terminal side of the strongly conserved GlyLeuPro or SerLeuPro sequence that flanks both sides of the cyclotide domain. Cyclotides have previously been assigned an antimicrobial function; here we describe a potent inhibitory effect on the growth and development of larvae from the Lepidopteran species Helicoverpa punctigera.
Naturally occurring circular proteins (i.e., proteins in which the N and C termini are linked via a peptide bond) were unknown before the mid 1990s; however, there have been increasing reports of both naturally occurring and synthetic circular proteins over the last few years. Of particular interest among naturally occurring examples have been a series of plant-derived proteins referred to as the cyclotides (1). These include kalata B1 (2), the circulins (3), cyclopsychotride (4), and several peptides from Viola species (5–7). The cyclotides comprise 29–31 amino acids, including six highly conserved Cys residues that form a cystine knot (8–9). In this structural motif, an embedded ring formed by two disulfide bonds and their connecting backbone segments is penetrated by a third disulfide bond. The combination of a cystine knot embedded in a cyclic backbone, referred to as a cyclic cystine knot, produces a unique protein fold that is topologically complex and has exceptional chemical and biological stability. These plant-derived circular proteins are quite different from the small cyclic peptides from microorganisms that have been well known for many years. The latter are typically 5–10 aa in length, lack disulfide bonds, and are not ribosomally synthesized. There are only limited examples of larger circular proteins from microbial sources and these include a 21-aa peptide from Escherichia coli (10) and a 70-aa protein from Enterococcus faecalis (11). Both lack disulfide bonds but appear to be gene products. Recently, two other small cyclic peptides have been reported in higher organisms. One is SFTI, a cyclic 14-aa trypsin inhibitor from sunflower seeds (12), and the other is RTD-1, an 18-aa tridisulfide peptide from rhesus monkey leukocytes (13). The latter is derived from the cotranslational processing of two related genes by a mechanism that is not yet known, and the biosynthetic origin of the former is unknown. We have recently reported the structures of both these molecules (14, 15).
The cyclotides contrast with the other circular proteins in that they have highly defined three-dimensional structures and, despite their small size, may be regarded as miniproteins. This attribute arises primarily from the knotted network of disulfide bonds that stabilizes the structures. The well-defined structures are associated with a range of biological activities. Indeed, the cyclotides were originally discovered either from screening programs or from anecdotal reports of their biological activity in traditional medicines. For example, in 1970, kalata B1was reported as the active ingredient in a tea used by women in the Congo region of Africa to accelerate childbirth (16, 17), although it was some 25 years later before the sequence and cyclic nature of the peptide were determined (2). The circulins were discovered in screens focusing on anti-HIV activity (3), cyclopsychotride for inhibition of neurotensin binding (4), and various Viola peptides for hemolytic activity (7). Thus, all of the known macrocyclic peptides have diverse biological activities, but their function in plants was not known. Tam et al. (18) have suggested a role in plant defense against microorganisms. Here we show a potential role in host defense against insects.
Experimental Procedures
RNA Isolation and Production of a Partial cDNA Clone Encoding Kalata B1.
RNA was isolated by using Trizol reagent and the protocol from GIBCO/BRL. Single-stranded cDNA was prepared from leaf RNA and amplified by the PCR by using one of two degenerate primers and oligo-dT (GeneWorks, Adelaide, Australia). The primers and the encoded protein sequence are shown in Fig. 1. The PCR fragments were isolated from agarose gels and cloned into pBluescript SK+ vector (Stratagene) for sequencing.
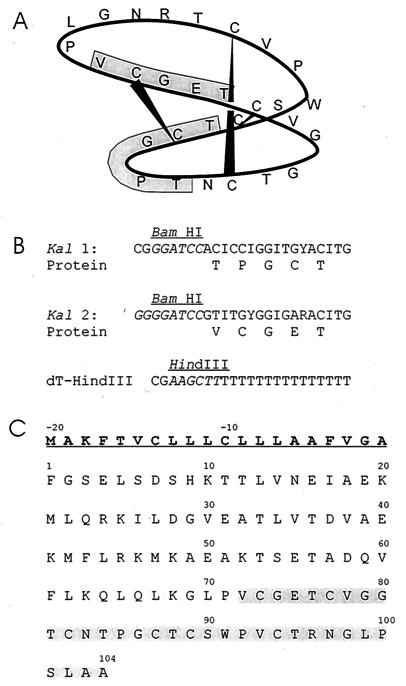
Figure 1.
(A) Schematic representation of kalata B1 showing the cyclic cystine knot, the amino acid sequence in single letter code, and the regions used for oligonucleotide primer design (shaded). (B) The primers used in the PCR reactions. I represents inosine, Y represents C or T, and R represents A, C, T, or G. The introduced restriction enzyme sites are in italics. (C) Amino acid sequence of the protein encoded by the Oak1 clone. The sequence corresponding to the PCR product obtained with the Kal2 and oligo-dT primers is shaded.
