Abstract
Rationale
Previous studies have suggested that there is an inverse genetic relationship between ethanol consumption (two-bottle choice, continuous access) and ethanol withdrawal (e.g., Metten et al., Behav Brain Res 95:113–122, 1998a).
Objectives
The current study used short-term selective breeding from heterogeneous stock (HS) animals to examine this relationship. The primary goal of the current study was to determine if reciprocal quantitative trait loci (QTLs) could be found in the selectively bred lines. The advantage of detecting QTLs in HS animals is that it is possible to extract a haplotype signature for the QTL, which in turn can be used to narrow the number of candidate genes generated from gene expression and sequence databases (see, e.g., Hitzemann et al., Mamm Genome 14:733–747, 2003).
Results
Seven reciprocal QTLs were detected on chromosomes (Chr) 1 (two), 3, 6, 11, 16, and 17 that exceeded the nominal LOD threshold of 10; genetic drift, which occurs during selection, dramatically increases the LOD threshold. The proximal Chr 1 QTL was examined in some detail. The haplotype structure of the QTL was such that the LP/J allele was associated with low withdrawal and high consumption. The QTL appears to be located in a gene-poor region between 170 and 173 Mbp. Based on available sequence data, two plausible candidate genes emerge—Nos1ap and Atf6α.
Conclusions
The data presented here confirm some aspects of the negative genetic relationship between acute ethanol withdrawal and ethanol consumption. The QTL data point to the potential involvement of NO signaling and/or the unfolded protein response.
Keywords: Ethanol, Withdrawal, Consumption, QTL, Mouse, Heterogeneous stock, Haplotype
Introduction
There is strong evidence, especially in mouse populations derived from the C57BL/6J (B6) and DBA/2J (D2) inbred strains, for a negative genetic relationship between ethanol consumption (two-bottle choice, continuous access) and both acute and chronic ethanol withdrawal (see Metten et al. 1998a). High ethanol withdrawal (scored as handling-induced convulsions [HICs]) is associated with low ethanol consumption and vice versa. Data supporting this relationship have been obtained in the BXD recombinant inbred (RI) panel, B6×D2 F2 intercrosses, two independent selections from B6×D2 F2 intercrosses, and in a BXD F1 (RIX) population. Putative reciprocal quantitative trait loci (QTLs), which appear to contribute to the negative correlation, are found on chromosomes (Chr) 1 (distal), 2 (mid), 4 (mid), and 15 (proximal; Metten et al. 1998a). Reciprocal QTLs are defined by alleles associated with high withdrawal/low consumption and vice versa. To date, the genes within the reciprocal QTL intervals that are driving the differential responses (the quantitative trait genes [QTGs]) have not been determined.
When moving beyond B6×D2 genotypes, the evidence supporting a reciprocal relationship between consumption and withdrawal has been mixed. Kosobud et al. (1988) found that withdrawal seizure-prone and seizure-resistant (WSP/WSR) selected lines, derived from a heterogeneous stock (HS/Ibg—a cross of eight inbred mouse strains), differed as expected in consumption; when compared to the WSP mice, the WSR mice showed a lower ethanol consumption. The WSP and WSR lines were selected for the severity of the HIC response after chronic ethanol exposure. However, a new selection for the same phenotype and from HS/Ibg animals failed to replicate these results (Metten et al. 1998a). When surveying panels of inbred mouse strains, the reciprocal relationship has been detected for chronic but not acute withdrawal (Crabbe et al. 1983; Belknap et al. 1993; Metten and Crabbe 1994). Chester et al. (2002, 2003) examined the relationship in the alcohol preferring (P) and the high alcohol drinking (HAD1 and HAD2) selected lines versus the non-alcohol preferring (NP) and the low alcohol drinking (LAD1 and LAD2) selected lines. The P and NP lines were derived from outbred Wistar rats, while the HAD and LAD lines were derived from an eight-strain rat HS. Acute withdrawal sensitivity was measured using the acoustic startle response. The P/NP and HAD1/LAD1 lines gave the expected acute withdrawal response—higher in the NP and LAD1 lines. However, there was no significant difference between the HAD2 and LAD2 lines. In a separate study and when white noise rather than a tone was used as the stimulus, the P/HAD1/HAD2 lines exhibited an enhanced withdrawal-induced startle response, whereas no change was found in the NP/LAD1/LAD2 lines (Chester et al. 2004). Recently, Chester and Barrenha (2007) examined the acute withdrawal-induced startle response in the high alcohol preferring (HAP1/HAP2) and low alcohol preferring (LAP1/LAP2) mouse lines, which were selectively bred from HS/Ibg mice (Grahame et al. 1999). Here, the male LAP lines, when compared with the male HAP lines, showed a withdrawal-induced suppression of the startle response; the effect in females was line dependent.
Metten et al. (1998b) used short-term selective breeding (STSB) from a B6×D2 F2 intercross to produce what they termed short-term drinking response high and low lines and high/low acute withdrawal lines; the lines showed the expected reciprocal response for acute withdrawal and consumption. The current study used a similar experimental design except that HS mice were used as the founding population. The HS population was formed by crossing the B6, D2, BALB/cJ (C), and LP/J (LP) inbred strains. This HS population (hereafter termed HS4) has approximately twice the genetic diversity of a B6×D2 F2 intercross and more than three times the genetic diversity of the standard BXD RI panel (see Roberts et al. 2007). The C strain is intermediate between the B6 and D2 strains in terms of ethanol consumption and acute withdrawal (Metten and Crabbe 1994; Fehr et al. 2002). The LP is similar to the D2 strain in terms of ethanol consumption (low; Yoneyama et al. 2008); to our knowledge, there are no reports describing ethanol withdrawal in the LP strain. However, acute ethanol withdrawal in the closely related 129S1/SvIJ strain (Beck et al. 2000) is moderate and similar to that seen in the B6 strain (Fehr et al. 2002).
The primary goal of the current study was to determine if reciprocal QTLs could be found in the selectively bred lines formed by short-term selective breeding from HS4. The advantage of detecting QTLs in HS animals is that it is possible to extract a more detailed haplotype signature for the QTL. The importance of extracting a more detailed haplotype signature can be seen in the following example. Consider that we have detected a HS4 QTL, but the only information available is that the genetic marker or markers differentiate the B6 and D2 strains; if the B6 strain has the A allele and the D2 strain has the B allele, the haplotype is simply AB. If we now add information about the C and LP strains, the QTL could have not one but four possible haplotypes (ABAA, ABBBB, ABAB, and ABBA). This increased information makes it possible to narrow the QTL interval and the number of candidate genes generated from gene expression and sequence databases (see, e.g., Hitzemann et al. 2003).
Finally, a secondary goal of these experiments was to determine if the reciprocal QTLs previously detected in various B6×D2 crosses (see above) would also be detected in the HS4 selected lines. If so, this would greatly expand the strategies that could be used to detect the underlying QTGs.
