Abstract
An important challenge in molecular assembly and hierarchical molecular engineering is to control and program the chiral directional self-assembly. Here, we present a versatile DNA surface adapter that can programmably self-assemble into various chiral supramolecular architectures, thereby regulating the chiral directional “bonding” of gold nanorods decorated by the surface adapter. Distinct optical chirality relevant to the ensemble conformation is demonstrated from the assembled novel stair-like and coil-like gold nanorod chiral metastructures, which is strongly affected by the spatial arrangement of neighboring nanorod pair. Our strategy provides new avenues for fabrication of tunable optical metamaterials by manipulating the directional self-assembly of nanoparticles using programmable surface adapters.
Keywords: DNA self-assembly, directional self-assembly, plasmonic, chiroptical activity, nanoparticle
Graphical Abstract
DNA adapter can be programmably assembled into different chiral supramolecular architectures following the distinct binding modality enabled by the selective interactions between specific binding domains. Gold nanorods site-specifically decorated on the DNA adapters can directionally self-assemble into new plasmonic chiral metastructures with facilely tunable configurations and handedness following the DNA chiral supramolecular matrix.
Directional self-assembly of nanoparticles makes it possible to construct colloidal analogues of molecules by mimicking “bond”-like interactions between “atom”-like particles, enabling the production of artificial materials with increased complexity and new collective properties.[1] In particular, chiral directional control of particle self-assembly allows breaking the mirror-symmetry of particle arrangements, producing marked optical activity with potentially far-reaching applications in optics,[2] magnetics,[3] and diagnostics.[4] Molecular chiral directional self-assembly may be dictated by directional hydrogen bonding or π-π stacking.[5] In contrast, particles often possess geometrical symmetry or surface isotropy, leading to symmetric interactions among them that result in highly-symmetric clusters[6] and periodic lattices.[7] Chiral directional self-assembly of particles requires breaking the surface symmetry, thus programmable anisotropic surface functionalization of particles is an interesting research direction.
For instance, the particle surface can be decorated with discrete domains of anisotropically-distributed synthetic polymers or biomolecules, called surface patches, that have molecular dimensions comparable to the sizes of the particles, leading to anisotropic assembly of particles.[8] DNA nanostructures have been utilized as fully programmable surface patches, providing both geometric and site-specific recognition of particles.[9] Especially, DNA origami-templated strategy proved to be successful on arbitury and deterministic positioning of nanoparticles, and various discrete chiral arrangements of nanoparticles have been achieved.[10] However, chiral supra-assembly of nanoparticles with tunable chiral directionality remains challenging.
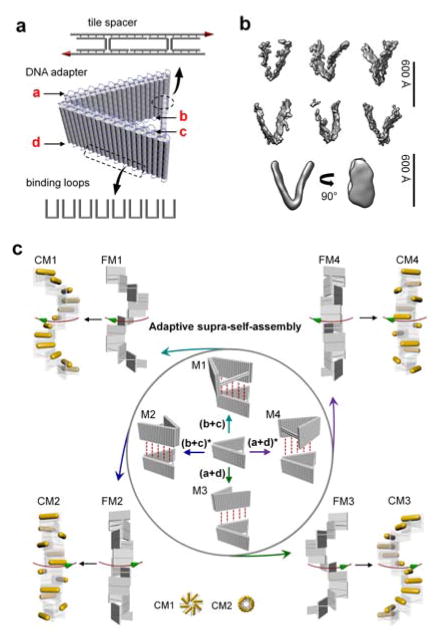
Here, we take the concept of DNA nanostructure as surface patches of nanoparticles a significant step further by creating a V-shaped “DNA adapter” (Figure 1a) that combines a particle-binding arm (a patch) and a structural control arm to allow self-assembly of gold nanorods into chiral super-structures that can be facilely tunable (Figure 1c). The DNA adaptors can be programmably connected into distinct stair-like or coil-like chiral supramolecular architectures with tuned handedness. Such chiral supramolecular assembly of DNA adaptor can be applied to govern the tunable directional self-assembly of gold nanorods decorated by the DNA adapter. As a result, plasmonic metastructures of new chiral forms, such as enantiomerically pure stair-like and coil-like gold nanorod helices that have never been reported before can be self-assembled, which are validated by cryo EM three-dimensional reconstructions, and they display interesting chiroptical properties that match well to theoretical predictions.