Preparation of the O. affinis cDNA Library.
Total RNA (1 mg) was prepared from leaves and stems, and mRNA was separated by using the PolyATract I (Promega) mRNA isolation system. Five micrograms of mRNA was used to produce the cDNA library with the Uni-Zap-cDNA synthesis kit and the GIGAPACKIII gold packaging extract from Stratagene. The Oak1 and Oak2 clones were obtained by screening the amplified library with a 32P-labeled DNA fragment of the Kal2 and oligo-dT PCR product (DNA-encoding amino acids 72–104 in Fig. 1C). The Oak3 and Oak4 clones were isolated by using the Oak1 clone as probe.
RNA and DNA Blots.
Total RNA (10 μg) was fractionated on 1.2% agarose gels in the presence of formaldehyde and transferred to HyBond N+ (Amersham Pharmacia) (19). Prehybridization and hybridization were performed at 42°C in 5× (SSPE)(3 M NaCl, 0·2 M NaH2PO4, 0·0.2 M EDTA)/1% (wt/vol) SDS/5× Denhardt's/50%(wt/vol) deionized formamide/200 μg/ml herring sperm DNA. The membrane was probed with P32-labeled Oak 1. Unbound probe was removed by washing three times with 2× SSPE and 1% SDS at 42°C for 10 min.
Genomic DNA was isolated from fresh leaf material (1.5 g), and 40 μg was digested with restriction enzymes; HindIII, BamH1, NdeI, and EcoRV (Promega, 5 units) before electrophoresis on a 0.7% agarose gel in the presence of ethidium bromide and TBE buffer (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3) (19). The DNA was transferred to Hybond N+ (19), and the blot was probed with Oak1 cDNA before it was prehybridized, hybridized, and washed as described for RNA blots.
Hybridizing RNA and DNA was visualized by using a model 400B PhosphorImager and imagequant software (Amersham Pharmacia).
Isolation of Cyclotides from O. affinis.
Kalata B1, B2, B6, and B7 were isolated from aerial parts of O. affinis by extraction with dichloromethane/methanol (50:50 vol/vol) and purified by using RP-HPLC [Vydac (Hesperia, CA) C18 column] (1). Kalata B6 and B7 have not been reported previously and were characterized by mass spectrometry and Edman sequencing.
Bioassays with Artificial Diets.
Helicoverpa punctigera larvae were raised on artificial diets based on haricot beans (20). The test diet was supplemented with the kalata B1 peptide (0.825 μmol/g of diet). The control diet contained casein in place of the inhibitor. Twenty neonates were added to each diet, and mortality was recorded every 2 days. Weight gain was recorded at the sixth day and every second day thereafter. The larvae were reared in 1.5-ml microfuge tubes (one larva/tube) until day eight, when they were transferred to individual plastic containers with lids [Solo (Urbana, IL) plastic portion cups, 28 ml]. Larvae were fed initially small amounts of diet (40 mg) that were replaced as required to provide a continuous supply. The larvae were kept in a temperature controlled room at 25 ± 1°C, 16:8 (light/dark).
Trypsin, Chymotrypsin, and α-Amylase Assays.
The midgut was dissected from 10 fourth instar larvae and homogenized in 2.5 ml of 10 mM Tris⋅HCl, pH8. The supernatant was collected after centrifugation (15,000 × g, 10 min, 4°C), and protein concentration was determined by using the Bradford method with reagents from Bio-Rad and BSA as standard. The supernatant was divided into 200-μl aliquots and stored at −80°C until use. Protease assays were essentially as described previously (21) and were performed in duplicate by using controls without enzyme. Substrates were 1 mM sodium-benzoyl-dl-arginine p-nitroanilide for trypsin and 1 mM N-succinyl-Ala-Ala-Pro-Phe p-nitroanilide (Sigma) for chymotrypsin. CAPS buffer (50 mM 3-[cyclohexylamino]-1-propane-sulfonic acid/75 mM NaCl/2.5 mM MgCl2, pH 10) was used for assays with gut extract, and 50 mM Tris⋅HCl, pH 8.0, was used for bovine trypsin and chymotrypsin. Enzyme and various amounts of kalata B1, kalata B2, or Nicotiana alata proteinase inhibitor (21) were mixed and preincubated in a 60-μl volume for 30 min at 30°C before the addition of substrate (40 μl). The reactions were continued for 30 min at 30°C before release of p-nitroanilide was recorded at 405 nm on a SpectraMax 250 microtiter plate reader (Molecular Devices). Trypsin activity was determined by using 4 μg (1.67 μM) bovine trypsin [Type XIII 1-tosylamido-2-phenylethyl chloromethyl ketone (TPCK) treated, Sigma] or 3.2 μg of gut protein. Chymotrypsin assays used 0.1 μg (0.04 μM) of bovine chymotrypsin (Type VII TLCK treated, Sigma) or 2.6 μg of gut protein.
