Abstract
Antimicrobial resistance against colistin has emerged worldwide threatening the efficacy of one of the last-resort antimicrobials used for the treatment of Enterobacteriaceae. To investigate the presence of the recently identified colistin resistance genes (mcr-4, mcr-5) in China, we established PCRs to detect mcr-4 and mcr-5 on 213 anal and 1,339 nasal swabs from apparently healthy pigs (n = 1,454) in nine provinces, and 1,696 cloacal and 1,647 oropharyngeal samples from poultry (n = 1,836) at live-bird markets in 24 provinces of China. The prevalence of the mcr-4 in swine swabs (41.4%; 642/1,552) was significantly higher than in swabs from poultry (11.5%; 384/3,343). The mcr-4 gene was found in geese (49.5%, 54/109), chickens (17.2%, 257/1,498), pigeons (17.2%, 17/99) and ducks (15.4%, 20/130). In a similar trend, the prevalence of the mcr-5 in swine swabs (33.1%; 514/1552) was significantly higher than in swabs from poultry (5.6%; 187/3,343). The mcr-5 was identified in geese (17.4%, 19/109), chickens (9.9%, 148/1,498), ducks (7.7%, 10/130) and pigeons (3%, 3/99). The mcr-4 prevalence in the nasal swabs from pigs (59.2%, 58/98) was significantly higher than that in anal swabs (29.6%, 29/98) (P<0.001). Similarly, the mcr-5 prevalence in the nasal swabs from pigs (61.2%, 60/98) was significantly higher than in anal swabs (44.9%, 44/98) (P = 0.02), and significantly higher in oropharyngeal swabs (7.2%, 109/1,507) than in the cloacal swabs (3.7%, 56/1,507) (P<0.001). This study further confirms the presence of the mcr-4 and mcr-5 in animals and indicates these genes are prevalent and widespread in food producing animals (pig and poultry) in China. Future studies are needed to characterize the bacteria carrying the mcr-4 and mcr-5 and their locations on plasmids and/or the bacterial chromosomes, and determine co-resistances in the mcr-4 and mcr-5 positive strains.
Introduction
Antimicrobial resistance is one of the most serious global threats to human health, especially the multiple drug resistant-pathogens of ESKAPE group (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter spp.) [1]. The reintroduction of the older and less user-friendly antibiotics such as colistin is an option for treatment of the infections with Gram-negative ESKAPE bacteria in humans [2]. However, the efficiency of colistin treatment is compromised by the presence of an increasing number of mobile colistin resistance (mcr) genes. After mcr-1 was first described in 2016, mcr-2 and mcr-3 genes have been found to occur very widely [3–22]. Up to the current writing of this manuscript, another two mcr genes (mcr-4 and mcr-5) were identified in Salmonella [23–26], but little is known about the prevalence of these two genes. In this study, PCRs were established to the presence of mcr-4 and mcr-5 genes in swine and poultry swab samples in China.
Materials and methods
Swab samples from swine and poultry
This study was reviewed and approved by the Institutional Animal Care and Use Committee of Yangzhou University College of Veterinary Medicine (YZU-CVM IACUC 2013#87, YZU-CVM IACUC 2015#57).
Nasal (n = 1,339) and anal (n = 213) swabs collected from apparently healthy pigs (n = 1,454) from nine provinces of China in 2016 (Table 1, S1 Table) [27] were used in this study to investigate the prevalence of mcr-4 and mcr-5. In addition, oropharyngeal and cloacal samples were obtained from poultry (n = 1,836) at 38 live-bird markets in 24 provinces in China between 2014 and 2015 (Table 1) [28], and 1,647 oropharyngeal and 1,696 cloacal samples from 1,836 birds were used in this investigation. The swabs from pigs and poultry were collected into tubes containing 400μl DNA/RNA Stabilization Buffer (Roche Molecular Biochemicals, IN, USA), and frozen at -80°C until DNA extraction. Swabs were centrifuged in the DNA/RNA Stabilization Buffer (3,000×g, 4°C for 5 min), and DNAs were extracted from the supernatants using the High Pure PCR Template Preparation Kit (Roche Diagnostic, USA) as described before [22, 28]. All samples from previous studies that had sufficient residual DNA extract were included in this investigation.