Figure 1.
a) Design of the DNA adapter. b) Cryo-ET subtomogram averaging of the DNA adapter. Subtomograms and the final average both show a ~ 45 degree inter-arm angle that is consistent with the design. c) The DNA adapters can be connected into different chiral supramolecular frameworks following the distinct chiral binding modality (M1–M4). Following the same mechanism, gold nanorods decorated with DNA adapter can be directionally self-assembled into different plasmonic chiral metastructures with distinct configurations and handedness following the DNA chiral supramolecular matrix. The cross sections of CM1 and CM2 exhibit pinwheel and coil morphologies, respectively.
Previously, chiral superstructures of DNA origami had been created, in which one had to insert or delete bases in parallel DNA helices to create a global twist or use the natural global twist of the DNA origami to begin with.[11] These structural torsions accumulate as DNA units further assemble into DNA superstructures. However, the chirality of such constructs is difficult and costly to tune, and the achievable structures are limited. Here, DNA adapter, which is rationally designed with distinct binding domains arranged in mirror-asymmetric manner, enables a facile new strategy for creating DNA chiral superstructures with facilely tuned handedness.
The DNA adapter is a 3D DNA origami nanostructure composed of two double-layered rectangular arms (both ~ 57 nm × 21 nm[12]) connected by a flexible hinge and 2-helix bundle (2-HB) spacer (~ 43 nm in length if straight). This 2-HB spacer fixes the inter-arm dihedral angle approximately 45 degrees (Figure 1a). Four binding domains, labeled “a”, “b”, “c”, and “d”, each consisting of 16-nucleotide (nt) single-stranded loops, enable intermolecular interactions through hybridization with a set of connecting strands (each 16 nt long). The reconstructed DNA adapter from cryo electron tomography shows two separated arms with inter-arm dihedral angles narrowly-distributed around 45 degrees, consistent with the design (Figure 1b). The 2-HB spacer is not observed in the averaged cryo-EM image, indicating that it is relatively flexible.
Figure 2a shows a large-area AFM image of the individual DNA adapters, which confirms the well formation and high purity of DNA adapter assembled as designed. The DNA adapters can self-assemble into four different chiral supramolecular frameworks (FMs) of helical morphology by selecting different combinations of the DNA connecting strands that link two specific binding domains (b+c or a+d) between the neighboring units (Figure 1c). The assemblies include the stair-like DNA frameworks (both left-handed FM1 and right-handed FM3) and the coil-like DNA frameworks (both right-handed FM2 and left-handed FM4). Similarly, gold nanorods decorated with DNA adapters can directionally self-assemble into four distinct chiral metastructures (CM), including two stair-like nanorod helices (CM1 and CM3) and two coil-like nanorod helices (CM2 and CM4), corresponding to their supramolecular scaffolds FM1 to FM4. CM1 has a side-by-side left-handed conformation, while CM2 has an end-to-end right-handed conformation. CM1 is mirror-symmetric to CM3, and CM2 is mirror-symmetric to CM4.
Figure 2.
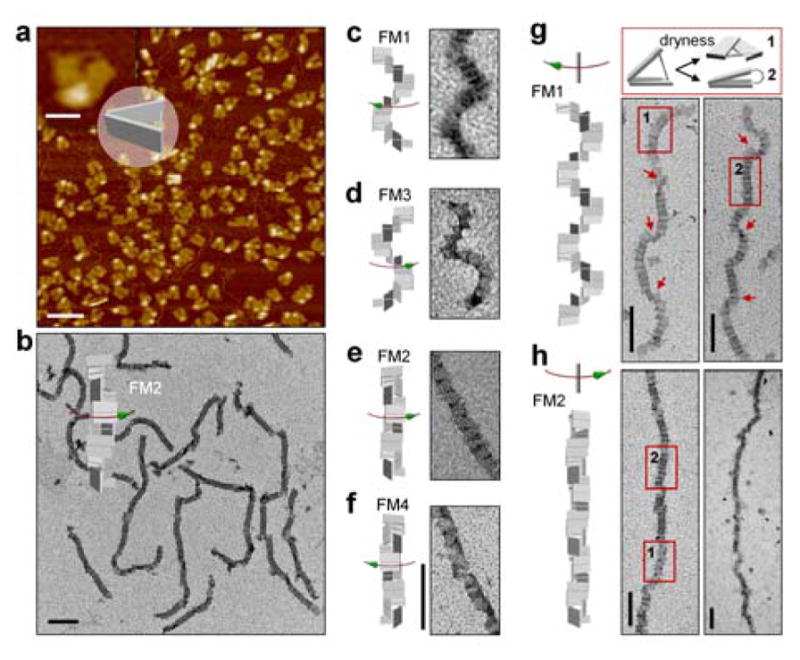
a) AFM image of the individual DNA adapter. The scale bar is 50 nm in the zoomed-in image and 200 nm in the zoomed-out image. b–h) Representative TEM images of the assembled DNA supramolecular structures. All scale bars are 200 nm. Red arrows in g indicate sharp twists in the superstructure due to flattening on the TEM grid. Numbered rectangular boxes in g and h represent the two flattened states (depicted in the top panel of g) of a short segment of a DNA superstructure after drying on the TEM grid.
