Abstract
In addition to its central role in energy metabolism, the mitochondrion has many other functions essential for cell survival. When stressed, the multifunctional mitochondria are expected to engender multifaceted cell stress with complex physiological consequences. Potential extra-mitochondrial proteostatic burdens imposed by inefficient protein import have been largely overlooked. Accumulating evidence suggests that a diverse range of pathogenic mitochondrial stressors that do not directly target the core protein import machinery, reduce cell fitness by disrupting the proteostatic network in the cytosol. The resulting stress, named mitochondrial Precursor Over-accumulation Stress (mPOS), is characterized by the toxic accumulation of unimported mitochondrial proteins in the cytosol. Here, we review our current understanding of how mitochondrial dysfunction can impact the cytosolic proteome and proteostatic signaling. We also discuss the intriguing possibility that the mPOS model may help untangle the cause-effect relationship between mitochondrial dysfunction and cytosolic protein aggregation, which are probably the two most prominent molecular hallmarks of neurodegenerative disease.
Keywords: mitochondria, mPOS, neurodegeneration
Introduction
Mitochondria are dynamic, double membrane-bound organelles that have evolved a wide range of functions. They are responsible for oxidative phosphorylation (OXPHOS), calcium signaling, reactive oxygen species production, cell death, phospholipid biosynthesis, fatty acid catabolism, and the synthesis of various metabolic intermediates and cofactors. To perform these functions, mitochondria depend on a proteome of >1,100 proteins, all but 13 of which are encoded in the nucleus, synthesized in the cytosol, and imported into the organelle [1–3]. Mitochondrial protein import is an intricate process, requiring several multi-subunit protein complexes located in the outer mitochondrial membrane (OMM), the intermembrane space (IMS), and the inner mitochondrial membrane (IMM) [4–6]. Import also requires the mitochondrial membrane potential, Δψm, which is generated by proton pumping from the matrix to the IMS by the respiratory chain (RC). Given the dependence of mitochondrial function on cytosolically synthesized proteins and the essential nature of mitochondrial functions, maintaining efficient mitochondrial protein import is clearly indispensable for cellular and organismal health.
Mitochondrial dysfunction causes many human diseases [7, 8]. Classic ‘mitochondrial diseases’ result from mutations in OXPHOS components encoded by both mitochondrial and nuclear genomes. It is generally accepted that mitochondrial diseases are caused by inadequate ATP production, and in many cases oxidative stress contributes as well. In mitochondrial DNA (mtDNA) diseases, pathologic phenotypes only develop when mutant mtDNA exceeds ~60% and ~90% for deletions and point mutations, respectively. This phenomenon is known as the “threshold effect” [9], which suggests that cells have some degree of tolerance to OXPHOS defects. When the threshold is exceeded, these diseases are typically childhood-onset and multisystem, although the neuromuscular system tends to be preferentially affected [10].
Mitochondrial dysfunction also causes late-onset neurodegenerative diseases, as exemplified by familial Parkinson’s disease[11], spinocerebellar ataxia[12], peripheral neuropathies [13], and several hereditary spastic paraplegias[14, 15]. In contrast to early-onset mitochondrial diseases, whether bioenergetic defect is the primary contributor to the pathogenesis of late-onset neurodegenerative disorders is debatable [16]. In many cases, these neurodegenerative diseases are caused by mitochondrial proteins that are not directly involved in OXPHOS. Although a mitochondrial etiology is well accepted for these diseases, the underlying mechanisms are poorly understood. Mitochondrial abnormalities are also frequently associated with some of the most common neurodegenerative diseases (e.g., Alzheimer’s disease and amyotrophic lateral sclerosis) that have no apparent genetic trigger of mitochondrial dysfunction. The mechanism(s) of mitochondrial involvement in these diseases are even less clear.
Recent studies in several model organisms have shown that the physiological consequences of mitochondrial damage extend far beyond bioenergetics. For example, mitochondria are involved in stress signaling, which plays a critical role in determining cell and organismal fitness and survival [17]. The effect of mitochondrial dysfunction on cytosolic proteostasis is another intriguing development, and is the focus of the present review. Early studies showed that improving global proteostasis robustly suppresses mitochondria-induced cellular degeneration[18]. It was later found that multiple pathways of mitochondrial damage can compromise protein import efficiency, which leads to cell death triggered by a novel mechanism named mitochondrial precursor over-accumulation stress (mPOS)[19]. Here, we will first review the latest developments on the impact of mitochondrial functionality on cytosolic proteostasis and proteostatic signaling. We will then discuss the potential implications of these studies for mitochondrial and neurodegenerative disease mechanisms, with particular emphasis on neurodegeneration marked by both mitochondrial dysfunction and cytosolic protein aggregation.
The discovery of mPOS
Mitochondrial protein import is the cornerstone of mitochondrial biogenesis and is essential for cell survival. However, OXPHOS is dispensable for many cell types (e.g., the baker’s yeast Saccharomyces cerevisiae) as glycolysis can compensate to maintain energy homeostasis. Growth inhibition in these cells by mutations directly affecting the protein import machineries would be expected to result from the loss of other essential cellular functions associated with mitochondria (e.g., the biosynthesis of iron sulfur cluster). Inconsistent with this expectation, recent genetic studies in yeast revealed that diverse mitochondrial stressors can lead to cell death due to proteostatic stress in the cytosol[18, 19]. Suppression of cytosolic proteostatic stress is sufficient to maintain viability in these cells. It appears that before mitochondrial damage reaches the threshold to severely affect essential mitochondrial functions, reduced protein import is a significant trigger of cell death due to increased proteostatic stress in the cytosol.
In early studies, the Chen group modeled mitochondrial stress in yeast by expressing a mutant allele of AAC2, aac2A128P [18, 20]. AAC2 encodes the major isoform of adenine nucleotide translocase that is involved in ATP/ADP exchange across the IMM. A128P is equivalent to the A114P allele in the human ANT1 gene that causes autosomal dominant Progressive External Ophthalmoplagia (adPEO) [21]. It first came as a surprise that expression of aac2A128P dominantly inhibited cell growth on a fermentable carbon source, a condition where mitochondrial ATP synthesis is dispensable [20]. It was subsequently found that Aac2A128P-induced cell death is suppressed by mutations that downregulate cytosolic protein synthesis [18]. This observation suggested that readjusting cytosolic proteostasis is important for accommodating mitochondrial damage.
Like several other clinically relevant variants of Aac2, the Aac2A128P protein is misfolded on the inner membrane and prone to aggregation [22]. Genetic and biochemical studies suggest that misfolded Aac2 causes significant proteostatic stress on the IMM [18]. Yeast cells expressing a misfolded Aac2 cannot tolerate the loss of proteins involved in IMM protein quality control including Yme1 and subunits of prohibitin[18, 23]. In addition, the biogenesis/stability of multiple IMM protein complexes is compromised in cells expressing aac2A128P [22]. Affected complexes include the respiratory complexes, and TIM22 and TIM23 protein translocases. TIM22 is required for the insertion of polytopic membrane proteins into the IMM and TIM23 promotes the transport of precursors across the IMM into the matrix. These deleterious effects may synergistically disrupt IMM integrity and reduce Δψm [20], which together are likely to diminish protein import.
To understand how Aac2A128P-induced IMM proteostatic stress kills cells, Wang and Chen screened for genes that suppress cell lethality when overexpressed [19]. Intriguingly, of the 40 suppressor clones characterized, none of them primarily functions in mitochondria. Instead, these genes are involved in proteostasis in the cytosol by participating in TOR signaling, ribosomal biogenesis, mRNA decay/silencing, tRNA modification, translational control, and protein folding/degradation. This finding led to the proposal that Aac2A128P does not kill cells by loss of a mitochondrial process critical for cell survival, but rather, by the loss of protein homeostasis in the cytosol. It was speculated that Aac2A128P-induced IMM damage reduces protein import efficiency. This results in a cytosolic stress, named mitochondrial Precursor Overaccumulation Stress (mPOS), characterized by the toxic accumulation and aggregation of unimported mitochondrial proteins (Fig. 1). The suppressor genes likely alleviate mPOS by globally reducing protein synthesis, preventing protein misfolding, increasing protein turnover, and stimulating the selective translation of potential stress-resistant proteins. Strong support for the mPOS model came from the analysis of the cytosolic proteome showing that many mitochondrial precursor proteins accumulate in the cytosol of cells expressing aac2A128P. The presence of these unimported mitochondrial proteins apparently represents a significant proteostatic burden for the cytosol. When it exceeds cell’s capacity to stabilize, sequestrate and remove these proteins, mPOS ensues and cell viability is compromised. These studies firmly established that mitochondrial damage that do not directly target the core protein import machinery and the OXPHOS apparatus is sufficient to compromise protein import into mitochondria and to cause cell death.