Amylase activity was assayed essentially as described by Morton et al. (22). Assays were performed at 30°C in 200 μl of reaction mixture containing 26 μg of gut protein and 0.5% (wt/vol) starch (Sigma) in CAPS buffer. Samples (20 μl) were removed at 10, 20, and 30 min for reducing sugar assay by using dinitrosalicyclic acid reagent. Potential inhibitory activity of kalata B1 and B2 was measured by mixing 20 μg with gut protein (26 μg) and preincubating at 30°C for 1 h in CAPS buffer before the enzyme assay was initiated by the addition of an equal volume of 1% (wt/vol) starch in the same buffer. The α-amylase inhibitor from wheat seeds (Sigma A1520) was used as a positive control.
Results
Isolation of cDNA Clones Encoding Cyclotides from O. affinis.
As kalata B1 is a cyclic protein of only 29 amino acids and the 5′ end of the coding region was unknown, two oligonucleotides were designed for PCR amplification in combination with oligo-dT to ensure PCR products were long enough for subcloning and library screening (see Fig. 1 A and B). A 400-bp fragment produced from primer Kal2 and oligo-dT had a 3′ untranslated region of 267 bp and a polyA tail of 32 bp together with what appeared to be the complete coding sequence for kalata B1. When used as a probe on RNA blots containing O. affinis leaf RNA, the partial kalata B1 clone hybridized to an RNA transcript of about 750 bases, suggesting that the cyclic peptide is derived from a larger precursor protein.
The cDNA library prepared from leaf and stem mRNA was screened initially by using the partial kalata B1 cDNA as probe. Two full-length clones were obtained; the first designated OaK1 for O. affinis kalata B1 was 725 bp in length and encodes a predicted protein of 124 amino acids (Fig. 1C). The 29-aa kalata B1 sequence is embedded in a precursor protein, which has a typical endoplasmic reticulum (ER) signal sequence of 20 amino acids. The B1 sequence is flanked by about 70 amino acids at the N terminus and four to seven amino acids at the C terminus. All six cysteines in the precursor are located in the B1 sequence. The predicted precursor has no potential N-glycosylation sites and, hence, has an expected mass of 11.18 kDa without the signal sequence.
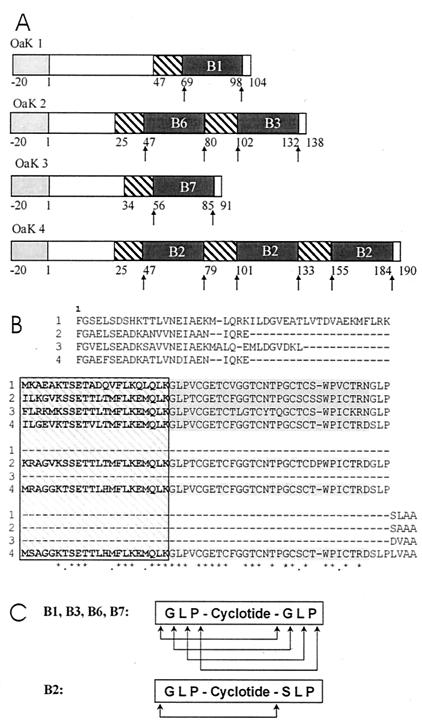
The second cDNA clone, designated Oak2, is 843 bp in length and, unlike the first clone, is predicted to encode a protein of 158 aa with two kalata B1-related sequences that we have called kalata B3 and B6 (Fig. 2). The predicted protein also has a typical ER signal sequence of 20 aa that is followed by a 46- to 49-aa sequence before the first kalata sequence (B6) is encountered. This peptide is separated from the kalata B3 sequence by about 25 aa. The B3 sequence is followed by four amino acids at the C terminus (SAAA) that are similar to those that flank B1 (SLAA) in the protein encoded by the Oak1 clone. Like the precursor encoded by the Oak1 clone, the precursor encoded by the Oak2 clone has no potential N-glycosylation sites, and all cysteine residues are confined to the putative mature peptide sequences. After removal of the potential ER signal sequence, the precursor encoded by the Oak 2 clone has a predicted mass of 14.56 kDa.
Figure 2.
(A) Block diagram of the precursor proteins predicted from the Oak1, 2, 3, and 4 clones showing the signal peptide (light shading), the regions corresponding to the mature kalata peptides (dark shading), the region of 22 aa on the N-terminal side of the kalata peptide sequence (N-T repeat, hatched). (B) Alignment of the amino acid sequence of the proteins encoded by the Oak1–Oak4 clones (labeled 1–4, respectively). The sequence begins with the first amino acid after the signal peptide. The N-T repeat sequences are boxed and hatched. The kalata sequences are shaded. Identical amino acids in the N-T repeat and kalata sequences are marked with an asterisk, and similar amino acids are marked with a dot. Gaps (−) were introduced to maximize the alignment. (C) The potential processing sites. The mature cyclic peptide retains one copy of the Gly-Leu-Pro sequence that could be derived entirely from one of the two flanking elements or partially from both depending on the initial cleavage sites for B1, B3, B6, and B7. The retention of Gly-Leu-Pro in the B2 cyclotide suggests cleavage before the Gly and Ser residues.
