Abstract
Artificial high-CO2 atmosphere (AHCA, 95% CO2 and 1% O2) has been widely applied as a postharvest de-astringency treatment for persimmon fruit. AHCA increases expression of transcription factors, including ethylene response factors (DkERF), that target de-astringency genes. Here, the promoter of DkERF9, a previously characterized AHCA-inducible and de-astringency regulator, was utilized to screen a cDNA library by yeast one hybrid assay. A novel NAC transcription factor, named DkNAC7, was identified. Dual-luciferase assay indicated that DkNAC7 could not only trans-activate the promoter of DkERF9, but also activated the previously identified deastringency-related gene DkPDC2. Real-time PCR analysis showed that DkNAC7 was up-regulated by AHCA treatment, in concert with the removal of astringency from persimmon fruit and subcellular localization showed DkNAC7 was located in the nucleus. Thus, these results indicate that DkNAC7 is a putative transcriptional activator involved in regulating persimmon fruit deastringency by trans-activition on both DkERF9 and DkPDC2, which encodes pyruvate decarboxylase.
Introduction
Persimmon (Diospyros kaki L.) is a worldwide crop, which originated in Southeast Asia. Persimmon fruit can be divided into astringent and non-astringent types, but most native cultivars in China are of the astringent types [1]. These astringent persimmon fruit have the unique feature of accumulating abundant amounts of condensed tannins (CT) [2]. Astringent persimmon accumulates abundant CTs in fruit even at maturity and soluble CTs (SCT) cause astringency [1,3], which severely affects the persimmon industry and consumer acceptance.
A range of artificial technologies have been developed to remove astringency, including high-CO2, ethylene and ethanol [4–8]. Among these, high CO2 (usually > 90%) is the most widely used treatment, in which the O2 level is reduced to 1%. In plants, hypoxia usually causes the accumulation of products from anaerobic metabolism [9], and these products (especially acetaldehyde) effectively reduce the SCTs and accelerate deastringency in persimmon fruit [5,7,10]. The activities of alcohol dehydrogenase (ADH, EC 1.1.1.1) and pyruvate decarboxylase (PDC, EC 4.1.1.1) and also their encoding genes (DkADH1, DkPDC1, DkPDC2 and DkPDC3) have been shown to increase in amount during deastringency [7,11]. Transient over-expression of DkPDC2 led to a lower level of SCTs in persimmon leaves [7], suggesting that DkPDC2 is a key gene for the de-astringency program of persimmon fruit. These results confirmed that CO2 driven astringency removal involves hypoxia-triggered acetaldehyde metabolism.
In the model plant Arabidopsis, a few ethylene response factors (ERFs) have been reported to be involved in the hypoxia response, including HRE1, HRE2, RAP2.2 and RAP2.12. These ERF genes could transcriptionally regulate ADH and PDC, and result in hypoxia tolerance [12–15]. As stated above, persimmon fruit deastringency by high CO2 treatment is considered to operate mainly through the hypoxia fermentation pathway. In persimmon, four DkERF were previously reported to be involved in persimmon fruit deastringency, including DkERF9/10/19/22 [7,8]. Of these, DkERF9 was characterized as an activator of the promoter of DkPDC2, a key gene for deastringency [7]. Due to the low oxygen in high CO2 treatment, these DkERFs were previously termed as hypoxia responsive [8]. But, high CO2 treatment is an atypical anoxia environment, with effects of both high CO2 and low O2, thus it could be induced either a high-CO2 or hypoxia response.