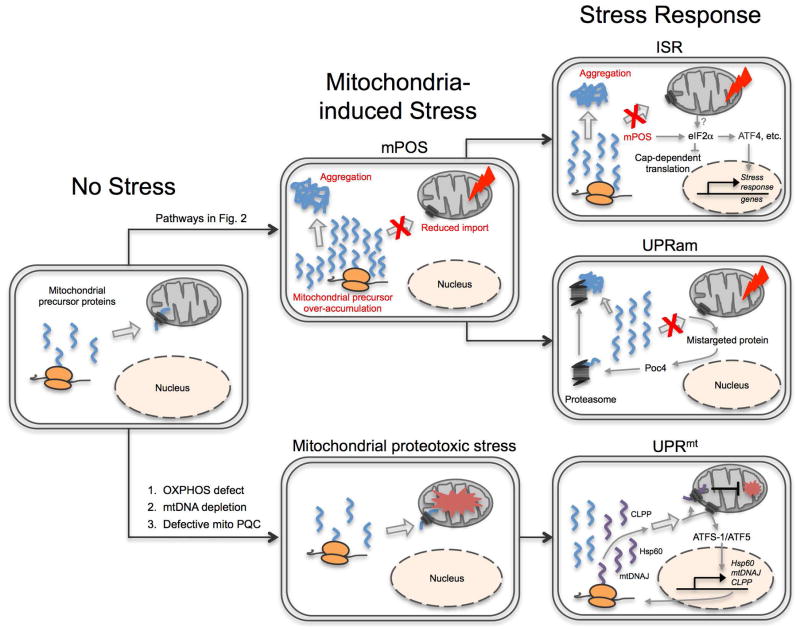
Fig. 1. Mitochondrial Precursor Over-accumulation Stress (mPOS) and its relationship to UPRam, UPRmt and ISR.
In unstressed cells, mitochondrial protein import is efficient and precursor proteins in the cytosol are kept to a minimum. Protein import can be reduced by intra- and extra-mitochondrial stressors (listed in Fig. 2) to cause mPOS. Unimported mitochondrial proteins can then activate the unfolded protein response activated by mistargeting of proteins (UPRam), which includes upregulation of proteasome activity. mPOS also activates the integrated stress response, characterized by the phosphorylation of eIF2α. This increases the translation of select transcriptional factors including ATF4 that in turn upregulates stress response genes. OXPHOS defect, mtDNA depletion and defective mitochondrial protein quality control (PQC) can cause mitochondrial proteotoxic stress. This triggers the mitochondrial unfolded protein response (UPRmt), driven by failure to import of ATFS-1 in worms and ATF5 in humans into mitochondria. ATFS-1/ATF5 translocates to the nucleus and drives transcription of mitochondria-destined stress response genes such as HSP60, mtDNAJ, and ClpP.
Conditions inducing mPOS
The physiological implications of the mPOS model are broad. The model entails that any condition that tips the balance between mitochondrial protein import and the cytosolic capacity to handle unimported proteins can compromise cell viability. Genetic studies in yeast showed that defects in many pathways can induce mPOS, albeit to variable degrees. These pathways include (1) mutations in the core mitochondrial protein import machinery, (2) IMM protein misfolding, (3) reduced IMM protein quality control, (4) mitochondrial DNA mutations, (5) Δψm dissipation, (6) mutations affecting precursor delivery, and (7) defects in stabilizing pre-imported or degrading unimported mitochondrial proteins (Fig. 2). In addition, (8) proteostatic stress from the cytosolic proteome can synergize with mPOS to further reduce cell viability. These studies are highlighted and discussed below.
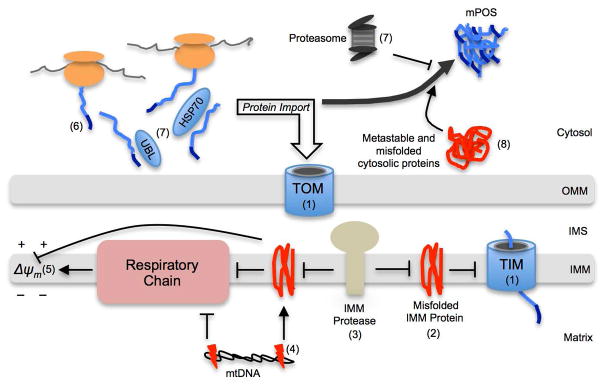
Fig. 2. Schematic of pathways inducing mPOS.
(1) Mutations in the core mitochondrial protein import machinery (TIM, translocase of the inner membrane; TOM, translocase of the outer membrane). (2) IMM protein misfolding. (3) Reduced IMM protein quality control, which can be caused by mutant IMM proteases. (4) mtDNA mutations, which disrupts the respiratory chain to reduce Δψm and increases protein misfolding through imbalanced mitochondrially and nuclear-encoded respiratory complex subunits. (5) Δψm dissipation by factors such as reduced respiration and increased proton leak. (6) Defects in precursor delivery to the OMM including mutations in the mitochondrial targeting sequences. (7) Defects in stabilizing (e.g., by mutant ubiquilin (UBL) or heat shock protein 70 kDa (HSP70)), or degrading (e.g., reduced proteasome function) unimported proteins. (8) Increased protein burden in the cytosol. OMM, outer mitochondrial membrane; IMS, intermembrane space; IMM, inner mitochondrial membrane.
There are many examples in the literature showing that defects in the core protein import machinery cause the cytosolic accumulation of unimported proteins (Fig. 2, pathway 1). For instance, defect in the mitochondrial intermembrane space import and assembly machinery (MIA) leads to the cytosolic accumulation of various unprocessed mitochondrial precursors [24]. Clearly, the cytosol has a limited capacity to degrade precursor proteins when there is a delay or block in protein import, and exceeding this capacity causes cell stress.
The prototypical trigger of mPOS is the misfolding of IMM proteins such as Aac2, as discussed above (Fig. 2, pathway 2) [19]. The mechanism by which IMM protein misfolding causes mPOS is not completely understood. The effects could be multiple. For example, the misfolded Aac2A128P could ectopically interact with other proteins to disrupt the assembly and reduce the stability of protein complexes in the IMM. The biogenesis and/or stability of the TIM22 and TIM23 complexes are clearly reduced in cells expressing misfolded variants of Aac2 [19]. Aac2A128P could also have increased retention time in the TIM22 channel thereby reducing the import of other substrate proteins. Lastly, it is also possible that the misfolded and aggregated Aac2A128P partially permeabilises the IMM, which would reduce Δψm and therefore protein import efficiency.
A contribution of mtDNA mutations and several other mitochondrial processes to mPOS was established through the study of cells depleted of mtDNA, known as ρ° cells. ρ° cells have nonfunctional RCs and low Δψm [25]. The remaining Δψm is maintained in the absence of RC by reversed ATP4−(cytosol)/ADP3− (matrix) exchange, as long as a robust free F1-ATPase is present in the matrix to convert the imported ATP into ADP [26]. It has been long believed that a low level of Δψm is sufficient to drive protein import, given that ρ° cells are viable in some cell types. However, genetic studies suggest that in other cell types, the ρ° condition can be lethal due to reduced Δψm and protein import. The first indication of this came from studies of the aerobic yeast, Kluyveromyces lactis, published more than two decades ago [27–29]. In this yeast, elimination of mtDNA is lethal. However, specific mutations in the free F1-ATPase that facilitate the conversion of ATP to ADP in the matrix can sustain the viability of ρ° cells by stimulating the electrogenic ATP4− (cytosol)/ADP3− (matrix) exchange [30]. The critical role of F1-ATPase-dependent Δψm maintenance in the survival of ρ° cells were then recapitulated in cultured human cells [31, 32]. Thus, a Δψm-dependent but OXPHOS-independent mechanism of cell survival is evolutionarily conserved upon mutation of mtDNA.
Subsequent studies in S. cerevisiae supported the idea that protein import in vivo is sensitive to Δψm reduction, based on the synthetic lethality between defective import machinery and the ρ° condition that reduces Δψm. In contrast to K. lactis and most higher eukaryotes, S. cerevisiae can naturally survive the ρ° condition, due to an intrinsically robust ATP hydrolyzing activity associated with the wild-type free F1-ATPase in this yeast [33]. Several studies showed that cells with decreased protein import cannot tolerate the ρ°-condition. As such, mutants defective in Tim9 and Tim10, components of an intermembrane chaperone complexe in the TIM22 protein import pathway, are ρ°-lethal [34]. Mutants of TIM12, TIM18, TIM54 and TOM70, encoding components of the TIM22 and TOM complexes, are also ρ°-lethal [34–36]. These genetic data support the idea that reduced Δψm diminishes protein import in vivo to kill cells.