Table 1. Prevalences of mcr-4 and mcr-5 in swabs from pigs and poultry.
| Province | pig | chicken | duck | goose | pigeon | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| mcr-4 | mcr-5 | mcr-4 | mcr-5 | mcr-4 | mcr-5 | mcr-4 | mcr-5 | mcr-4 | mcr-5 | |
| Anhui | 4/34 | 0/34 | ||||||||
| Fujian | 6/35 | 1/35 | 2/33 | 1/33 | 0/13 | 0/13 | ||||
| Gansu | 2/57 | 0/57 | ||||||||
| Guangdong | 8/40 | 11/40 | 24/65 | 10/65 | 0/4 | 0/4 | ||||
| Guangxi | 23/130 | 12/130 | 3/10 | 1/10 | ||||||
| Hainan | 6/70 | 15/70 | ||||||||
| Hebei | 26/96 | 3/96 | 2/6 | 0/6 | 10/34 | 1/34 | ||||
| Heilongjiang | 15/60 | 4/60 | ||||||||
| Henan | 35/63 | 8/63 | 11/56 | 12/56 | 1/7 | 0/7 | 3/7 | 1/7 | ||
| Hubei | 13/64 | 3/64 | 4/6 | 0/6 | ||||||
| Hunan | 0/70 | 8/70 | ||||||||
| Inner Mongolia | 9/65 | 6/65 | 0/5 | 0/5 | ||||||
| Jiangsu | 238/590 | 109/590 | 11/154 | 20/154 | 3/31 | 5/31 | 5/9 | 7/9 | 5/46 | 1/46 |
| Jiangxi | 18/49 | 24/49 | 1/11 | 3/11 | 1/9 | 2/9 | ||||
| Jilin | 33/63 | 2/63 | 24/70 | 8/70 | ||||||
| Liaoning | 10/37 | 3/37 | 0/7 | 0/7 | 2/6 | 1/6 | ||||
| Shaanxi | 0/70 | 0/70 | ||||||||
| Shandong | 3/60 | 8/60 | 1/59 | 0/59 | 1/3 | 0/3 | 1/8 | 0/8 | ||
| Shanghai | 51/53 | 6/53 | 44/70 | 9/70 | ||||||
| Shanxi | 20/20 | 4/20 | ||||||||
| Sichuan | 9/70 | 3/70 | ||||||||
| Tibet | 5/30 | 0/30 | ||||||||
| Xinjiang | 17/70 | 4/70 | ||||||||
| Yunnan | 41/130 | 27/130 | 10/70 | 7/70 | ||||||
| Zhejiang | 197/395 | 303/395 | 8/57 | 5/57 | 3/12 | 0/12 | 0/1 | 0/1 | ||
| Total | 621/1454 | 478/1454 | 257/1498 | 148/1498 | 20/130 | 10/130 | 54/109 | 19/109 | 17/99 | 3/99 |
| 42.7% | 32.9% | 17.2% | 9.9% | 15.4% | 7.7% | 49.5% | 17.4% | 17.2% | 3.0% | |
Real Time PCRs for mcr-4 and mcr-5
The representing nucleotide sequences for mcr-4 (MF543359) and mcr-5 (KY807920, KY807921) were obtained from the NCBI, and were aligned using the Clustal Multiple Alignment Algorithm. Based on the alignment, two mcr-4 PCRs were designed, one with a short 206-bp amplicon (forward: 5’-AGGTTTAGTGTTCGGGTTACGACTGG-3’; reverse: 5’-GCATTGGGATAGTCGCCTTTTTTTACTA-3’) and another with a long 1,165-bp amplicon (forward: 5’-AATTGTCGTGGGAAAAGCCGC-3’; reverse: 5’-CTGCTGACTGGGCTATTACCGTCAT-3’). The short amplicon PCR was used to establish prevalence data and positive samples were then tested with the long amplicon qPCR for phylogenetic studies.
Similarly, two mcr-5 PCRs were designed, the one producing a short 271-bp amplicon (forward: 5’-GTGAAACAGGTGATCGTGACTTACCG-3’, reverse: 5’-CGTGCTTTACACCGATCATGTGCT-3’) and the other a long 1,251-bp amplicon (forward: 5’-ACTCGACTGCCACCAGATCATCG-3’, reverse: 5’-CGCTGGAGTGTCAAGCCACTACTG-3’). The short amplicon PCR was used to establish prevalence data and positive samples were then tested with the long amplicon PCR for phylogenetic studies.
