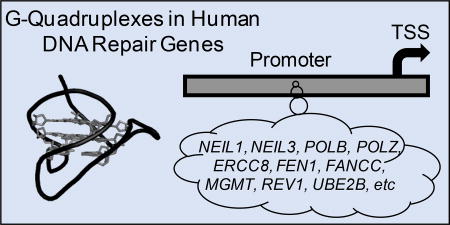
Abstract
The cellular response to oxidative stress includes transcriptional changes, particularly for genes involved in DNA repair. Recently, our laboratory demonstrated that oxidation of 2`-deoxyguanosine (G) to 8-oxo-7,8-dihydro-2`-deoxyguanosine (OG) in G-rich potential G-quadruplex sequences (PQSs) in gene promoters impacts the level of gene expression up or down depending on the position of the PQS in the promoter. In the present report, bioinformatic analysis found that the 390 human DNA repair genes in the genome ontology initiative harbor 2,936 PQSs in their promoters and 5`-untranslated regions (5`-UTRs). The average density of PQSs in human DNA repair genes was found to be nearly twofold greater than the average density of PQSs in all coding and non-coding human genes (7.5 vs. 4.3 per gene). The distribution of the PQSs in the DNA repair genes on the non-transcribed (coding) vs. transcribed strands reflects that of PQSs in all human genes. Next, literature data were interrogated to select 30 PQSs to catalog their ability to adopt G-quadruplex (G4) folds in vitro using five different experimental tests. The G4 characterization experiments concluded that 26 of the 30 sequences could adopt G4 topologies in solution. Last, four PQSs were synthesized into the promoter of a luciferase plasmid and co-transfected with the G4-specific ligands pyridostatin, Phen-DC3, or BRACO-19 in human cells to determine whether the PQSs could adopt G4 folds. The cell studies identified changes in luciferase expression when the G4 ligands were present, and the magnitude of the expression changes dependent on the PQS and the coding vs. template strand on which the sequence resided. Our studies demonstrate PQSs exist at a high density in human DNA repair gene promoters and a subset of the identified sequences fold in vitro and in vivo.
TOC Graphic
Introduction
Oxidative stress represents a shift in the redox balance toward the oxidative state.1 During oxidative stress there is an increase in reactive oxygen species (ROS) that are electron deficient and can readily oxidize biomolecules.2,3 Oxidative damage to the genome is particularly troublesome because it can result in mutations that can impact the phenotype of parent and daughter cells.4 The cellular damage imposed by ROS, particularly on the genome, has been implicated in the initiation and propagation of cancer, neurological disease, cardiovascular disease, and the aging process.5–7 Nature has evolved a DNA repair system to counteract ROS-mediated damage to the genome.8,9 The DNA repair pathways are divided into subgroups that include base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR), homologous recombination (HR), non-homologous end joining (NHEJ), cross-link repair (XLR), translesion synthesis (TLS), and a group of proteins associated with DNA repair (e.g., DNA damage signaling, ubiquitinases, etc). To date, the gene ontology initiative has classified 390 genes to be involved in human DNA repair.10 Transcriptome profiling before and after oxidative stress have identified many DNA repair genes that are activated, repressed, or remain the same after exposure to the stress.11,12 The molecular details that drive the changes in expression levels during oxidative stress are ongoing questions for deeper inquiry.
Our laboratory has recently begun efforts to understand the interplay between oxidative stress and gene regulation controlled by potential G-quadruplex sequences (PQSs) in gene promoters.13,14 The experiments we have conducted to date have found oxidation of 2`-deoxyguanosine (G) to 8-oxo-7,8-dihydro-2`-deoxyguanosine (OG) in PQSs located in either the VEGF or NTHL1 gene promoters guide the DNA repair process to the regulatory region.13 By directing DNA repair on the gene promoter in the PQS context, formation of a G-quadruplex (G4) can occur for transcriptional induction or repression depending on the position of the PQS.13,15 Other laboratories have documented oxidation, most likely at G, in the VEGF,16 BCL2,17 SIRT1,18 and TNFα19 promoters also facilitates an interplay of DNA repair and gene induction. Thus, we asked whether DNA repair genes harbor PQSs for gene regulation during oxidative stress.
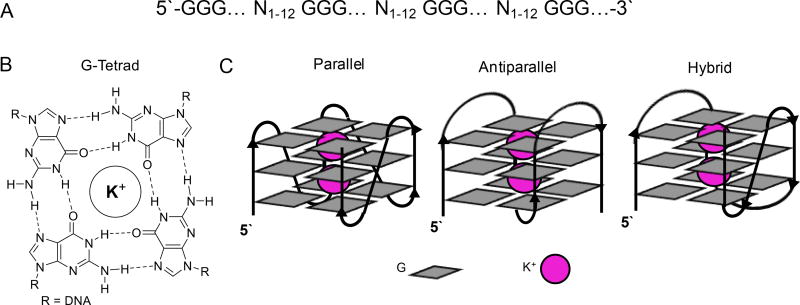
Sequences of DNA with four or more runs of G with each run possessing at least three G nucleotides and with intervening sequences (loops) between the G runs of generally ≤12 nucleotides have the potential to adopt G4 folds (Figure 1A).20,21 The G4 structure is comprised of at least three tetrads of four Gs each participating in Hoogsteen base pairs, in which each run of Gs provides one of the members to the tetrads (Figure 1B). The three tetrads stack on one another to allow coordination to metal ions in the interior of the channel formed by the tetrad stack (Figure 1C).20,21 The major monovalent cation in human cells is K+ (~140 mM), which is also more tightly bound by the G4 than the lower concentration of Na+ ions (~12 mM) found in human cells.21 The nucleotides between the G runs provide the loops that hold the structure together. A fascinating aspect regarding G4s is that they adopt many different folding types with dependency on the sequence, nature of the cation, and/or the analysis conditions (Figure 1C).20,21
Figure 1.
(A) Generalized PQS, (B) structure of a G-tetrad, and (C) typical G4 folding topologies.
Bioinformatic algorithms have been developed to inspect genomes of interest for PQSs.22–25 Bioinformatic analysis of the whole human genome has identified >375,000 classically defined PQSs with four G runs and loops of up to seven nucleotides between the G tracks.22,26 Next-generation sequencing results obtained from human genomic DNA identified >700,000 sequences that might adopt G4 folds.27 The discrepancy between these numbers represents the large number of non-classical G4s with loops >7 nucleotides long and/or structures possessing bulges in the G-tetrad core that natural human genomic sequences can adopt. Interestingly, the PQSs observed are biased toward gene promoters, UTRs, and intron-exon boundaries, particularly in the first intron.28 The Balasubramanian laboratory demonstrated G4 folding in human cells by immunofluorescence,29 and recently they identified ~10,000 PQSs fold to G4s and regulate transcription in human keratinocytes by G4 ChIP-Seq.30 The Maizels laboratory conducted ChIP-Seq for the G4-specific helicases XPB and XPD to locate folded G4s in cells that preferentially are found in promoters, UTRs, and introns.31 Our understanding of how G4s regulate transcription is best described by work in the Hurley laboratory on the c-MYC oncogene.32 Additional genes reported to be regulated by a promoter PQS include VEGF,33 PDGF-A,34 KRAS,35 SRC,36 and many others;37,38 additionally, in some instances the C-rich complementary strand may adopt an i-motif structure for gene regulation as was recently reported by Hurley and co-workers for the BCL2 gene promoter.39 Regulation of transcription via G4 folds from sequences in the 5`-UTR at the RNA level has been demonstrated.40 Roles for G4s in epigenetic changes to chromatin,41 stimulation of retrotransposition,42 participation in stress granule assembly,43,44 HIV infections,45 and causing polymerase stalling and DNA instability have also been reported.46 There continues to be considerable interest in understanding the role of G4s in regulating biological processes. In the present study, we consider the possibility that many human DNA repair genes may be regulated by promoter PQSs. The first step to support this possibility includes identifying all PQSs in these genes by a bioinformatic approach, followed by cataloging a subset of these PQSs for their ability to adopt G4 folds in solution.
Materials and Methods
Bioinformatic Analysis
The human genome assembly GRCh38 was used to obtain all genomic sequences studied, and the gene annotations were obtained from the UCSC genome table browser.47 Specifically, the sequences studied were 2,000 nucleotides (nt) upstream and 1,000 nt downstream of the transcription start site (TSS) for all human genes (coding and non-coding); these were downloaded and combined for study. The PQSs from the whole human genome were identified using Quadparser22 running a modified set of parameters: the loop lengths were ≤12 nt, and 4 or more G tracks with three or more Gs per track were inspected (Figure 1A). The human genome ontology initiative using the AmiGO tool (Oct. 2017) was inspected to identify the classified genes (e.g., DNA repair, oncogenes).10 The functional classification of each gene was determined using PANTHER (v12).48 The genes with PQSs in their promoters and 5`-UTRs were extracted with the BEDTools intersect command,49 and determination of PQS distances to the TSS, as well as counts of PQSs in random gene samples were calculated using custom script run in Python v2.7 that can be found on GitHub at the following address https://github.com/dychangfeng/DNA_repair_promoter_G4/blob/master/G4_DNArepair_gene_notebook.ipynb.