Materials and methods
Animals
All mice were maintained in a temperature-controlled colony room (21–23°C) on a 12-h light–dark cycle and were allowed free access to food and water. Details concerning the formation of the HS4 colony are found elsewhere (Malmanger et al. 2006). Briefly, the colony is maintained as 48 families; the standard circle design is used for breeding. G19 animals were used as the founding population for STSB. Three to four males and females from each of 48 families were used for selection; details of the selection phenotypes (acute ethanol withdrawal and ethanol consumption) follow. The HS4 animals were divided into two cohorts of approximately equal size; one cohort was used to select for ethanol withdrawal, and the other was used to select for ethanol consumption. However, all animals were phenotyped for both traits, with ethanol consumption always preceding acute withdrawal. The order of testing is important; in the B6, D2, and C inbred strains, testing first for ethanol consumption does not affect the withdrawal response. However, the reverse is not true; ethanol consumption is significantly decreased in the B6 and C strains after exposure to acute withdrawal (unpublished observations).
For both phenotypes, animals were selected to form ten High- and ten Low-response families; brother–sister mating was avoided even if it meant using an animal that was not in the highest or lowest tier. To the extent possible, animals used for acute withdrawal selection were mated to balance differences in baseline withdrawal scores. A similar selection procedure was followed for each subsequent generation. Selection was stopped at S5 and S4 for acute withdrawal and ethanol consumption, respectively. All animal care, breeding, and testing procedures were approved by the Laboratory Animal Users Committees at the Veterans Affairs Medical Center, Portland, OR, USA and Oregon Health & Science University, Portland, OR, USA.
Behavioral testing
The withdrawal response was measured using the HIC, which is elicited by lifting the animal by the tail and looking for convulsive signs (Crabbe et al. 1991). Signs are rated from 0 (absent) to 7 (severe) and in part determined by whether the sign was elicited by simply lifting the animal or by spinning the animal in a 180–360° arc. At T−30min and T−10 min, baseline withdrawal measures were taken. At T0, animals were administered 4.0 g/kg ethanol, an anesthetic dose in all animals. Withdrawal signs were measured beginning at T+2 h and continuing every hour until T+12 h. Peak withdrawal signs are generally seen 6–8 h after ethanol administration (Metten et al. 1998a, b). Data are reported either as the corrected area under the curve (CAUC) or the corrected maximum response (CMAX); the correction is obtained by subtracting the average baseline value from the post-drug values.
Ethanol consumption was measured in individually housed animals presented with a continuous choice between tap water and an ethanol solution. The duration of the procedure was 10 days. For the first 2 days, animals were presented with two water bottles, and on each day, the volume of water consumed from each bottle was measured. On day 3, 3% ethanol in water (v/v) was substituted for one of the water bottles. On day 7, 10% ethanol was substituted for the 3% ethanol solution. The position of the ethanol bottle was changed on days 5, 7, and 9. Ethanol consumption was measured as the average consumption on days 4 and 6 (3% ethanol) and on days 8 and 10 (10% ethanol). Data are reported as grams per kilogram ethanol consumed. The consumption of 10% ethanol was the selection phenotype. Data were also collected for total fluid volume consumed, which were used to measure ethanol preference.
All behavioral data were analyzed using a standard factorial or repeated measures analysis of variance (ANOVA); Tukey’s HSD test was used for the post hoc analysis. These analyses were run using Statistica (StatSoft, Tulsa, OK, USA).
Genotyping the selected lines
DNA was extracted as described elsewhere (Malmanger et al. 2006). Mice were genotyped using a custom single nucleotide polymorphism (SNP) array and the Illumina Golden Gate Assay (San Diego, CA, USA). A description of the array is found in Hitzemann et al. (2008). Samples were run locally using procedures exactly as described by the manufacturer. The SNP data for the High and Low lines were analyzed using a marker-by-marker C2 analysis. In the absence of a correction for drift, the nominal LOD threshold (4.2) is obtained by correcting for the number of multiple comparisons. The drift correction was patterned after that of Belknap et al. (1997); details of how the correction was calculated are found in Hitzemann et al. (2008). For the experiments described here, the estimated −LogP threshold for a significant QTL was 10.5.
Results
Ethanol withdrawal and consumption in G19 HS4 mice
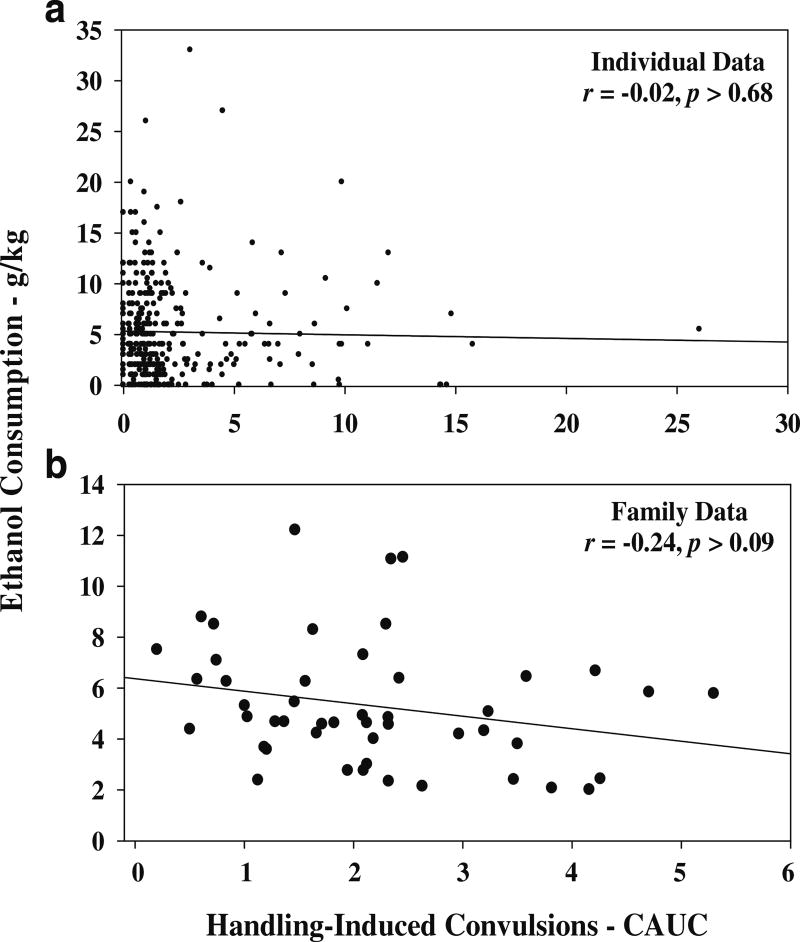
The data in Fig. 1a illustrate that there was no significant correlation between ethanol withdrawal and consumption in individual G19 HS4 mice (r=−0.02, p>0.68). The metrics being compared are grams per kilogram of ethanol consumed per 24 h and the CAUC for HIC scores (see “Materials and methods” for details on the calculation of the CAUC). The data in Fig. 1b illustrate that when the data are grouped according to family, there is a trend toward a negative correlation (r=−0.24, p>0.09). Among individual animals, males compared to females showed a significantly greater withdrawal score (6.25±0.43 vs 4.28±0.30—relative CAUC units, p<0.0003). In contrast, females as compared to males showed significantly greater ethanol consumption (2.84±0.29 vs 1.55±0.16 g/kg/24 h, p<0.0002). These data suggested that it would be of value to analyze the family data separately for males and females; the correlation between withdrawal and consumption was essentially unchanged (−0.16Males and −0.26Females).