To distinguish the connections between different binding domains, native DNA crossovers involving 40 connecting strands and mutated crossovers involving 20 connecting strands were designed for the binding domains (b+c) and domains (a+d), respectively. Therefore, FM1 and FM2 are expected to grow to longer extent than FM3 and FM4 due to the stronger connections. As shown in Figure 2b–h, the lengths of most FM1 and FM2 structures were several hundred nanometers, with FM2 approaching the micrometer scale. The lengths of FM3 and FM4 were relatively shorter than FM1 and FM2 (Figures S18 and S19). It is noted that FM3 and FM4 can also grow much longer by modifying the connections to form native DNA crossovers at the binding domains between the DNA adapters. The stair-like and coil-like DNA chiral supramolecular frameworks show clear morphological differences. The characteristic winding and twisting of the superstructures were observed in the stair-like FM1 (Figure 2c and g), while relatively straight conformations arose from the coil-like FM2 (Figure 2b, e and h). The same is true when comparing the stair-like FM3 to the coil-like FM4. Abrupt structural twists (for FM1) or opening of the edges (for FM2) were observed due to collapsing of the helical superstructures on the surface of the TEM grid during sampling (Figure 2g and h).
Having validated and characterized the tunable self-assembly of the DNA adapters into various chiral supramolecular structures, we proceeded to study the chiral directional self-assembly of gold nanorods templated on the self-assembled DNA suprastructures. As illustrated in Figure 3a, the optical module that incorporate a gold nanorod on the patch arm of the DNA adapter can self-assemble into distinct stair-like and coil-like plasmonic chiral metastructures with addition of different sets of DNA connecting strands. Cryo-ET tomograms demonstrated the expected chiral assembly of the gold nanorods (Figure 3b). The overall metastructure conformation was affected by the limited rigidity of the 2-HB adapter spacer and the integration of the gold nanorods,[13] in which the observed periodicity (~8–12 units per helical turn) of the helical assemblies deviated from the design value (8 units per helical turn). Although there are slight periodicity deviations, the observed chiral conformation (stair- or coil-like) and handedness agreed to the chiral metastructure design. For example, as shown in the CM1 tomograms, the observed structure exhibited the expected left-handed stair-like conformation. TEM images (Figure S26) showed that CM1 and CM2 are generally several hundred nanometers long, by average longer than CM3 and CM4, which are consistent to the sizes of their corresponding DNA supramolecular templates.
Figure 3.
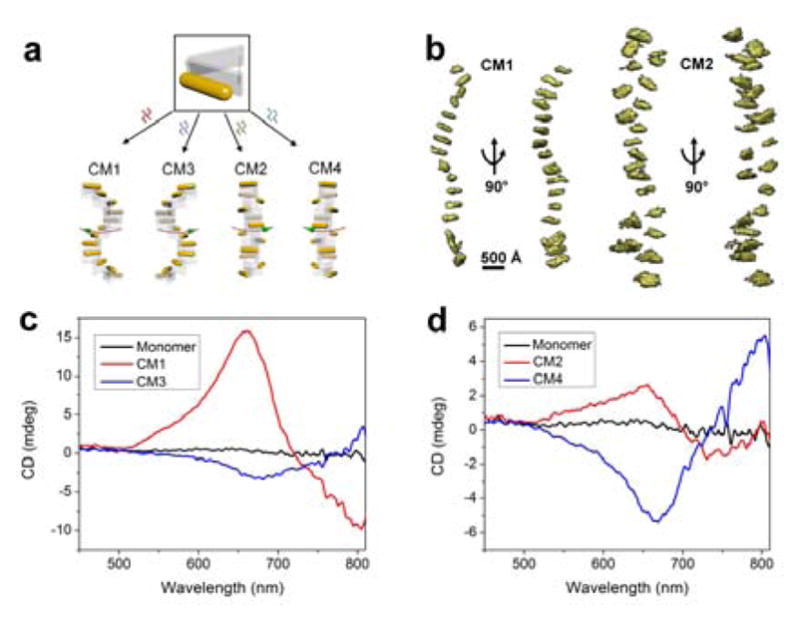
a) Tunable chiral directional self-assembly of nanorods by use of four different sets of DNA connecter strands. b) Tomograms from cryo-ET for individual metastructures CM1 and CM2, respectively. c–d) CD spectra of the chiral metastructures CM1-CM4.