Oligomer Preparation
The oligomers were synthesized by the DNA/Peptide core facility at the University of Utah using commercially available phosphoramidites and a standard solid-phase synthesis protocol. The crude oligomers were purified using a semi-preparative, anion-exchange HPLC column running line A = 1:9 ddH2O:MeCN, and line B = 20 mM LiOAc (pH 7) with 1 M LiCl in 1:9 ddH2O:MeCN and a flow rate = 3 mL/min while monitoring the elution via the absorbance at 260 nm. After purification, the oligomers were dialyzed against ddH2O for 36 h while changing the water three times to remove the purification salts. The dialyzed samples were lyophilized to dryness and resuspended in ddH2O. The concentrations were determined by the absorbance at 260 nm using the primary sequence to estimate the extinction coefficients. All oligomers were stored at −20 °C when not being studied. The G4 strands were annealed in the desired salt and buffer by heating them to 90 °C for 5 min and then slowly cooling the samples to room temperature over ~4 h. After reaching room temperature, the samples were stored at 4 °C for at least 24 h prior to their study.
1H-NMR Analysis
The PQS samples were annealed in 300 µL at a 300 µM concentration in 20 mM KPi (pH 7.0) and 50 mM KCl in 9:1 H2O:D2O. The annealed samples were placed in a D2O-matched Shigemi NMR tube. The samples were analyzed on an 800-MHz NMR spectrometer (Varian, Inc.) with the temperature set to 24 °C. Each sample was scanned 2,048 times using the Watergate solvent suppression pulse sequence. The data were analyzed and plotted using the instrument’s software.
Circular Dichroism Analysis
The PQS samples were annealed at 10 µM concentration in 20 mM lithium cacodylate buffer (pH 7.4) with 140 mM KCl and 12 mM NaCl. The samples were placed in a 0.2-cm quartz cuvette for circular dichroism (CD) analysis at 20 °C (Jasco J-815 circular dichroism spectrometer). The recorded data were solvent background subtracted and then normalized on the y-axis to units of molar ellipticity ([Θ]) for plotting and comparative purposes.
Thermal Melting Analysis
The thermal melting (Tm) values were determined on samples of 5 µM oligomer in buffered solutions with physiological K+ and Na+ concentrations (20 mM lithium cacodylate pH 7.4, 140 mM KCl, and 12 mM NaCl). The melting experiments were initiated by thermally equilibrating the samples at 20 °C for 10 min followed by heating at 0.5 °C/min and equilibrating at each 1 °C increment for 1 min. Readings at 260 and 295 nm were taken after each 1 °C change in the temperature starting at 20 °C up to 95 °C. Plots of absorbance at 295 nm vs. temperature were constructed, and the Tm values were determined by a two-point analysis protocol using the instrument’s software (Shimadzu Scientific UV-1800 spectrometer).
Thermal Difference Spectra Analysis
Measurement of the thermal difference spectra (TDS) for each PQS was achieved by annealing the strands at 3 µM concentration in 20 mM lithium cacodylate buffer (pH 7.4) with 140 mM KCl and 12 mM NaCl. The samples were placed in a 1.0-cm cuvette for UV-vis analysis initially at 20 °C (Shimadzu Scientific UV-1800 spectrometer). After recording the low temperature spectra, the samples were heated to 90 °C followed by thermal equilibration for 20 min, after which the UV-vis spectra were re-recorded. The arithmetic difference of the two spectra was calculated (Abs90 °C − Abs20 °C) and plotted to obtain the TDS plots.
Thioflavin-T Fluorescence Analysis
The PQS samples were annealed at 4 µM concentrations in 20 mM lithium cacodylate buffer (pH 7.4) with 140 mM KCl and 12 mM NaCl. The fluorescence assay was conducted by diluting the G4 stocks to a final concentration of 1 µM with 0.5 µM thioflavin T (3,6-dimethyl-2-(4-dimethylaminophenyl) benzothiazolium cation) present in the same buffer they were annealed. The samples were placed in a 0.2-cm quartz cuvette with the spectrometer (Hitachi, F-7000 fluorescence spectrometer) set to excite the thioflavin T at 425 nm. The emission spectra were collected over the range of 440 to 700 nm at 2 nm intervals. The experimental spectra were corrected by subtracting the buffer and thioflavin-T background spectra prior to plotting the data reported. The c-MYC G4 sequence (5`-GG GTG GGG AGG GTG GGG-3`) was studied as a positive control, and the sequence 5`-TGT TCA TCA TGC GTC GTC GGT ATA TCC CAT-3` was used as a single-stranded control and addition of the complementary strand to this final strand provided the double-stranded control.
Native Polyacrylamide Gel Electrophoresis Analysis
To conduct the native polyacrylamide gel electrophoresis (PAGE) analysis, 2 pmoles of oligomer were 5` labeled with 32P following a literature protocol.50 Next, to a 10 µM solution of each non-radiolabeled PQS was added 20,000 cpm of the same sequence that was 5` labeled with 32P in 20 µL final volume of buffer (20 mM Tris (pH 7.8), 100 mM KOAc). The samples were annealed as stated above. To the samples were loaded dye and then they were electrophoresed in comparison to a ladder of 5`-32P radiolabeled poly-dCn (n = 10, 15, 20, 30, and 40 nt) on a 20% native PAGE doped with 100 mM KOAc. The gel was run at either 4 °C or 20 °C at 25 W.
Plasmid Preparation and Dual Glo Luciferase Assay
The PQSs were inserted into the SV40 promoter regulating the Renilla luciferase gene in the psiCHECK2 plasmid (Promega) via an approach we previously reported.13 The full details and PCR primer sequences for insertion of the BLM, MGMT, NEIL1, and NEIL3 PQSs can be found in the Supplementary Information file. Wild-type glioblastoma cells (U87 MG) were grown in Dulbecco’s Modified Eagle Medium supplemented with 10% FBS, 20 µg/mL gentamicin, 1× glutamax, and 1× non-essential amino acids. The cells were grown at 37 °C with 5% CO2 at ~80% relative humidity and were split when they reached 70–80% confluence. The transfection experiments were conducted in white, 96-well plates by seeding 2 × 104 cells per well and then allowing them to grow for 24 h. After 24 h, the cells were transfected with 200–400 ng of plasmid using X-tremeGene HP DNA transfection agent (Roch) following the manufacturer’s protocol in Opti-MEM media. All transfection experiments were conducted at least 4 times. Next, 48 h post transfection, the Dual-Glo luciferase (Promega) assay was conducted following the manufacturer’s protocol.
The G-quadruplex specific ligands pyridostatin, Phen-DC3, and BRACO-19 were obtained from commercial sources and stock solutions of these ligands were made in DMSO. For the titration studies, pyridostatin was added at 1 or 5 µM directly to the media at the same time the plasmid was transfected into the cells. For the titration studies with Phen-DC3 or BRACO-19, they were added to the media at 10 or 20 µM concentrations directly to the media at the same time the plasmids were transfected into the cells. A control experiment was conducted with DMSO and the original psiCHECK2 plasmid to verify that DMSO did not impact the expression levels observed.
Results and Discussion
Bioinformatic analysis of human DNA repair gene promoters and 5`-UTRs for PQSs
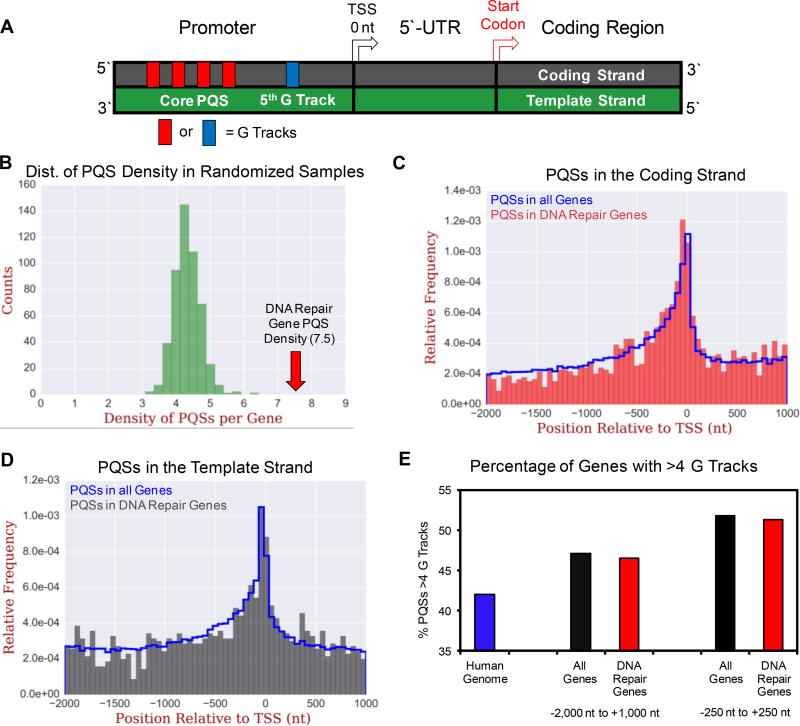
The human genome assembly GRCh38 provided the promoter and 5`-UTR sequences for the 390 human DNA repair genes found in the human genome ontology initiative (Figure 2A);10 additionally, the same sequence space was also obtained for all annotated genes in the human genome. The two different sequence data sets were analyzed with Quadparser22 (settings were ≥4 G tracks with ≥3 Gs per track and loops ≤12 nts) to locate PQSs in these regulatory regions. From the DNA repair genes, the sequence analysis found 2,936 PQSs in 353 of the 390 possible genes (Table S1). There were 37 genes that did not have a PQS in their promoter or 5`-UTR (Table S2). The analysis on all annotated genes (20,338 coding + 22,521 non-coding = 42,859) identified 194,172 PQSs (Table S3), while the entire genome has 708,572 PQSs with the consensus sequence searched in all regions except the telomere. These sequence data sets were used for the following analyses.
