Abstract
Brain tumor segmentation is a fundamental step in surgical treatment and therapy. Many hand-crafted and learning based methods have been proposed for automatic brain tumor segmentation from MRI. Studies have shown that these approaches have their inherent advantages and limitations. This work proposes a semantic label fusion algorithm by combining two representative state-of-the-art segmentation algorithms: texture based hand-crafted, and deep learning based methods to obtain robust tumor segmentation. We evaluate the proposed method using publicly available BRATS 2017 brain tumor segmentation challenge dataset. The results show that the proposed method offers improved segmentation by alleviating inherent weaknesses: extensive false positives in texture based method, and the false tumor tissue classification problem in deep learning method, respectively. Furthermore, we investigate the effect of patient’s gender on the segmentation performance using a subset of validation dataset. Note the substantial improvement in brain tumor segmentation performance proposed in this work has recently enabled us to secure the first place by our group in overall patient survival prediction task at the BRATS 2017 challenge.
Keywords: Brain Tumor Segmentation, Deep Learning, Convolutional Neural Network, Texture Features, Random Forest, Label Fusion
1. INTRODUCTION
Glioblastoma (GB) is the most invasive variant of brain tumors and is often fatal [1–3] with a median survival of 12.1 to 14.6 months even with radiotherapy [4]. Therefore, accurate and timely detection and segmentation of the tumor is imperative for effective treatment planning and follow-up evaluations. The analysis and diagnosis of brain tumors are performed by a radiologist utilizing Magnetic Resonance Imaging (MRI). The segmentation of gliomas in MRI is typically performed manually, which is quite tedious and susceptible to human errors [5]. This necessitates automatic tumor segmentation which is faster, less prone to human errors and may assist radiologists. Nevertheless, the development of such computer-based methods is quite challenging due to the large variations in tumor shape, size, location, structure, and tissue types. Additionally, The MRI imaging technology may pose further problems in terms of intensity inhomogeneity, aliasing, and changes in intensity range within and between sequences of MRI.
Many automatic tumor segmentation algorithms [5–11] have been proposed in the literature that can be broadly divided into two categories: feature-based and probabilistic atlas-based. The atlas-based method essentially incorporates a probabilistic atlas that is typically estimated using MRI [10–12]. The feature-based methods can be again divided into two groups: classical hand-crafted feature based techniques and artificial neural network (ANN) based deep learning methods. Studies of both these categories have shown varying degrees of success in tumor segmentation. Feature-based techniques [6, 7, 9] involve classical image analysis and machine learning methods in which a set of features are extracted from the MRI and then fed into a conventional classifier to perform the segmentation task. Many hand-crafted methods prefer voxel-wise features such as voxel intensity values, local intensity differences, and intensity distributions to segment tumor tissue types in MRI [9, 13–15]. Additionally, our prior work [9] on multi-class abnormal brain tissue segmentation introduces a novel set of multi-resolution-fractal based texture features such as fractal piecewise triangular prism surface area (PTPSA) [16] and texton [17] to capture the variations in tumor tissue in MRI. The hand-crafted methods are often susceptible to variations in the MRI as the chosen features may be highly sensitive to a particular intensity range. Furthermore, hand-engineered features may not always be optimal for a specific task. For example, though the tumor tissues are segmented accurately, further analysis of our work in [9] demonstrates an inherent degree of over-sensitivity, with increased false positive voxel classifications.