Fig. 1.
Ethanol consumption and acute ethanol withdrawal in HS4, generation 19, mice. For acute withdrawal, animals were first tested at T−30 min and T−10 min for baseline withdrawal scores. At T−0, animals were injected with 4 g/kg of ethanol. Testing for HICs began at T+2 h and continued for every hour up to 12 h. Animals were always phenotyped for consumption first. a The relationship between consumption and withdrawal for individual mice (N=348). b The relationship when the data are collapsed across families (N=48). Consumption is expressed as grams of ethanol consumed per 24 h. Withdrawal is expressed as the CAUC for HICs. To calculate the CAUC, the average baseline score was subtracted from the postethanol scores
Animals were identified on the basis of six different coat colors—black (86), brown (22), dilute (27), albino (80), agouti (78), and piebald (30). The ANOVA revealed no significant effect for coat color on ethanol withdrawal (p>0.8). There was a modestly significant coat color effect for consumption (F5,318=2.5, p<0.03); however, the post hoc analysis (Tukey HSD for unequal N) revealed no significant difference among groups. The trend was for higher ethanol consumption in the dilute animals. There was no significant family effect for consumption (p>0.60); however, the family effect for withdrawal was significant (F47,274=1.66, p<0.006). The range of family variation is illustrated in Fig. 1b.
Short-term selective breeding (STSB) for ethanol withdrawal
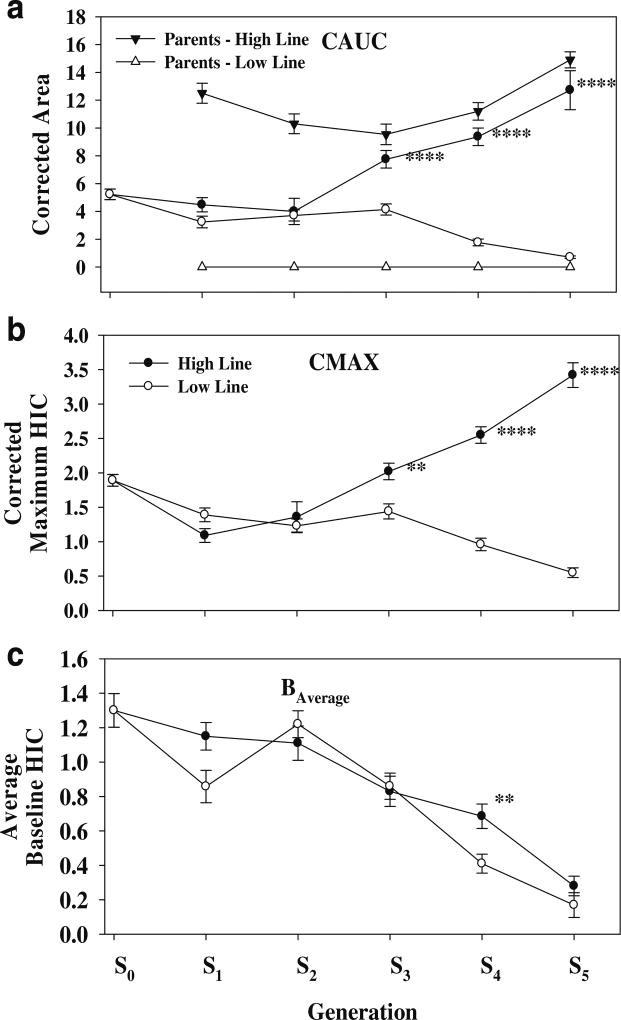
The selection for acute withdrawal is illustrated in Fig. 2a. The ANOVA revealed a significant effect for line (F1,861=19, p<1.3 × 10−5), generation (F4,861=10, p<7 × 10−8), and the line × generation interaction (F4,861= 32, p<9 × 10−25). At S5, the difference in the CAUC between the High and Low lines was 12.7±1.4 vs 0.7± 0.9 (relative CAUC units). Over generations S3 to S5, the High Line was significantly different from the Low Line at p<1 × 10−4 or better (Tukey HSD for unequal N). Within the High Line, generations S3 to S5 differed from generations S1 and S2 at p<1 × 10−3 or better. Within the Low line, generation S3 differed significantly from generations S4 and S5 at p<0.02 and 0.01, respectively. Paralleling the segregation for the CAUC, there was also a segregation for the CMAX (Fig. 2b); the ANOVA revealed a significant line × generation interaction (F4,861=43, p<9 × 10−25). At S5, the difference between the High and Low lines was (in relative units) 3.4±0.2 vs 0.6±0.01 (p<1.2 × 10−5). During selection, there was a significant generation effect for the average baseline withdrawal score (F4,861=40, p<1 × 10−30; Fig. 2c); the average baseline score (relative units) decreased from 1.0± 0.06 to 0.22±0.05 (p<6 × 10−5). At S5, there was no significant difference in baseline withdrawal score between the High and Low lines (0.28±0.06 vs 0.17±0.07).
Fig. 2.