Apart from ERFs, some other transcription factors were reported as high-CO2/hypoxia responsive in persimmon fruit, such as DkMYB6 [16] and DkTGA1 [17]. NAC genes are the main transcription factors reported to be involved in the plant hypoxia response. In Arabidopsis, more than 100 NAC genes have been characterized [18] that share highly conserved consensus in the N-terminal region of a Petunia gene (NAM), Arabidopsis ATAF1/2 and CUC2 proteins [19]. Among these genes, hypoxia-responsive NAC genes have rarely been reported, and the results from studies on ANAC102 also indicate that additional NAC genes might exist for the hypoxia response, as ANAC102 knockout lines did not show altered ADH gene transcription in Arabidopsis [9]. In persimmon, six NAC genes have been characterized, among which DkNAC1/3/5/6 were high-CO2/hypoxia responsive, however their regulatory roles in persimmon deastringency remain unclear [20]. Thus, the potential role of NAC genes in regulating persimmon deastringency still lacks experimental evidence.
Here, a novel NAC transcription factor (DkNAC7) was obtained as a result of yeast one hybrid screening by using the promoter of DkERF9 as bait and the regulatory role of DkNAC7 in persimmon de-astringency was investigated using yeast one-hybrid assay, dual-luciferase, real-time PCR and subcellular localization.
Materials and methods
Plant materials and treatment
‘Mopanshi’ (astringent cultivar) persimmon (D. kaki) fruit were obtained from a commercial orchard at Fangshan (Beijing, China) in 2012. Fruit without disease or mechanical wounding were selected and treated with artificial high-CO2 atmosphere (AHCA, 95% CO2 and 1% O2) or air in air-tight containers for 1 d. The physiological data and sampling information were described in Wang et al. [21].
RNA extraction and cDNA synthesis
Total RNAs were extracted from frozen fruit flesh (2.0 g) and the cDNA synthesis carried out according to the method used previously [6].
Gene isolation and sequence analysis
A NAC transcription factor was obtained based on the Matchmaker Gold Yeast One-hybrid Library Screening System (Clontech, USA), using the promoter of deastringency-related DkERF9 [7,16] as the bait DNA sequence. The full-length NAC gene was isolated with a SMART RACE cDNA Amplification Kit (Clontech). The sequences of primers are described in Table 1. For phylogenetic analysis, the NAC genes in persimmon and methods were as described in Min et al [20]
Table 1. Sequences of the primers used for RACE, full-length amplification, real time PCR and vector construction.
| Gene | Methods used | Primers (5’-3’) |
|---|---|---|
| DkNAC7 | 3’RACE (Primary PCR) | CAAGCCCTTCCGATTCGATGCCAT |
| DkNAC7 | 3’RACE (Secondary PCR) | GGAAGACAACAGGAAAGGACAGGCC |
| DkNAC7 | 5’RACE (Primary PCR) | CTCGTCATCCTCCCATTCCTCCTCAAC |
| DkNAC7 | 5’RACE (Secondary PCR) | CTCGTGCATCACCCAGTTGGTCCTCT |
| DkNAC7 | Full-length clone (FP) | CATCGGCGGTGACCAAAACGG |
| DkNAC7 | Full-length clone (RP) | CACAAAGTCCCTAGATCTCAGA |
| DkNAC7 | Y1H constructs (FP) | CGCGAATTCATGGGCCTCGATCCATCGTC |
| DkNAC7 | Y1H constructs (RP) | GATGGATCCCTACCTCGATGCATTTCCCG |
| DkNAC7 | SK vector construction (FP) | CGCGCGGCCGCATGGGCCTCGATCCATCGTC |
| DkNAC7 | SK vector construction (RP) | GATGGATCCCTACCTCGATGCATTTCCCG |
| DkNAC7 | Q-PCR (FP) | TGAGTTTCAAAATTGGGAGT |
| DkNAC7 | Q-PCR (RP) | CCCTAGATCTCAGATGGTGA |
| DkNAC7 | GFP vector construction (FP) | CGCGGTACCATGGGCCTCGATCCATCGTC |
| DkNAC7 | GFP vector construction (RP) | CATGTCGACCCTCGATGCATTTCCCG |
| DkERF9 | pAbAi vector construction (FP) | CGCGAGCTCAAATAATTTAATTAAAGATA |
| DkERF9 | pAbAi vector construction (RP) | CGCGTCGACATACACAGGAAAACAGGATT |
Note: underlined sequences show cutting sites for restriction enzymes
Yeast one-hybrid assay (Y1H)
According to library screening results, the protein-DNA interaction was verified with DkNAC7-AD and DkERF9 promoter, individually. Meanwhile, interaction between DkNAC7 and DkPDC2 promoter was also investigated by Y1H. The promoter of DkERF9 was constructed into pAbAi vector (primers are listed in Table 1). The DkPDC2-pAbAi was constructed by Min et al. [8]. The full-length sequence of transcription factor DkNAC7 was subcloned into pGADT7 AD vector (AD) (primers are listed in Table 1).