On the other hand, the recent ANN methods utilize deep machine learning based approaches [5, 8, 18] in which both the features and the classifier are learned directly from the raw MRI data for the segmentation task. Deep learning essentially involves the development of architectures capable of hierarchical feature learning, instead of hand-engineering feature extraction. This often results in better performance as the features that are learned from examples tend to be superior to the features that are hand-crafted. Deep neural networks, specifically convolutional neural networks (CNN), have shown state-of-the-art performance in many object recognition tasks [18–20]. Several studies have proposed the use of CNN for brain tumor segmentation using MRI. An early study in [21] proposes the use of a rather shallow three-layer convolutional network for a 2D patch-wise MRI based tumor segmentation task. A recent study in [18] proposes a more complex cascaded CNN architecture with varying filter sizes and two-stage training for improved scale invariance and to overcome the problems in imbalanced tumor tissue representations in training data. The increased availability of training data has enabled the development of deeper CNN architectures with improved versatility in learning. Accordingly, Pereira et al. [5] propose a CNN that utilizes small 3 × 3 convolutional kernels in each layer, allowing for a deeper architecture of 11 layers. Our experiments on a similar CNN architecture for brain tumor segmentation with a larger training and validation dataset from the latest BRATS 2017 challenge show impressive accuracy in whole tumor localization and segmentation. However, we also observe that our CNN-based method may further be improved in multiclass tumor tissue segmentation.
Consequently, in this work, we propose a semantic label fusion algorithm by combining two representative state-of-the-art segmentation algorithms: texture based hand-crafted, and deep learning based methods to obtain robust tumor segmentation. Note the substantial improvement in brain tumor segmentation performance proposed in this work has recently facilitated to securing the first place by our group in overall patient survival prediction task at the BRATS 2017 challenge. In addition, we investigate the effect of patient’s gender on the segmentation performance using a subset of validation dataset. The remainder of this paper is organized as follows. Section 2 discusses the deep CNN architecture and training, the multi-fractal texture based segmentation model and training, and the proposed label fusion methodology. Section 3 provides the tumor segmentation results obtained using the proposed pipeline on the BRATS 2017 training, validation, and testing datasets. Section 4 concludes the findings.
2. METHODS
2.1 Deep convolutional neural network based segmentation
The recent developments in deep learning have enabled new avenues in various medical image processing research. Several recent studies [5, 8] apply CNN-based deep learning techniques to obtain brain tumor segmentation. The state-of-the-art deep CNN architecture proposed by Pereira et al. [5] has been reported to achieve the highest performance in the BRATS 2013, and the second highest in the BRATS 2015 challenges, respectively [5]. MRI based tumor segmentation involves volumetric data processing, in which each voxel in the MRI volume is classified into five types of tissues such as background, enhanced tumor, edema, necrosis and non-enhanced tumor, respectively. For the deep learning brain tumor segmentation in this study, we configure our CNN architecture based on the specifications provided in [5]. The CNN based processing is performed for each MRI slice in the volume. The input for each pixel in the MRI slice is an image patch of size (33 × 33 × 4) which captures a 16 pixel-wide local neighborhood surrounding the pixel in question. The third dimension of the image patch is comprised of the four MRI modalities: T1, T1Gd, T2, and FLAIR. The output of CNN is the classification of the corresponding middle pixel of input patch of each MRI slice as one of the tissue types mentioned above.