Figure 2.
Bioinformatic analysis of PQSs in all human genes compared to human DNA repair genes. (A) Diagram of a basic gene structure. (B) Average density of PQSs per gene from 500 randomized samples of 390 human genes per sample compared to the average density of PQSs in human DNA repair genes. (C) Location of PQSs in all human genes and human DNA repair genes on the coding strand or (D) template strand. (E) Counts of PQSs with more than four G tracks in all human genes (coding + non-coding) vs. human DNA repair genes.
To understand if human DNA repair genes possess a greater average density of PQSs than expected by chance, we took a random sample of 390 human genes, equal to the number in the DNA repair gene ontology, and determined the average density of PQSs per gene. By running this random selection process 500 times, we found the random samples to have 1,680 PQSs per 390 genes on average, which yields an average density of ~4.3 PQSs per gene (Figure 2B). In contrast, the 390 DNA repair genes possessed 2,936 PQSs with a density of 7.5 PQSs per gene (Figure 2B). These randomization plots found human DNA repair genes have ~1.8-times more PQSs in their promoters and 5`-UTRs than expected by chance (P = 4.8 × 10−20). An interesting finding was the MTA1 gene had the largest number of unique PQS forming regions (some regions could fold to more than one G4) with 67. Furthermore, the MTA1 protein is involved in signaling the response to oxidative stress, and the expression levels of this protein change following a burst of ROS.51 The comparisons provided show that DNA repair genes are two-fold enriched in PQSs in their promoters and 5`-UTRs relative to a random sample of other genes from the human genome.
Similarly to the human DNA repair genes, proto-oncogenes were found by the Maizels laboratory to be significantly enriched in PQSs by bioinformatic analysis; further, their studies also found tumor suppressor genes had a low density of PQSs.25 This finding was supported by G4 ChIP-Seq results looking for folded G4s in human keratinocytes under normal growth conditions along with ChIP-Seq for the G4-specific helicases XPB/XPD.30,31 The ChIP-Seq studies also concluded that highly transcribed genes yielded greater numbers of possibly folded G4s in cells. All of these studies and our initial findings with DNA repair genes suggest a non-random occurrence of PQSs in the human genome with respect to gene type. The leading hypothesis is that some of these are involved in gene regulation possibly when folded to non-B-form structures such as G4s. Why these sequences were selected for regulation of certain types of genes and not others is not well understood. The molecular-level details for a few genes have been described,52 but many questions still remain. Additional bioinformatic and experimental studies are required to further develop our knowledge regarding regulatory PQSs.
In our previous studies, we found that the coding (non-template) vs. template strand in which the PQS resides in a gene promoter alters the direction in which gene expression changes when a G is oxidized in the sequence.15 Thus, plots were made of the coding or template strand vs. the location of the PQSs in human DNA repair genes relative to the TSS (Figures 2A, 2C, and 2D). Overlaid on the DNA repair gene data is a plot of the PQS distribution for all coding and non-coding genes found in the known human gene table (UCSC table browser).47 For the coding strand, there is a slight increase in the number of PQSs on the promoter side of the TSS compared to the whole genome (Figure 2C); otherwise, the PQS distributions mirrored one another. The plot of the template strand distribution of PQSs in the promoter and 5`-UTRs between the DNA repair genes and the whole genome are very similar (Figure 2D). Thus, human DNA repair genes are enriched in PQSs relative to other genes (~1.8-fold; Figure 2B), and the PQSs are distributed similarly to the whole genome with a slight preference for them being located on the coding strand on the promoter side of the TSS (Figures 2C and 2D).
In a final analysis of the PQSs found in human DNA repair genes, we inspected for PQSs that had more than four G tracks. The reason for this analysis is twofold: (1) Our sequencing work on the mammalian genome for the G oxidation product OG identified a preference for OG formation in PQSs.53 This observation is consistent with chemical studies demonstrating sequences with runs of G are more prone to oxidative modification via transfer of an electron hole through the DNA stack.54 When Gs are oxidized to OG or other products, the modified sites cannot participate in G:G Hoogsteen base pairing in tetrads resulting in poor G4 formation;55 however, we demonstrated if the PQS has more than four G runs, the additional run allows extrusion of the oxidized G track to maintain the fold.56 (2) The presence of more than four G runs maximized the transcriptional output in our studies demonstrating the VEGF gene was upregulated when the G oxidation product OG was in the regulatory PQS.13,56 Thus, we seek to understand the frequency in which these additional G runs exist in DNA repair genes.
The percentage of genes with more than four G tracks was quantified (Figure 2E). First, the PQSs throughout the whole genome, which includes intergenic regions but not telomeres, found 42.1% of the PQSs have more than four G tracks. When analyzing for >4 G track sequences between −2,000 to +1,000 nt flanking the TSS, 46.5% and 47.1% of human DNA repair and all genes had >4 track PQSs, respectively. Next, we inspected for the percent of PQSs with >4 G tracks between −250 to +250 nt flanking the TSS because this region had the greatest enrichment in PQSs (Figures 2C and 2D). The percentage of PQSs with >4 G tracks in the smaller region flanking the TSS was 51.8% for the DNA repair genes and 51.3% for all genes (Figure 2E). From these values, the DNA repair genes compared to all genes had nearly identical numbers of five G track or greater PQSs in their promoters and 5`-UTRs when inspecting the larger and smaller regions flanking the TSS. More interestingly, when moving from the whole genome to regions flanking the TSS we observed an increase in the PQSs with >4 G tracks. This was most pronounced when inspecting the region 250 nt flanking the TSSs. For all genes and DNA repair genes, ~51% had >4 G tracks showing a greater frequency of >4 G track PQSs than the whole genome (~42%; Figure 2E).
Initial structural characterization of 30 PQSs to determine G-quadruplex formation
The number of sequences identified by the bioinformatic analysis was far too large to characterize all of them by initial methods; therefore, the sequence population was reduced for an initial structural inquiry to those that have the greatest potential to fold in cells. Reduction of the PQS population was achieved by inspecting the sequences found in the DNA repair genes (Table S1) against data from human cells that identified folded G4s by G4 ChIP-Seq30 and ChIP-Seq for the G4-specific helicases XPB and XPD.31 In the G4 ChIP-Seq data, 316 human DNA repair PQSs were found, and the G4-specific helicase ChIP-Seq data possessed 233 of the PQSs (Tables S4 and S5). There exist 63 sequences common to both data sets (Table S6). On an additional note, 18 out of the 63 PQSs in the intersected ChIP-Seq data sets are stress response genes (PRKDC, TAOK3, RAD9A, UBE2B, ORAOV1, UBE2V1, RAD51, POLD1, POLD4, MSH5, MBD4, NSMCE1, TDG, PNKP, SOD1, HMGA2, USP28, and HMGA1). From this population, we picked PRKDC, RAD9A, and UBE2B for the structural studies described below.
Overall, we chose 30 sequences from these cellular data sets for initial characterization of G4 folding (Table 1). The sequences selected favored those with short loop lengths because established regulatory PQSs typically have loop lengths of 1–4 nt, and these generally have greater stability resulting in an increased ability to impact genomic processes such as polymerase bypass.20,38,57 This approach may cause loss of some potentially important PQSs; however, this provides us a place to start understanding whether PQSs found in human DNA repair genes can adopt G4 folds. Many of the sequences had five or more G runs, and for the initial characterization, the four G runs calculated by QGRS mapper58 to be the most stable were studied. Lastly, all sequences studied had 2-nt overhangs of the natural sequence on the 5` and 3` ends to maintain a more relevant sequence context. Previous studies with the human telomere sequence have found omission of the natural 5` and 3` overhangs impacts the structures observed.59
Table 1.