Auto-activation and the interaction analysis were conducted according to the manufacturer’s protocol.
Dual luciferase assay
Dual-luciferase assay was used as a rapid and efficient method to detect in vivo trans-activation or trans-repression effects of transcription factors [22]. Full-length DkNAC7 was inserted into pGreen II 0029 62-SK vector (SK), using the primers listed in Table 1. The dual luciferase assay was carried out with Nicotiana benthamiana leaves, using the protocol described by Min et al. [7]. Three independent experiments (with minimum five replicates) were performed to verify the results.
Real-time PCR analysis
For real-time PCR, gene specific oligonucleotide primers were designed and are shown in Table 1. The quality and specificity of primers were checked by melting curve and PCR products resequencing. The DkACT was chosen as a housekeeping gene to monitor the abundance of mRNA [7].
Real-time PCR reactions were carried out on a CFX96 instrument (Bio-Rad). The PCR protocols were according to our previous reports, using Ssofast EvaGreen Supermix Kit (Bio-Rad) [6]. The relative expression of this NAC gene was calibrated with values for day 0 fruit set as 1.
Subcellular localization analysis
35S-DkNAC7-GFP was transiently expressed in tobacco leaves by Agrobacterium-mediated infiltration (GV3101) according to previous reports [23,24]. The green fluorescent protein (GFP) fluorescence in tobacco leaves was imaged 2 d after infiltration using a Zeiss LSM710NLO confocal laser scanning microscope. Primers used for GFP construction are described in Table 1.
Statistical analysis
Least Significant Difference (LSD) test was used to compare the statistical significance differences among treatments by using DPS 7.05 or Student’s t-test. The figures were drawn with Origin 8.0.
Results and discussion
Y1H based library screening discovered a novel NAC gene, which targeted the DkERF9 promoter
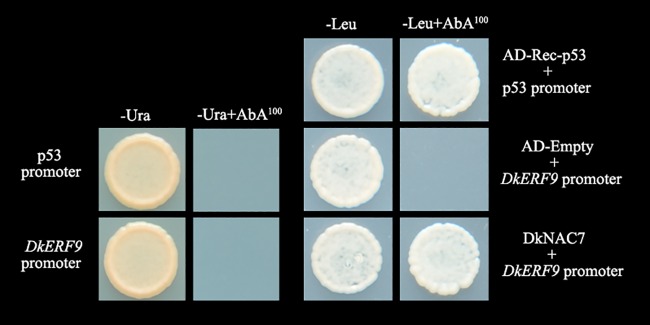
In our previous reports, DkERF9 transcription factor was shown to be involved in persimmon de-astringency via regulation of the DkPDC2 promoter [7]. In order to obtain further information about the transcriptional regulatory mechanism controlling persimmon fruit deastringency, Y1H based screening was employed to screening the potential interacting transcription factors, using the DkERF9 promoter as bait. A total of 150 PCR products were obtained, among which only one NAC transcription factor gene was characterized. Individual verifications with Y1H indicated that the NAC transcription factor could bind to DkERF9 promoter (Fig 1). As six DkNAC genes were reported previously in persimmon fruit [20], this NAC transcription factor was named as DkNAC7 (GenBank no.MG792350) (Fig 1).