Short-term selection for acute ethanol withdrawal. a The response to selection in the High and Low responding lines. The average CAUC for the parents at each generation of selection is also illustrated. b The effects of selection on the corrected maximum HIC score (CMAX). c The effects of selection on the average baseline score. Asterisks indicate High line significantly different from respective Low line value. **p<10−2; ****p<10−4
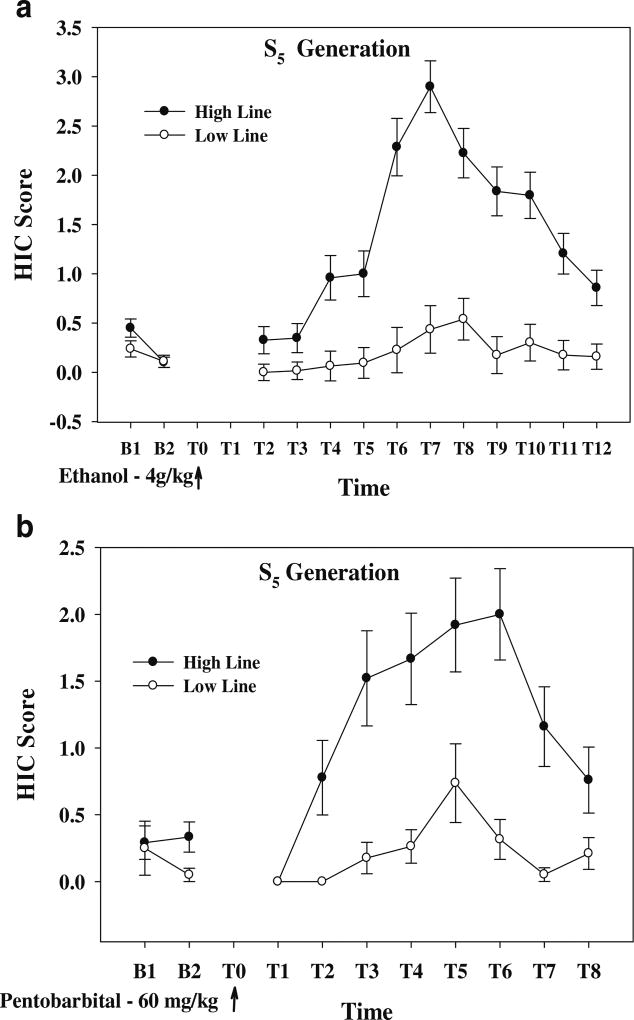
The data in Fig. 3a illustrate the time course of the ethanol withdrawal response for the High and Low lines at S5. The peak response in the High line occurred 6–8 h after the injection of ethanol (4 g/kg, IP). Naive S5 mice were challenged with 60 mg/kg (IP) of pentobarbital. The time course of the withdrawal response is illustrated in Fig. 3b. The difference in the CAUC between the High and Low lines was 7.0±1.3 vs 1.6±0.56 (p<6 × 10−4); the CMAX was also significantly different (2.5±0.31 vs 0.82±0.31, p< 1 × 10−3). Ethanol naive S5 animals were also tested for ethanol consumption using the standard protocol (see “Materials and methods”). Average consumption in the High and Low lines (10% ethanol, two-bottle choice, 24-h continuous access) was 0.98±0.29 vs 1.2±0.31 g/kg, p>0.5; data not shown).
Fig. 3.
Time course for acute ethanol and acute pentobarbital withdrawal in animals selected for acute ethanol withdrawal at the S5 generation. a The time course for acute ethanol withdrawal. b The time course for acute pentobarbital withdrawal. All animals were drug naïve until testing. N=16 to 20 animals per line. Data are reported as the uncorrected HIC score
Short-term selective breeding (STSB) for ethanol consumption
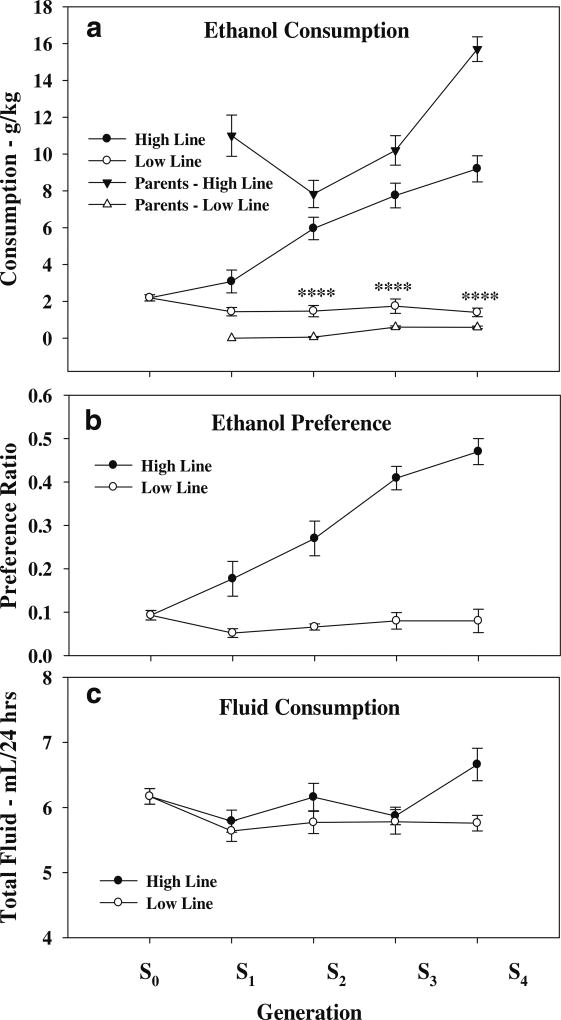
The segregation of the High and Low lines for ethanol consumption is illustrated in Fig. 4. The ANOVA revealed a significant effect for line (F3,414=12, p<3 × 10−7), generation (F3,414=13, p<3 × 10−8), and the line × generation interaction (F3,414=11, p<1 × 10−6). The post hoc analysis revealed a significant difference between the High and Low lines at S2 (5.1±0.62 vs 1.5±0.23 g/kg, p<8 × 10−3), at S3 (7.8±0.67 vs 1.7±0.39 g/kg, p<3 × 10−5), and at S4 (9.1±0.71 vs 1.4±0.22 g/kg, p<3 × 10−5). Segregation for preference paralleled the segregation for consumption—the ANOVA revealed a significant line × generation effect interaction (F3,414=14, p<1 × 10−8); at S4, preference (volume of 10% alcohol consumed/total fluid consumed) was significantly different between the High and Low lines (0.47±0.032 vs 0.081±0.027, p<3 × 10−5); although to a lesser degree, preference was also significantly different (p<5 × 10−4 or better) at S2 and S3. The ANOVA for total volume consumed revealed a significant effect for line (F3,414=8.2, p<3 × 10−5) but not generation or the line × generation interaction. On average, the difference in total fluid consumption between the High and Low lines was 6.10±0.15 vs 5.81±0.07 ml, p<0.01).
Fig. 4.
Short-term selection for ethanol consumption (two-bottle choice, water vs 10% ethanol, continuous access). a The response to selection in the High and Low responding lines. Consumption in the parents at each generation of selection is also illustrated. b The effects of selection on ethanol preference. Preference is measured as the ratio of ethanol fluid consumed/total fluid consumed. c The effects of selection on total fluid consumed. Asterisks indicate High line significantly different from respective Low line value. ****p<10−4
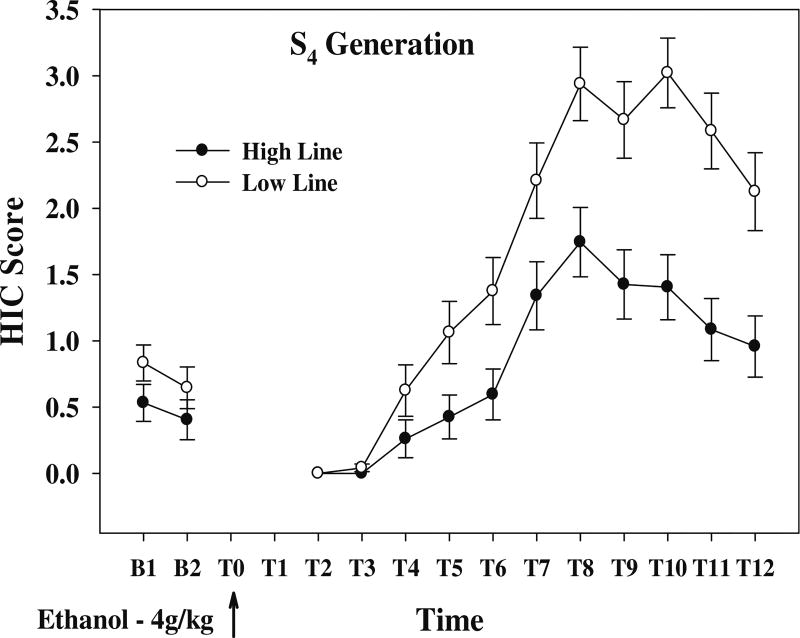
The data in Fig. 5 illustrate acute ethanol withdrawal at S4 in the STSB lines for ethanol consumption. The CAUC (relative units) for the High and Low lines was 5.91±1.14 vs 12.1±1.27 (p<3 × 10−4). The CMAX was also significantly different—2.18±0.24 vs 3.28±0.20 (p<5 × 10−4). The difference in the baseline scores was not significant (p>0.4).