PQSs selected for biophysical characterization, the coding vs. template strand for the PQSs, their G4 score, and the ChIP-Seq data sets from which the sequences were identified.
| Gene | Promoter PQSs Characterized | Coding/ Template Strand |
G4 Scorea |
G4 ChIP- Seqb |
G4-Specific Helicase ChIP-Seqc |
|---|---|---|---|---|---|
| BLM | 5`-GA GGG A GGGG C GGG A GGG AA | Coding | 41 | X | X |
| ERCC8 | 5`-AA GGG CTAGAA GGG CCA GGGG A GGGG TT | Template | 39 | X | |
| FAAP24 | 5`-GT GGGG CTCTGT GGGG CC GGG AATTA GGGG TA | Coding | 39 | X | X |
| FANCA | 5`-GC GGG CTC GGG CGCA GGG AGCCGCCGCC GGGG CT | Coding | 35 | X | X |
| FANCC | 5`-TC GGG TCCGT GGGG C GGGG C GGG CG | Template | 39 | X | X |
| FEN1 | 5’-TT GGG CTGAA GGG T GGGG AA GGGGG AA | Template | 39 | X | |
| GTF2H1 | 5`-GC GGG AACCCGT GGGGG A GGG A GGG AA | Coding | 36 | X | X |
| LIG4 | 5`-TT GGGGG TCT GGGGG ATCCGGTCGT GGGGG TGTCT GGG AC | Coding | 69* | X | |
| MGMT | 5`-CC GGGGG C GGGG CC GGGG CGCGC GGGGG CG | Template | 60 | X | |
| MMS19 | 5`-AA GGG AGA GGGG CCGGCCT GGGGG C GGGG TT | Template | 39 | X | |
| NEIL1 | 5`-CC GGG CCT GGGGG A GGG AAA GGG CC | Template | 42 | X | |
| NEIL3 | 5`-TT GGG C GGGG CCT GGG C GGGG CC | Coding | 41 | X | |
| NTHL1 | 5`-GT GGG CGC GGG TGA GGG CCC GGG AC | Coding | 42 | X | |
| PARP3 | 5`-AA GGG CT GGGG AA GGG CC GGG AC | Coding | 41 | X | |
| PCNA | 5`-CA GGG CGAC GGGGG C GGGG C GGGG CG | Coding | 41 | X | X |
| PMS1 | 5`-CT GGG TGC GGG TGC GGG TGC GGGG TT | Coding | 42 | X | X |
| POLB | 5`-CT GGG C GGGG C GGGG C GGGG CT | Template | 42 | X | |
| POLH | 5`-CT GGGG CT GGG AGA GGG TGTC GGG AC | Coding | 41 | X | X |
| POLL | 5’-CC GGG AA GGG CCTCA GGG CCT GGG TT | Template | 39 | X | |
| POLZ (REV3L) | 5`-GA GGG AA GGG C GGG C GGG CG | Template | 41 | X | |
| PRKDC | 5`-TA GGGG CATTTCC GGG TCC GGG CCGAGC GGG CG | Coding | 38 | X | |
| RAD9A | 5`-AT GGGG AGGC GGGGG C GGGG CCGGCA GGGG CG | Coding | 59 | X | |
| RAD17 | 5`-CC GGG A GGG ACT GGG CT GGGG CA | Template | 40 | X | |
| RAD54L | 5`-GA GGGG C GGGG C GGGGG C GGGGG TG | Template | 62 | X | |
| RECQL | 5`-CA GGG T GGG AAGCTGAGTC GGG AGAAATGAAGCC GGG AA | Coding | 61* | X | |
| REV1 | 5`-CA GGG C GGGG CC GGGG A GGGG AG | Coding | 42 | X | X |
| RPA1 | 5`-GC GGG CGCT GGG A GGG AGACCA GGG CG | Template | 37 | X | X |
| UBE2B | 5`-GC GGG TTTAAGA GGG TGA GGG C GGG TA | Template | 36 | X | X |
| WRN | 5`-GA GGGG A GGG AA GGGG AGGC GGGG AG | Coding | 40 | X | |
| XAB2 | 5`-GT GGG TT GGG AGGCT GGG CA GGGG AT | Template | 39 | X |
The G4 scores were calculated with the QGRS mapper algorithm using the default settings,58 with the exception of the sequences marked with *, in which the sequence length was increased to 45 nt to accommodate the longer sequences.
Sequences identified with an X were found in the G4 ChIP-Seq data.30
Sequences identified with an X were found in the XPB/XPD G4-specific helicase ChIP-Seq data.31
The 30 promoter PQSs selected are found in many subsets of DNA repair genes including BER (NEIL1, NEIL3, and NTHL1), HR (RAD54L), NER (ERCC8, GTF2H1, MMS19, and RPA1), XLR (FAAP24, FANCA, and FANCC), repair associated nucleases (FEN1), MMR (PMS1), NHEJ (LIG4 and PRKDC), direct DNA repair (MGMT), TLS (PARP3, PCNA, POLB, POLH, POLL, POLZ, and REV1), and other proteins associated with DNA repair (BLM, RAD9A, RAD17, RECQL, UBE2B, WRN, and XAB2; Tables 1 and S1). The sequences were scored via the QGRS mapper algorithm58 using the default settings (length = 35 nt) to yield scores ranging from 35–69 (Table 1); two PQSs were too long to be analyzed by the default settings, and thus the length setting was increased to 45 nt to accommodate the sequences. To give some meaning to these scores, the human telomere sequence,59 c-MYC regulatory G4,38 and VEGF regulatory G438 have QGRS mapper scores of 42, 41, and 41, respectively. The sequences selected for characterization spanned a range of scores with 16 of them scoring >40 by this approach. Lastly, all sequences studied were identified in at least one ChIP-Seq data set from the literature (Table 1).30,31
The 30 chosen sequences were then synthesized, HPLC purified, and annealed in NMR buffer containing K+ cation (20 mM KPi pH 7.0, 50 mM KCl, and 22 °C) at 300 µM DNA concentration. The NMR analysis conditions at lower than physiological ionic strength were selected on the basis of literature precedence to achieve the best possible NMR spectra,60 and the DNA concentration had to be 300 µM to maximize the signals. Previous studies with other promoter G-quadruplexes found increased concentrations used for NMR analysis did not impact the molecularity of the G-quadruplex folds;61 additionally, the 2-nt tails added to the ends were previously shown to strongly favor unimolecular G-quadruplex folds (i.e., prevent concatenation).62 We took a subset of these sequences and analyzed them by native PAGE (see below) at 300 µM concentration and did not find any major bands that suggest major multimolecular folds were monitored during the NMR experiments.
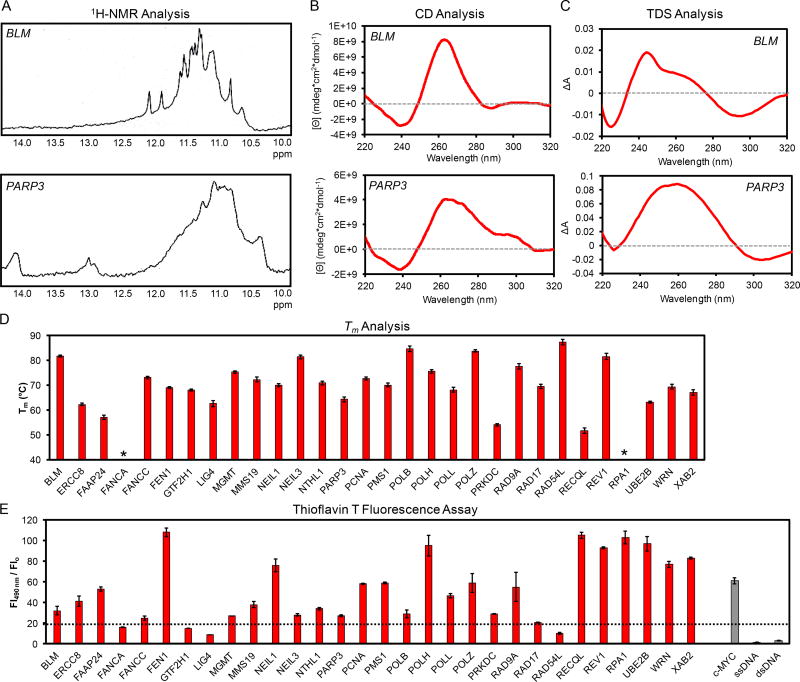
The 30 sequences were analyzed by 1H-NMR to identify whether imino protons were present that are diagnostic of G:G Hoogsteen base pairs (10–12 ppm)60 found in G-tetrad building blocks of G4 structures (Figures 3A and S1). All sequences produced characteristic imino proton peaks except FANCA and RPA1. Inspection of these two sequences suggest FANCA has the potential to adopt a possible five base-paired hairpin that can compete with the G4 fold (5`-GCG GGC TCG GGC GCA GGG AGC CGC CGC CGG GGC T-3`, underline = hairpin), while the RPA1 sequence cannot adopt a competitive hairpin. The only trend observed in the data was that sequences with long G runs that have the potential to adopt many possible structures furnished spectra with broad imino peaks, while those with fewer possible structures generally provided better resolved imino spectra (Figures 3A and S1). Hoogsteen base pairs between two Gs can be found in other secondary structures, such as hairpins or triplexes;63,64 thus, caution is strongly warranted when interpreting the 1H-NMR results. To further establish G4 folding, additional methods of structural analysis were pursued.