A formal connection between ρ°-lethality and mPOS was established in a study showing that manipulation of the cytosolic proteostatic network suppresses ρ°-lethality triggered by various mitochondrial stressors[19]. S. cerevisiae cells expressing misfolded Aac2 are ρ°-lethal [23]. Loss of protein homeostasis on the IMM by disrupting IMM protein quality control genes such as YME1, and PHB1 and PHB2 encoding subunits of the prohibitin complex, also leads to ρ°-lethality [36, 37]. As mentioned above, null mutant of TOM70 is ρ°-lethal [35]. It was found that many genes involved in cytosolic proteostasis that suppress aac2A128P-induced mPOS also suppress the ρ°-lethal phenotype of yme1Δ, phb1Δ, phb2Δ and tom70Δ [18, 19]. Several of these genes have been previously known to suppress the ρ°-lethal phenotype of a tim18 mutant[35]. These observations strongly support the idea that mtDNA damage (Fig. 2, pathway 4), low Δψm (Fig. 2, pathway 5), mutations in the core protein import machinery (Fig. 2, pathway 1), IMM protein misfolding (Fig. 2, pathway 2) and reduced protein quality control (Fig. 2, pathway 3) can converge to induce mPOS and ultimately, cell death.
Extra-mitochondrial processes can also cause mPOS. Efficient targeting of precursor proteins to mitochondria is critical for preventing mPOS. In yeast, nuclear-encoded mitochondrial proteins modified to impair targeting to mitochondria accumulate in the cytosol and confer significant toxicity when proteasome activity was sub-optimal for their degradation (Fig. 2, pathway 6) [24]. In addition, mPOS may also be caused by failure of cytosolic chaperones to hold and deliver nascent precursors to the import machinery (Fig. 2, pathway 7). The cytosolic Hsp70, Hsp90 and their cochaperones participate in the stabilization and delivery of precursors onto the OMM for import [38]. Interestingly, the ribosome–associated chaperones, Ssb1, Ssb2 and Zuo1, were found to suppress ρ°-lethality and mPOS-induced cell death in tim18Δ, yme1Δ, tom70Δ, atp1Δ and aac2A128P cells, and loss of these chaperones renders the cells ρ°-lethal[19, 35]. One possible explanation for these findings is that these proteins play a “holdase” function to prevent the folding of the mitochondrial precursor proteins in the cytosol. In human cells, recent studies showed that ubiquitin-like proteins (ubiquilins) play roles in handling pre-imported mitochondrial membrane proteins, preventing their aggregation and targeting them to proteasome for degradation [39]. Membrane proteins may be particularly susceptible to cytosolic aggregation due to high hydrophobicity. Indeed, genetic ablation of specific ubiquilins, including ubiquilin-2, results in cytosolic accumulation and aggregation of mitochondrial membrane proteins[39, 40].
There is strong evidence suggesting that the proteasome is critical for degrading unimported mitochondrial proteins. Early studies showed that proteasome inhibition results in cytosolic accumulation of mitochondrial proteins in neuronal cells [41, 42]. This is corroborated by the observation in yeast that loss of proteasomal chaperones (e.g., Blm10 and Poc4) and reduced expression of proteasomal genes in rpn4Δ cells are synthetically lethal with mitochondrial stress induced by yme1Δ [19]. Loss of the proteasomal chaperones, Poc3 and Poc4, leads to hypersensitivity to the cytosolic accumulation of mitochondrial precursors[24]. As previously proposed, there may exist a basal, innocuous level of mitochondrial protein import failure [43]. mPOS may develop when there is a shift in the balance between mitochondrial precursor import efficiency, precursor handling and delivery, and proteasome activity (Fig. 2, pathway 7).
Finally, cytosolic proteome stress and mPOS can synergize to kill cells (Fig. 2, pathway 8). As described above, defective proteasomal function synergizes with mitochondrial damage to induced cell death[19]. This may be at least partially explained by its effect on the stability of the cytosolic proteome in addition to its role in processing unimported mitochondrial proteins. Similarly, the Dunn group found that a dot6Δ tod6Δ double mutant is ρ°-lethal [44]. DOT6 and TOD6 are transcriptional repressors of genes required for ribosomal biogenesis and protein synthesis in the TOR-signaling pathway. Disruption of these two genes likely upregulates global protein synthesis and increase the proteostatic burden in the cytosol, which synergizes with the ρ° condition to enhance mPOS. The role of DOT6 and TOD6 in modulation of mPOS is also highlighted by their ability to suppress mPOS in aac2A128P, yme1Δ and tom70Δ cells when overexpressed [19].
Interplay between mPOS, UPRam, UPRmt and ISR
The cellular response to mitochondria-induced proteostatic stress is an emerging field. mPOS is defined as a mitochondria-induced cytosolic stress, which triggers multiple adaptive stress response pathways in yeast including reduction of global protein synthesis [19, 24]. mPOS also upregulates the levels of Nog2 and Gis2, two highly conserved proteins involved in ribosomal reconfiguration and cap-independent protein translation, respectively [19]. How these proteostatic activities suppress mPOS and ameliorate cell survival are yet to be worked out.
Additional response pathways triggered by mutations in the protein import machinery were discovered by the Chacinska group. Particularly interesting is the activation of proteasomal function, as part of a process named the unfolded protein response activated by mistargeting of proteins (UPRam) [24] (Fig. 1). It would be expected that mPOS and UPRam are intimately related. Failure to import mitochondrial proteins causes precursor overaccumulation stress (i.e. mPOS), and the mistargeted precursors then activate the proteasome to alleviate that stress (i.e. the UPRam). A closer look at these two yeast studies solidifies this relationship. Poc4 is part of a proteasome assembly chaperone complex[45], and was identified as a potent mPOS suppressor by Wang and Chen[19]. The Chacinska group found that Poc4 is upregulated when protein import is compromised, and that it is required for proteasomal activation in these settings [24]. These data suggest that, at least in yeast, mPOS may activate the UPRam, and that the UPRam is directed to alleviate mPOS. How Poc4 is upregulated in response to mPOS is yet to be investigated.
mPOS may also interact with UPRmt (Fig. 1). The UPRmt is a mitochondrial retrograde signaling pathway that transcriptionally regulates a set of genes designed to restore mitochondrial proteostasis (e.g., Hsp60, mtDNAJ and ClpP) [17, 46]. It is interesting that the UPRmt can be experimentally induced in Caenorhabditis elegans and cultured human cells by some of the same conditions that cause mPOS including mitochondrial protein misfolding, mtDNA depletion and OXPHOS disruption [47–51]. Additionally, the best-characterized UPRmt activation pathway depends on inefficient mitochondrial protein import of the transcription factor ATFS-1 in worms (or ATF5 in humans), resulting in its cytosolic accumulation and nuclear translocation. Is some level of mPOS required for the activation of the UPRmt, or is there a distinct mechanism to specifically reduce ATFS-1/ATF5 import in vivo? It is important to note that the UPRmt directs upregulation of nuclear-encoded mitochondrial proteins. As such, its activation would be predicted to exacerbate mPOS. An important area of research moving forward will be to understand the interactions between mPOS and the UPRmt under various stress conditions.
Finally, mPOS may intersect with the integrated stress response (ISR). A key player in ISR is eIF2α. When phosphorylated, eIF2α increases the translation of stress-activated transcription factors including ATF4, while inhibiting cap-dependent translation and global protein synthesis. Pharmacological inhibition of protein import in human cells (i.e. mPOS) induces phosphorylation of eIF2α and increases ATF4 expression [52]. Studies from multiple model systems demonstrate that diverse mitochondrial stress activates the ISR [53–58]. Interestingly, ISR activation precedes bioenergetic defects in human neurons treated with a RC inhibitor [59]. We propose that mPOS activates the ISR, an idea that requires further exploration.
Implications of mPOS for human diseases
The induction of mPOS by the clinically relevant aac2A128P mutation raises the possibility that mitochondrially induced cytosolic proteostatic stress may occur under pathophysiological conditions. A priori, defects in any step listed in Fig. 2 would trigger mPOS and potentially contribute to disease. On the other hand, reduced cytosolic anti-mPOS activity could also predispose to cell death and disease development. Wang and Chen showed that mPOS can be suppressed by manipulating cytosolic processes [19]. The mechanisms by which some of these pathways exert their anti-mPOS function are unclear. Nonetheless, several genes in the pathways are clinically relevant. For example, the Pbp1 protein is a ortholog of human ATXN2, a RNA-processing protein associated with Spinocerebellar Ataxia Type 2, amyotrophic lateral sclerosis, fronto-temporal lobar dementia, and levodopa-responsive Parkinson’s disease [60]. The potential implication of the mPOS model for mitochondria-induced pathologies could be broad. As hypothesized by Wang and Chen[19], the mPOS model may provide a conceptual framework to understand the enigmatic association between mitochondrial dysfunction and cytosolic proteostatic stress in mitochondria-induced diseases. Here, we will focus on classic mitochondrial diseases and ageing-related neurodegenerative disorders.
mPOS and mitochondrial disease
Classic mitochondrial diseases are primarily caused by energy deficiency and are often accompanied by oxidative stress. However, potential non-bioenergetic factors in the pathogenesis of these diseases are understudied. Here we discuss tissue culture, mouse and human patient data that suggest a possible involvement of mPOS in mitochondrial diseases.