The specificity of the primers for the mcr-4 and mcr-5 PCRs was verified by BLASTN and DNA sequencing of the amplicons obtained using synthesized plasmids containing portions of the target mcr-4 and mcr-5 that were cloned into the SacI site (Takara Biothechnology, Dalian, China). The sensitivity of the mcr-4 PCRs and mcr-5 PCRs was determined by amplifying dilutions of the synthesized plasmids. The PCRs were quantified using the PicoGreen DNA fluorescence assay (Molecular Probes, Eugene, OR, USA) with standards prepared with the synthesized plasmids (104, 103, 102, 101, and 100 copies/reaction) (Genscript, Nanjing, China).
The PCRs were performed on a Roche LightCycler 480 II PCR instrument. The PCRs with short amplicons were SYBR based, and were used to determine the presence of mcr-4 and mcr-5 in swabs in this study. The positive samples determined by PCRs were further amplified by PCRs with long amplicons. The PCR products of both short and long amplicons were sequenced using forward and reverse primers (BGI, Shanghai, China).
Phylogenetic analysis
The mcr sequences obtained from this study and those from GenBank for the mcr-4 and mcr-5 were aligned used the MEGA 6.0 software to compare their similarities.
Statistical analysis
Multiple Pearson Chi-square test was used to compare differences between animal species as well as between oropharyngeal/anal and oral/nasal swabs with Bonferroni adjusted p-values. P value below 0.05 was considered significantly different.
Results
Establishment of PCRs for mcr-4 and mcr-5
The SYBR-based real-time PCRs detected the positive control plasmids containing the target mcr-4 and mcr-5 sequences with a detection limit of one gene copy per reaction. The detection limit was 10 copies per reaction for the mcr-4 and mcr-5 PCRs with long amplicons. The specificity of the PCRs was verified by gel electrophoresis and DNA sequencing.
Prevalence of mcr-4
Overall, the prevalence of the mcr-4 as detected by the short amplicon Real Time PCR in swine swabs (41.4%; 642/1,552) was significantly higher than in swabs from poultry (11.5%; 384/3,343) (P<0.01). The mcr-4 positivity determined by PCR was 17.4% for the anal (37/213) and 45.2% of the nasal swabs (605/1,339) in pigs, and 11.5% for the cloacal (195/1,696) and 11.5% of the oropharyngeal swabs (189/1,647) in poultry (Table 2, S2–S6 Tables).
Table 2. Comparison of the prevalence of mcr-4 and mcr-5 in nasal/oropharyngeal and anal/cloacal swabs of pigs and poultry.
| Positivity of mcr | pig | chicken | duck | goose | pigeon | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nasal | anal | total* | Oro** | cloacal | total | oro | cloacal | total | oro | cloacal | total | oro | cloacal | total | |
| (1339) | (213) | (1454) | (1350) | (1383) | (1498) | (122) | (122) | (130) | (109) | (109) | (109) | (66) | (82) | (99) | |
| mcr-4 | 605 | 37 | 621 | 137 | 143 | 257 | 9 | 11 | 20 | 37 | 30 | 54 | 6 | 11 | 17 |
| 45.2% | 17.4% | 42.7% | 10.1% | 10.3% | 17.2% | 7.4% | 9.0% | 15.4% | 33.9% | 27.5% | 49.5% | 9.1% | 13.4% | 17.2% | |
| mcr-5 | 455 | 59 | 478 | 93 | 58 | 148 | 8 | 3 | 10 | 16 | 6 | 19 | 1 | 2 | 3 |
| 34.0% | 27.7% | 32.9% | 6.9% | 4.2% | 9.9% | 6.6% | 2.5% | 7.7% | 14.7% | 5.5% | 17.4% | 1.5% | 2.4% | 3.0% | |
| mcr-4 and mcr-5 | 255 | 27 | 266 | 28 | 6 | 33 | 3 | 1 | 4 | 9 | 2 | 10 | 1 | 0 | 1 |
| 19.0% | 12.7% | 18.3% | 2.1% | 0.4% | 2.2% | 2.5% | 0.8% | 3.1% | 8.3% | 1.8% | 9.2% | 1.2% | 0.0% | 1.0% | |
* Total means the total number of the assayed animals. Under the column of Total, when one of the Nasal/Oral and Anal/cloacal swabs was positive, this animal was considered to be positive
** oro indicates oropharyngeal swab
Overall, poultry in 21 of the 24 provinces of China we studied were positive for the mcr-4. The mcr-4 gene was found in swabs all four of the poultry species we studied, geese (49.5%, 54/109), chickens (17.2%, 257/1,498), pigeons (17.2%, 17/99), and ducks (15.4%, 20/130). The prevalence of the mcr-4 in cloacal swabs from geese (27.5%, 30/109) was significantly higher than that from pigeons (13.4%, 11/82), chickens (10.3%, 143/1,383), and ducks (9%, 11/122). Similarly, the oropharyngeal swabs from geese (33.9%, 37/109) were most commonly positive by mcr-4 PCR than those from chickens (10.1%; 137/1,350), pigeons (9.1%; 6/66) and ducks (7.4%; 9/122) (Table 1, S3–S6 Tables).