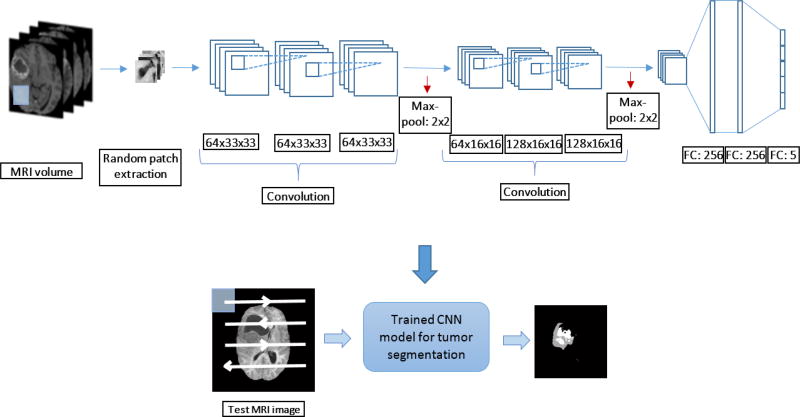
The training of the CNN is conducted using the training dataset of 210 high-grade MRI volumes provided in the BRATS 2017 segmentation and survival prediction challenges. All the MRI volumes are pre-processed prior to training patch extraction as follows: 1) bias field correction on T1 modality MRI following the N4ITK algorithm [22], 2) intensity normalization following [23] is applied to all four modalities for inter-volume consistency. The training set for the CNN is 900,000 image patches randomly obtained from the BRATS 2017 training MRI volume set, representing 500,000 normal tissue and 100,000 samples for each abnormal tissue type. The imbalanced set of training samples are obtained in order to sufficiently represent the inherent imbalance in MRI pixel presence for each tissue type for the CNN. The training set is subsequently enhanced with data augmentation by applying affine rotation in (0°, 90°, 180°, 270°) for each patch to obtain a final dataset of 3,600,000 patches. The trained CNN is then used to obtain the segmentation mask for the fusion step. The CNN based segmentation pipeline is shown in Figure 1.
Figure 1.
CNN based tumor segmentation pipeline. The detailed CNN architecture is provided with patch-wise training and testing
In addition, the outputs of each convolutional and fully connected layers of the CNN is activated using the leaky rectified linear (ReLu) function with the leak factor of 0.3. Dropout regularization is utilized with a rate of 0.1 to all the fully connected layers to prevent over-fitting. The CNN is trained by the stochastic gradient descent method in a batch-wise training scheme with 100 image patches per batch.
2.2 Multi-fractal texture features and Random Forest (RF) based segmentation
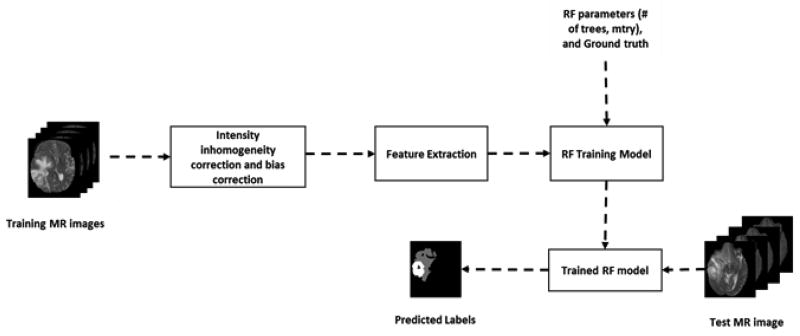
Our previous work [24, 25] on multiclass (edema, necrosis, enhance tumor, and non-enhance tumor) brain tumor segmentation in multimodality MRI (T1, T1C, T2, and FLAIR) have shown success [12]. The RF-based segmentation starts with preprocessing the MR images. The preprocessing steps comprise registration, resampling, re-slicing, skull stripping, bias correction and intensity inhomogeneity correction [26]. Then spatially varying texture features such as PTPSA [27], multifractal Brownian motion (mBm) [16], regular texture (texton) [28] and intensity are extracted from each preprocessed MRI modality. These features capture both global and local characteristics of different tissues. We then combine all the features into a 3D matrix and use this feature matrix for training a Random Forest (RF) for classification [29]. The RF model is trained on a training dataset with known tissue label (ground truth) used as the target. The trained model is subsequently used to predict the tissue labels in a testing dataset. Figure 2 shows the complete pipeline of the RF multiclass brain tumor segmentation method (henceforth this pipeline referred as texture+RF).
Figure 2.
Flow diagram of multiclass abnormal brain tissue segmentation using texture+RF.
The training and testing datasets for the texture+RF is the same BRATS 2017 high-grade MRI volumes used for the deep CNN method discussed above. However, the training methodology is quite different as the feature extraction in texture+RF is done on the whole MRI slice, with no patch-wise processing. Despite this, the final output in both cases is slice-wise segmentation masks, which enables straightforward label fusion.