It appears that OXPHOS defects triggers proteostatic stress in cultured human cells. Pharmacological RC inhibition causes cytosolic protein aggregation in cell lines [61], cultured primary neurons[62], and in mouse and human neurons in vivo [63–65]. Increased expression of a cytosolic chaperone (HSP70) that participates in the stabilization and targeting of mitochondrial precursors robustly suppresses aggregate formation and cell death caused by RC-inhibition [66]. Other studies showed that a wide range of OXPHOS stressors trigger cellular responses to improve cytosolic proteostasis [52, 67–69]. This may be interpreted as a response to mPOS.
More direct evidence for mPOS in mitochondrial disease came from studies in mice [70, 71]. For example, Johnson et al. showed that the neurological symptoms in a mouse model for Leigh Syndrome, which is deficient in complex I subunit Ndufs4 (NADH dehydrogenase (ubiquinone) Fe-S protein 4), were partially alleviated by rapamycin treatment [70]. Rapamycin binds and inhibits mammalian TOR (mTOR), thereby partially inhibiting protein synthesis. Interestingly, brain lesions in the complex I-deficient mice were prevented and neurological symptoms and premature death were delayed by rapamycin treatment in the absence of a corresponding rescue in mitochondrial respiration. This suggests that at least a portion of the pathomechanism is cytosolic and independent of respiratory deficiency.
Studies from humans also implicate mPOS in mitochondrial diseases. A recent transcriptome analysis of skeletal muscle from a heterogeneous group of 12 mitochondrial disease patients with confirmed RC deficiencies revealed upregulation of the proteasome and RNA processing proteins, and the downregulation of cytosolic ribosomal proteins [72]. Alterations to these pathways have been shown to suppress mPOS in yeast [18, 19, 24]. It is important to note that cytosolic aggregates are not typically detected in mitochondrial disease patients. This suggests that if mPOS occurs, either it is effectively contained in vivo, or it contributes to disease without the formation of detectable aggregates.
How mtDNA mutations affect cytosolic proteostasis is not well understood. It is conventionally thought that ATP depletion and oxidative stress decrease ATP-dependent protein degradation and increase protein damage in the cytosol, respectively. The mPOS model would provide a direct link between mitochondrial dysfunction and cytosolic proteostasis. It can be speculated that blocking the electron transport chain may reduce Δψm and potentialize mPOS, consistent with the observation that inhibition of the electron transport chain synergizes with misfolded Aac2 to kill yeast cells [22]. In addition, deletion of genes encoding OXPHOS components affects the assembly of respiratory complexes. This may lead to the accumulation and misfolding of their unassembled partner subunits, including those that are nuclear-encoded. For example, knockout of individual accessory subunits of complex I decreases the stability of other complex I subunits [73]. Excessive protein misfolding may collapse the proteostatic network on the IMM to affect the biogenesis, stability and functionality of protein complexes including those required for protein import. In summary, accumulating evidence suggests that mPOS is present in human cells with mtDNA mutations and respiratory deficiency. Its potential implications for pathogenesis in mitochondrial diseases need further investigation.
Neurodegeneration caused by mutations in the core mitochondrial protein import machinery
Studies in yeast strongly suggest that mutations in mitochondrial protein import complex subunits cause ρ°-lethality via mPOS [34–36]. It is therefore reasonable to speculate that mPOS may contribute to diseases caused by mutations affecting the core protein import machinery. The TIM23 complex transports precursor proteins across the IMM into the matrix. Heterozygous TIM23 knockout mice develop neurodegenerative symptoms and have a shortened lifespan [74], supporting a link between diminished protein import and neurodegeneration. In humans, defective assembly of the DDP1/TIMM8a-TIMM13 complex, which facilitates the transport of TIM22 substrates [75], causes deafness dystonia syndrome [76, 77]. It remains to be investigated whether mPOS contributes to the pathogenesis of this disease.
Neurodegenerative diseases affecting IMM proteostasis
IMM stress is a significant inducer of mPOS in yeast (Fig. 2). A hypersusceptibility of the nervous system to IMM stress is strongly supported by the discoveries of several neuropathogenic mutations affecting protein quality control on the membrane. IMM protein quality is maintained by a network of chaperones and proteases that hold, fold, and degrade misfolded proteins to prevent their toxic accumulation [78, 79]. Mutations in IMM quality control chaperones and proteases cause or are implicated in many neurodegenerative diseases, including Parkinson’s disease [80], amyotrophic lateral sclerosis [81], spastic ataxia-neuropathy syndrome [82], spastic paraplegia 7 [83], and spinocerebellar ataxia [12]. Does mPOS contribute to neurodegeneration in these diseases? Altered cytosolic proteostasis as a result of IMM stress is not unprecedented. The Langer group has shown that neuron-specific depletion of prohibitin, a chaperone complex on the IMM, leads to severe neurodegeneration [84]. Interestingly, prohibitin depletion in these mice also causes hyperphosphorylation and cytosolic aggregation of the microtubule-associated protein tau in neurons, a hallmark of Alzheimer’s disease. The mutant mice exhibit behavioral impairment, cognitive deficiencies, and death usually by 5 months old. Histopathologically, cytosolic tau aggregates appear concurrently with neuronal damage at 6 weeks old. Both pathological markers occur long before OXPHOS defects and oxidative stress can be detected [84]. These observations suggest that mitochondrial damage can cause neurodegeneration independent of OXPHOS deficiency and oxidative stress. In a more recent study, Kondadi and coworkers showed that depletion of AFG3L2, which codes for a subunit in the IMM quality control m-AAA protease, also leads to tau hyperphosphorylation and cytosolic aggregation in mice[85]. Mutations in AFG3L2 cause spinocerebellar ataxia 28[12]. This study thus provides a clinically relevant example for a causative role of IMM stress in affecting cytosolic proteostasis.
Numerous neuropathogenic mutations have been found in IMM proteins that have no apparent link with membrane proteostasis. For instance, missense mutations in CHCHD2, CHCHD10, and ATAD3A cause or associate with Parkinson’s disease [86], frontotemporal dementia/amyotrophic lateral sclerosis [87, 88], and dominant hereditary spastic paraplegia[14], respectively. While there is no evidence that these mutant proteins are misfolded or directly affect protein homeostasis on the IMM, the detection of cytosolic stress and/or a cytosolic stress response suggests that mPOS may occur. For example, CHCHD2 mutations are associated with neuronal cytosolic protein aggregation in Parkinson’s disease patients [89]. Pathogenic CHCHD10 mutations cause cytosolic protein aggregation in multiple model organisms (discussed in detail below) [90]. Furthermore, fibroblasts from ATAD3A-induced hereditary spastic paraplegia patients exhibit reduced mTOR signaling, possibly indicating a response to increased proteostatic stress in the cytosol [14]. Further work is required to evaluate whether mPOS plays a role in the pathogenesis of these diseases.
Common ageing-associated neurodegenerative diseases
Despite obvious differences in pathology, many ageing-related neurodegenerative diseases share two key hallmarks: dysfunctional mitochondria and cytosolic protein aggregation [91, 92]. Both hallmarks are widely believed to contribute to neurodegeneration, but whether and how these two seemingly unrelated pathways interact remain unclear. Often these two pathogenic pathways have been investigated independently. Given the consistency of their concurrence within and across neurodegenerative diseases, there is the possibility that mitochondrial dysfunction and cytosolic protein aggregation are related by a single pathogenic mechanism. There is significant evidence to suggest that protein aggregates can cause mitochondrial dysfunction [93–98]. For example, mouse models expressing pathogenic aggregate-prone proteins, including mutant SOD1, TDP-43, α-synuclein and Huntingtin, tend to develop dysfunctional mitochondria [95, 99–101]. Here, we offer mPOS as a novel mechanism for the interaction between the two neurodegeneration pathways.
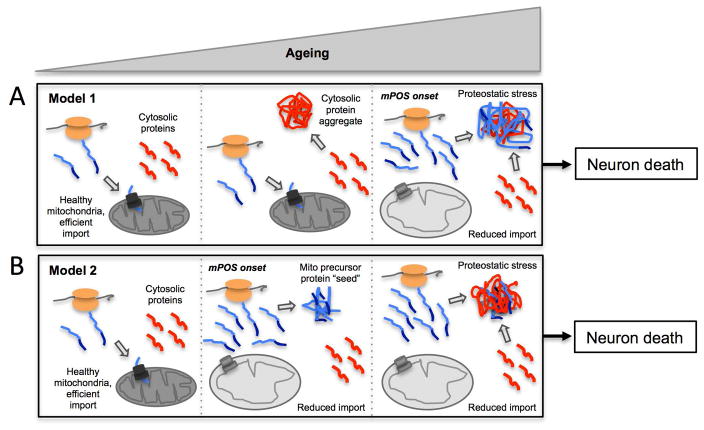
We propose that there are at least two feasible routes by which mPOS may contribute to pathological protein aggregation in the cytosol of neurons. First, protein aggregates of non-mitochondrial proteins may precede mitochondrial dysfunction, and unimported mitochondrial precursors add to the proteostatic burden once mitochondrial degeneration ensues during ageing (Fig. 3A). mPOS would therefore contribute to disease progression, not onset. Second, unimported precursors may serve as ‘seeding structures’ that challenge the proteostatic network and induce secondary aggregation of cytosolic proteins (Fig. 3B). Importantly, this model may not require severe defects in mitochondrial protein import. A basal level of precursor accumulation that would normally be tolerated in young cells may be amplified during ageing to seed cytosolic aggregates in neurons “preconditioned” by the presence (or increased concentration) of aggregation-prone proteins. Here, mPOS would be implicated in disease onset and progression. Neither of these models are mutually exclusive with aggregate-induced mitochondrial dysfunction. If aggregates cause mitochondrial dysfunction, then both models would portend a feed-forward loop that would cause protein aggregation and mitochondrial dysfunction to accelerate, presumably until neuronal death.