The third clone, designated Oak3, was 677 bp long and encodes a predicted protein of 111 aa (Fig. 2). It has only one kalata B1-related sequence, designated here as kalata B7. The fourth clone, Oak4, has a 993-bp insert and encodes a predicted protein of 210 aa (Fig. 2). This protein has three identical kalata-like sequences that we have called kalata B2, because the sequence corresponds to a peptide given this name but not fully identified in an early study of O. affinis by Gran (16). The B1 and B3 sequences from the Oak1 and Oak2 clones also correspond to peptides with the same name that have been extracted from plant tissues (1). The B6 and B7 peptides predicted from the Oak2 and Oak3 clones, however, did not correspond to any of the known kalata molecules. Further isolation of peptides from O. affinis leaves resulted in the discovery of two new cyclic peptides with the same mass and sequence predicted for B6 and B7.
Genes Encoding Kalata-Like Peptides Belong to a Multigene Family in O. affinis.
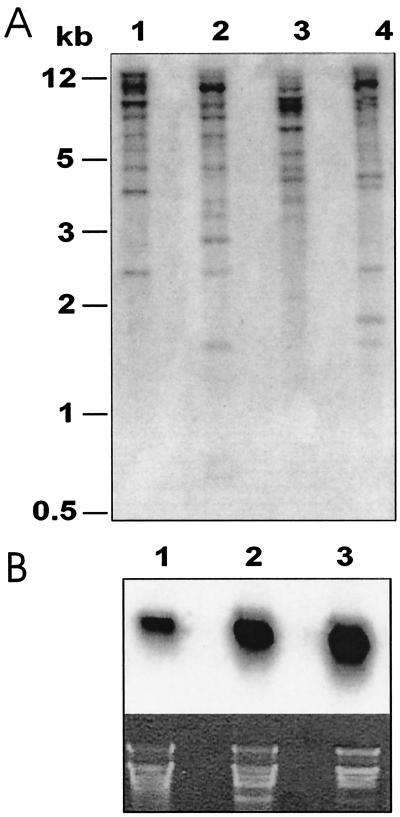
Craik et al. (1) have isolated three cyclotides (kalata B4, B5, and kalata S) from O. affinis that are not encoded by the cDNA clones described here, consistent with expression of more than four cyclotide genes. The size of the cyclotide gene family was estimated by digesting genomic DNA with HindIII, BamHI, NdeI, and EcoRV and subjecting it to DNA blot analysis with the Oak1 cDNA as probe. Up to 12 hybridizing bands were obtained in all of the digests (Fig. 3A), suggesting the cyclotides are derived from a multigene family with up to 12 related genes.
Figure 3.
Gel blot analysis of RNA and genomic DNA from O. affinis. (A) Blot of genomic DNA digested with: (1) HindIII, (2) BamHI, (3) Nde1, and (4) EcoRV probed with radiolabeled Oak1. (Upper) (i) Blot of total RNA from 1 roots; 2, leaves; 3, shoots probed with radiolabeled Oak1. (Lower) Identical gel to (Upper) stained with ethidium bromide.
Cyclotide Genes Are Expressed in Roots, Leaves, and Stems of O. affinis.
Cyclotides have been isolated from all tissues of O. affinis, but the leaves are the best source on a dry-weight basis (1). We asked whether this was because of differences in extraction efficiency, peptide stability, or gene expression. RNA blots were performed by using the Oak1 cDNA as probe. Expression was higher in young leaves than mature leaves and was relatively low in roots (Fig. 3B).
Effect of Kalata B1 on the Growth and Development of H. punctigera Larvae.
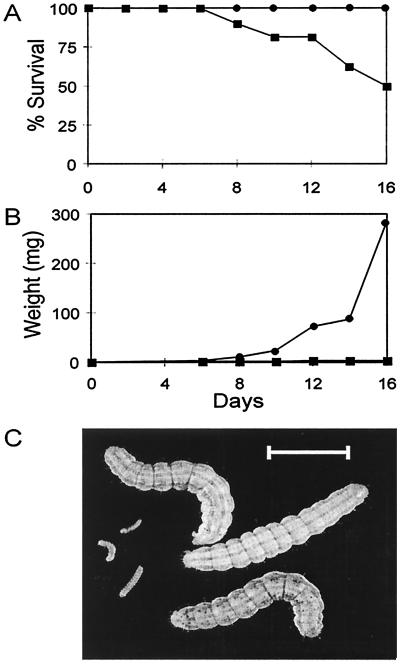
Although antimicrobial activity has been described for kalata B1 (18), a potential role in insect defense has not been examined. Purified kalata B1 was incorporated into an artificial diet and fed to larvae for 16 days postemergence. Kalata B1 had a significant effect on the development of larvae compared with larvae on a diet free of kalata. No mortality was observed in the first 6 days, although 50% failed to survive past day sixteen (Fig. 4A). After 16 days, none of the 12 survivors had progressed past the first instar stage of development and weighed 3.3 mg (±1.1 mg standard deviation). Most larvae on the control diet, however, had achieved fifth instar, and the average mass was 284 mg (±96.1 mg standard deviation; Fig. 4 B and C).