2.3 Semantic label fusion using texture-based RF and CNN for segmentation
This section discusses our proposed semantic label fusion pipeline for improved brain tumor segmentation. The CNN architecture utilized in this study has shown to perform well in BRATS 2013 and BRATS 2015 competitions, respectively [5]. Our experimentations with this CNN method on the latest BRATS 2017 training and validation data shows that though the method captures the whole tumor quite well, the classification between abnormal tissue types is not satisfactory. Accurate segmentation of abnormal tissue types is vital for many tasks including that of patient survival prediction, in which most relevant features are based on the abnormal tissue volume statistics.
In comparison, the texture+RF method classifies each tumor tissue type fairly well, albeit with a considerable amount of false positives. This is, in turn, detrimental to the task of survival prediction due to the addition of false information in the segmentation step. Therefore, we combine the segmentation outcome from CNN with texture+RF to overcome these limitations inherent to each approach. Specifically, the segmented output of the RF method is fused with the binarized outcomes from the deep learning based segmentation method. The binarization is a pixel-wise operation where the output pixel of the slice k in MRI volume V is obtained as follows:
| (1) |
where is the CNN output segmentation mask value at (i, j) of slice k in MRI volume V.
Subsequently, the label fusion is performed using the binarized output of the CNN and the multi-class output of the texture+RF model in an elementwise multiplication as follows:
| (2) |
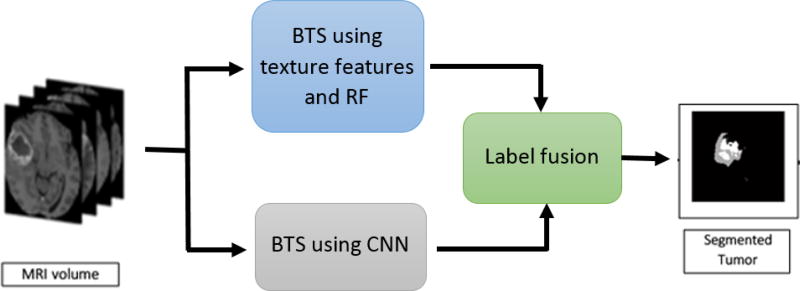
where is the fused segmentation mask of slice k in MRI volume V, and is the multi-class output segmentation mask of slice k in MRI volume V obtained by the texture+RF model. Equation (2) essentially combines multiclass prediction from the RF segmentation with the whole tumor segmentation determined by the deep CNN, effectively suppressing the performance shortcomings observed in each individual technique. The proposed algorithm for obtaining tumor segmentation via label fusion is shown in Algorithm 1 and the overall label fusion pipeline is shown in Figure 3, respectively.
Algorithm 1.
Multiclass deep CNN+RF segmentation with label fusion
| /* Initialization */ |
| - Pre-process Training and testing datasets for CNN, and texture+RF |
| - Initialize CNN and RF parameters |
| /* Training */ |
| Train the CNN architecture |
| - Extract random patches as discussed in section 2.1 |
| - Train CNN using batch-wise stochastic gradient descent |
| Train the texture+RF model |
| - Extract multi-fractal texture features from each slice, and accumulate in a feature matrix |
| - Train RF classifier with training feature matrix |
| /* Segmentation */ |
| For each testing MRI Volume V |
| For each MRI slice k |
| - Extract patches corresponding to each non-zero pixel |
| - Perform patch-wise classification with trained CNN to obtain: |
| - Apply Equation (1) to obtain the corresponding binary output: |
| - Perform multi-class segmentation using texture+RF to obtain: |
| - Apply label fusion in Equation (2) to obtain the final segmentation mask: |
| - Use to obtain the segmentation of slice k |
| End |
| End |
Figure 3.