Fig. 3. Models by which mPOS may reconcile mitochondrial dysfunction and cytosolic protein aggregation in neurodegeneration.
(A) Model 1, cytosolic aggregates precede mitochondrial damage and mPOS during ageing. (B) Model 2, unimported proteins induced by mPOS provide a seeding structure with which cytosolic proteins co-aggregate during ageing. Blue, mitochondrial precursor proteins; red, cytosol-derived protein aggregates; orange, ribosomes.
An mPOS contribution to neurodegenerative diseases co-hallmarked by mitochondrial dysfunction and cytosolic protein aggregation is supported by numerous studies. Several mitochondrial proteins co-aggregate with synthetic amyloidogenic proteins expressed in human cells in the absence of apparent mitochondrial stress [102]. These same mitochondrial proteins, as well as many OXPHOS components, are inherently supersaturated in the cellular environment; that is, they are expressed at concentrations higher than their solubility[103]. Such proteins are referred to as ‘metastable’, and the metastable proteome is thought to drive protein aggregation in neurodegenerative diseases [104]. Therefore, mitochondrial proteins are implicated in cytosolic aggregate formation in a disease-non-specific manner. Below is a description of additional evidence that potentially implicates mPOS in three neurodegenerative diseases co-hallmarked by mitochondrial dysfunction and cytosolic protein aggregation: Parkinson’s disease (PD), Alzheimer’s disease (AD), and amyotrophic lateral sclerosis (ALS). These diseases share other key characteristics that make the prospect of a pathogenic mPOS contribution more likely and more exciting: the etiology of the majority of patients is unknown and there is a disease dependence on ageing
A causative contribution of mitochondrial dysfunction has been established in PD [105], as evidenced by two main observations: (1) mutations in mitochondrial proteins cause familial PD [80, 106, 107], and (2) environmental toxins that inhibit complex I can cause PD [64, 65, 108]. These familial and environment-induced PD patients often share a key pathological hallmark with sporadic PD patients: intraneuronal cytosolic α-synuclein protein aggregates known as Lewy bodies (LBs). LBs are also thought to cause PD[109], but consideration of mitochondria-induced PD with concurrent LB pathology suggests mitochondrial dysfunction can cause cytosolic protein aggregation in these patients. One possible way mitochondrial dysfunction could cause cytosolic protein aggregation is through mPOS. Consistent with this, the two mutant mitochondrial proteins that cause familial PD, PTEN-induced putative kinase 1 (PINK1) [106] and Htra2 [80, 107], can reasonably be expected to induce mPOS. PINK1 plays a role in mitophagy important for mitochondrial quality control [11]. Reduced mitochondrial quality (e.g., membrane depolarization) in the absence of PINK function may decrease protein import. Loss of PINK1 in mice has been clearly shown to cause a progressive reduction in mitochondrial protein import [110]. In addition, Htra2 is an IMM-associated protease, and its deletion in mice results in Parkinsonism and accumulation of misfolded IMM proteins [58], a known inducer of mPOS. Consistently, transcriptome analysis from Htra2 knockout mice indicated a general response to rescue cytosolic proteostasis, including an upregulation of HSPs and activation of the ISR, which is an adaptive responses to cytosolic proteostatic stresses like mPOS.
There is accumulating evidence that mPOS may also contribute to sporadic PD. Reduced activity of complex I is consistently observed in sporadic PD brains [111], consistent with complex I inhibition-induced PD noted above. Furthermore, it is well established that complex I inhibition induces cytosolic α-synuclein aggregation [61–63], which we propose may occur through mPOS (see above for a discussion of possible mechanisms). Support for a potential involvement of mPOS in PD comes from the detection of mitochondrial proteins in LBs isolated from the brains of PD patients. Using immunohistochemistry, mitochondrial proteins have been shown to colocalize with α-synuclein in intraneuronal cytoplasmic inclusions in sporadic PD patient brains [112–114]. A proteomic analysis of laser capture microdissected LBs from human brains identified additional mitochondrial proteins[115]. This set of mitochondrial components had a clear bias towards IMM proteins, and some of which overlap with the aggregation-prone metastable proteome [102, 103]. It should be noted that one of these studies showed an association of LBs with whole mitochondria [112]. The reason for such an association is unknown. Nevertheless, whether mitochondrial dysfunction contributes to cytosolic α-synuclein aggregation in PD via the mPOS mechanism warrants further investigation.
The link between mitochondrial dysfunction and AD is less understood compared with PD. The neuropathological hallmarks of AD include neuron loss, extracellular deposits of amyloid beta and neurofibrillary tangles (NFTs). NFTs are intraneuronal cytosolic protein aggregates marked by the presence of hyperphosphorylated, microtubule-associated protein, tau. Mitochondrial dysfunction is also observed in AD, which occurs early in disease progression [16, 116–119]. Both mitochondrial dysfunction and NFTs are thought to contribute to AD pathogenesis, but the relationship between the two is unclear. Multiple lines of evidence suggest that dysfunctional mitochondria may contribute to NFTs, possibly via mPOS. First, mPOS-related mitochondrial dysfunction can induce tau hyperphosphorylation and cytosolic aggregation (i.e. NFTs). We present three examples of this: (1) as discussed above, IMM proteostatic stress induced by defective IMM protein quality control can induce intraneuronal tau aggregation in two different mouse models [84, 85]; (2) inhibition of complex I induces cytosolic tau hyperphosphorylation and aggregation in rats and cultured neurons [120, 121]; and (3) the mitochondrial membrane uncoupler CCCP, which dissipates Δψm, also causes tau pathology in cultured neurons [121]. Second, genetic variants in the core protein import machinery, specifically in the TOMM40 gene, predict AD age of onset [122]. This suggests that protein import efficiency contributes to disease onset, possibly through mPOS. Third, the metastable mitochondrial proteins discussed above, specifically IMM OXPHOS proteins, are downregulated in AD patient brains [123]. Ciryam et al. interpreted this result as a compensatory response to prevent aggregation of these susceptible metastable mitochondrial proteins, which would be consistent with mPOS occurring in AD neurons. Fourth, mitochondrial proteins are found in NFTs. An early study raised antibodies against NFTs and found that the anti-NFT antibodies were specific for the α-subunit of the F1Fo-ATP synthase [124]. Proteomic analysis of laser capture microdissected NFTs confirmed the presence of the α-subunit of ATP synthase, and detected other ATP synthase subunits, several IMM carrier proteins, a complex IV subunit and a metabolic matrix protein [125]. Taken together, the evidence warrants the study of mPOS as a potential neuronal stressor in AD.
Finally, it is possible that mPOS contributes to ALS as well, although the evidence for this is also less strong than for PD. ALS is marked by coincident mitochondrial dysfunction and cytosolic protein aggregation, both of which are thought to be causative [126, 127]. These two hallmarks are observed in both familial and sporadic ALS. Two genes linked to familial ALS may be related to mPOS. (1) CHCHD10 is an IMM protein involved in cristae morphology maintenance, and mutations in the gene cause familial ALS [87, 88]. Interestingly, ALS-linked mutations in CHCHD10 were shown to induce cytosolic protein aggregation of TDP-43 in multiple model systems [90]. TDP-43 is a nuclear transcription factor that commonly mislocalizes to and aggregates in the cytosol of sporadic and familial ALS neurons [127]. It is tempting to speculate that CHCHD10 mutations induce mPOS and the resulting unimported mitochondrial precursors seed TDP-43 aggregation in these patients. (2) Mutations in ubiquilin-2 cause ALS [128]. Ubiquilin-2, along with ubiquilin-1 and -4, functions as a chaperone to keep mitochondrial membrane proteins unfolded and soluble before import [39]. When pre-imported membrane proteins linger in the cytosol too long, these ubiquilins deliver the precursor proteins to the proteasome for degradation. If pathogenic ubiquilin-2 mutations cause mitochondrial membrane precursor proteins to be unchaperoned in the cytosol and unable to be delivered to the proteasome, this would likely cause mPOS.
There is little evidence so far to suggest that mPOS contributes to sporadic ALS, other than concurrent mitochondrial dysfunction and cytosolic protein aggregation. However, this could be a fruitful avenue of exploration, especially considering the connection between mPOS and familial ALS subtypes.