Circular dichroism (CD) measurements of the stair-like CM1 and CM3 exhibited opposite CD signals (Figure 3c), agreed with their mirror-image structures. The characteristic peak-dip and dip-peak spectral line shapes are consistent with the left-handedness of CM1 and the right-handedness of CM3. However, for the coil-like CM2 and CM4, the right-handed CM2 presented a peak-dip CD signal, while the left-handed CM4 showed a dip-peak CD signal (Figure 3d), indicating that the optical chirality was opposite to that predicted by the ensemble handedness of the assemblies. This marked difference between stair-like and coil-like metastructures suggests that the nature of the optical chirality cannot be simply predicted from the apparent overall handedness of the superstructure, which was not emphasized in the previous studies.[10a]
CM1 showed a rapid increase in CD signal with the increase of the assembly time (Figure 4a), while the CD increase rate was significantly slower for CM2. DLS measurements indicated that the sizes of both CM1 and CM2 metastructures increased with assembly time following a similar trend (Figure S28), and they grew into metastructures of similar average lengths (Figure S25–26). We further used the discrete dipole approximation (DDA) method to calculate the theoretical CD spectra of the four chiral metastructures with varied numbers of nanorod layers, from 2 to 12 layers (Figures S31 and S32). The calculations produced the same bisignated CD spectral line shapes as those we experimentally measured (Fiures 3d–d). The theoretical chiral signals of both CM1 and CM2 increased linearly with increasing number of nanorod layers, with a much attenuated slope for CM2 (Figure 4b). These calculations are consistent with the experimental results of the dependence of CD signal on the assembly time (Figure 4a).
Figure 4.
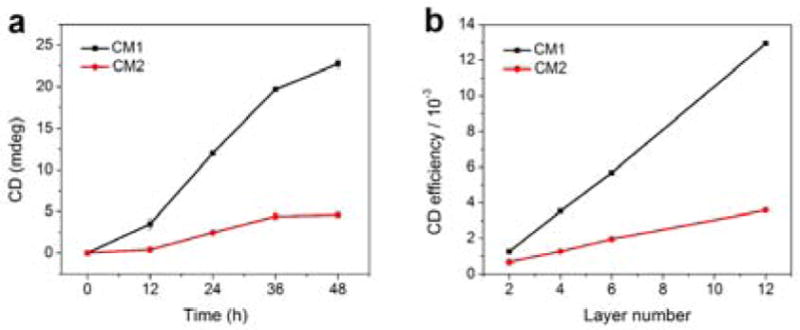
a) CD maximal signals of CM1 and CM2 experimentally measured after various annealing times. b) Theoretical peak values of CM1 and CM2 CD signals depend on the number of nanorod layers.