Fig 1. Protein-DNA interaction between DkNAC7 and the promoter of DkERF9 using yeast one hybrid analysis.
Interaction was confirmed on SD medium lacking Leu in the presence of aureobasidin A (-Leu+AbA100). AD-Rec-p53 and p53-AbAi were used as a positive control; AD-empty and pDkERF9-AbAi were used as a negative control.
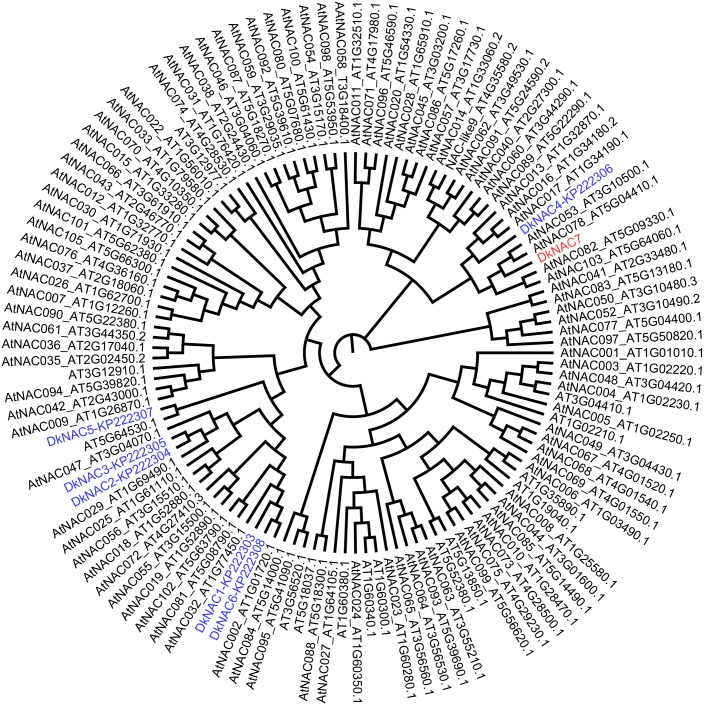
Phylogenetic tree analysis indicated that DkNAC7 was close to DkNAC4, but not the other five previously reported DkNAC genes (Fig 2) [20]. Compared to Arabidopsis NAC transcription factors, DkNAC7 was close to AtNAC078, which was reported to regulate flavonoid biosynthesis under high light in Arabidopsis [25], while it was not clustered with ANAC102, which was shown to be induced by low oxygen (0.1%) in Arabidopsis [9].
Fig 2. Phylogenetic analysis of DkNAC7 and NAC members from persimmon and Arabidopsis.
Persimmon DkNAC is highlighted in red (DkNAC7, newly isolated) and blue (previously reported). The amino acid sequences of NAC transcription factors were obtained from the Arabidopsis Information Resource or National Center for Biotechnology Information, and their accession numbers are included in the diagram. The phylogenetic tree was constructed with Figtree (v 1.3.1).
In vivo regulatory roles of DkNAC7 on deastringency related genes (DkERF, DkADH1 and DkPDC2)
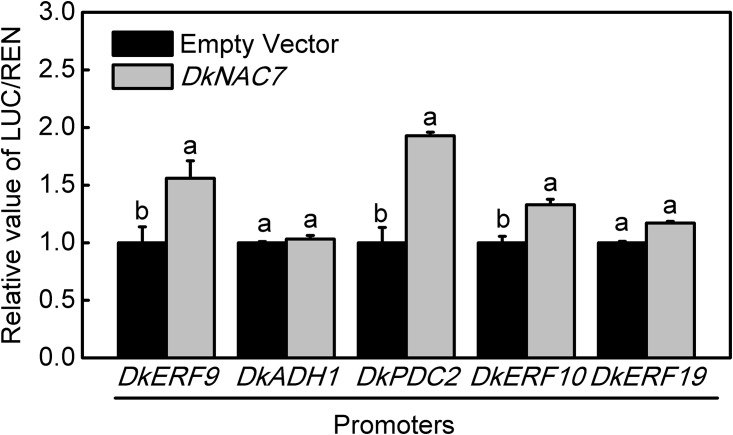
Further investigations on the possible transcriptional regulatory linkage between DkNAC7 and deastringency related genes were carried out. Three previously studied DkERF genes (DkERF9/10/19) and two structural genes (DkADH1 and DkPDC2) were selected for test. Dual luciferase assay indicated that DkNAC7 could significantly trans-activate the promoters of DkERF9 and DkPDC2 with 1.55 and 1.92-fold enhancement, respectively (Fig 3). The effect of DkNAC7 on the DkERF10 promoter also reached statistical significance, but the response was very limited (about 1.32-fold) and the DkNAC7 gene had no significant effects on the promoters of DkADH1 and DkERF19 (Fig 3).