Figure 3.
Examples of (A) 1H-NMR, (B) CD, and (C) TDS spectra for the BLM and PARP3 PQSs, as well as the (D) Tm and (E) thioflavin-T fluorescence intensities for all PQSs with comparisons to a known G4, dsDNA, and ssDNA controls. Data for all of the sequences are provided in Figure S1. *These sequences failed to provide thermal transitions while monitoring at 295 nm. The 1H-NMR, CD, and Tm data for the NTHL1 PQS were reported in our previous work.13
After 1H-NMR analysis, the PQSs were analyzed by spectroscopic methods requiring more dilute samples. Support that the following analyses were inspecting predominantly intramolecular G4 folds, the sequences were interrogated by native PAGE at 4 and 20 °C in comparison to a single-stranded DNA ladder. Intramolecular structures (i.e., G-quadruplexes and hairpins) migrate faster on a native PAGE than a single-stranded control of the same length, and in contrast, multi-molecular structures generally migrate slower than a single-stranded control of the same length as the monomer strand.65 When the sequences were annealed at the highest concentration used in the spectroscopic methods (10 µM), most of the sequences when folded provided a band that migrated faster than the single-stranded control of a similar length at 20 °C; in contrast, when the gel was run at the traditional 4 °C a significant amount of material failed to migrate out of the wells and could not be interpreted (Figure S2). Similarly, when the native PAGE was used to interrogate a subset of the samples at 300 µM used in the NMR studies, some of the material did not migrate out of the wells and could not be interpreted. One exception was LIG4 that migrated as a single band in line with the single-stranded control of the same length. Many of the sequences migrated as more than one band. If the two bands migrated faster than the single-stranded control, we concluded that the sequences likely adopted more than one possible G4 structure in solution. If one band migrated similarly to the control and one was faster than the control, a mixture of G4 and single-stranded folds likely existed; alternatively, this result could suggest a monomer and multi-molecular structure were present in solution. A major limitation of native PAGE results from the low resolution of the data and the many hours of electrophoresis required to achieve separation, and as a consequence, we cannot differentiate these possibilities; however, with this limitation in mind, the analysis does suggest all sequences adopted a G4 fold with the exception of LIG4.
The CD spectrum for each PQS was recorded in a buffered solution designed to mimic the K+ and Na+ concentrations inside human cells (20 mM lithium cacodylate pH 7.0, 140 mM KCl, 12 mM NaCl) at 10 µM strand concentration. The spectra recorded fall into four general categories: (1) Ones that have a λmax = 262 nm and λmin ~245 nm, consistent with literature sources for a parallel-stranded G4 (Figure 1C); (2) those having a λmax = 262 and 290–295 nm and λmin ~245 nm that is consistent with a mixture of parallel- and antiparallel-stranded conformations or a mixed-hybrid conformation (i.e., mixture of folds; Figure 1C); (3) those showing a λmax = 295 nm and λmin ~260 nm indicative of a antiparallel conformation (Figure 1C); or (4) spectra with a λmax = 265–280 nm and λmin ~240 nm that supports the conclusion that sequence did not adopt a known G4 structure (Figures 3B and S1).66–68 Sequences that adopt parallel-stranded G4 topologies include BLM, ERCC8, FAAP24, FANCC, FEN1, GTF2H1, NEIL1, PCNA, PMS1, POLB, POLH, POLL, POLZ, RAD9A, RAD54L, REV1, UBE2B, and WRN. Sequences that gave CD spectra consistent with a mixture of folds include MGMT, MMS19, NEIL3, NTHL1, PARP3, PRKDC, and XAB2. The RAD17 PQS provided a CD spectrum nearly identical to that observed for a hybrid human telomere sequence (λmax ~ 295 nm, λshoulder ~ 264 nm, and λmin ~245 nm).69 Lastly, those that did not adopt known G4s on the basis of CD spectroscopy were FANCA, LIG4, RECQL, and RPA1, in which FANCA and RPA1 had λmax = 270–280 nm and λmin = 240 nm suggestive of single-stranded DNA. The single-stranded nature of FANCA and RPA1 were further supported by these sequences failing to provide G:G Hoogsteen imino signatures in the 1H-NMR experiments (Figure S1). In contrast, LIG4 and RECQL had λmax = 265 nm and λmin = 240 nm that in tandem with poor G:G imino protons in the 1H-NMR experiment does not support a known G4 fold (Figure S1). To further support whether each sequence adopts a G4, further analyses were undertaken.
Thermal difference spectra (TDS) provide another method to assess if a new PQS can adopt a G4 fold.70 The UV-vis spectrum for a given sequence was recorded while folded at 20 °C and then denatured at 90 °C, followed by taking the arithmetic difference of the spectra to obtain the TDS. The temperatures selected ensure the sequences were in the folded and unfolded states, which will be discussed below for the thermal melting experiments. Sequences that adopt G4 folds yield TDS with a negative peak at 295 nm and positive peaks at 240 and 270 nm.70 The most consistent G4-specific peak in a TDS is the negative band around 295 nm that was preferentially used to assign G4 folding.70 Most sequences gave G4-consistent TDS (Figures 3C and S1). The sequences that failed to yield TDS indicative of G4 folding include FAAP24, LIG4, PRKDC, RECQL, RPA1, and XAB2. On the basis of comparisons to the previous results, it was expected that LIG4, RECQL, and RPA1 would fail this test for G4 folding; however, FAAP24, PRKDC, and XAB2 were not expected to give inconsistent G4 TDS. These four sequences do not share any sequence or CD spectral similarities to explain why they do not yield a G4-type TDS. It must be noted that this method is low resolution and has been shown to yield false positive and negative results.70 Even with this limitation, the TDS support G4 folding for most of these sequences.
The Tm values for the 30 sequences were then measured by following the decay in absorbance at 295 nm as a function of increasing the temperature from 20 to 95 °C. All of the sequences furnished Tm values for the denaturing processes greater than physiological temperature except FANCA and RPA1, which are the two sequences that did not appear to adopt G4 folds, as discussed above (Figures 3D and S1). Trends in the Tm values failed to emerge when making comparisons to the fold type on the basis of CD spectra or the quality of 1H-NMR imino peaks. A study by Piazza, et al. found that G4s with Tm values >60 °C are capable of stalling polymerase bypass in cellulo;57 therefore, taking this polymerase study as a guide, all sequences studied have the potential to impact biological processes except FAAP24, FANCA, PRKDC, RECQL, and RPA1 (Figure 3D).
The final study to establish G4 folding monitored the fluorescence emission enhancement of the G4-specific fluorophore thioflavin T (ThT) following a literature protocol.71 The ThT assay for the 30 PQSs were compared to the fluorescence changes observed for the c-MYC G4-forming sequence used as a positive control and dsDNA and ssDNA structures utilized as negative controls (Figures 3E and S1). Literature sources have found enhancement of ThT fluorescence emission of >20 Fl490nm/Flo in the presence of a folded G4;71 therefore, utilizing this metric leads to the conclusion that all PQSs studied adopt G4 folds except FANCA, GTF2H1, LIG4, RAD54L, RECQL1, and RPA1. The sequences FANCA, LIG4, RECQL, and RPA1 were anticipated to fail this test on the basis of the previous results; however, the reasons GTF2H1 and RAD54L failed are not immediately clear. This final confirmation provided additional support that many of the selected PQSs can adopt G4 folds.
The six biophysical methods provided data (Figures 3A–E and S1) to establish whether the 30 PQSs found in human DNA repair gene promoters can adopt G4 folds (Figure 3). The data were collectively evaluated to bin the sequences into high, medium, or low probability to adopt a G4 structure. The Mergny laboratory followed a similar approach for assigning G4 folding probability in an attempt to minimize false positive sequences that are claimed to adopt G4 folds.23 The data were equally weighted in the process of assigning folding probability; however, 1H-NMR and CD spectroscopy provide the most direct readout of whether a PQS has adopted a G4 fold. A high classification equals a G4 positive result in 4 or more of the experiments, a medium classification equals a G4 positive result in 3 of the experiments, and a low classification equals 2 or fewer positive results out of the experiments (Figure S3). The sequences with the lowest probability of folding include FANCA, LIG4, RECQL, and RPA1, while the sequence PRKDC has a medium probability of adopting a G4 structure under the conditions studied. All remaining sequences have a high probability of adopting a G4 fold under the conditions of the present studies. The approach of looking at folding probability by many complementary methods provides an excellent way to establish more confidently whether a sequence can adopt a G4 structure. For instance, if only 1H-NMR was used to establish folding, the LIG4 sequence would appear to have adopted a G4 fold; however, the additional methods suggest that LIG4 does not adopt a G4 fold. Even though the studies looked at short sequences outside of their biological context, these biophysical analyses are a necessary first step to understand the possibility of G4 formation in a cell. With this limitation in mind, the utilization of many complementary G4 characterization methods allows us to identify the best possible sequences for future study in a biological setting.