Concluding remarks
The present review demonstrates that the mPOS model can rationalize many observations that suggest a causative link of mitochondrial dysfunction to cytosolic proteostatic stress. Although some evidence suggests that mPOS occurs in the classic mitochondrial diseases, its contribution to pathogenesis needs to be investigated. It appears that strong evidence exists to support a role of mPOS in contributing to the development of ageing-dependent neurodegenerative diseases. Wang et al. captured an ageing-dependent trait that accelerates mPOS-induced cell degeneration in yeast [18]. In mice, ageing-dependent deficiency in mitochondrial protein import has been clearly documented [129]. The interplay between ageing and mPOS adds another dimension for investigating the pathogenic mechanism of neurodegenerative diseases. Further studies are certainly warranted. Looking forward, these studies may help to identify key drivers of neurodegeneration during ageing. Study of anti-mPOS pathways will also be an important field of investigation. Elucidation of these pathways may offer novel mechanisms of disease and new therapeutic targets for treating neurodegeneration.
Acknowledgments
This work was supported by the National Institutes of Health grants AG023731 and AG047400.
References
- 1.Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schonfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci USA. 2003;100:13207–12. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44:D1251–7. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vafai SB, Mootha VK. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491:374–83. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- 4.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–44. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dudek J, Rehling P, van der Laan M. Mitochondrial protein import: common principles and physiological networks. Biochim Biophys Acta. 2013;1833:274–85. doi: 10.1016/j.bbamcr.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 6.Wiedemann N, Pfanner N. Mitochondrial machineries for protein import and assembly. Annu Rev Biochem. 2017;86:685–714. doi: 10.1146/annurev-biochem-060815-014352. [DOI] [PubMed] [Google Scholar]
- 7.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–59. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rossignol R, Faustin B, Rocher C, Malgat M, Mazat JP, Letellier T. Mitochondrial threshold effects. Biochem J. 2003;370:751–62. doi: 10.1042/BJ20021594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lax NZ, Gorman GS, Turnbull DM. Review: Central nervous system involvement in mitochondrial disease. Neuropathol Appl Neurobiol. 2017;43:102–118. doi: 10.1111/nan.12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 2015;85:257–73. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Bella D, Lazzaro F, Brusco A, Plumari M, Battaglia G, Pastore A, Finardi A, Cagnoli C, Tempia F, Frontali M, Veneziano L, Sacco T, Boda E, Brussino A, Bonn F, Castellotti B, Baratta S, Mariotti C, Gellera C, Fracasso V, Magri S, Langer T, Plevani P, Di Donato S, Muzi-Falconi M, Taroni F. Mutations in the mitochondrial protease gene AFG3L2 cause dominant hereditary ataxia SCA28. Nat Genet. 2010;42:313–21. doi: 10.1038/ng.544. [DOI] [PubMed] [Google Scholar]
- 13.Chen H, Chan DC. Critical dependence of neurons on mitochondrial dynamics. Curr Opin Cell Biol. 2006;18:453–9. doi: 10.1016/j.ceb.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 14.Cooper HM, Yang Y, Ylikallio E, Khairullin R, Woldegebriel R, Lin KL, Euro L, Palin E, Wolf A, Trokovic R, Isohanni P, Kaakkola S, Auranen M, Lonnqvist T, Wanrooij S, Tyynismaa H. ATPase-deficient mitochondrial inner membrane protein ATAD3A disturbs mitochondrial dynamics in dominant hereditary spastic paraplegia. Hum Mol Genet. 2017;26:1432–1443. doi: 10.1093/hmg/ddx042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen JJ, Durr A, Cournu-Rebeix I, Georgopoulos C, Ang D, Nielsen MN, Davoine CS, Brice A, Fontaine B, Gregersen N, Bross P. Hereditary spastic paraplegia SPG13 is associated with a mutation in the gene encoding the mitochondrial chaperonin Hsp60. Am J Hum Genet. 2002;70:1328–32. doi: 10.1086/339935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schon EA, Przedborski S. Mitochondria: the next (neurode)generation. Neuron. 2011;70:1033–53. doi: 10.1016/j.neuron.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pellegrino MW, Nargund AM, Haynes CM. Signaling the mitochondrial unfolded protein response. Biochim Biophys Acta. 2013;1833:410–6. doi: 10.1016/j.bbamcr.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang XW, Zuo XM, Kucejova B, Chen XJ. Reduced cytosolic protein synthesis suppresses mitochondrial degeneration. Nature Cell Biology. 2008;10:1090–1097. doi: 10.1038/ncb1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Chen XJ. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature. 2015;524:481–484. doi: 10.1038/nature14859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen XJ. Induction of an unregulated channel by mutations in adenine nucleotide translocase suggests an explanation for human ophthalmoplegia. Hum Mol Genet. 2002;11:1835–43. doi: 10.1093/hmg/11.16.1835. [DOI] [PubMed] [Google Scholar]
- 21.Kaukonen J, Juselius JK, Tiranti V, Kyttala A, Zeviani M, Comi GP, Keranen S, Peltonen L, Suomalainen A. Role of adenine nucleotide translocator 1 in mtDNA maintenance. Science. 2000;289:782–5. doi: 10.1126/science.289.5480.782. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Wang X, Chen XJ. Misfolding of mutant adenine nucleotide translocase in yeast supports a novel mechanism of Ant1-induced muscle diseases. Mol Biol Cell. 2015;26:1985–1994. doi: 10.1091/mbc.E15-01-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Salinas K, Zuo X, Kucejova B, Chen XJ. Dominant membrane uncoupling by mutant adenine nucleotide translocase in mitochondrial diseases. Hum Mol Genet. 2008;17:4036–44. doi: 10.1093/hmg/ddn306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wrobel L, Topf U, Bragoszewski P, Wiese S, Sztolsztener ME, Oeljeklaus S, Varabyova A, Lirski M, Chroscicki P, Mroczek S, Januszewicz E, Dziembowski A, Koblowska M, Warscheid B, Chacinska A. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature. 2015;524:485–8. doi: 10.1038/nature14951. [DOI] [PubMed] [Google Scholar]
- 25.Appleby RD, Porteous WK, Hughes G, James AM, Shannon D, Wei YH, Murphy MP. Quantitation and origin of the mitochondrial membrane potential in human cells lacking mitochondrial DNA. Eur J Biochem. 1999;262:108–16. doi: 10.1046/j.1432-1327.1999.00350.x. [DOI] [PubMed] [Google Scholar]
- 26.Chen XJ, Clark-Walker GD. The petite mutation in yeasts: 50 years on. Int Rev Cytol. 2000;194:197–238. doi: 10.1016/s0074-7696(08)62397-9. [DOI] [PubMed] [Google Scholar]
- 27.Chen XJ, Clark-Walker GD. Mutations in MGI genes convert Kluyveromyces lactis into a petite-positive yeast. Genetics. 1993;133:517–25. doi: 10.1093/genetics/133.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen XJ, Clark-Walker GD. Specific mutations in alpha- and gamma-subunits of F1-ATPase affect mitochondrial genome integrity in the petite-negative yeast Kluyveromyces lactis. EMBO J. 1995;14:3277–86. doi: 10.1002/j.1460-2075.1995.tb07331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen XJ, Clark-Walker GD. The mitochondrial genome integrity gene, MGI1, of Kluyveromyces lactis encodes the beta-subunit of F1-ATPase. Genetics. 1996;144:1445–54. doi: 10.1093/genetics/144.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark-Walker GD, Hansbro PM, Gibson F, Chen XJ. Mutant residues suppressing ρ°-lethality in Kluyveromyces lactis occur at contact sites between subunits of F1-ATPase. Biochim Biophys Acta. 2000;1478:125–37. doi: 10.1016/s0167-4838(00)00003-0. [DOI] [PubMed] [Google Scholar]
- 31.Buchet K, Godinot C. Functional F1-ATPase essential in maintaining growth and membrane potential of human mitochondrial DNA-depleted ρ° cells. J Biol Chem. 1998;273:22983–9. doi: 10.1074/jbc.273.36.22983. [DOI] [PubMed] [Google Scholar]
- 32.Chen WW, Birsoy K, Mihaylova MM, Snitkin H, Stasinski I, Yucel B, Bayraktar EC, Carette JE, Clish CB, Brummelkamp TR, Sabatini DD, Sabatini DM. Inhibition of ATPIF1 ameliorates severe mitochondrial respiratory chain dysfunction in mammalian cells. Cell Rep. 2014;7:27–34. doi: 10.1016/j.celrep.2014.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen XJ, Clark-Walker GD. Alpha and beta subunits of F1-ATPase are required for survival of petite mutants in Saccharomyces cerevisiae. Mol Gen Genet. 1999;262:898–908. doi: 10.1007/s004380051156. [DOI] [PubMed] [Google Scholar]