Fig 3. Regulatory effects of DkNAC7 on the promoters of deastringency-related genes (DkERF9/10/19, DkADH1, DkPDC2) using the dual-luciferase assay.
The ratio of LUC/REN of the empty vector (SK) plus promoter was used as calibrator (set as 1). Error bars indicate SEs from five replicates. Different letters above the columns indicate significant differences (P<0.05).
In persimmon, twenty-two ethylene response factors (DkERFs) were differently expressed in response to high CO2 treatment. Of these 22 genes, only four ERFs (DkERF9/10/19/22) were fund to be involved in persimmon de-astringency [7,8], via the interaction with promoters of de-astringent related target genes (e.g. DkADH1, DkPDC2 and DkPDC3). Furthermore, a MYB transcription factor (DkMYB6) and a bZIP transcription factor (DkTGA1) were also characterized as regulators of persimmon fruit astringency removal, respectively [16,17]. Thus, these finding with DkNAC7 unveiled a new transcription factor that participates in regulation of persimmon fruit deastringency. Furthermore, DkNAC7 is not closely related to the low oxygen-induced ANAC102 gene [9] from phylogenetic result (Fig 2) which suggests that more than one various type NAC transcription factor may contribute to the hypoxia response.
Cascade regulations of DkNAC7-DkERF9-DkPDC2
Comparing the effects DkNAC7 and the previously characterized TFs on the DkPDC2 promoter, DkNAC7 was shown to have only a relatively limited action, which was only slightly stronger than DkTGA1 [17]. Y1H analyses indicated that DkNAC7 cannot bind to and is therefore an indirect regulator for hypoxia responsive DkPDC2 (S1 Fig). As the present results indicated that interaction between DkNAC7 and DkERF9 promoter (Fig 1) and our previous study indicated that DkERF9 could physically bind to DkPDC2 promoter [17]. Thus, it could be proposed that a regulatory cascade involving DkNAC7-DkERF9-DkPDC2 contributes to persimmon fruit deastringency. The regulatory roles of NAC transcription factors in hierarchical interactions with ERFs have also been reported in other fruits, for instance MdNAC029/MdNAP, an apple NAC gene, was reported to directly repressed the expression of two ERF genes (MdCBF1 and MdCBF4) by binding to their promoters, thus negatively regulating cold tolerance via the CBF-dependent pathway [26]. These finding from persimmon not only partial explain the transcriptional regulations during deastringency, but also provided a new example of NAC-ERF regulation. Moreover, since the NAC-ERF cascade contributes to persimmon deastringency (high-CO2/hypoxia response) and apple cold tolerance, this raises the question whether other NAC-ERF may be involved in abiotic stress responses.