Prevalence of mcr-5
Overall, the mcr-5 short amplicon PCR were significantly more commonly positive with swabs from pigs (nasal swabs: 34.0%, 455/1,339; anal swabs: 27.7%, 59/213) than with swabs from poultry (oropharyngeal swabs: 7.2%, 118/1,647; cloacal swabs: 4.1%, 69/1,696) (Table 1, Table 2, S2–S6 Tables).
Positive mcr-5 PCR were obtained from poultry sampled in 19 of the 24 provinces with all four species being positive, mainly geese (17.4%, 19/109), chickens (9.9%, 148/1,498), ducks (7.7%, 10/130), and pigeons (3%, 3/99). The oropharyngeal swabs from geese were most commonly mcr-5 positive (14.7%, 16/109) followed by those from chickens (6.9%, 93/1,350), ducks (6.6%, 8/122), and pigeons (1.5%, 1/66). The prevalence of the mcr-5 in cloacal swabs was 5.5% in geese (6/109), 4.2% in chickens (58/1,383), 2.5% in ducks (3/122), and 2.4% in pigeons (2/82) (Table 1, S3–S6 Tables).
Co-occurrence of mcr-4 and mcr-5
We identified both the mcr-4 and mcr-5 in swabs from 48 poultry (2.6%, 48/1,836), including 33 chickens (oropharyngeal: 28; cloacal: 6), four ducks (oropharyngeal: 3; cloacal: 1), ten geese (oropharyngeal: 9; cloacal: 2) and one pigeon (oropharyngeal: 1; cloacal: 0) (Table 2). Both the mcr-4 and mcr-5 were detected in 18.3% (266/1,454) of the pigs with 27 anal swabs (27/213) and 255 nasal swabs (255/1,339) found positive for both genes (Table 2).
Comparison of the presence of mcr in swabs from the upper and lower alimentary tract
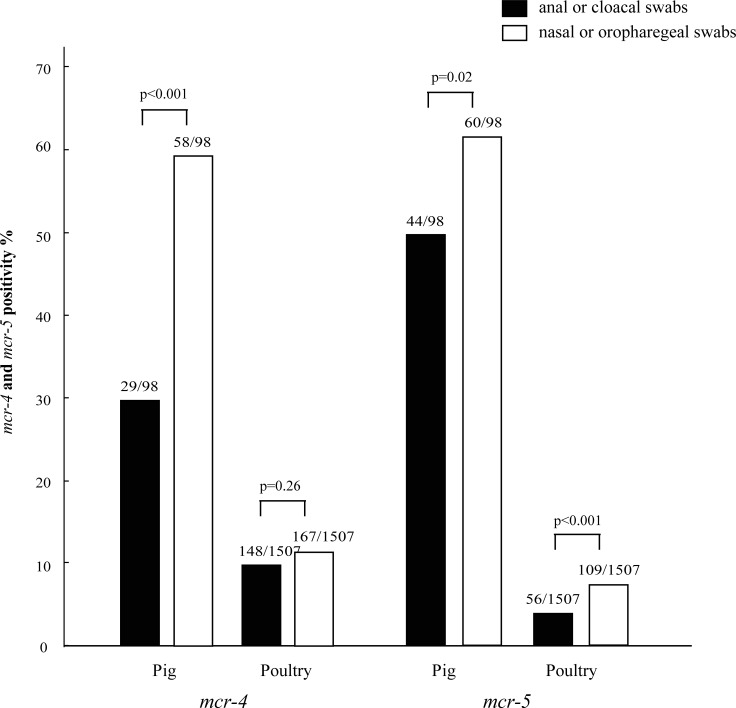
Swabs from both the upper (represented by nasal and oropharyngeal swabs) and lower alimentary tract (represented by anal and cloacal swabs) were available for each of 98 pigs and of 1,507 poultry we studied. Both of the swabs were positive for mcr-4 in 21 (21.4%, 21/98) of the pigs but only the anal swab was mcr-4 positive in eight pigs (8.2%, 8/98), and only the nasal swab was mcr-4 positive in 37 pigs (37.8%, 37/98) (Fig 1, S3 Table). The prevalence of mcr-4 in the nasal swabs from pigs (59.2%, 58/98) was significantly higher (p<0.001) than that in anal swabs (29.6%, 29/98) (Fig 1).