- 34.Senapin S, Chen XJ, Clark-Walker GD. Transcription of TIM9, a new factor required for the petite-positive phenotype of Saccharomyces cerevisiae, is defective in spt7 mutants. Curr Genet. 2003;44:202–10. doi: 10.1007/s00294-003-0437-9. [DOI] [PubMed] [Google Scholar]
- 35.Dunn CD, Jensen RE. Suppression of a defect in mitochondrial protein import identifies cytosolic proteins required for viability of yeast cells lacking mitochondrial DNA. Genetics. 2003;165:35–45. doi: 10.1093/genetics/165.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dunn CD, Lee MS, Spencer FA, Jensen RE. A genomewide screen for petite-negative yeast strains yields a new subunit of the i-AAA protease complex. Mol Biol Cell. 2006;17:213–26. doi: 10.1091/mbc.E05-06-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thorsness PE, White KH, Fox TD. Inactivation of YME1, a member of the ftsH-SEC18-PAS1-CDC48 family of putative ATPase-encoding genes, causes increased escape of DNA from mitochondria in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:5418–26. doi: 10.1128/mcb.13.9.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoseini H, Pandey S, Jores T, Schmitt A, Franz-Wachtel M, Macek B, Buchner J, Dimmer KS, Rapaport D. The cytosolic cochaperone Sti1 is relevant for mitochondrial biogenesis and morphology. FEBS J. 2016;283:3338–52. doi: 10.1111/febs.13813. [DOI] [PubMed] [Google Scholar]
- 39.Itakura E, Zavodszky E, Shao S, Wohlever ML, Keenan RJ, Hegde RS. Ubiquilins chaperone and triage mitochondrial membrane proteins for degradation. Mol Cell. 2016;63:21–33. doi: 10.1016/j.molcel.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whiteley AM, Prado MA, Peng I, Abbas AR, Haley B, Paulo JA, Reichelt M, Katakam A, Sagolla M, Modrusan Z, Lee DY, Roose-Girma M, Kirkpatrick DS, McKenzie BS, Gygi SP, Finley D, Brown EJ. Ubiquilin1 promotes antigen-receptor mediated proliferation by eliminating mislocalized mitochondrial proteins. eLife. 2017:6. doi: 10.7554/eLife.26435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X, Zhang Y, Xie P, Piao J, Hu Y, Chang M, Liu T, Hu L. Proteomic characterization of an isolated fraction of synthetic proteasome inhibitor (PSI)-induced inclusions in PC12 cells might offer clues to aggresomes as a cellular defensive response against proteasome inhibition by PSI. BMC Neurosci. 2010;11:95. doi: 10.1186/1471-2202-11-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muqit MM, Abou-Sleiman PM, Saurin AT, Harvey K, Gandhi S, Deas E, Eaton S, Payne Smith MD, Venner K, Matilla A, Healy DG, Gilks WP, Lees AJ, Holton J, Revesz T, Parker PJ, Harvey RJ, Wood NW, Latchman DS. Altered cleavage and localization of PINK1 to aggresomes in the presence of proteasomal stress. J Neurochem. 2006;98:156–69. doi: 10.1111/j.1471-4159.2006.03845.x. [DOI] [PubMed] [Google Scholar]
- 43.Bragoszewski P, Turek M, Chacinska A. Control of mitochondrial biogenesis and function by the ubiquitin-proteasome system. Open Biol. 2017:7. doi: 10.1098/rsob.170007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akdogan E, Tardu M, Garipler G, Baytek G, Kavakli IH, Dunn CD. Reduced glucose sensation can increase the fitness of Saccharomyces cerevisiae lacking mitochondrial DNA. PLoS One. 2016;11:e0146511. doi: 10.1371/journal.pone.0146511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Le Tallec B, Barrault MB, Courbeyrette R, Guerois R, Marsolier-Kergoat MC, Peyroche A. 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol Cell. 2007;27:660–74. doi: 10.1016/j.molcel.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 46.Schulz AM, Haynes CM. UPR(mt)-mediated cytoprotection and organismal aging. Biochim Biophys Acta. 2015;1847:1448–56. doi: 10.1016/j.bbabio.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21:4411–9. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aldridge JE, Horibe T, Hoogenraad NJ. Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PLoS ONE. 2007;2:e874. doi: 10.1371/journal.pone.0000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martinus RD, Garth GP, Webster TL, Cartwright P, Naylor DJ, Hoj PB, Hoogenraad NJ. Selective induction of mitochondrial chaperones in response to loss of the mitochondrial genome. Eur J Biochem. 1996;240:98–103. doi: 10.1111/j.1432-1033.1996.0098h.x. [DOI] [PubMed] [Google Scholar]
- 50.Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337:587–90. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fiorese CJ, Schulz AM, Lin YF, Rosin N, Pellegrino MW, Haynes CM. The transcription factor ATF5 mediates a mammalian mitochondrial UPR. Curr Biol. 2016;26:2037–43. doi: 10.1016/j.cub.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quiros PM, Prado MA, Zamboni N, D’Amico D, Williams RW, Finley D, Gygi SP, Auwerx J. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J Cell Biol. 2017;216:2027–2045. doi: 10.1083/jcb.201702058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baker BM, Nargund AM, Sun T, Haynes CM. Protective coupling of mitochondrial function and protein synthesis via the eIF2alpha kinase GCN-2. PLoS Genet. 2012;8:e1002760. doi: 10.1371/journal.pgen.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khan NA, Nikkanen J, Yatsuga S, Jackson C, Wang L, Pradhan S, Kivela R, Pessia A, Velagapudi V, Suomalainen A. mTORC1 regulates mitochondrial integrated stress response and mitochondrial myopathy progression. Cell Metab. 2017;26:419–428 e5. doi: 10.1016/j.cmet.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 55.Michel S, Canonne M, Arnould T, Renard P. Inhibition of mitochondrial genome expression triggers the activation of CHOP-10 by a cell signaling dependent on the integrated stress response but not the mitochondrial unfolded protein response. Mitochondrion. 2015;21:58–68. doi: 10.1016/j.mito.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 56.Bao XR, Ong SE, Goldberger O, Peng J, Sharma R, Thompson DA, Vafai SB, Cox AG, Marutani E, Ichinose F, Goessling W, Regev A, Carr SA, Clish CB, Mootha VK. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. eLife. 2016:5. doi: 10.7554/eLife.10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsuyama T, Tsubouchi A, Usui T, Imamura H, Uemura T. Mitochondrial dysfunction induces dendritic loss via eIF2alpha phosphorylation. J Cell Biol. 2017;216:815–834. doi: 10.1083/jcb.201604065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moisoi N, Klupsch K, Fedele V, East P, Sharma S, Renton A, Plun-Favreau H, Edwards RE, Teismann P, Esposti MD, Morrison AD, Wood NW, Downward J, Martins LM. Mitochondrial dysfunction triggered by loss of HtrA2 results in the activation of a brain-specific transcriptional stress response. Cell Death Differ. 2009;16:449–64. doi: 10.1038/cdd.2008.166. [DOI] [PubMed] [Google Scholar]
- 59.Krug AK, Gutbier S, Zhao L, Poltl D, Kullmann C, Ivanova V, Forster S, Jagtap S, Meiser J, Leparc G, Schildknecht S, Adam M, Hiller K, Farhan H, Brunner T, Hartung T, Sachinidis A, Leist M. Transcriptional and metabolic adaptation of human neurons to the mitochondrial toxicant MPP+ Cell Death Dis. 2014;5:e1222. doi: 10.1038/cddis.2014.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Auburger G, Sen NE, Meierhofer D, Basak AN, Gitler AD. Efficient prevention of neurodegenerative diseases by depletion of starvation response factor Ataxin-2. Trends Neurosci. 2017;40:507–516. doi: 10.1016/j.tins.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 61.Lee HJ, Shin SY, Choi C, Lee YH, Lee SJ. Formation and removal of alpha-synuclein aggregates in cells exposed to mitochondrial inhibitors. J Biol Chem. 2002;277:5411–7. doi: 10.1074/jbc.M105326200. [DOI] [PubMed] [Google Scholar]
- 62.Chaves RS, Melo TQ, Martins SA, Ferrari MF. Protein aggregation containing beta-amyloid, alpha-synuclein and hyperphosphorylated tau in cultured cells of hippocampus, substantia nigra and locus coeruleus after rotenone exposure. BMC Neurosci. 2010;11:144. doi: 10.1186/1471-2202-11-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, Greenamyre JT. Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat Neurosci. 2000;3:1301–6. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- 64.Singer TP, Castagnoli N, Jr, Ramsay RR, Trevor AJ. Biochemical events in the development of parkinsonism induced by 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine. J Neurochem. 1987;49:1–8. doi: 10.1111/j.1471-4159.1987.tb03384.x. [DOI] [PubMed] [Google Scholar]