Human DNA repair PQSs are targets for G4-specific molecules in human cells
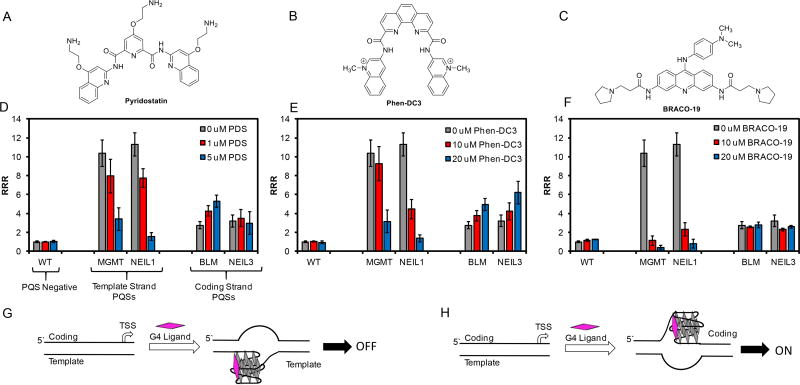
In the final studies, we set out to determine whether some of the human DNA repair PQSs could possibly fold within the context of a human cell. To achieve this goal, we incorporated the BLM and NEIL3 PQSs found in the coding strand (Table 1) of the promoter and the MGMT and NEIL1 PQSs located in the template strand of the promoter (Table 1) into the SV40 promoter sequence in a plasmid expressing the Renilla luciferase gene. The PQSs were inserted following a method we previously reported.13 The plasmid selected also contained the firefly luciferase gene that was not modified and was therefore used as an internal standard. To determine if the PQSs could possibly adopt G4 folds in human cells, the modified plasmids were transfected into human glioblastoma cells (U87) and then titrated with G4-specific compounds. The luciferase expression levels were quantified and normalized to a relative response ratio (RRR) and compared across the titration series to determine if the G4-specific compounds alter expression of the Renilla luciferase gene containing the PQSs. The G4-specific compounds pyridostatin (PDS; Figure 4A),27 Phen-DC3 (Figure 4B),72 and BRACO-19 (Figure 4C)73 were chosen for study because they have all been previously validated to selectively bind G4 sequences over duplex or single-stranded DNA in the cellular context. Lastly, the concentrations for the compounds were selected on the basis of literature precedence.36,57,74
Figure 4.
Demonstration that the BLM, MGMT, NEIL1, and NEIL3 human DNA repair gene promoter PQSs can fold in human glioblastoma cells when targeted with G4-specific ligands. The structures of (A) pyridostatin, (B) Phen-DC3, and (C) BRACO-19. (D–F) The changes in Renilla luciferase expression relative to firefly luciferase (i.e., RRR) expression measured for the four PQSs targeted with the three G4 ligands. Models to understand how gene expression decreased when the PQS is in the (G) template strand or increased when the PQS is in the (H) coding strand.
The G4 ligand studies aided in demonstration that the PQSs from human DNA repair genes are targeted by the G4-specific ligands and alter gene expression on the basis of the following observations (Figures 4D–4F). For the MGMT and NEIL1 PQSs naturally located in the template strand, when PDS, Phen-DC3, or BRACO-19 were titrated in the media along with the plasmid, reduction in Renilla luciferase expression was observed. As a control, the wild-type (WT) plasmid with the native SV40 promoter regulating Renilla luciferase that does not have a PQS was studied with the G4 ligands, and in each case the expression was not impacted by the ligands (Figures 4D–4F). Therefore, this supports the changes in luciferase expression measured result from the ligands binding the G4 folded state of the PQSs in the modified plasmids. The decrease in luciferase expression is consistent with the ligands stabilizing the G4 fold on the template strand and decreasing RNA pol II preinitiation leading to a decrease in mRNA synthesis (Figure 4G). Further, these observations are consistent with other cellular studies that showed that these ligands, when added to cell culture media, result in a decrease in gene expression.36,73,75 The observation of suppression of Renilla luciferase expression when regulated by either the MGMT or NEIL1 PQSs demonstrate a strong possibility for these PQSs to fold in the cellular context. These two examples do not reflect all of the PQSs identified in the bioinformatic studies (Figure 2) or those initially characterized (Figure 3), but these cellular results suggest it is likely that human DNA repair PQSs can fold to G4s in human cells.
In the studies with the BLM or NEIL3 PQSs located in the coding strand of the promoter regulating the Renilla luciferase gene, we observed either no change in gene expression or an increase in luciferase expression (Figures 4D–4F) when the G4 ligands were added. First, the relative changes in expression observed when the PQSs were located in the coding strand vs. the template strand were lower in the coding strand cases. Because each study targeted a different PQS, we cannot rule out the suggestion that different sequences caused the difference in expression change rather than the location on the coding vs. template strand. Even with this limitation in mind, when the PQSs are in the coding strand, transcription was induced for both PQSs when Phen-DC3 was added to the cell cultures. These results are consistent with previous literature findings,75 and they suggest that when PQSs fold in the coding strand, transcription can be enhanced (Figure 4H). In contrast, pyridostatin (PDS) addition to the cell cultures increased expression for the BLM PQS and had no impact when the NEIL3 PQS was present. Lastly, BRACO-19 did not alter luciferase expression when either the BLM or NEIL3 PQSs were present.
Regardless of all these ligand differences, these studies aid in validating that the human DNA repair PQSs in the coding strand of the promoters can possibly impact transcription when folded by increasing gene expression. We must caution that all of these studies placed the PQSs −24 nts from the TSS, and therefore, how the location of the PQS in the natural promoter may impact gene expression when possibly folded was not studied and cannot be addressed with the present data. Furthermore, these plasmid studies make a good attempt at understanding chemistry inside cells, but they do not completely model the genomic context.
As stated in the introduction, our long-term goal is to better understand the interplay between oxidatively derived DNA damage in gene promoters with PQSs and the impact this has on gene expression. Our goal is not related to understanding G4 ligands and how these molecules impact gene expression. Outside of our main goal, these cellular studies do identify some interesting points that may be of interest to the G4 community. First, PDS, Phen-DC3, and BRACO-19 targeted both template PQSs (i.e., MGMT and NEIL1) leading to a reduction in transcription. There did not appear to be any selectivity for these sequences by these three ligands. The lack of sequence selectivity should be of concern to the G4 community. When the PQSs were in the coding strand, gene expression was generally increased. Therefore, treatment of cells with these ligands will decrease and increase gene expression with dependency on the promoter sequences and the coding vs. template strand in which the targeted PQS resides. Selective targeting of different sequences will be very challenging to tune with large planar molecules that do not have built-in sequence selectivity. We do realize the four different sequences inspected in the present study do not paint the complete picture of the genome, but the observations made should be of importance to researchers trying to synthesize and understand G4 ligands for pharmacological applications.
Conclusions
In the present report, we evaluated 390 human DNA repair gene promoters and 5`-UTRs for PQSs utilizing the Quadparser algorithm.22 We identified 2,936 PQSs in 353 of the DNA repair genes that have an average density of 7.5 PQSs per gene (Figure 2B). By evaluating 500 random populations of 390 human genes, we found the average PQS density was 4.3 per gene in the human genome; thus, human DNA repair genes possess ~1.8-times more PQSs than the human genes on average. The finding that DNA repair genes have a greater density of PQSs than expected by chance is an observation that, in tandem with oncogenes having high PQSs,25,30,31 leads to many intriguing future questions. The distribution of the PQSs in the DNA repair genes was similar to the whole genome when looking at their presence on non-transcribed (coding) or transcribed strands (Figures 2C and 2D). The number of PQSs in the DNA repair genes reflects that of the whole genome when evaluating the percentage of sequences that have more than four G tracks (Figure 2E). Further, we found that PQSs near TSSs (±250 nt) are enriched with >4 G track PQSs relative to the whole genome. This final observation supports our previous observation that regulatory PQSs favor >4 tracks of G runs for maintaining their folds when damaged by ROS.56
We then intersected the PQSs identified with ChIP-Seq data for folded sequences in human cells,30,31 that found hundreds of DNA repair sequences that have strong potential to fold in cellulo (Tables S4–S6). We cataloged the ability of 30 of these PQSs to adopt G4 topologies in vitro by five different tests for G4 folding (Table 1 and Figures 3A–E). These experiments determined 26 of the sequences have a medium to high potential to adopt G4 folds in solution (Figures 3A–E and S3). These initial structural characterization results suggest a possibility that these sequences can adopt G4 folds in cells. As a step forward in understanding whether a subset of these human DNA repair promoter PQSs could fold in cells and alter gene expression, four PQSs (i.e., BLM, MGMT, NEIL1, and NEIL3) were synthesized in the promoter of a luciferase gene in a plasmid. We demonstrated in human glioblastoma cells that when the plasmids were co-transfected with G4 specific ligands, luciferase expression was altered (Figure 4). Furthermore, the up or down change in gene expression was dependent on the coding (up) vs. template (down) strand in which the PQS was synthesized. However, studies outside of the genomic context cannot fully reproduce the complexity of nucleosomes and other higher order chromatin structure; thus, caution is warranted when interpreting these results. Lastly, these data provide the first steps in our efforts to understand whether DNA repair genes harbor PQSs in their regulatory regions that aid in transcriptional regulation during oxidative stress. The present results are critical in guiding our future cellular studies to understand how this class of genes may be regulated by G-rich sequences during cellular oxidative stress.14
Supplementary Material
Acknowledgments
This work was supported by a National Cancer Institute grant (R01 CA090689). The authors greatly appreciate the advice of Dr. Peter Flynn (University of Utah) for the NMR measurements. The oligonucleotides were provided by the DNA/Peptide core facility at the University of Utah that is supported in part by the NCI Cancer Center Support grant (P30 CA042014).