Figure 4.
Effect of kalata B1 on growth and development of H. punctigera larvae. (A) Survival of larvae fed an artificial diet containing kalata B1 (■) and the control (●) diet. (B) Average mean weight of larvae fed on kalata B1 (■) and control (●) diet. (C) Size of larvae after 16 days on artificial diet containing kalata B1 (Left) or control diet (Right). (Bar = 1 cm.)
Effect of Kalata B1 and B2 on H. punctigera Digestive Enzymes.
Inhibition of endogenous digestive proteinases is one mechanism by which small disulfide-rich defense molecules exert their effect, as exemplified by the proteinase inhibitors from N. alata (23). This possibility was tested for kalata by using a series of enzyme inhibition assays. Kalata B1 and B2 had no effect on bovine or H. punctigera trypsins and chymotrypsins, whereas the N. alata proteinase inhibitors abolished all trypsin and most chymotrypsin activity under the same conditions (Table 1). Similarly, the kalata peptides failed to inhibit α-amylases from Helicoverpa gut (Table 1).
Table 1.
Effect of kalata B1 and B2 on trypsin, chymotrypsin and α-amylase activity
| Assay | Potential inhibitor | Highest concentration tested, μM | IC50, μM |
|---|---|---|---|
| Trypsin | |||
| BT | NaPI | 5 | 0.62 |
| BT | B1/B2 | 20 | n.i. |
| GE | NaPI | 0.5 | 0.02 |
| GE | B1/B2 | 20 | n.i. |
| Chymotrypsin | |||
| BC | NaPI | 5 | 0.045 |
| BC | B1/B2 | 34 | n.i. |
| GE | NaPI | 20 | 15 |
| GE | B1/B2 | 34 | n.i. |
| α-amylase | |||
| GE | Wheat AI | 4.2 | 0.83 |
| GE | B1/B2 | 34 | n.i. |
NaPI, Nicotiana alata proteinase inhibitor; Wheat AI, α-amylase inhibitor from wheat seeds; B1/B2, kalata B1 and B2; BT, bovine trypsin; BC, bovine chymotrypsin; GE, H. punctigera gut extract; IC50, concentration of inhibitor required for 50% inhibition of activity; n.i., not inhibited.
Discussion
In this paper, we have described a series of precursor proteins for the plant cyclotides. Although they are small (29–31 aa), the cyclotides adopt the features of larger proteins, including elements of secondary structure, a tightly intertwined set of disulfide bonds, and a well defined three-dimensional fold. As far as we are aware, the cyclotides are the only known naturally occurring cyclic proteins in higher organisms, apart from SFTI and RTD-1, which are half the size of the cyclotides at 14–18 aa. The finding here that the cyclotides are true gene products distinguishes them from a range of smaller cyclic peptides (5–12 aa) that are found in fungi and bacteria. The latter are typically synthesized by multienzyme complexes and include examples such as gramicidin and cyclosporin. The unique circular nature of cyclotides makes their biosynthesis and function of much interest. Processing mechanisms for the natural production of circular proteins have not been reported so far.
The plant cyclotides are produced by cleavage and cyclization from precursor proteins incorporating one, two, or three cyclotide domains. The predicted precursors have typical endoplasmic reticulum signal sequences. Thus, it is likely that the precursors enter the secretory pathway where folding and disulfide bond formation occurs before the cleavage and cyclization events that release the mature cyclic peptides. Peptides that are the apparent products of all of the described clones have been isolated from plant tissues (1).
The modular structure of the precursors has several interesting features. The four precursor proteins have a relatively long N-terminal domain that is not tightly conserved in sequence or in length. This domain is followed by a relatively well-conserved sequence of 25 aa that precedes the cyclotide domain. We have called this sequence the N-terminal repeat fragment, because it is repeated together with the cyclotide sequence in precursors with more than one cyclotide domain. The function of the sequence is not known, but it may have an essential role in folding or cyclization. We have previously noted that the folded structures of the cyclotides are interesting in that they incorporate a hydrophobic patch on the surface of the molecule (2). The exposure of hydrophobic residues appears to be a consequence of the tightly packed disulfide core of the cyclotides that leaves no room for internalization of hydrophobic residues. The surface exposure probably leads to the relatively long retention times on RP-HPLC that characterize the cyclotide family (1). We speculate that the N-terminal repeat fragment may be a folding auxiliary that helps to stabilize the development of the hydrophobic surface during protein folding. Formation of disulfide bonds presumably provides the stabilizing features that subsequently hold the structure in place.