A systematic tuning of the chiral metastructure configuration could be accomplished by varying the length of the spacer between the two DNA adapter arms to adjust the adapter angle. This would tune the chiral optical responses in a manner similar to that of a DNA ruler. As illustrated in Figure 5a, a new way of generating tunable gold nanorod chiral metastructures could be designed. We calculated the theoretical chiral responses of CM1 and CM2 metastructures produced with DNA adapter angles varying from 15 to 60 degrees (Figures 5b–e). The higher-order chiral metastructures (6-layer) exhibit the same sign of optical chirality as a single pair of neighboring nanorods (2-layer), the chiral repeating unit, but a larger amplitude. According to the conceptual frame of plasmon hybridization theory,[14] the resonance energy of a nanorod twisted pair is split under left-handed and right-handed circularly polarized light (LCP and RCP) as a result of the generation of a mixture of antibonding and bonding resonance modes from the plasmon hybridizations between nanorods. For CM1 and CM2 with similar dihedral angle (model in Figure 1) between neighboring nanorods, LCP excites plasmon modes slightly blue-shifted with respect to RCP because of the more prominent antibonding mode excitation (Figure S33).[15] The differentiated absorptions for LCP and RCP of these plasmon modes caused the peak-dip CD line shape via reduction in RCP absorption relative to LCP absorption (Figure S31). Due to a similar mechanism, the pair of neighboring nanorods in CM3 and CM4 show a reversed dip-peak CD line shape. These results are in agreement with the experimentally measured optical chirality (Figure 3 in main text) and suggest that the optical chirality of these metastructures is strongly affected by intrinsic plasmon couplings within the chiral repeating units. Moreover, CM1 and CM2 exhibit distinct ensemble configurations and helical periodicity, which further account for the difference in the the generated CD signals. For examples, if the orientations of the rods in CM2 are examined from bottom to top, rods 1 and 2 form a left-handed structure as do rods 2 and 3; rods 1 and 3, however, form a right-handed structure. In contrast, neighboring rod pairs in CM1 always form left-handed structures. Therefore, the overall CD signal of CM2 may be diminished relative to that of CM1 due to a cancelling out effect. Besides, CM1 showed a stronger plasmon coupling compared to CM2 due to the closer inter-rod distances perpendicular to the axis of the helix, thus it partially explains the stronger dependence of the CD intensity on the number of layers of the gold nanorods in the metastructure (Figures S33 and S35) (For more theoretical calculations and optical discussions, please see the Section 7 in the supporting information).
Figure 5.
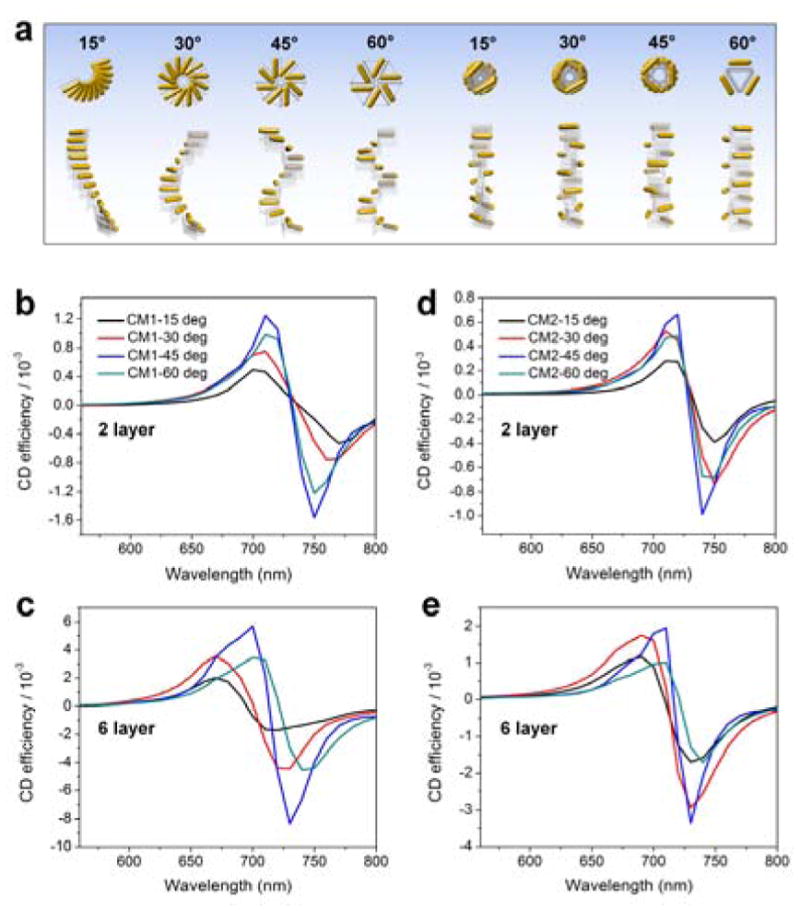
a) Left 4 and right 4 chiral metastructures are stair-like helices and coil-like helices resulting from the M1 and M2 binding modality, respectively, with angle of the DNA adapter varied from 15 to 60 degrees. b–e) Theoretical CD spectra of CM1 (b, c) and CM2 (d, e) chiral metastructures in panel a. Note the different Y-axis scales used in b–e.