Semantic label fusion pipeline using convolutional neural network (CNN) and random forest (RF) for brain tumor segmentation
3. RESULTS
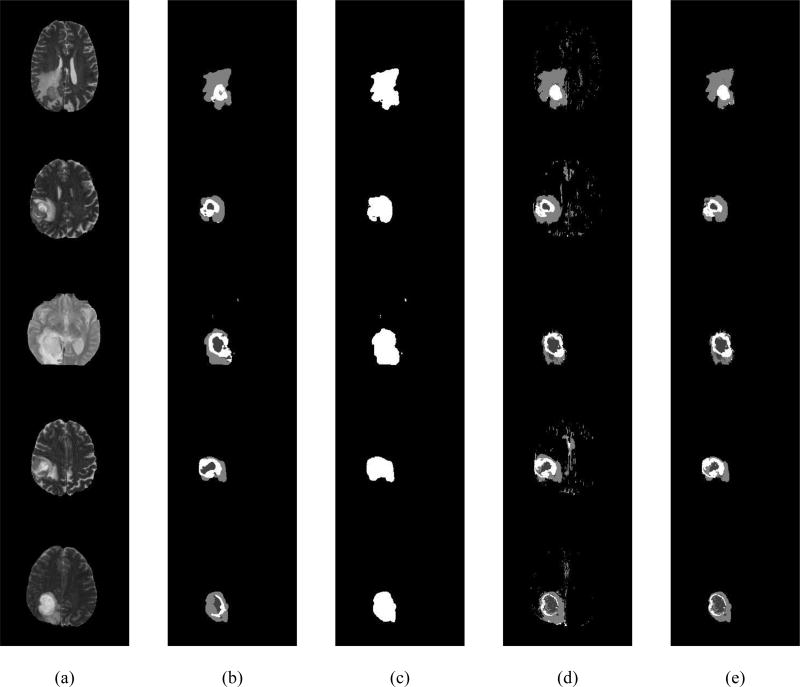
This section reports the performance of the proposed deep CNN+RF fusion-based segmentation method using high-grade GBM tumor MRI in the latest BRATS 2017 dataset [7, 31]. First, we assess the performance using the training and validation dataset split provided by the BRATS 2017 challenge organizing committee. For comparison, we utilize the above mentioned state-of-the-art hand-crafted texture+RF [9] segmentation method and the state-of-the-art deep CNN based brain tumor segmentation pipeline [5]. Figure 4 shows example segmentation results from the deep CNN based pipeline, the binary mask of the CNN outcome, the texture+RF segmentation outcome, and the proposed deep CNN+RF fusion outcome in (b), (c), (d), and (e), respectively for each MRI slice (shown in (a)) from five randomly chosen validation data.
Figure 4.
Example input slices and segmentation outcomes: (a) A slice from T2 MRI from five randomly chosen validation data, (b) the segmentation outcome of CNN, (c) The binary mask of CNN outcome, (d) Segmentation outcome from texture+RF (e) Proposed deep CNN+RF fusion model segmentation outcome.
Figure 4 (d) demonstrates that the outcome of the texture+RF method contains a significant amount of false positives. On the other hand, the outcome of the CNN in Figure 4 (b) contains a well localized whole tumor with no false positives. However, the tumor tissue classification for deep CNN does not appear to be as accurate compared to the texture+RF outcome. Consequently, the proposed deep CNN+RF fusion outcome alleviates both these drawbacks to obtain an improved segmentation result as evident in Figure 4 (e).
Next, we show a quantitative comparison between the above-mentioned segmentation techniques using dice similarity coefficient computed as follows,
| (3) |
where, “a” indicates the number of pixels where both the segmentation decision and ground truth label confirms the presence of tumor and “b” denotes the number of pixels where the decision mismatch. The dice coefficient is computed on three tumor regions: Dice_ET is computed considering only the enhanced tumor tissue class, DICE_WT is computed on the whole tumor that includes all the tissue classes and Dice_TC is computed on the core tumor tissue class, respectively.