Fig. 5.
Time course of acute ethanol withdrawal in S4 animals selected for High and Low ethanol consumption. Data are reported as the uncorrected HIC score. N=20/line
Detection of reciprocal quantitative trait loci (QTL) in the selection lines
Forty-seven to 56 each of the selection lines for acute withdrawal and consumption were genotyped using a panel of 768 SNPs. Because of the genetic drift that occurs during selection, it was estimated that the threshold for a significant association was LOD> 10. Seven reciprocal QTLs were detected (Table 1). The table illustrates the marker with the peak LOD score. The founding HS4 population was genotyped to determine if the reciprocal QTLs would also be detected. All of the QTLs were detected with the appropriate haplotype structure (see Table 1), with the exception of the reciprocal QTL on Chr 6. The data in Table 2 illustrate the allele segregation for the reciprocal QTLs. For some of the QTLs, there was a complete loss of heterozygosity (see the QTLs on Chr 1, 3, and 16).
Table 1.
Detection of coincident reciprocal QTLs for acute ethanol withdrawal and ethanol consumption
| SNP | Phenotype | Chr | Mbp | Haplotype B6:D2:C:LP |
−LogP |
|---|---|---|---|---|---|
| rs13476229 | Acute withdrawal | 1 | 171.14 | 1:1:1:2 | 20.30 |
| Consumption | 24.30 | ||||
| rs6157620 | Acute withdrawal | 1 | 185.26 | 1:1:2:2 | 17.40 |
| Consumption | 11.00 | ||||
| rs6376008 | Acute withdrawal | 3 | 86.74 | 1:1:1:2 | 12.40 |
| Consumption | 14.20 | ||||
| rs3718735 | Acute withdrawal | 6 | 101.08 | 1:2:2:2 | 13.30 |
| Consumption | 17.80 | ||||
| rs6386362 | Acute withdrawal | 11 | 106.79 | 1:2:2:2 | 12.90 |
| Consumption | 12.00 | ||||
| rs4165467 | Acute withdrawal | 16 | 25.32 | 1:1:1:2 | 18.60 |
| Consumption | 18.70 | ||||
| rs6390174 | Acute withdrawal | 17 | 38.59 | 1:2:2:1 | 16.60 |
| Consumption | 25.10 |
Short-term selective breeding was used to produce High and Low lines for acute ethanol withdrawal and ethanol consumption. At S5 for acute withdrawal and S4 for consumption, 47 to 56 each of the lines were genotyped using a SNP panel (see “Materials and methods”). Each SNP was analyzed using a marker-by-marker analysis (χ2). Because of genetic drift, the threshold for a significant QTL was set a −LogP≥10.5. Data in the table indicate the SNP associated with each QTL with the highest −LogP and the haplotype structure of each QTL
Table 2.
Allele segregation of the coincident reciprocal QTLs for acute ethanol withdrawal and ethanol consumption
| Phenotype | Chr | Mbp | Line–allele | |||
|---|---|---|---|---|---|---|
|
|
||||||
| High | Low | |||||
|
|
|
|||||
| A | B | A | B | |||
| Acute withdrawal | 1 | 171.14 | 88 | 6 | 25 | 69 |
| Consumption | 37 | 69 | 112 | 0 | ||
| Acute withdrawal | 1 | 185.26 | 14 | 80 | 72 | 20 |
| Consumption | 80 | 26 | 32 | 78 | ||
| Acute withdrawal | 3 | 86.74 | 94 | 0 | 53 | 41 |
| Consumption | 59 | 45 | 112 | 0 | ||
| Acute withdrawal | 6 | 101.08 | 4 | 90 | 50 | 42 |
| Consumption | 60 | 46 | 3 | 109 | ||
| Acute withdrawal | 11 | 106.79 | 4 | 90 | 49 | 43 |
| Consumption | 60 | 46 | 12 | 98 | ||
| Acute withdrawal | 16 | 25.32 | 37 | 57 | 92 | 0 |
| Consumption | 103 | 3 | 44 | 66 | ||
| Acute withdrawal | 17 | 38.59 | 10 | 84 | 66 | 26 |
| Consumption | 100 | 6 | 27 | 85 | ||
Experimental details are found in the legend to Table 1. The data presented in this table show the allele segregation at each of the SNPs shown in Table 1. The “A” allele is always the B6 strain allele
The proximal Chr 1 QTL associated with marker rs13476229 was examined in additional detail. As noted in Table 1, the haplotype for this SNP was LP:B6=D2=C. The Mouse Phenome SNP database (www.jax.org/phenome/snp.html) was queried for the distribution of SNPs with this haplotype over the interval from 160 to 180 Mbp. The number of SNPs that had data for all four strains was small (N=295), primarily due to the lack of data for the LP and C strains. However, the data suggested that the LP:B6=D2=C haplotype was found only between ~170 and 173 Mbp. Substituting the closely related (see Beck et al. 2000) 129S1/SvImJ (129) and the BALB/cByJ (CBy) strains for the LP and C strains, respectively, and focusing on the region between 168 and 178 Mbp, 7449 SNPs with the haplotype of interest were reported. Only one of these SNPs was found in the interval between 168 and 170 Mbp, and only 16 were found between 173 and 178 Mbp. The 170- to 173-Mbp interval is a relatively gene-poor region, containing only 20 known or predicted genes. These are Pbx1, Nuf2, Rgs5, Rgs4, 1700084C01Rik, Ddr2, Hsd17b7, Uhmk1, Uap1, Sh2d1b1, 1700015E13Rik, Nos1ap, Olmfml2b, Atf6, Dusp12, Fcrla, Fcrlb, Fcgr3, Fcgr2b, and Fcgr4. Two of these genes (Nos1ap {nitric oxide synthase [neuronal] adaptor protein} and Atf6 {activating transcription factor 6}) can be linked to ethanol response (Spanagel et al. 2002; Chen et al. 2008). Both genes have been reported to have coding non-synonymous SNPs (http://phenome.jax.org/pub-cgi/phenome/) with the129:B6=D2=CBy haplotype structure. The LP and C strains were sequenced to confirm that these SNPs also had the LP:B6=D2=C haplotype structure.