In conclusion, we developed a new approach for tunable self-assembly of DNA chiral supramolecular architectures by creating a versatile DNA origami adapter. The chiral directional self-assembly of plasmonic nanorods decorated by the DNA adapter can be conveniently manipulated and tuned. The assembled stair-like and coil-like gold nanorod chiral metastructures demonstrated distinctive chiral optical responses. The demonstrated chiral metastructures with easily tuned hierarchy and handedness could be potentially used for DNA sensing devices and reconfigurable optically-active metamaterials. The tunable self-assembly of DNA chiral superstructures opens new possibilities for the programmable realization of a variety of supramolecular constructs that could function as chiral frameworks for heterogeneous nanocomponent self-assembly, thus lead to a host of new topographical and functional architectures.
Supplementary Material
Acknowledgments
This work was supported by grants from the Army Research Office, National Institute of Health, Office of Naval Re-search and National Science Foundation to H.Y. and Y.L. Q.W. thanks to financial support from National Natural Science Foundation of China (No. 21425103, 21673280).
Footnotes
Supporting information for this article is given on Wiley website.
Contributor Information
Dr. Xiang Lan, Biodesign Center for Molecular Design and Biomimetics, The Biodesign Institute, Arizona State University, Tempe, Arizona 85287, United States. School of Molecular Sciences, Arizona State University, Tempe, Arizona 85287, United States
Dr. Zhaoming Su, National Center for Macromolecular Imaging, Verna and Marrs McLean Department of Biochemistry and Molecular Biology, Baylor College of Medicine, Houston, Texas 77030, United States
Dr. Yadong Zhou, Chemistry Department, University of Central Florida, 4111 Libra Drive Orlando, FL 32816-2366, United States
Mr. Travis Meyer, Wallace H. Coulter Department of Biomedical Engineering at Georgia Institute of Technology and Emory University, 1760 Haygood Drive Health Sciences Research Bldg E186, Atlanta, GA 30322, United States
Prof. Yonggang Ke, Wallace H. Coulter Department of Biomedical Engineering at Georgia Institute of Technology and Emory University, 1760 Haygood Drive Health Sciences Research Bldg E186, Atlanta, GA 30322, United States
Prof. Qiangbin Wang, CAS Key Laboratory of Nano-Bio Interface, Division of Nanobiomedicine and i-Lab, Suzhou Institute of Nano-Tech and Nano-Bionics, Chinese Academy of Sciences, Suzhou 215123, China
Prof. Wah Chiu, National Center for Macromolecular Imaging, Verna and Marrs McLean Department of Biochemistry and Molecular Biology, Baylor College of Medicine, Houston, Texas 77030, United States
Prof. Na Liu, Max Planck Institute for Intelligent Systems, Heisenbergstrasse 3, 70569 Stuttgart, Germany. Kirchhoff Institute for Physics, University of Heidelberg, Im Neuenheimer Feld 227, 69120 Heidelberg, Germany
Prof. Shengli Zou, Chemistry Department, University of Central Florida, 4111 Libra Drive Orlando, FL 32816-2366, United States
Prof. Hao Yan, Biodesign Center for Molecular Design and Biomimetics, The Biodesign Institute, Arizona State University, Tempe, Arizona 85287, United States. School of Molecular Sciences, Arizona State University, Tempe, Arizona 85287, United States
Prof. Yan Liu, Biodesign Center for Molecular Design and Biomimetics, The Biodesign Institute, Arizona State University, Tempe, Arizona 85287, United States. School of Molecular Sciences, Arizona State University, Tempe, Arizona 85287, United States
References