The isolation of the cDNA clones together with the B1, B2, B3, B6, and B7 cyclotides from plant tissues provides some insight into the potential processing sites. Each of the kalata B1, B3, B6, and B7 sequences in the predicted proteins from the Oak1, Oak2, and Oak3 clones (Fig. 2B) is flanked on both sides by the highly conserved sequence -Gly-Leu-Pro-. The circularization process thus appears to involve cleavage at a homologous site within both flanking sequences and ligation of the new N and C termini. The mature cyclic peptide retains one copy of the Gly-Leu-Pro sequence, which may be derived entirely from one of the original flanking elements, or partially from both depending on the initial cleavage sites (Fig. 2C).
The discovery of the Oak4 clone together with the corresponding B2 peptide from the plant assisted with further refinement of the potential processing site. Unlike the proteins encoded by the other three clones, the Oak4 protein has three copies of a kalata-like sequence. This sequence (B2) is flanked by Gly-Leu-Pro at the N terminus and Ser-Leu-Pro at the C terminus (Fig. 2B). The B2 peptide isolated from the plant (1) incorporates the Gly-Leu-Pro sequence, indicating that N- and C-terminal processing has occurred at the peptide bonds preceding the Gly and Ser, respectively.
Comparison of the flanking sequences for all of the precursors shown in Fig. 2 indicates that cleavage at the N terminus of the cyclotide domain probably occurs between the Lys and Gly, and cleavage at the C terminus occurs between Asp/Asn and Gly/Ser residues. Enzymes that accommodate both Lys and Asp/Asn residues in the P1 position have not been described in plants, although proteases specific for Asn residues are common and are located in the secretory pathway (24–26). Indeed, an asparaginyl endopeptidase has been implicated in the maturation of Con A within the secretory pathway of the plant Canavalia ensiformis (Jack bean) (27, 28). Con A maturation involves cleavage next to three Asn residues within the precursor accompanied by transposition and ligation (by formation of a peptide bond) of fragments released from the N and C termini. Processing intermediates can be isolated after the first two cleavage events but not after the third, suggesting that the cleavage and formation of the new peptide bond occurs simultaneously. Indeed, purified asparaginyl endopeptidase is sufficient for production of mature Con A from the precursor (28). The observation that asparaginyl endopeptidases are common in plants, but posttranslational peptide bond formation is rare, led Sheldon et al. (28) to suggest that the ligation event is a consequence of the structure of the Con A precursor rather than a simple transpeptidation event. That is, the residues that are ultimately connected by the new peptide bond in mature Con A are located close together in the precursor. It is possible that a similar asparaginyl endopeptidase is involved in production of the cyclotides, because the C-terminal cleavage site has a conserved Asn or Asp residue. However, unlike Con A, the second cleavage involves a Lys residue, suggesting looser specificity if a single enzyme is involved or requirement for a second protease.
We have also considered the possibility that the cyclotides are produced by a nonenzymic process. For example, it has been demonstrated that intein-based mechanisms, which are normally used for the autocatalytic splicing of two linear protein domains (29), can be adapted for the production of artificial circular proteins, both in vitro and in vivo (30–32). Inspection of the sequences of the precursor proteins described here does not reveal any homologies with known intein sequences, all of which are substantially larger. Nevertheless, the possibility of a new type of autocatalytic processing cannot be excluded at this stage.
The function of cyclotides in plants has not been elucidated previously, but the high level of expression in leaves (17), as well as production of several isoforms within a single plant, is consistent with a role in recognition or defense. Furthermore, plant defense molecules are often small cysteine-rich proteins (33) that are trafficked through the secretory pathway. These defense molecules include insecticidal molecules such as proteinase and α-amylase inhibitors as well as potent antimicrobial molecules such as thionins and defensins. There is also precedent for precursors containing repeated domains of small defense-related proteins. The proteinase inhibitor precursors from tobacco species have up to six or eight repeated proteinase inhibitor domains (34, 35) that are released after transit through the secretory pathway. Precursors with eight cysteine proteinase inhibitor domains have been identified in potato (36), and Impatiens balsamina has a precursor that is processed into six small cysteine-rich antimicrobial peptides (37).