Discussion
The results presented here illustrate that selection for ethanol consumption produced lines with the predicted reciprocal relationship to acute withdrawal, but the selection for acute withdrawal did not produce lines that differed in ethanol consumption. In fact, both the High and Low lines for acute withdrawal had lower levels of consumption than the average for the progenitor HS4 population. The failure to demonstrate differences in consumption between the withdrawal lines may simply reflect the unique genetics of the HS4 animals, e.g., epistatic interactions that are present in the HS4 but not in B6×D2 intercrosses where the reciprocal relationship is readily observed (Metten et al. 1998a, b).
The question may be raised as to whether or not the selection for acute withdrawal was for a fundamentally different phenotype than the STSB selection reported by Metten et al. (1998b). This selection has been described in some detail. The lines were found to also differ in withdrawal convulsions after the administration of diazepam, nitrous oxide, zolpidem, and pentobarbital; the lines did not differ in threshold convulsant sensitivity to pentylenetetrazol, N-methyl-d-aspartate, or kainic acid (Metten et al. 1998b). These data were taken to indicate that the selection was not simply for a general difference in CNS activation but rather for some features associated with the mechanisms of action of the depressant drugs. Given the mechanisms of action of these drugs, a GABAergic mechanism is strongly implicated (see, e.g., Mihic et al. 1997; Buck and Finn 2001). In the current study, the High and Low acute withdrawal lines were also found to differ (and in the appropriate direction) for withdrawal after acute pentobarbital administration (Fig. 3).
The use of STSB has many advantages, including the ability to rapidly detect whether a particular phenotype is heritable, providing a strategy for confirming QTLs detected by some other independent experiment, and the ability to produce essentially unlimited numbers of animals that can be used for gene-expression analyses (see, e.g., Palmer et al. 2005). The major disadvantage of STSB is genetic drift and the stochastic fixation of some alleles that have no relevance to the phenotype of interest. Thus, one expects that many of the QTLs detected in STSB experiments will be false positives. To some extent, this problem can be dealt with by adjusting the LOD threshold (upward) for a significant QTL. However, this adjustment will always be an estimate and may well in some cases underestimate the needed correction. One common solution to this problem is to have a replicate selection—the idea here being that the chance of randomly fixing the same alleles in two independent selections is very small. The experimental design used in the current study extends this design by using two independent selections, which were predicted to generate reciprocal QTLs. Given the size of the SNP panel used to genotype animals, it was expected that one reciprocal QTL would be detected by chance. However, the chance probability of detecting seven reciprocal QTLs is very small (~10−21), and thus, we assume that the majority of these QTLs are real. Although not an independent experiment, the observation that we were able to confirm six of the seven QTLs in the founding HS4 population supports this position.
One goal of the current study was to determine if QTLs detected elsewhere for withdrawal and consumption would be confirmed in the HS4 selected lines. Metten et al. (1998a) noted that for B6×D2 crosses, there was evidence ranging from provisional to significant for reciprocal QTLs on Chr 1, 2, 4, and 15. Subsequent studies, focusing on the acute withdrawal phenotype, have confirmed QTLs on Chr 1 and 4 and an additional QTL on Chr 11 (Buck and Finn 2001; Fehr et al. 2002; Shirley et al. 2004; Hood et al. 2006). The Chr 4 QTL has been the best characterized; the associated QTG appears to be Mpdz (multiple PDZ domain protein; Fehr et al. 2002; Shirley et al. 2004). Focusing on the consumption/preference phenotype, QTLs have been confirmed on Chr 2, 3, 4, and 9 (Belknap and Atkins 2001). Of these, the Chr 9 QTL has been best characterized; putative QTGs are Hyou1 (Orp150; hypoxia upregulated protein 1) and Scn4b (sodium channel subunit beta-4; Mulligan et al. 2006). The overlap of the two data-sets is on Chr 4; however, the Chr 4 congenic strain, which captured the withdrawal QTL (see Fehr et al. 2002), did not differ in ethanol consumption (Buck, unpublished observation). Of the seven reciprocal QTLs detected in the current study, only three (Chr 6, 11, and 17) have a haplotype where the B6 differs from the D2 strain. Thus, the only potential overlap with the QTLs noted above is for the withdrawal QTL on Chr 11; however, the reciprocal Chr 11 QTL is distal to the QTL detected in the B6×D2 crosses. The question arises as to whether or not we detected any of the B6×D2 withdrawal or consumption QTLs. Although we have not looked at this issue extensively, we have been unable to detect the Chr 4 withdrawal QTL in either the withdrawal selected lines or in the parental HS4 population; in contrast, we have reliably detected the Chr 9 QTL for consumption. Although, overall, the QTL data collected in this and previous studies appear generally discordant, this may well not be the case. For example, a QTL detected in one genotype may be silent in another, but the loci may have strong epistatic effects. To examine these and related issues, we are currently in the process of phenotyping a much larger HS4 population and densely genotyping these animals across the regions of interest.
The more proximal reciprocal Chr 1 QTL lying between 170 and 173 Mbp was examined in some detail. The interval contains 20 known and predicted genes, several of which are highly expressed in the brain (http://symatlas.gnf.org/SymAtlas/). These include Rgs4 and Rgs5 (regulators of g-protein signaling 4 and 5), Umhk1 (U2AF homology motif [UHM] kinase), and Nos1ap (nitric oxide synthase [neuronal] adaptor protein); variants in each of these genes have been implicated as having a role in schizophrenia (e.g., Bakker et al. 2007; Campbell et al. 2008; Puri et al. 2007; Brzustowicz et al. 2004). To our knowledge, none of these genes has been directly implicated as having a role in ethanol-related behaviors. Indirectly, one could strongly infer a role for Nos1ap. Nos1ap (also known as carboxy-terminal PDZ domain ligand of nNos; neuronal nitric oxide synthase) competes with PSD95 for the nNos PDZ domain and prevents the coupling of nNos activation with NMDA-receptor-mediated calcium influx (Jaffrey et al. 1998). nNos knockout mice have been shown to have an increase in ethanol consumption (Spanagel et al. 2002). Direct sequencing confirmed that a non-synonymous SNP in Nos1ap has the correct haplotype structure. The question of whether this SNP is functionally relevant remains to be determined.
Atf6 emerged as another candidate gene in the proximal Chr 1 QTL interval. Here, one is actually referring to Atf6α, which is distinct from Atf6β (Hai and Hartman 2001; Thuerauf et al. 2007). For Atf6α, two non-synonymous coding SNPs with the appropriate haplotype structure were confirmed. One of these, H77P occurs in the N-terminus region known to be key for transcriptional activation and proteasomal degradation (Thuerauf et al. 2007). ATF6α is a 670-amino-acid transmembrane endoplasmic reticulum (ER) protein that is cleaved in response to ER stress; the cytoplasmic fragment is translocated to the nucleus where it activates the expression of a variety of genes, some of which are involved in the unfolded protein response (UPR). Chen et al. (2008) have shown that under in vitro conditions in both SH-SY5Y neuroblastoma cells and primary cerebellar cultures, ethanol can markedly potentiate the ER stress response induced by tunicamycin or thapsigargin; the mechanism of this ethanol effect appears to involve the production of reactive oxygen species. It also should be noted that, in addition to UPR-associated genes, ATF6α also regulates the expression of calmodulin 1, which in turn is involved in the regulation of nNOS (e.g., Spratt et al. 2007). Finally, while it may be only a coincidence, two additional genes involved in the UPR (Ire1 and Atf6β; Lin et al. 2007) lie within the haplotype-defined intervals for the Chr 11 and 17 QTLs, respectively.