Table 1 provides segmentation performance comparison among three methods: deep CNN, texture+RF, and deep CNN+RF fusion using a subset of the BRATS 2017 validation dataset consisting of 33 high-grade tumor patients. The mean values of Dice_ET, Dice_WT, and Dice_TC in Table 1 are shown as reported by the BRATS 2017 challenge organizers. The results show a huge improvement in the enhanced tumor (ET); and small improvements in the whole tumor (WT), and tumor core (TC) segmentation in comparison to deep CNN, and significant improvements in all the performance metrics compared to the texture+RF methods, respectively.
Table 1.
Performance comparison between Texture+RF, CNN, and proposed deep CNN+RF fusion based segmentation methods using BRATS 2017 dataset (mean dice values)
| Methods | Dice_ET | Dice_WT | Dice_TC |
|---|---|---|---|
| Texture+RF | 0.53242 | 0.50775 | 0.53085 |
| Deep CNN | 0.64423 | 0.81228 | 0.68326 |
| Proposed Deep CNN+RF Fusion | 0.74579 | 0.81460 | 0.69797 |
In addition, we perform a gender-based analysis on the performance of the proposed pipeline. We first determine the patients’ gender subject to the availability of information from BRATS 2017 validation dataset. Accordingly, we identify 12 MRI volumes with gender information, of which 3 volumes are from female patients and 9 from male patients. Then, we compute the mean dice similarity coefficients for each criteria to perform gender based analysis. The tumor segmentation performance for patients with M-F gender difference is shown in Table 2.
Table 2.
Brain tumor segmentation performance of the Deep CNN+RF fusion model on Male VS. Female Subjects (mean dice values)
| Methods | Dice_ET | Dice_WT | Dice_TC |
|---|---|---|---|
| Male | 0.81772 | 0.87028 | 0.78511 |
| Female | 0.82345 | 0.87785 | 0.73945 |
Table 2 shows that segmentation performance of the proposed method is quite similar between male and female subjects for whole tumor tissue. However, the reruslts also indicate that the pateints’ gender has discernible influence on the segmentation performance of enhancing and tumor core tissue, respectively.
Finally, for completeness, we report the overall segmentation results of our proposed deep CNN+RF fusion based method using the BRATS 2017 test dataset. Table 2 shows the mean, standard deviation (StdDev) and median values of Dice_ET, Dice_WT and Dice_TC, respectively obtained from 95 high-grade test patients. Note that the reported values shown in Table 3 are provided by the BRATS 2017 challenge organizing committee. However, we are unable to perform gender based analysis on the testing dataset due to the unavaliability of necessary gender information.
Table 3.
Segmentation performance of the proposed deep CNN+RF fusion based method on BRATS 2017 high-grade test dataset
| Label | Dice_ET | Dice_WT | Dice_TC |
|---|---|---|---|
| Mean | 0.73 | 0.83 | 0.72 |
| StdDev | 0.16 | 0.08 | 0.17 |
| Median | 0.78 | 0.85 | 0.78 |
Table 3 shows that the mean Dice values of the test dataset are comparable to the mean Dice values of the validation dataset (as shown in Table 1) obtained by the proposed deep CNN+RF fusion based segmentation method. This validates the consistency of the proposed deep CNN+RF fusion based segmentation method. As mentioned above, the overall improvements in segmentation accuracy of abnormal tissue types such as tumor core (TC), and enhanced tumor (ET) has contributed to a substantial enhancement in our performance of the patient survival prediction task at the recent BRATS 2017 challenge, yielding the highest estimation accuracy among all global contestants.