Tam et al. (18) have reported on the potent and specific antimicrobial activity of the cyclotides, kalata B1, circulin A and B, and cyclopsychotride, against several bacterial and fungal species, although no plant pathogens were tested. Interestingly the antimicrobial activity is salt dependent, suggesting the initial interaction between the cyclotides and the microbial surface is electrostatic, similar to that described for defensins (18). Here we report a major effect of kalata B1 on the growth and development of larvae from the Lepidopteran species H. punctigera. Caterpillars fed artificial diets containing 0.15% wt/vol (520 μM) kalata B1 failed to progress past the first instar stage of development. Kalata B1 has a greater effect on larval growth than most serine proteinase inhibitors (23, 38) that are produced by many plants for defense against insect pests. We do not know whether failure to thrive was caused by a toxic activity or an antifeedant effect that ultimately led to death by starvation. Small Cys-rich plant peptides are often proteinase or α-amylase inhibitors that can retard insect growth by blocking digestion of protein or starch. Kalata B1 and B2, however, had no effect on trypsins, chymotrypsins, or α-amylases from Helicoverpa gut and thus must have a different mechanism of action. The kalata peptides have hemolytic activity (18), raising the possibility that the insecticidal activity results from damage to membranes within the insect gut. Whatever the mode of action, it is clear that the stable framework provided by the cyclic cystine knot that makes up the core of the plant cyclotide provides an exciting template for potential agricultural and pharmaceutical applications.
Acknowledgments
We thank David Norman (University of Dundee) for providing a sample of O. affinis seed, Chelsea Clavarino and Jackie Stevens for technical support. and Liz Johnson for critical reading of the manuscript. This work was supported by a grant from the Australian Research Council (ARC). D.C. is an ARC Senior Research Fellow. The Institute for Molecular Bioscience is a Special Research Centre of the ARC. C.J. was supported by an Australian Postgraduate Research Award. J.W. was supported by a La Trobe University postgraduate award.
Footnotes
References
- 1.Craik D J, Daly N L, Bond T, Waine C. J Mol Biol. 1999;294:1327–1336. doi: 10.1006/jmbi.1999.3383. [DOI] [PubMed] [Google Scholar]
- 2.Saether O, Craik D J, Campbell I D, Sletten K, Juul J, Norman D G. Biochemistry. 1995;34:4147–4158. doi: 10.1021/bi00013a002. [DOI] [PubMed] [Google Scholar]
- 3.Gustafson K R, Sowder R C, II, Henderson L E, Parsons I C, Kashman Y, Cardellina J H, II, McMahon J B, Buckheit R W, Jr, Pannell L K, Boyd M R. J Am Chem Soc. 1994;116:9337–9338. [Google Scholar]
- 4.Witherup K M, Bogusky M J, Anderson P S, Ramjit H, Ransom R W, Wood T, Sardana M. J Nat Prod. 1994;57:1619–1625. doi: 10.1021/np50114a002. [DOI] [PubMed] [Google Scholar]
- 5.Schöpke T, Hasan Agha M I, Kraft R, Otto A, Hiller K. Sci Pharm. 1993;61:145–153. [Google Scholar]
- 6.Claeson P, Göransson U, Johansson S, Luijendijk T, Bohlin L. J Nat Prod. 1998;61:77–81. doi: 10.1021/np970342r. [DOI] [PubMed] [Google Scholar]
- 7.Göransson U, Luijendijk T, Johansson S, Bohlin L, Claeson P. J Nat Prod. 1999;62:283–286. doi: 10.1021/np9803878. [DOI] [PubMed] [Google Scholar]
- 8.Isaacs N W. Curr Opin Struct Biol. 1995;5:391–395. doi: 10.1016/0959-440x(95)80102-2. [DOI] [PubMed] [Google Scholar]
- 9.McDonald N Q, Hendrickson W A. Cell. 1993;73:421–424. doi: 10.1016/0092-8674(93)90127-c. [DOI] [PubMed] [Google Scholar]
- 10.Blond A, Peduzzi J, Goulard C, Chiuchiolo M J, Barthelemy M, Prigent Y, Salomon R A, Farias R N, Moreno F, Rebuffat S. Eur J Biochem. 1999;259:747–755. doi: 10.1046/j.1432-1327.1999.00085.x. [DOI] [PubMed] [Google Scholar]
- 11.Martinezbueno M, Maqueda M, Galvez A, Samyn B, Vanbeeumen J, Coyette J, Valdivia E. J Bacteriol. 1994;176:6334–6339. doi: 10.1128/jb.176.20.6334-6339.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luckett S, Santiago Garcia R, Barker J J, Konarev A V, Shewry P R, Clarke A R, Brady R L. J Mol Biol. 1999;290:525–533. doi: 10.1006/jmbi.1999.2891. [DOI] [PubMed] [Google Scholar]