Fig 1. Prevalence of mcr-4 and mcr-5 in the upper and lower alimentary system of pigs and poultry.
In swabs collected from both locations in 98 pigs and 1,507 poultry, the prevalences of the mcr-4 and mcr-5 in pigs and the mcr-5 in poultry were significantly higher in nasal/oropharyngeal swabs than in the anal/cloacal swabs.
Both the oropharyngeal and cloacal swabs were mcr-4 positive in 36 poultry (2.4%: 23 chickens, 13 geese). The cloacal swab was the only positive swab for 112 birds (7.4%: 82 chickens, 10 ducks; 17 geese, 3 pigeons) and the oropharyngeal swab was the only positive swab for 131 birds (8.7%: 98 chickens, 5 ducks, 24 geese, 4 pigeons) (Fig 1, S3–S6 Tables). The prevalence of the mcr-4 in the oropharyngeal swabs (11.1%, 167/1,507) of poultry was not significantly different from that in the cloacal swabs (9.8%, 148/1,507) (Fig 1).
With respect to the mcr-5 PCR, 36 of the 98 (36.7%) pigs, from which both nasal and anal swabs were taken, were positive for both swabs; only the anal swab was positive for 8 pigs (8.2%, 8/98) and only the nasal swab positive for 24 pigs (24.5%, 24/98) (Fig 1, S2 Table). The prevalence of the mcr-5 in the nasal swabs from these pigs (61.2%, 60/98) was significantly higher (p = 0.02) than in anal swabs (44.9%, 44/98) (Fig 1).
Of the 1,507 poultry from which both oropharyngeal and cloacal swabs were collected, 7 birds (0.5%: 3 chickens, 1 duck, 3 geese) were mcr-5 positive in both swabs. Forty-nine birds (3.3%: 43 chickens; 2 ducks, 3 geese, 1 pigeons) were positive to mcr-5 only for cloacal swabs, and 102 birds (6.8%: 82 chickens, 6 ducks, 13 geese, 1 pigeon) were positive to mcr-5 in only oropharyngeal swabs (Fig 1, S3–S6 Tables). The prevalence of the mcr-5 in oropharyngeal swabs (7.2%, 109/1,507) was significantly higher (p<0.001) than in the cloacal swabs (3.7%, 56/1,507) (Fig 1).
Phylogenetic comparison
We successfully sequenced the products of 26 of the mcr-4 long amplicon PCRs (9 pigs, 6 chickens, 10 geese, 1 pigeon) and 24 of the mcr-5 long amplicon PCRs (15 pigs, 6 chickens, 3 geese).
The nucleotide sequences of the mcr-4 (1,165-bp) we obtained were all identical to one another, and to a sequence obtained from a vaginal swab from a woman in China (MG520404). Further, they had 99.8% similarity (two mismatches) with the first mcr-4 sequence, reported from a Salmonella strain in Italy in 2013 (MF543359). The similarity at the amino acid level (388-aa) with the Salmonella pMCR_R3445 strain from Italy was 99.5% (two mismatches).
The nucleotide sequences (1,251-bp) of the mcr-5 identified in our study were identical to one another and to the first mcr-5, sequenced from Salmonella enterica subsp. pSE12-02541 (KY807920), as well as the mcr-5 of Salmonella enterica subsp. pSE13-SA01718 (KY807921), and the mcr-5 in the vaginal swab from a woman in China (MG520405).
All nucleotide sequences were submitted to GenBank with accession numbers MG586909 to MG586912 for mcr-4 and MG586913 to MG586915 for mcr-5.
Discussion
The usefulness of colistin, the last-resort antibiotic used to treat multidrug resistant Gram-negative bacterial infections, is being compromised with the recent identification of the mobile colistin resistance gene, mcr-1 [3], and the subsequent finding of mcr-2, mcr-3, mcr-4 and mcr-5 [4–5, 23–26]. The mcr-1, mcr-2 and mcr-3 have been detected in bacteria or swabs from a variety of hosts in China and elsewhere in the world [3–22] However, there are yet only few reports on mcr-4 and mcr-5 [23–26]. Our study shows the mcr-4 and mcr-5 occur widely in pigs and poultry in China (Table 1, S2–S6 Tables). The prevalences of the genes we detected using PCR of swabs from animals were considerably higher than those obtained with studies that relied on bacterial isolates [23, 24]. The sensitive and specific PCR we used to detect the mcr-4 and mcr-5 directly in swabs avoided the limitations introduced by bacterial isolation and the associated underestimation of the prevalence of the mcr’s, the so-called ‘phantom resistome’ [29]. Although bacterial isolation for resistance testing is expensive, laborious, time consuming, and limits the resistant strains detected in a sample, it is an important adjunct to detection by molecular methods and enables a more complete understanding of colistin resistance and its epidemiology. Our molecular study might have provided more accurate data on the true prevalence of the mcr. However, the data did not enable us to determine the bacterial species that carried the resistance genes, or the location of the mcr in plasmids or in the bacterial chromosomes.