4. CONCLUSION
This study proposes a semantic label fusion method for brain tumor segmentation that combines the outcomes of a texture+RF based hand-crafted technique with a state-of-the-art deep CNN for improved performance. Accurate segmentation of brain tumors in MRI is vital for effective treatment planning, follow-up evaluations, and in potentially fatal conditions, estimating the patient’s survival duration. Our study shows that the state-of-the-art texture+RF method generates significant number of false positives in the segmentation results, while providing good tumor tissue classification. In comparison, a state-of-the-art deep CNN based algorithm provides good tumor localization with inadequate tissue labelling. Consequently, the proposed segmentation algorithm with label fusion achieves superior performance by minimizing the above shortcomings inherent to the texture+RF and CNN based techniques, respectively. We also find that the performance of the proposed model shows discernable influence of gender of the patient in segmentation of enhancing tumor and tumor core tissues, respectively. However, the effect of gender difference will need to be further investigated with larger patient cases. Finally, the substantial improvement in brain tumor segmentation performance proposed in this work has recently enabled us to secure the first place by our group in overall patient survival prediction task at the BRATS 2017 challenge.
Acknowledgments
The authors would like to acknowledge partial funding of this work by the National Institute of Health (NIH) through NIBIB/NIH grant# R01 EB020683.
References
- 1.Louis David N, Ohgaki Hiroko, Wiestler Otmar D, Cavenee Webster K, Burger Peter C, Jouvet Anne, Scheithauer Bernd W, Kleihues Paul. The 2007 WHO Classification of Tumours of the Central Nervous System. Acta Neuropathologica. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kleihues Paul, Sobin Leslie H. WHO Classification of Tumours: Pathology and Genetics of Tumours of the Nervous System. Lyon, France: International Agency for Research on Cancer; 2000. Glioblastoma; pp. 29–39. [Google Scholar]
- 3.Claes An, Idema Albert J, Wesseling Pieter. Diffuse Glioma Growth: a Guerilla War. Acta Neuropathologica. 2007;114(5):443–458. doi: 10.1007/s00401-007-0293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stupp Roger, Mason Warren P, Van Den Bent Martin J, Weller Michael, Fisher Barbara, Taphoorn Martin JB, Belanger Karl, et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. New England Journal of Medicine. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 5.Pereira S, Pinto A, Alves V, Silva CA. Brain Tumor Segmentation Using Convolutional Neural Networks in MRI Images. IEEE transactions on medical imaging. 2016;35(5):1240–1251. doi: 10.1109/TMI.2016.2538465. [DOI] [PubMed] [Google Scholar]
- 6.Farahani K, Menze B, Reyes M. Brats 2014 Challenge Manuscripts (2014) URL http://www.braintumorsegmentation.org.
- 7.Menze BH, et al. The multimodal brain tumor image segmentation benchmark (BRATS) IEEE transactions on medical imaging. 2015;34(10):1993–2024. doi: 10.1109/TMI.2014.2377694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Havaei M, et al. Brain tumor segmentation with deep neural networks. Medical Image Analysis. 2016 doi: 10.1016/j.media.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Reza S, Iftekharuddin K. Multi-class Abnormal Brain Tissue Segmentation Using Texture. Multimodal Brain Tumor Segmentation. 2013:38. [Google Scholar]