Expression and subcellular localization analyses for DkNAC7
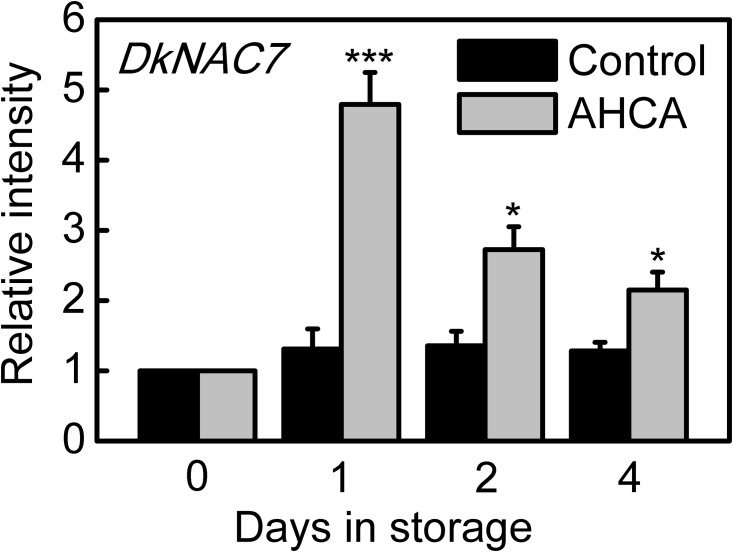
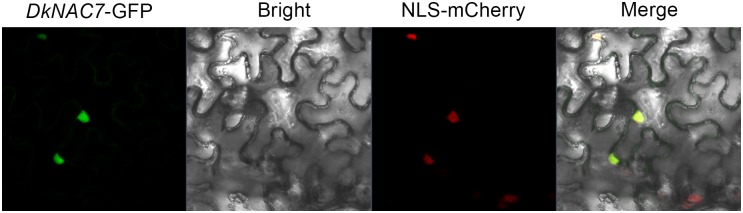
The above-mentioned regulatory effects of DkNAC7 on deastringency related genes encouraged us to study the response of DkNAC7 to deastringency treatment. From previous results, AHCA treatment (also called CO2 treatment or high CO2 treatment: 95% CO2 and 1% O2) was very effective in removing astringency in various persimmon [5,7,21]. Therefore, using previously described materials [21], the expression of DkNAC7 was analyzed. The DkNAC7 gene exhibited a sharp increase in expression in response to AHCA treatment, with the highest level at 1 d (Fig 4). After removal of CO2 treatment, transcripts of DkNAC7 decreased concomitantly, but remained statistically significantly higher than in control fruits. Such expression was similar to most of the previously identified deastringency related transcription factors. Furthermore, subcellular localization analysis of DkNAC7 in tobacco leaves using GFP tagging, showed strong signals in the nucleus (Fig 5).
Fig 4. Expression of DkNAC7 in response to AHCA treatment (95% CO2, 1% O2, 1 day).
Relative mRNA abundance was evaluated by real-time PCR. Day 0 fruit values were set as 1. Error bars represent ± SE from three replicates (*p < 0.05; ***p < 0.001).
Fig 5. Subcellular localization of DkNAC7-GFP in tobacco leaves transformed by agroinfiltration.
GFP fluorescence of DkNAC7 is indicated. Bars = 25 μm.
Taken all the results of Y1H, dual-luciferase assay, expression and subcellular localization together, we propose DkNAC7 as a novel regulator of persimmon fruit deastringency, acting via direct regulation of the DkERF9. Again, DkNAC7 was not closest homolog to the low oxygen-induced ANAC102 gene [9], indicating either potential differences between species or organs, or the complexity of NAC-regulatory roles. Another possible explanation would be the differences between experimental treatments, as in model plant or crops, anoxia treatments were generally low O2, but AHCA treatment in persimmon involves high CO2 and low O2. Thus, the deastringency related DkNAC7, as well as the previously characterized transcription factors, could be termed as high-CO2/hypoxia responsive.
Supporting information
(TIF)
Acknowledgments
This research was supported by the National Key Research and Development Program (2016YFD0400102), the National Natural Science Foundation of China (31722042; 31672204), the Natural Science Foundation of Zhejiang Province, China (LR16C150001), the Fundamental Research Funds for the Central Universities, and the 111 Project (B17039).