- 13.Tang Y-Q, Yuan J, Ösapay G, Ösapay K, Tran D, Miller C J, Ouellette A J, Selsted M E. Science. 1999;286:498–502. doi: 10.1126/science.286.5439.498. [DOI] [PubMed] [Google Scholar]
- 14.Korsinczky M L J, Schirra H J, Rosengren K J, West J, Wade J D, Condie B A, Otvos L, Anderson M A, Craik D J. J Mol Biol. 2001;311:579–591. doi: 10.1006/jmbi.2001.4887. [DOI] [PubMed] [Google Scholar]
- 15.Trabi M, Schirra H J, Craik D J. Biochemistry. 2001;40:4211–4221. doi: 10.1021/bi002028t. [DOI] [PubMed] [Google Scholar]
- 16.Gran L. Medd Nor Farm Selsk. 1970;12:173–180. [Google Scholar]
- 17.Gran L. Lloydia. 1973;36:174–178. [PubMed] [Google Scholar]
- 18.Tam J P, Lu Y-A, Yang J-L, Chiu K-W. Proc Natl Acad Sci USA. 1999;96:8913–8918. doi: 10.1073/pnas.96.16.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E, Maniatis T. In: Molecular Cloning: A Laboratory Manual. Nolan C, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 20.Teakle R E, Jensen J M, Giles J E. J Invert Pathol. 1985;46:166–173. [Google Scholar]
- 21.Heath R L, Barton P A, Simpson R J, Reid G E, Lim G, Anderson M A. Eur J Biochem. 1995;230:250–257. doi: 10.1111/j.1432-1033.1995.tb20558.x. [DOI] [PubMed] [Google Scholar]
- 22.Morton R L, Schroeder H E, Bateman K S, Chrispeels M J, Armstrong E, Higgins T J V. Proc Natl Acad Sci USA. 2000;97:3820–3825. doi: 10.1073/pnas.070054597. . (First Published April 4, 2000; 10.1073/pnas.070054597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heath R L, McDonald G, Christeller J T, Lee M, Bateman K, West J, van Heeswijck R, Anderson M A. J Insect Physiol. 1997;43:833–842. doi: 10.1016/s0022-1910(97)00026-7. [DOI] [PubMed] [Google Scholar]
- 24.Scott M P, Jung R, Muntz K, Nielson N C. Proc Natl Acad Sci USA. 1992;89:658–662. doi: 10.1073/pnas.89.2.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hara-Nishimuri I, Takeuchi Y, Inoue K, Nishimura M. Plant J. 1993;4:793–800. doi: 10.1046/j.1365-313x.1993.04050793.x. [DOI] [PubMed] [Google Scholar]
- 26.Takeda O, Miura Y, Mitta M, Matsushita H, Kato I, Abe Y, Yokosawa H, Ishii S-i. J Biochem (Tokyo) 1994;116:541–546. doi: 10.1093/oxfordjournals.jbchem.a124559. [DOI] [PubMed] [Google Scholar]
- 27.Carrington D M, Auffret A, Hanke D E. Nature (London) 1985;313:64–67. doi: 10.1038/313064a0. [DOI] [PubMed] [Google Scholar]
- 28.Sheldon P S, Keen J N, Bowles D J. Biochem J. 1996;320:865–870. doi: 10.1042/bj3200865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perler F B, Xu M-Q, Paulus H. Curr Opin Chem Biol. 1997;1:292–299. doi: 10.1016/s1367-5931(97)80065-8. [DOI] [PubMed] [Google Scholar]
- 30.Camarero J A, Muir T W. J Am Chem Soc. 1999;121:5597–5598. [Google Scholar]
- 31.Evans T C, Martin D, Kolly R, Panne D, Sun L, Ghosh I, Chen L X, Benner J, Liu X Q, Xu M Q. J Biol Chem. 2000;275:9091–9094. doi: 10.1074/jbc.275.13.9091. [DOI] [PubMed] [Google Scholar]
- 32.Scott C P, Abel-Santos E, Wall M, Wahnon D C, Benkovic S J. Proc Natl Acad Sci USA. 1999;96:13638–13643. doi: 10.1073/pnas.96.24.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Broekaert W F, Cammue B P A, DeBolle M F C, Thevissen K, De Samblanx G W, Osborn R W. Crit Rev Plant Sci. 1997;16:297–323. [Google Scholar]
- 34.Lee M C S, Scanlon M J, Craik D J, Anderson M A. Nat Struct Biol. 1999;6:526–530. doi: 10.1038/9293. [DOI] [PubMed] [Google Scholar]
- 35.Choi D, Park J-A, Seo Y S, Chun Y J, Kim W T. Biochim Biophys Acta. 2000;1492:211–215. doi: 10.1016/s0167-4781(00)00073-7. [DOI] [PubMed] [Google Scholar]
- 36.Waldron C, Wegrich L M, Owens Merlo P A, Walsh T A. Plant Mol Biol. 1993;23:801–812. doi: 10.1007/BF00021535. [DOI] [PubMed] [Google Scholar]
- 37.Tailor R H, Acland D P, Attenborough S, Cammue B P A, Evans I J, Osborn R W, Ray J A, Rees S B, Broekaert W F. J Biol Chem. 1997;272:24480–24487. doi: 10.1074/jbc.272.39.24480. [DOI] [PubMed] [Google Scholar]
- 38.Jongsma M A, Bolter C. J Insect Physiol. 1997;43:885–895. doi: 10.1016/s0022-1910(97)00040-1. [DOI] [PubMed] [Google Scholar]