The mcr-1 gene has spread to most continents, and has been detected in various bacterial isolates from animals, human and the environment, including Escherichia coli, Klebsiella pneumoniae, Enterobacter cloacae and Enterobacter aerogenes [30]. The mcr-2 gene was found on rarely occasion, in Escherichia. coli isolates from porcine of Belgium and in flies of China [4, 22]. After its first characterization on a IncHI2-type plasmid, pWJ1, from Escherichia coli isolated from a Chinese pig [5], the mcr-3 gene was shown to be present in bacteria isolated from humans in Denmark, chickens and flies in China, pigs in Japan and cattle in Spain [20–22, 26, 31].
As far as we know, the mcr-4 gene was first detected in two Salmonella enterica serovar Typhimurium strains isolated from human fecal samples and in Salmonella and E. coli isolated from pigs in Italy, Spain and Belgium [25]. After the initially discovery of the mcr-5 gene in Salmonella enterica subsp. enterica serovar Paratyphi B, the gene was detected in Escherichia coli from diseased pigs and healthy pigs in Japan [26].
Our findings of very high prevalences of the mcr-4 and mcr-5 in pigs and poultry from large areas in China are most likely associated with the prolonged and widespread use of colistin as a growth promoter in food animals in China. However, it should be noted that these two genes might also be more prevalent in other countries as few studies looked for them.
It is noticeable that the prevalences of the mcr-4 and mcr-5 were generally significantly higher in the nasal/oropharyngeal swabs than in the anal/cloacal swabs in both pigs and poultry. This suggests that bacteria in saliva and respiratory secretions might play important roles in the maintenance and transmission of colistin resistance genes in pigs and poultry. Further comparative studies are needed to determine the bacterial species carrying the mcr-4 and mcr-5 in the upper and lower alimentary tract and how there might be transmission of the resistance genes between these populations.
Compared to the reported sequences (MF543359; MG581979) [23, 25], the mcr-4 identified in this study demonstrated nucleotide mutations (MF543359: 2/1,165; MG581979: 3/1,165), resulting in change in amino acids (MF543359: 2/338; MG581979: 3/338). The alignment of mcr-5 genes in this study with the initial sequence from Salmonella species in German and E. coli in Japan [24, 26] demonstrated that they were identical.
In conclusions, our study further confirms the presence of the mcr-4 and mcr-5 in bacteria from animals and indicates that these genes are widespread in food producing animals (pigs and poultry) in China. Future studies are needed to characterize the bacteria carrying the mcr-4 and mcr-5 and their locations on plasmids and/or the bacterial chromosomes.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
This work was supported by a grant from the National Key R & D Program of China (2016YFD0500804), a grant from the National Natural Science Foundation of China (NO: 31472225), the Priority Academic Program Development of Jiangsu Higher Education Institutions, and the Jiangsu Co-Innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses.
Data Availability
All relevant data are within the paper and its Supporting Information files. All the nucleotide sequences were submitted to GenBank with accession numbers MG586909 to MG586912 for mcr-4 and MG586913 to MG586915 for mcr-5.