Footnotes
The Supporting Information is available free of charge on the ACS Publications website at DOI:XXX. Complete list of genes analyzed and the PQSs identified in human gene promoters and 5`-UTRs, and all data to support G4 folding for the sequences selected for study by 1H-NMR, CD, Tm, TDS, ThT analysis, and native PAGE, as well as a list of G4 folding probabilities.
Conflict of interest statement. The authors declare no conflicts of interest in this work.
References
- 1.Trachootham D, Lu W, Ogasawara MA, Nilsa RD, Huang P. Redox regulation of cell survival. Antioxid. Redox Signal. 2008;10:1343–1374. doi: 10.1089/ars.2007.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cadet J, Wagner JR, Shafirovich V, Geacintov NE. One-electron oxidation reactions of purine and pyrimidine bases in cellular DNA. Int. J. Radiat. Biol. 2014;90:423–432. doi: 10.3109/09553002.2013.877176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.West JD, Marnett LJ. Endogenous reactive intermediates as modulators of cell signaling and cell death. Chem. Res. Toxicol. 2006;19:173–194. doi: 10.1021/tx050321u. [DOI] [PubMed] [Google Scholar]
- 4.Roberts SA, Gordenin DA. Hypermutation in human cancer genomes: footprints and mechanisms. Nat. Rev. Cancer. 2014;14:786–800. doi: 10.1038/nrc3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lonkar P, Dedon PC. Reactive species and DNA damage in chronic inflammation: reconciling chemical mechanisms and biological fates. Int. J. Cancer. 2011;128:1999–2009. doi: 10.1002/ijc.25815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurian GA, Rajagopal R, Vedantham S, Rajesh M. The role of oxidative stress in myocardial ischemia and reperfusion injury and remodeling: Revisited. Oxid. Med. Cell Longev. 2016;2016:1656450. doi: 10.1155/2016/1656450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Markesbery WR. The role of oxidative stress in Alzheimer disease. Arch. Neurology. 1999;56:1449–1452. doi: 10.1001/archneur.56.12.1449. [DOI] [PubMed] [Google Scholar]
- 8.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallace SS. Base excision repair: A critical player in many games. DNA Repair. 2014;19:14–26. doi: 10.1016/j.dnarep.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S, Ami GOH Web Presence Working Group. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mangerich A, Knutson CG, Parry NM, Muthupalani S, Ye W, Prestwich E, Cui L, McFaline JL, Mobley M, Ge Z, Taghizadeh K, Wishnok JS, Wogan GN, Fox JG, Tannenbaum SR, Dedon PC. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E1820–E1829. doi: 10.1073/pnas.1207829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aguilera-Aguirre L, Hosoki K, Bacsi A, Radak Z, Sur S, Hegde ML, Tian B, Saavedra-Molina A, Brasier AR, Ba X, Boldogh I. Whole transcriptome analysis reveals a role for OGG1-initiated DNA repair signaling in airway remodeling. Free Radic. Biol. Med. 2015;89:20–33. doi: 10.1016/j.freeradbiomed.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleming AM, Ding Y, Burrows CJ. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. U. S. A. 2017;114:2604–2609. doi: 10.1073/pnas.1619809114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleming AM, Burrows CJ. 8-Oxo-7,8-dihydroguanine, friend and foe: Epigenetic-like regulator versus initiator of mutagenesis. DNA Repair (Amst) 2017;56:75–83. doi: 10.1016/j.dnarep.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleming AM, Zhu J, Ding Y, Burrows CJ. 8-Oxo-7,8-dihydroguanine in the context of a promoter G-quadruplex is an on-off switch for transcription. ACS Chem. Biol. 2017;12:2417–2426. doi: 10.1021/acschembio.7b00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pastukh V, Roberts JT, Clark DW, Bardwell GC, Patel M, Al-Mehdi AB, Borchert GM, Gillespie MN. An oxidative DNA "damage" and repair mechanism localized in the VEGF promoter is important for hypoxia-induced VEGF mRNA expression. Am. J. Physiol. Lung Cell Mol. Physiol. 2015;309:L1367–1375. doi: 10.1152/ajplung.00236.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 18.Antoniali G, Lirussi L, D'Ambrosio C, Dal Piaz F, Vascotto C, Casarano E, Marasco D, Scaloni A, Fogolari F, Tell G. SIRT1 gene expression upon genotoxic damage is regulated by APE1 through nCaRE-promoter elements. Mol. Biol. Cell. 2014;25:532–547. doi: 10.1091/mbc.E13-05-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan L, Zhu B, Hao W, Zeng X, Vlahopoulos SA, Hazra TK, Hegde ML, Radak Z, Bacsi A, Brasier AR, Ba X, Boldogh I. Oxidized guanine base lesions function in 8-oxoguanine DNA glycosylase1-mediated epigenetic regulation of nuclear factor kappaB-driven gene expression. J. Biol. Chem. 2016;291:25553–25566. doi: 10.1074/jbc.M116.751453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5'-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gray RD, Chaires JB. Kinetics and mechanism of K+- and Na+-induced folding of models of human telomeric DNA into G-quadruplex structures. Nucleic Acids Res. 2008;36:4191–4203. doi: 10.1093/nar/gkn379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bedrat A, Lacroix L, Mergny JL. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016;44:1746–1759. doi: 10.1093/nar/gkw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahakyan AB, Chambers VS, Marsico G, Santner T, Di Antonio M, Balasubramanian S. Machine learning model for sequence-driven DNA G-quadruplex formation. Sci. Rep. 2017;7:14535. doi: 10.1038/s41598-017-14017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chambers VS, Marsico G, Boutell JM, Di Antonio M, Smith GP, Balasubramanian S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015;33:877–881. doi: 10.1038/nbt.3295. [DOI] [PubMed] [Google Scholar]
- 28.Maizels N, Gray LT. The G4 genome. PLoS Genet. 2013;9:e1003468. doi: 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansel-Hertsch R, Beraldi D, Lensing SV, Marsico G, Zyner K, Parry A, Di Antonio M, Pike J, Kimura H, Narita M, Tannahill D, Balasubramanian S. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016;48:1267–1272. doi: 10.1038/ng.3662. [DOI] [PubMed] [Google Scholar]
- 31.Gray LT, Vallur AC, Eddy J, Maizels N. G quadruplexes are genomewide targets of transcriptional helicases XPB and XPD. Nat. Chem. Biol. 2014;10:313–318. doi: 10.1038/nchembio.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brooks TA, Hurley LH. Targeting MYC expression through G-quadruplexes. Genes Cancer. 2010;1:641–649. doi: 10.1177/1947601910377493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun D, Liu WJ, Guo K, Rusche JJ, Ebbinghaus S, Gokhale V, Hurley LH. The proximal promoter region of the human vascular endothelial growth factor gene has a G-quadruplex structure that can be targeted by G-quadruplex-interactive agents. Mol. Cancer Ther. 2008;7:880–889. doi: 10.1158/1535-7163.MCT-07-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin Y, Rezler EM, Gokhale V, Sun D, Hurley LH. Characterization of the G-quadruplexes in the duplex nuclease hypersensitive element of the PDGF-A promoter and modulation of PDGF-A promoter activity by TMPyP4. Nucleic Acids Res. 2007;35:7698–7713. doi: 10.1093/nar/gkm538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez R, Miller KM, Forment JV, Bradshaw CR, Nikan M, Britton S, Oelschlaegel T, Xhemalce B, Balasubramanian S, Jackson SP. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012;8:301–310. doi: 10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rigo R, Palumbo M, Sissi C. G-quadruplexes in human promoters: A challenge for therapeutic applications. Biochim. Biophys. Acta. 2017;1861:1399–1413. doi: 10.1016/j.bbagen.2016.12.024. [DOI] [PubMed] [Google Scholar]