Overall, the current study illustrates that STSB from HS4 animals resulted in the detection of reciprocal QTLs for acute ethanol withdrawal and ethanol consumption. The coincident and reciprocal QTLs detected in the current study appear to be different from those previously described (Metten et al. 1998a). Finally, the advantage of mapping in HS4-selected animals is illustrated: The proximal Chr 1 QTL was readily refined in terms of interval size and in terms of generating plausible candidate genes.
Acknowledgments
This study was supported in part by AA 10760, 11034, and 13484.
The authors wish to thank Dr. Kristin Demarest for help in reviewing and editing the manuscript. The authors also want to thank Dr. Pam Metten for help with the handling-induced convulsion assay.
Footnotes
All of the authors agree that there was no actual or potential conflict of interest in relation to this article.
Contributor Information
R. Hitzemann, Department of Behavioral Neuroscience, Oregon Health & Science University, 3181 S.W. Sam Jackson Park Road, Portland, OR 97239-3098, USA Research Service, Veterans Affairs Medical Center, Portland, OR 97239, USA.
S. Edmunds, Research Service, Veterans Affairs Medical Center, Portland, OR 97239, USA
W. Wu, Department of Behavioral Neuroscience, Oregon Health & Science University, 3181 S.W. Sam Jackson Park Road, Portland, OR 97239-3098, USA
B. Malmanger, Department of Behavioral Neuroscience, Oregon Health & Science University, 3181 S.W. Sam Jackson Park Road, Portland, OR 97239-3098, USA
N. Walter, Department of Behavioral Neuroscience, Oregon Health & Science University, 3181 S.W. Sam Jackson Park Road, Portland, OR 97239-3098, USA
J. Belknap, Department of Behavioral Neuroscience, Oregon Health & Science University, 3181 S.W. Sam Jackson Park Road, Portland, OR 97239-3098, USA Research Service, Veterans Affairs Medical Center, Portland, OR 97239, USA.
P. Darakjian, Department of Behavioral Neuroscience, Oregon Health & Science University, 3181 S.W. Sam Jackson Park Road, Portland, OR 97239-3098, USA
S. McWeeney, Department of Preventive Medicine and Epidemiology, Oregon Health & Science University, Portland, OR 97239-3098, USA
References
- Bakker SC, Hoogendoorn ML, Hendriks J, Verzijlbergen K, Caron S, Verduijn W, Selten JP, Pearson PL, Kahn RS, Sinke RJ. The PIP5K2A and RGS4 genes are differentially associated with deficit and non-deficit schizophrenia. Genes Brain Behav. 2007;6:113–119. doi: 10.1111/j.1601-183x.2006.00234.x. [DOI] [PubMed] [Google Scholar]
- Beck JA, Lloyd S, Hafezparast M, Lennon-Pierce M, Eppig JT, Festing MF, Fisher EM. Genealogies of mouse inbred strains. Nat Genet. 2000;24(1):23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Atkins AL. The replicability of QTLs for murine alcohol preference drinking behavior across eight independent studies. Mamm Genome. 2001;12(12):893–899. doi: 10.1007/s00335-001-2074-2. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Crabbe JC, Young ER. Voluntary consumption of ethanol in 15 inbred mouse strains. Psychopharmacology (Berl) 1993;112:503–510. doi: 10.1007/BF02244901. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Richards SP, O’Toole LA, Helms ML, Phillips TJ. Short-term selective breeding as a tool for QTL mapping: ethanol preference drinking in mice. Behav Genet. 1997;27:55–66. doi: 10.1023/a:1025615409383. [DOI] [PubMed] [Google Scholar]
- Brzustowicz LM, Simone J, Mohseni P, Hayter JE, Hodgkinson KA, Chow EW, Bassett AS. Linkage disequilibrium mapping of schizophrenia susceptibility to the CAPON region of chromosome 1q22. Am J Hum Genet. 2004;74:1057–1063. doi: 10.1086/420774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck KJ, Finn DA. Genetic factors in addiction: QTL mapping and candidate gene studies implicate GABAergic genes in alcohol and barbiturate withdrawal in mice. Addiction. 2001;96:139–149. doi: 10.1046/j.1360-0443.2001.96113910.x. [DOI] [PubMed] [Google Scholar]
- Campbell DB, Lange LA, Skelly T, Lieberman J, Levitt P, Sullivan PF. Association of RGS2 and RGS5 variants with schizophrenia symptom severity. Schizophr Res. 2008;101:67–75. doi: 10.1016/j.schres.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Ma C, Bower KA, Shi X, Ke Z, Luo J. Ethanol promotes endoplasmic reticulum stress-induced neuronal death: involvement of oxidative stress. J Neurosci Res. 2008;86:937–946. doi: 10.1002/jnr.21540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester JA, Barrenha GD. Acoustic startle at baseline and during acute alcohol withdrawal in replicate mouse lines selectively bred for high or low alcohol preference. Alcohol Clin Exp Res. 2007;31:1633–1644. doi: 10.1111/j.1530-0277.2007.00462.x. [DOI] [PubMed] [Google Scholar]
- Chester JA, Price CS, Froehlich JC. Inverse genetic association between alcohol preference and severity of alcohol withdrawal in two sets of rat lines selected for the same phenotype. Alcohol Clin Exp Res. 2002;26:19–27. [PubMed] [Google Scholar]
- Chester JA, Blose AM, Froehlich JC. Further evidence of an inverse genetic relationship between innate differences in alcohol preference and alcohol withdrawal magnitude in multiple selectively bred rat lines. Alcohol Clin Exp Res. 2003;27:377–387. doi: 10.1097/01.ALC.0000056619.98553.50. [DOI] [PubMed] [Google Scholar]
- Chester JA, Blose AM, Froehlich JC. Acoustic startle reactivity during acute alcohol withdrawal in rats that differ in genetic predisposition toward alcohol drinking: effect of stimulus characteristics. Alcohol Clin Exp Res. 2004;28:677–687. doi: 10.1097/01.alc.0000125345.19665.09. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Jr, Young ER, Kosobud A. Genetic correlations with ethanol withdrawal severity. Pharmacol Biochem Behav. 1983;18(Suppl 1):541–547. doi: 10.1016/0091-3057(83)90233-2. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Merrill CD, Belknap JK. Effects of convulsants on handling-induced convulsions in mice selected for ethanol withdrawal severity. Brain Res. 1991;550:1–6. doi: 10.1016/0006-8993(91)90397-e. [DOI] [PubMed] [Google Scholar]