- 1.Wang YF, Wang Y, Breed DR, Manoharan VN, Feng L, Hollingsworth AD, Weck M, Pine DJ. Nature. 2012;491:51–55. doi: 10.1038/nature11564. [DOI] [PubMed] [Google Scholar]
- 2.a) Lv JW, Hou K, Ding DF, Wang DW, Han B, Gao XQ, Zhao M, Shi L, Guo J, Zheng YL, Zhang X, Lu CG, Huang L, Huang W, Tang ZY. Angew Chem Int Ed. 2017;56:5055–5060. doi: 10.1002/anie.201701512. [DOI] [PubMed] [Google Scholar]; b) Guerrero-Martinez A, Auguie B, Alonso-Gomez JL, Dzolic Z, Gomez-Grana S, Zinic M, Cid MM, Liz-Marzan LM. Angew Chem Int Ed. 2011;50:5499–5503. doi: 10.1002/anie.201007536. [DOI] [PubMed] [Google Scholar]
- 3.a) Barron LD. Nature. 2000;405:895–896. doi: 10.1038/35016183. [DOI] [PubMed] [Google Scholar]; b) Barron LD. Nat Mater. 2008;7:691–692. doi: 10.1038/nmat2263. [DOI] [PubMed] [Google Scholar]
- 4.a) Wu X, Xu L, Liu L, Ma W, Yin H, Kuang H, Wang L, Xu C, Kotov NA. J Am Chem Soc. 2013;135:18629–18636. doi: 10.1021/ja4095445. [DOI] [PubMed] [Google Scholar]; b) Ma W, Kuang H, Xu LG, Ding L, Xu CL, Wang LB, Kotov NA. Nat Commun. 2013;4:2689. doi: 10.1038/ncomms3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Lin YY, Pashuck ET, Thomas MR, Amdursky N, Wang ST, Chow LW, Stevens MM. Angew Chem Int Ed. 2017;56:2361–2365. doi: 10.1002/anie.201610976. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Liu MH, Zhang L, Wang TY. Chem Rev. 2015;115:7304–7397. doi: 10.1021/cr500671p. [DOI] [PubMed] [Google Scholar]
- 6.a) Chen Q, Bae SC, Granick S. J Am Chem Soc. 2012;134:11080–11083. doi: 10.1021/ja303434d. [DOI] [PubMed] [Google Scholar]; b) Fan JA, Wu CH, Bao K, Bao JM, Bardhan R, Halas NJ, Manoharan VN, Nordlander P, Shvets G, Capasso F. Science. 2010;328:1135–1138. doi: 10.1126/science.1187949. [DOI] [PubMed] [Google Scholar]
- 7.a) Jones MR, Macfarlane RJ, Lee B, Zhang JA, Young KL, Senesi AJ, Mirkin CA. Nat Mater. 2010;9:913–917. doi: 10.1038/nmat2870. [DOI] [PubMed] [Google Scholar]; b) Lu F, Yager KG, Zhang YG, Xin HL, Gang O. Nat Commun. 2015;6:6912. doi: 10.1038/ncomms7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choueiri RM, Galati E, Therien-Aubin H, Klinkova A, Larin EM, Querejeta-Fernandez A, Han LL, Xin HL, Gang O, Zhulina EB, Rubinstein M, Kumacheva E. Nature. 2016;538:79–83. doi: 10.1038/nature19089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu WY, Halverson J, Tian Y, Tkachenko AV, Gang O. Nat Chem. 2016;8:867–873. doi: 10.1038/nchem.2540. [DOI] [PubMed] [Google Scholar]
- 10.a) Kuzyk A, Schreiber R, Fan Z, Pardatscher G, Roller EM, Hogele A, Simmel FC, Govorov AO, Liedl T. Nature. 2012;483:311–314. doi: 10.1038/nature10889. [DOI] [PubMed] [Google Scholar]; b) Lan X, Chen Z, Dai G, Lu X, Ni W, Wang Q. J Am Chem Soc. 2013;135:11441–11444. doi: 10.1021/ja404354c. [DOI] [PubMed] [Google Scholar]; c) Urban MJ, Dutta PK, Wang PF, Duan XY, Shen XB, Ding BQ, Ke YG, Liu N. J Am Chem Soc. 2016;138:5495–5498. doi: 10.1021/jacs.6b00958. [DOI] [PubMed] [Google Scholar]
- 11.Dietz H, Douglas SM, Shih WM. Science. 2009;325:725–730. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai XC, Martin TG, Scheres SHW, Dietz H. P Natl Acad Sci USA. 2012;109:20012–20017. doi: 10.1073/pnas.1215713109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma J, Chhabra R, Cheng A, Brownell J, Liu Y, Yan H. Science. 2009;323:112–116. doi: 10.1126/science.1165831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prodan E, Radloff C, Halas NJ, Nordlander P. Science. 2003;302:419–422. doi: 10.1126/science.1089171. [DOI] [PubMed] [Google Scholar]
- 15.Yin XH, Schaferling M, Metzger B, Giessen H. Nano Lett. 2013;13:6238–6243. doi: 10.1021/nl403705k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.