- 38.Kendrick S, Hurley LH. The role of G-quadruplex/i-motif secondary structures as cis-acting regulatory elements. Pure Appl. Chem. 2010;82:1609–1621. doi: 10.1351/PAC-CON-09-09-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kendrick S, Kang HJ, Alam MP, Madathil MM, Agrawal P, Gokhale V, Yang D, Hecht SM, Hurley LH. The dynamic character of the BCL2 promoter i-motif provides a mechanism for modulation of gene expression by compounds that bind selectively to the alternative DNA hairpin structure. J. Am. Chem. Soc. 2014;136:4161–4171. doi: 10.1021/ja410934b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bugaut A, Balasubramanian S. 5'-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012;40:4727–4741. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guilbaud G, Murat P, Recolin B, Campbell BC, Maiter A, Sale JE, Balasubramanian S. Local epigenetic reprogramming induced by G-quadruplex ligands. Nat. Chem. 2017;9:1110–1117. doi: 10.1038/nchem.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahakyan AB, Murat P, Mayer C, Balasubramanian S. G-quadruplex structures within the 3' UTR of LINE-1 elements stimulate retrotransposition. Nat. Struct. Mol. Biol. 2017;24:243–247. doi: 10.1038/nsmb.3367. [DOI] [PubMed] [Google Scholar]
- 43.Byrd AK, Zybailov BL, Maddukuri L, Gao J, Marecki JC, Jaiswal M, Bell MR, Griffin WC, Reed MR, Chib S, Mackintosh SG, MacNicol AM, Baldini G, Eoff RL, Raney KD. Evidence that G-quadruplex DNA accumulates in the cytoplasm and participates in stress granule assembly in response to oxidative stress. J. Biol. Chem. 2016;291:18041–18057. doi: 10.1074/jbc.M116.718478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lyons SM, Gudanis D, Coyne SM, Gdaniec Z, Ivanov P. Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs. Nat. Commun. 2017;8:1127. doi: 10.1038/s41467-017-01278-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Nicola B, Lech CJ, Heddi B, Regmi S, Frasson I, Perrone R, Richter SN, Phan AT. Structure and possible function of a G-quadruplex in the long terminal repeat of the proviral HIV-1 genome. Nucleic Acids Res. 2016;44:6442–6451. doi: 10.1093/nar/gkw432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piazza A, Cui X, Adrian M, Samazan F, Heddi B, Phan AT, Nicolas AG. Non-Canonical G-quadruplexes cause the hCEB1 minisatellite instability in Saccharomyces cerevisiae. Elife. 2017;6:e26884. doi: 10.7554/eLife.26884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.UCSC Genome Browser. http://genome.ucsc.edu/
- 48.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fleming AM, Burrows CJ. G-Quadruplex folds of the human telomere sequence alter the site reactivity and reaction pathway of guanine oxidation compared to duplex DNA. Chem. Res. Toxicol. 2013;26:593–607. doi: 10.1021/tx400028y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang R-A. MTA1-a stress response protein: a master regulator of gene expression and cancer cell behavior. Cancer Metastasis Rev. 2014;33:1001–1009. doi: 10.1007/s10555-014-9525-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brooks TA, Hurley LH. The role of supercoiling in transcriptional control of MYC and its importance in molecular therapeutics. Nat. Rev. Cancer. 2009;9:849–861. doi: 10.1038/nrc2733. [DOI] [PubMed] [Google Scholar]
- 53.Ding Y, Fleming AM, Burrows CJ. Sequencing the mouse genome for the oxidatively modified base 8-oxo-7,8-dihydroguanine by OG-Seq. J. Am. Chem. Soc. 2017;139:2569–2572. doi: 10.1021/jacs.6b12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Delaney S, Barton JK. Long-range DNA charge transport. J. Org. Chem. 2003;68:6475–6483. doi: 10.1021/jo030095y. [DOI] [PubMed] [Google Scholar]
- 55.Zhou J, Fleming AM, Averill AM, Burrows CJ, Wallace SS. The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic Acids Res. 2015;43:4039–4054. doi: 10.1093/nar/gkv252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fleming AM, Zhou J, Wallace SS, Burrows CJ. A role for the fifth G-track in G-quadruplex forming oncogene promoter sequences during oxidative stress: Do these "spare tires" have an evolved function? ACS Cent. Sci. 2015;1:226–233. doi: 10.1021/acscentsci.5b00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Piazza A, Adrian M, Samazan F, Heddi B, Hamon F, Serero A, Lopes J, Teulade-Fichou MP, Phan AT, Nicolas A. Short loop length and high thermal stability determine genomic instability induced by G-quadruplex-forming minisatellites. EMBO J. 2015;34:1718–1734. doi: 10.15252/embj.201490702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kikin O, D'Antonio L, Bagga PS. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Phan AT, Kuryavyi V, Luu KN, Patel DJ. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007;35:6517–6525. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adrian M, Heddi B, Phan AT. NMR spectroscopy of G-quadruplexes. Methods. 2012;57:11–24. doi: 10.1016/j.ymeth.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 61.Agrawal P, Hatzakis E, Guo K, Carver M, Yang D. Solution structure of the major G-quadruplex formed in the human VEGF promoter in K+: insights into loop interactions of the parallel G-quadruplexes. Nucleic Acids Res. 2013;41:10584–10592. doi: 10.1093/nar/gkt784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sengar A, Heddi B, Phan AT. Formation of G-quadruplexes in poly-G sequences: structure of a propeller-type parallel-stranded G-quadruplex formed by a G15 stretch. Biochemistry. 2014;53:7718–7723. doi: 10.1021/bi500990v. [DOI] [PubMed] [Google Scholar]
- 63.Cerofolini L, Amato J, Giachetti A, Limongelli V, Novellino E, Parrinello M, Fragai M, Randazzo A, Luchinat C. G-triplex structure and formation propensity. Nucleic Acids Res. 2014;42:13393–13404. doi: 10.1093/nar/gku1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mooers BH, Eichman BF, Ho PS. The structures and relative stabilities of d(G × G) reverse Hoogsteen, d(G × T) reverse wobble, and d(G × C) reverse Watson-Crick base-pairs in DNA crystals. J. Mol. Biol. 1997;269:796–810. doi: 10.1006/jmbi.1997.1100. [DOI] [PubMed] [Google Scholar]
- 65.Sun D, Hurley LH. Biochemical techniques for the characterization of G-quadruplex structures: EMSA, DMS footprinting, and DNA polymerase stop assay. Methods Mol. Biol. 2010;608:65–79. doi: 10.1007/978-1-59745-363-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karsisiotis AI, Hessari NM, Novellino E, Spada GP, Randazzo A, Webba da Silva M. Topological characterization of nucleic acid G-quadruplexes by UV absorption and circular dichroism. Angew. Chem., Int. Ed. 2011;50:10645–10648. doi: 10.1002/anie.201105193. [DOI] [PubMed] [Google Scholar]
- 67.Kypr J, Kejnovska I, Renciuk D, Vorlickova M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gray RD, Buscaglia R, Chaires JB. Populated intermediates in the thermal unfolding of the human telomeric quadruplex. J. Am. Chem. Soc. 2012;134:16834–16844. doi: 10.1021/ja307543z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mergny JL, Li J, Lacroix L, Amrane S, Chaires JB. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 2005;33:e138. doi: 10.1093/nar/gni134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Renaud de la Faverie A, Guedin A, Bedrat A, Yatsunyk LA, Mergny JL. Thioflavin T as a fluorescence light-up probe for G4 formation. Nucleic Acids Res. 2014;42:e65. doi: 10.1093/nar/gku111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Piazza A, Boule J-B, Lopes J, Mingo K, Largy E, Teulade-Fichou M-P, Nicolas A. Genetic instability triggered by G-quadruplex interacting Phen-DC compounds in Saccharomyces cerevisiae. Nucleic Acids Res. 2010;38:4337–4348. doi: 10.1093/nar/gkq136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Burger AM, Dai F, Schultes CM, Reszka AP, Moore MJ, Double JA, Neidle S. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005;65:1489–1496. doi: 10.1158/0008-5472.CAN-04-2910. [DOI] [PubMed] [Google Scholar]
- 74.Perrone R, Butovskaya E, Daelemans D, Palu G, Pannecouque C, Richter SN. Anti-HIV-1 activity of the G-quadruplex ligand BRACO-19. J. Antimicrob. Chemother. 2014;69:3248–3258. doi: 10.1093/jac/dku280. [DOI] [PubMed] [Google Scholar]
- 75.Halder R, Riou J-F, Teulade-Fichou M-P, Frickey T, Hartig JS. Bisquinolinium compounds induce quadruplex-specific transcriptome changes in HeLa S3 cell lines. BMC Res. Notes. 2012;5:138–138. doi: 10.1186/1756-0500-5-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.