Funding Statement
This work was supported by a grant from the National Key R & D Program of China (2016YFD0500804), a grant from the National Natural Science Foundation of China (NO: 31472225), the Priority Academic Program Development of Jiangsu Higher Education Institutions, and the Jiangsu Co-Innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pogue JM, Kaye KS, Cohen DA, Marchaim D. Appropriate antimicrobial therapy in the era of multidrug-resistant human pathogens. Clin Microbiol Infect. 2015;21: 302–312. doi: 10.1016/j.cmi.2014.12.025 [DOI] [PubMed] [Google Scholar]
- 2.Temkin E, Adler A, Lerner A, Carmeli Y. Carbapenem-resistant Enterobacteriaceae: biology, epidemiology, and management. Ann N Y Acad Sci. 2014;1323: 22–42. doi: 10.1111/nyas.12537 [DOI] [PubMed] [Google Scholar]
- 3.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16: 161–168. doi: 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- 4.Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 2016;21 doi: 10.2807/1560-7917 [DOI] [PubMed] [Google Scholar]
- 5.Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, et al. Novel Plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. MBio. 2017;8. pii: e00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prim N, Turbau M, Rivera A, Rodríguez-Navarro J, Coll P, Mirelis B. Prevalence of colistin resistance in clinical isolates of Enterobacteriaceae: A four-year cross-sectional study. J Infect. 2017. September 14 pii: S0163-4453(17)30297-9. doi: 10.1016/j.jinf.2017.09.008 [DOI] [PubMed] [Google Scholar]
- 7.Venditti C, Nisii C, D'Arezzo S, Vulcano A, Di Caro A. Letter to the Editor: Surveillance of mcr-1 and mcr-2 genes in Carbapenem-resistant Klebsiella pneumoniae strains from an Italian Hospital. Euro Surveill. 2017;22 pii: 30604. doi: 10.2807/1560-7917.ES.2017.22.35.30604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Liu Y, Qi X, Wang R, Jin L, Zhao M, et al. Molecular epidemiology of colistin-resistant Enterobacteriaceae in inpatient and avian isolates from China: high prevalence of mcr-negative Klebsiella pneumoniae. Int J Antimicrob Agents. 2017;50: 536–541. doi: 10.1016/j.ijantimicag.2017.05.009 [DOI] [PubMed] [Google Scholar]
- 9.Zrfluh K, Stephan R, Widmer A, Poirel L, Nordmann P, Nüesch HJ, et al. Screening for fecal carriage of MCR-producing Enterobacteriaceae in healthy humans and primary care patients. Antimicrob Resist Infect Control. 2017;6: 28 doi: 10.1186/s13756-017-0186-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pragasam AK, Shankar C, Veeraraghavan B, Biswas I, Nabarro LE, Inbanathan FY, et al. Molecular Mechanisms of Colistin Resistance in Klebsiella pneumoniae Causing Bacteremia from India-A First Report. Front Microbiol. 2017;7: 2135 doi: 10.3389/fmicb.2016.02135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Terveer EM, Nijhuis RHT, Crobach MJT, Knetsch CW, Veldkamp KE, Gooskens J, et al. Prevalence of colistin resistance gene (mcr-1) containing Enterobacteriaceae in feces of patientsattending a tertiary care hospital and detection of a mcr-1 containing, colistin susceptible E. coli. PLoS One. 2017;12: e0178598 doi: 10.1371/journal.pone.0178598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baron S, Bardet L, Dubourg G, Fichaux M, Rolain JM. mcr-1 plasmid-mediated colistin resistance gene detection in an Enterobacter cloacae clinicalisolate in France. J Glob Antimicrob Resist. 2017;10: 35–36. doi: 10.1016/j.jgar.2017.05.004 [DOI] [PubMed] [Google Scholar]
- 13.Saly M, Jayol A, Poirel L, Megraud F, Nordmann P, Dubois V. Prevalence of faecal carriage of colistin-resistant Gram-negative rods in a university hospital in western France, 2016. J Med Microbiol. 2017;66: 842–843. doi: 10.1099/jmm.0.000497 [DOI] [PubMed] [Google Scholar]
- 14.Hu YY, Wang YL, Sun QL, Huang ZX, Wang HY, Zhang R, et al. Colistin resistance gene mcr-1 in gut flora of children. Int J Antimicrob Agents. 2017;50: 593–597. doi: 10.1016/j.ijantimicag.2017.06.011 [DOI] [PubMed] [Google Scholar]
- 15.Tada T, Uechi K, Nakasone I, Shimada K, Nakamatsu M, Kirikae T, et al. Emergence of a colistin-resistant Escherichia coli clinical isolate harboring mcr-1 in Japan. Int J Infect Dis. 2017;63: 21–22. doi: 10.1016/j.ijid.2017.07.023 [DOI] [PubMed] [Google Scholar]