- Fehr C, Shirley RL, Belknap JK, Crabbe JC, Buck KJ. Congenic mapping of alcohol and pentobarbital withdrawal liability loci to a <1 centimorgan interval of murine chromosome 4: identification of Mpdz as a candidate gene. J Neurosci. 2002;22:3730–3738. doi: 10.1523/JNEUROSCI.22-09-03730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahame NJ, Li TK, Lumeng L. Selective breeding for high and low alcohol preference in mice. Behav Genet. 1999;29:47–57. doi: 10.1023/a:1021489922751. [DOI] [PubMed] [Google Scholar]
- Hai T, Hartman MG. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene. 2001;273:1–11. doi: 10.1016/s0378-1119(01)00551-0. [DOI] [PubMed] [Google Scholar]
- Hitzemann R, Malmanger B, Reed C, Lawler M, Hitzemann B, Coulombe S, Buck K, Rademacher B, Walter N, Polyakov Y, Sikela J, Gensler B, Burgers S, Williams RW, Manly K, Flint J, Talbot C. A strategy for the integration of QTL, gene expression, and sequence analyses. Mamm Genome. 2003;14:733–747. doi: 10.1007/s00335-003-2277-9. [DOI] [PubMed] [Google Scholar]
- Hitzemann R, Malmanger B, Belknap J, Darakjian P, McWeeney S. Short-term selective breeding for High and Low prepulse inhibition of the acoustic startle response; pharmacological characterization and QTL mapping in the selected lines. Pharmacol Biochem Behav. 2008;90:525–533. doi: 10.1016/j.pbb.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood HM, Metten P, Crabbe JC, Buck KJ. Fine mapping of a sedative-hypnotic drug withdrawal locus on mouse chromosome 11. Genes Brain Behav. 2006;5:1–10. doi: 10.1111/j.1601-183X.2005.00122.x. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Snowman AM, Eliasson MJ, Cohen NA, Snyder SH. CAPON: a protein associated with neuronal nitric oxide synthase that regulates its interactions with PSD95. Neuron. 1998;20:115–124. doi: 10.1016/s0896-6273(00)80439-0. [DOI] [PubMed] [Google Scholar]
- Kosobud A, Bodor AS, Crabbe JC. Voluntary consumption of ethanol in WSP, WSC and WSR selectively bred mouse lines. Pharmacol Biochem Behav. 1988;29:601–607. doi: 10.1016/0091-3057(88)90026-3. [DOI] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmanger B, Lawler M, Coulombe S, Murray R, Cooper S, Polyakov Y, Belknap J, Hitzemann R. Further studies on using multiple-cross mapping (MCM) to map quantitative trait loci. Mamm Genome. 2006;17:1193–1204. doi: 10.1007/s00335-006-0070-2. [DOI] [PubMed] [Google Scholar]
- Metten P, Crabbe JC. Common genetic determinants of severity of acute withdrawal from ethanol, pentobarbital and diazepam in inbred mice. Behav Pharmacol. 1994;5:533–547. doi: 10.1097/00008877-199408000-00014. [DOI] [PubMed] [Google Scholar]
- Metten P, Phillips TJ, Crabbe JC, Tarantino LM, McClearn GE, Plomin R, Erwin VG, Belknap JK. High genetic susceptibility to ethanol withdrawal predicts low ethanol consumption. Mamm Genome. 1998a;9:983–990. doi: 10.1007/s003359900911. [DOI] [PubMed] [Google Scholar]
- Metten P, Belknap JK, Crabbe JC. Drug withdrawal convulsions and susceptibility to convulsants after short-term selective breeding for acute ethanol withdrawal. Behav Brain Res. 1998b;95:113–122. doi: 10.1016/s0166-4328(97)00216-7. [DOI] [PubMed] [Google Scholar]
- Mihic SJ, Ye Q, Wick MJ, Koltchine VV, Krasowski MD, Finn SE, Mascia MP, Valenzuela CF, Hanson KK, Greenblatt EP, Harris RA, Harrison NL. Sites of alcohol and volatile anaesthetic action on GABA(A) and glycine receptors. Nature. 1997;389:385–389. doi: 10.1038/38738. [DOI] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov YA, Grahame NJ, Phillips TJ, Finn DA, Hoffman PL, Iyer VR, Koob GF, Bergeson SE. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci U S A. 2006;103(16):6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AA, Verbitsky M, Suresh R, Kamens HM, Reed CL, Li N, Burkhart-Kasch S, McKinnon CS, Belknap JK, Gilliam TC, Phillips TJ. Gene expression differences in mice divergently selected for methamphetamine sensitivity. Mamm Genome. 2005;16:291–305. doi: 10.1007/s00335-004-2451-8. [DOI] [PubMed] [Google Scholar]
- Puri V, McQuillin A, Choudhury K, Datta S, Pimm J, Thirumalai S, Krasucki R, Lawrence J, Quested D, Bass N, Moorey H, Morgan J, Punukollu B, Kandasami G, Curtis D, Gurling H. Fine mapping by genetic association implicates the chromosome 1q23.3 gene UHMK1, encoding a serine/threonine protein kinase, as a novel schizophrenia susceptibility gene. Biol Psychiatry. 2007;61:873–879. doi: 10.1016/j.biopsych.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Roberts A, Pardo-Manuel de Villena F, Wang W, McMillan L, Threadgill DW. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mamm Genome. 2007;18:473–481. doi: 10.1007/s00335-007-9045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley RL, Walter NA, Reilly MT, Fehr C, Buck KJ. Mpdz is a quantitative trait gene for drug withdrawal seizures. Nat Neurosci. 2004;7:699–700. doi: 10.1038/nn1271. [DOI] [PubMed] [Google Scholar]
- Spanagel R, Siegmund S, Cowen M, Schroff KC, Schumann G, Fiserova M, Sillaber I, Wellek S, Singer M, Putzke J. The neuronal nitric oxide synthase gene is critically involved in neurobehavioral effects of alcohol. J Neurosci. 2002;22:8676–8683. doi: 10.1523/JNEUROSCI.22-19-08676.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt DE, Taiakina V, Guillemette JG. Calcium-deficient calmodulin binding and activation of neuronal and inducible nitric oxide synthases. Biochim Biophys Acta. 2007;1774:1351–1358. doi: 10.1016/j.bbapap.2007.07.019. [DOI] [PubMed] [Google Scholar]
- Thuerauf DJ, Marcinko M, Belmont PJ, Glembotski CC. Effects of the isoform-specific characteristics of ATF6 alpha and ATF6 beta on endoplasmic reticulum stress response gene expression and cell viability. J Biol Chem. 2007;282:22865–22878. doi: 10.1074/jbc.M701213200. [DOI] [PubMed] [Google Scholar]
- Yoneyama N, Crabbe JC, Ford MM, Murillo A, Finn DA. Voluntary ethanol consumption in 22 inbred mouse strains. Alcohol. 2008;42:149–160. doi: 10.1016/j.alcohol.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]