Abbreviations
- AbA
aureobasidin A
- AD
pGADT7
- ADH
alcohol dehydrogenase
- cv
Cultivar
- ERF
ethylene response factor
- PDC
pyruvate decarboxylase
- SCT
soluble condensed tannins
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by the National Key Research and Development Program (2016YFD0400102 to KC), the National Natural Science Foundation of China (31722042; 31672204 to XY), the Natural Science Foundation of Zhejiang Province, China (LR16C150001 to XY), the Fundamental Research Funds for the Central Universities, and the 111 Project (B17039 to XY). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Akagi T, Ikegami A, Tsujimoto T, Kobayashi S, Sato A., Kono A., et al. DkMyb4 is a Myb transcription factor involved in proanthocyanidin biosynthesis in persimmon fruit. Plant Physiol. 2009; 4, 2028–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo C, Zhang QL, Luo ZR. Genome-wide transcriptome analysis of Chinese pollination-constant nonastringent persimmon fruit treated with ethanol. BMC Genomics 2014; 15, 112 doi: 10.1186/1471-2164-15-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yonemori K, Suzuki Y. Differences in three-dimensional distribution of tannin cells in flesh tissue between astringent and non-astringent type persimmon. Acta Hortic. 2008; 833, 119–124. [Google Scholar]
- 4.Ikegami A, Eguchi S, Kitajima A, Inoue K, Yonemori K. Identification of genes involved in proanthocyanidin biosynthesis of persimmon (Diospyros kaki) fruit. Plant Sci. 2007; 172, 1037–1047. [Google Scholar]
- 5.Salvador A, Arnal L, Besada C, Larrea V, Quiles A, Pérez-Munuera I. Physiological and structural changes during ripening and deastringency treatment of persimmon fruit cv. ‘Rojo Brillante’. Postharvest Biol Technol. 2007; 46, 181–188. [Google Scholar]
- 6.Yin XR, Shi YN, Min T, Luo ZR, Yao YC, Xu Q, et al. Expression of ethylene response genes during persimmon fruit astringency removal. Planta. 2012; 235, 895–906. doi: 10.1007/s00425-011-1553-2 [DOI] [PubMed] [Google Scholar]
- 7.Min T, Yin XR, Shi YN, Luo ZR, Yao YC, Grierson D, et al. Ethylene-responsive transcription factors interact with promoters of ADH and PDC involved in persimmon (Diospyros kaki) fruit de-astringency. J Exp Bot. 2012; 63, 6393–6405. doi: 10.1093/jxb/ers296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Min T, Fang F, Ge H, Shi YN, Luo ZR, Yao YC, et al. Two novel anoxia-induced ethylene response factors that interact with promoters of deastringency-related genes from persimmon. PLOS One 2014; 9, e97043 doi: 10.1371/journal.pone.0097043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christianson JA, Wilson IW, Llewellyn DJ, Dennis ES. The low-oxygen-induced NAC domain transcription factor ANAC102 affects viability of Arabidopsis seeds following low-oxygen treatment. Plant Physiol. 2009; 149, 1724–1738. doi: 10.1104/pp.108.131912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuo T, Ito S. On mechanisms of removing astringency in persimmon fruits by carbon dioxide treatment. I. Some properties of the two processes in the de-astringency. Plant Cell Physiol. 1997; 18, 17–25. [Google Scholar]
- 11.Tamura F, Tanabe K, Itai A, Hasegawa M. Characteristics of acetaldehyde accumulation and removal of astringency with ethanol and carbon dioxide treatments in ‘Saijo’ persimmon fruit. J. Jpn. Soc. Hortic. Sci. 1999; 68, 1178–1183. [Google Scholar]
- 12.Licausi F, van Dongen JT, Giuntoli B, Novi G, Santaniello A, Geigenberger P, et al. HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J. 2010; 62, 302–315. doi: 10.1111/j.1365-313X.2010.04149.x [DOI] [PubMed] [Google Scholar]