- 16.Kim ES, Chong YP, Park SJ, Kim MN, Kim SH, Lee SO, et al. Detection and genetic features of MCR-1 producing plasmid in human Escherichia coli infection in South Korea. Diagn Microbiol Infect Dis. 2017;89: 158–160. doi: 10.1016/j.diagmicrobio.2017.06.020 [DOI] [PubMed] [Google Scholar]
- 17.Newton-Foot M, Snyman Y, Maloba MRB, Whitelaw AC. Plasmid-mediated mcr-1 colistin resistance in Escherichia coli and Klebsiella spp. clinical isolates from the Western Cape region of South Africa. Antimicrob Resist Infect Control. 2017;6: 78 doi: 10.1186/s13756-017-0234-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohsin J, Pál T, Petersen JE, Darwish D, Ghazawi A, Ashraf T, et al. Plasmid-Mediated Colistin Resistance Gene mcr-1 in an Escherichia coli ST10 Bloodstream Isolate in the Sultanate of Oman. Microb Drug Resist. 2017. August 11 doi: 10.1089/mdr. 2017.0131 [DOI] [PubMed] [Google Scholar]
- 19.Zurfluh K, Nüesch-Inderbinen M, Klumpp J, Poirel L, Nordmann P, Stephan R. Key features of mcr-1 Bearing plasmids from Escherichia coli isolated from humans and food. Antimicrob Resist Infect Control. 2017;6: 91 doi: 10.1186/s13756-017-0250-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Litrup E, Kiil K, Hammerum AM, Roer L, Nielsen EM, Torpdahl M. Plasmid borne colistin resistance gene mcr-3 in Salmonella isolates from human infections, Denmark, 2009–17. Euro Surveill. 2017;22 pii: 30587. doi: 10.2807/1560 7917.ES.2017.22.31.30587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roer L, Hansen F, Stegger M, Sönksen UW, Hasman H, Hammerum AM. Novel mcr-3 variant, encoding mobile colistin resistance, in an ST131 Escherichia coli isolate from bloodstream infection, Denmark, 2014. Euro Surveill. 2017;22. 22846. [PubMed] [Google Scholar]
- 22.Zhang J, Wang J, Chen L, Yassin AK, Kelly P, Butaye P, et al. Housefly (Musca domestica) and blow fly (Protophormia terraenovae) as vectors of bacteria carrying Colistin resistance genes. Appl Environ Microbiol. 2017;84 pii: e01736-17. doi: 10.1128/AEM.01736-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carattoli A, Villa L, Feudi C, Curcio L, Orsini S, Luppi A, et al. Novel plasmid mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill. 2017;22 pii: 30589. doi: 10.2807/1560-7917.ES.2017.22.31.30589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borowiak M, Fischer J, Hammerl JA, Hendriksen RS, Szabo I, Malorny B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother. 2017. September 18 doi: 10.1093/jac/dkx327 [DOI] [PubMed] [Google Scholar]
- 25.Carretto E, Brovarone F, Nardini P, Russello G, Barbarini D, Pongolini S, et al. Detection of mcr-4 positive Salmonella enterica serovar Typhimurium in clinical isolates of human origin, Italy, October to November 2016. Eurosurveillance. 2018;23(2):17–00821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fukuda A, Sato T, Shinagawa M, Takahashi S, Asai T, Yokota S-i, et al. High prevalence of mcr-1, mcr-3, and mcr-5 in Escherichia coli derived from diseased pigs in Japan. International journal of antimicrobial agents. 2017. [DOI] [PubMed] [Google Scholar]
- 27.Li M, Jelocnik M, Yang F, Gong J, Kaltenboeck B, Polkinghorne A, et al. Asymptomatic infections with highly polymorphic Chlamydia suis are ubiquitous in pigs. BMC Vet Res. 2017;13: 370 doi: 10.1186/s12917-017-1295-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo W, Li J, Kaltenboeck B, Gong J, Fan W, Wang C. Chlamydia gallinacea, not C. psittaci, is the endemic chlamydial species in chicken (Gallus gallus). Sci Rep. 2016;6: 19638 doi: 10.1038/srep19638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Zhang R, Li J, Wu Z, Yin W, Schwarz S, et al. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat Microbiol 2017;2: 16260 doi: 10.1038/nmicrobiol.2016.260 [DOI] [PubMed] [Google Scholar]
- 30.Skov RL, Monnet DL. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Eurosurveillance. 2016;21(9). [DOI] [PubMed] [Google Scholar]
- 31.Hernández M, Iglesias MR, Rodríguez-Lázaro D, Gallardo A, Quijada NM, Miguela-Villoldo P, et al. Co-occurrence of colistin-resistance genes mcr-1 and mcr-3 among multidrug-resistant Escherichia coli isolated from cattle, Spain, September 2015. Eurosurveillance. 2017;22(31). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. All the nucleotide sequences were submitted to GenBank with accession numbers MG586909 to MG586912 for mcr-4 and MG586913 to MG586915 for mcr-5.