- 10.Menze Bjoern H, Van Leemput Koen, Lashkari Danial, Weber Marc-André, Ayache Nicholas, Golland Polina. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; Berlin, Heidelberg: 2010. A generative model for brain tumor segmentation in multi-modal images; pp. 151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gooya A, et al. GLISTR: Glioma Image Segmentation and Registration. IEEE transactions on medical imaging. 2012 Aug 13;31(10):1941–1954. doi: 10.1109/TMI.2012.2210558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon D, Shinohara RT, Akbari H, Davatzikos C. Combining generative models for multifocal glioma segmentation and registration; International Conference on Medical Image Computing and Computer-Assisted Intervention; 2014. pp. 763–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geremia E, Menze BH, Clatz O, Konukoglu E, Criminisi A, Ayache N. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2010. Spatial decision forests for MS lesion segmentation in multi-channel MR images; pp. 111–118. [DOI] [PubMed] [Google Scholar]
- 14.Zikic D, et al. Decision forests for tissue-specific segmentation of high-grade gliomas in multi-channel MR; International Conference on Medical Image Computing and Computer-Assisted Intervention; 2012. pp. 369–376. [DOI] [PubMed] [Google Scholar]
- 15.Bauer S, Nolte L-P, Reyes M. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2011. Fully automatic segmentation of brain tumor images using support vector machine classification in combination with hierarchical conditional random field regularization; pp. 354–361. [DOI] [PubMed] [Google Scholar]
- 16.Islam A, Reza SM, Iftekharuddin KM. Multifractal texture estimation for detection and segmentation of brain tumors. IEEE transactions on biomedical engineering. 2013;60(11):3204–3215. doi: 10.1109/TBME.2013.2271383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung T, Malik J. Representing and recognizing the visual appearance of materials using three-dimensional textons. International journal of computer vision. 2001;43(1):29–44. [Google Scholar]
- 18.Havaei M, et al. Brain tumor segmentation with Deep Neural Networks. Medical Image Analysis. 35(Supplement C):18–31. doi: 10.1016/j.media.2016.05.004. 2017/01/01/ 2017. [DOI] [PubMed] [Google Scholar]
- 19.Dieleman S, Willett KW, Dambre J. Rotation-invariant convolutional neural networks for galaxy morphology prediction. Monthly notices of the royal astronomical society. 2015;450(2):1441–1459. [Google Scholar]
- 20.Zhong Z, Jin L, Xie Z. Document Analysis and Recognition (ICDAR), 2015 13th International Conference on. IEEE; 2015. High performance offline handwritten chinese character recognition using googlenet and directional feature maps; pp. 846–850. [Google Scholar]
- 21.Zikic D, Ioannou Y, Brown M, Criminisi A. Segmentation of brain tumor tissues with convolutional neural networks. Proceedings MICCAI-BRATS. 2014:36–39. [Google Scholar]
- 22.Tustison NJ, et al. N4ITK: improved N3 bias correction. IEEE transactions on medical imaging. 2010;29(6):1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nyúl LG, Udupa JK, Zhang X. New variants of a method of MRI scale standardization. IEEE transactions on medical imaging. 2000;19(2):143–150. doi: 10.1109/42.836373. [DOI] [PubMed] [Google Scholar]
- 24.Reza S, Iftekharuddin KM. SPIE, Medical Imaging 2014: Computer-Aided Diagnosis. San Diego, California: 2014. Multi-fractal texture features for brain tumor and edema segmentation; p. 903503-903503-10. [Google Scholar]
- 25.Ahmed S, Iftekharuddin KM, Vossough A. Efficacy of Texture, Shape, and Intensity Feature Fusion for Posterior-Fossa Tumor Segmentation in MRI. IEEE Transactions on Information Technology in Biomedicine. 2011;15:206–213. doi: 10.1109/TITB.2011.2104376. [DOI] [PubMed] [Google Scholar]
- 26.Kleesiek Jens, Biller Armin, Urban Gregor, Kothe U, Bendszus Martin, Hamprecht F. ilastik for Multi-modal Brain Tumor Segmentation. MICCAI BraTS (Brain Tumor Segmentation) Challenge. 2014 [Google Scholar]
- 27.Iftekharuddin Khan M, Jia Wei, Marsh Ronald. Fractal analysis of tumor in brain images. Machine Vision and Applications. 2003;13:352–362. [Google Scholar]
- 28.Leung Thomas, Malik Jitendra. Representing and Recognizing the Visual Appearance of Materials using Three-dimensional Textons. International Journal of Computer Vision. 2001;43:29–44. [Google Scholar]
- 29.Breiman L. Machine Learning. Kluwer Academic Publishers; 2001. Random Forests; pp. 5–32. [Google Scholar]