- 13.Hinz M, Wilson IW, Yang J, Buerstenbinder K, Llewellyn D, Dennis ES, et al. Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 2010; 153, 757–772. doi: 10.1104/pp.110.155077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang CY, Hsu FC, Li JP, Wang NN, Shih MC. The AP2/ERF transcription factor AtERF73/HRE1 modulates ethylene responses during hypoxia in Arabidopsis. Plant Physiol. 2011; 156, 202–212. doi: 10.1104/pp.111.172486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papdi C, Pérez-Salamó I, Joseph MP, Giuntoli B, Bögre L, Koncz C, et al. The low oxygen, oxidative and osmotic stress responses synergistically act through the ethylene response factor VII genes RAP2.12, RAP2.2 and RAP2.3. Plant J. 2015; 82, 772–784. doi: 10.1111/tpj.12848 [DOI] [PubMed] [Google Scholar]
- 16.Fang F, Wang MM, Zhu QG, Min T, Grierson D, Yin XR, et al. DkMYB6 is involved in persimmon fruit deastringency, via transcriptional activation on both DkPDC and DkERF. Postharvest Biol Technol. 2016; 111, 161–167. [Google Scholar]
- 17.Zhu QG, Wang MM, Gong ZY, Fang F, Sun NJ, Li X, et al. Involvement of DkTGA1 transcription factor in anaerobic response leading to persimmon fruit postharvest de-astringency. PLOS One. 2016; 11, e0155916 doi: 10.1371/journal.pone.0155916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nuruzzaman M, Manimekalai R, Sharon AM, Satoh K, Kondoh H, Ooka H, et al. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010; 465, 30–44. doi: 10.1016/j.gene.2010.06.008 [DOI] [PubMed] [Google Scholar]
- 19.Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell. 1997; 9, 841–857. doi: 10.1105/tpc.9.6.841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Min T, Wang MM, Wang HX, Liu XF, Fang F, Grierson D, et al. Isolation and expression of NAC genes during persimmon fruit postharvest astringency removal. Int J Mol Sci. 2015; 16, 1894–1906. doi: 10.3390/ijms16011894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang MM, Zhu QG, Deng CL, Luo ZR, Sun NJ, Grierson D, et al. Hypoxia-responsive ERFs involved in postdeastringency softening of persimmon fruit. Plant Biotechno. J. 2017; 15,1409–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin XR, Allan AC, Chen KS, Ferguson IB. Kiwifruit EIL and ERF genes involved in regulating fruit ripening. Plant Physiol. 2010; 153, 1280–1292. doi: 10.1104/pp.110.157081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Q, Yin XR, Zeng JK, Ge H, Song M, Xu CJ, et al. Activator-and repressor-type MYB transcription factors are involved in chilling injury induced flesh lignification in loquat via their interactions with the phenylpropanoid pathway. J Exp Bot. 2014; 65, 4349–4359. doi: 10.1093/jxb/eru208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li SJ, Yin XR, Wang WL, Liu XF, Zhang B, Chen KS. Citrus CitNAC62 cooperates with CitWRKY1 to participate in citric acid degradation via up-regulation of CitAco3. J Exp Bot. 2017; 68, 3419–3426. doi: 10.1093/jxb/erx187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morishita T, Kojima Y, Maruta T, Nishizawa-Yokoi A, Yabuta Y, Shigeoka S. Arabidopsis NAC transcription factor, ANAC078, regulates flavonoids biosynthesis under high-light. Plant and Cell Physiol. 2009; 50, 2210–2222. [DOI] [PubMed] [Google Scholar]
- 26.An JP, Li R, Qu FJ, You CX, Wang XF, Hao YJ. An apple NAC transcription factor negatively regulates cold tolerance via CBF-dependent pathway. J Plant Physiol. 2017; 221, 74–80. doi: 10.1016/j.jplph.2017.12.009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.