- 65.Langston JW, Ballard P, Tetrud JW, Irwin I. Chronic Parkinsonism in humans due to a product of meperidine-analog synthesis. Science. 1983;219:979–80. doi: 10.1126/science.6823561. [DOI] [PubMed] [Google Scholar]
- 66.Zhou Y, Gu G, Goodlett DR, Zhang T, Pan C, Montine TJ, Montine KS, Aebersold RH, Zhang J. Analysis of alpha-synuclein-associated proteins by quantitative proteomics. J Biol Chem. 2004;279:39155–64. doi: 10.1074/jbc.M405456200. [DOI] [PubMed] [Google Scholar]
- 67.Mineri R, Pavelka N, Fernandez-Vizarra E, Ricciardi-Castagnoli P, Zeviani M, Tiranti V. How do human cells react to the absence of mitochondrial DNA? PLoS One. 2009;4:e5713. doi: 10.1371/journal.pone.0005713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Waveren C, Sun Y, Cheung HS, Moraes CT. Oxidative phosphorylation dysfunction modulates expression of extracellular matrix--remodeling genes and invasion. Carcinogenesis. 2006;27:409–18. doi: 10.1093/carcin/bgi242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Behan A, Doyle S, Farrell M. Adaptive responses to mitochondrial dysfunction in the ρ° Namalwa cell. Mitochondrion. 2005;5:173–93. doi: 10.1016/j.mito.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 70.Johnson SC, Yanos ME, Kayser EB, Quintana A, Sangesland M, Castanza A, Uhde L, Hui J, Wall VZ, Gagnidze A, Oh K, Wasko BM, Ramos FJ, Palmiter RD, Rabinovitch PS, Morgan PG, Sedensky MM, Kaeberlein M. mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science. 2013;342:1524–8. doi: 10.1126/science.1244360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peng M, Ostrovsky J, Kwon YJ, Polyak E, Licata J, Tsukikawa M, Marty E, Thomas J, Felix CA, Xiao R, Zhang Z, Gasser DL, Argon Y, Falk MJ. Inhibiting cytosolic translation and autophagy improves health in mitochondrial disease. Hum Mol Genet. 2015;24:4829–47. doi: 10.1093/hmg/ddv207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Z, Tsukikawa M, Peng M, Polyak E, Nakamaru-Ogiso E, Ostrovsky J, McCormack S, Place E, Clarke C, Reiner G, McCormick E, Rappaport E, Haas R, Baur JA, Falk MJ. Primary respiratory chain disease causes tissue-specific dysregulation of the global transcriptome and nutrient-sensing signaling network. PLoS One. 2013;8:e69282. doi: 10.1371/journal.pone.0069282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stroud DA, Surgenor EE, Formosa LE, Reljic B, Frazier AE, Dibley MG, Osellame LD, Stait T, Beilharz TH, Thorburn DR, Salim A, Ryan MT. Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature. 2016;538:123–126. doi: 10.1038/nature19754. [DOI] [PubMed] [Google Scholar]
- 74.Ahting U, Floss T, Uez N, Schneider-Lohmar I, Becker L, Kling E, Iuso A, Bender A, de Angelis MH, Gailus-Durner V, Fuchs H, Meitinger T, Wurst W, Prokisch H, Klopstock T. Neurological phenotype and reduced lifespan in heterozygous Tim23 knockout mice, the first mouse model of defective mitochondrial import. Biochim Biophys Acta. 2009;1787:371–6. doi: 10.1016/j.bbabio.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 75.Roesch K, Curran SP, Tranebjaerg L, Koehler CM. Human deafness dystonia syndrome is caused by a defect in assembly of the DDP1/TIMM8a-TIMM13 complex. Hum Mol Genet. 2002;11:477–86. doi: 10.1093/hmg/11.5.477. [DOI] [PubMed] [Google Scholar]
- 76.Tranebjaerg L, Schwartz C, Eriksen H, Andreasson S, Ponjavic V, Dahl A, Stevenson RE, May M, Arena F, Barker D, et al. A new X linked recessive deafness syndrome with blindness, dystonia, fractures, and mental deficiency is linked to Xq22. J Med Genet. 1995;32:257–63. doi: 10.1136/jmg.32.4.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jin H, May M, Tranebjaerg L, Kendall E, Fontan G, Jackson J, Subramony SH, Arena F, Lubs H, Smith S, Stevenson R, Schwartz C, Vetrie D. A novel X-linked gene, DDP, shows mutations in families with deafness (DFN-1), dystonia, mental deficiency and blindness. Nat Genet. 1996;14:177–80. doi: 10.1038/ng1096-177. [DOI] [PubMed] [Google Scholar]
- 78.Rugarli EI, Langer T. Mitochondrial quality control: a matter of life and death for neurons. EMBO J. 2012;31:1336–49. doi: 10.1038/emboj.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bohovych I, Chan SS, Khalimonchuk O. Mitochondrial protein quality control: the mechanisms guarding mitochondrial health. Antioxid Redox Signal. 2015;22:977–94. doi: 10.1089/ars.2014.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Strauss KM, Martins LM, Plun-Favreau H, Marx FP, Kautzmann S, Berg D, Gasser T, Wszolek Z, Muller T, Bornemann A, Wolburg H, Downward J, Riess O, Schulz JB, Kruger R. Loss of function mutations in the gene encoding Omi/HtrA2 in Parkinson’s disease. Hum Mol Genet. 2005;14:2099–111. doi: 10.1093/hmg/ddi215. [DOI] [PubMed] [Google Scholar]
- 81.Daoud H, Valdmanis PN, Gros-Louis F, Belzil V, Spiegelman D, Henrion E, Diallo O, Desjarlais A, Gauthier J, Camu W, Dion PA, Rouleau GA. Resequencing of 29 candidate genes in patients with familial and sporadic amyotrophic lateral sclerosis. Arch Neurol. 2011;68:587–93. doi: 10.1001/archneurol.2010.351. [DOI] [PubMed] [Google Scholar]
- 82.Pierson TM, Adams D, Bonn F, Martinelli P, Cherukuri PF, Teer JK, Hansen NF, Cruz P, Mullikin JC, Blakesley RW, Golas G, Kwan J, Sandler A, Fuentes Fajardo K, Markello T, Tifft C, Blackstone C, Rugarli EI, Langer T, Gahl WA, Toro C. Whole-exome sequencing identifies homozygous AFG3L2 mutations in a spastic ataxia-neuropathy syndrome linked to mitochondrial m-AAA proteases. PLoS Genet. 2011;7:e1002325. doi: 10.1371/journal.pgen.1002325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Casari G, De Fusco M, Ciarmatori S, Zeviani M, Mora M, Fernandez P, De Michele G, Filla A, Cocozza S, Marconi R, Durr A, Fontaine B, Ballabio A. Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell. 1998;93:973–83. doi: 10.1016/s0092-8674(00)81203-9. [DOI] [PubMed] [Google Scholar]
- 84.Merkwirth C, Martinelli P, Korwitz A, Morbin M, Bronneke HS, Jordan SD, Rugarli EI, Langer T. Loss of prohibitin membrane scaffolds impairs mitochondrial architecture and leads to tau hyperphosphorylation and neurodegeneration. PLoS Genet. 2012;8:e1003021. doi: 10.1371/journal.pgen.1003021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kondadi AK, Wang S, Montagner S, Kladt N, Korwitz A, Martinelli P, Herholz D, Baker MJ, Schauss AC, Langer T, Rugarli EI. Loss of the m-AAA protease subunit AFG(3)L(2) causes mitochondrial transport defects and tau hyperphosphorylation. EMBO J. 2014;33:1011–26. doi: 10.1002/embj.201387009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Funayama M, Hattori N. CHCHD2 and Parkinson’s disease--authors’ reply. Lancet Neurol. 2015;14:682–3. doi: 10.1016/S1474-4422(15)00097-6. [DOI] [PubMed] [Google Scholar]
- 87.Bannwarth S, Ait-El-Mkadem S, Chaussenot A, Genin EC, Lacas-Gervais S, Fragaki K, Berg-Alonso L, Kageyama Y, Serre V, Moore DG, Verschueren A, Rouzier C, Le Ber I, Auge G, Cochaud C, Lespinasse F, N’Guyen K, de Septenville A, Brice A, Yu-Wai-Man P, Sesaki H, Pouget J, Paquis-Flucklinger V. A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain. 2014;137:2329–45. doi: 10.1093/brain/awu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Johnson JO, Glynn SM, Gibbs JR, Nalls MA, Sabatelli M, Restagno G, Drory VE, Chio A, Rogaeva E, Traynor BJ. Mutations in the CHCHD10 gene are a common cause of familial amyotrophic lateral sclerosis. Brain. 2014;137:e311. doi: 10.1093/brain/awu265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ogaki K, Koga S, Heckman MG, Fiesel FC, Ando M, Labbe C, Lorenzo-Betancor O, Moussaud-Lamodiere EL, Soto-Ortolaza AI, Walton RL, Strongosky AJ, Uitti RJ, McCarthy A, Lynch T, Siuda J, Opala G, Rudzinska M, Krygowska-Wajs A, Barcikowska M, Czyzewski K, Puschmann A, Nishioka K, Funayama M, Hattori N, Parisi JE, Petersen RC, Graff-Radford NR, Boeve BF, Springer W, Wszolek ZK, Dickson DW, Ross OA. Mitochondrial targeting sequence variants of the CHCHD2 gene are a risk for Lewy body disorders. Neurology. 2015;85:2016–25. doi: 10.1212/WNL.0000000000002170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Woo JA, Liu T, Trotter C, Fang CC, De Narvaez E, LePochat P, Maslar D, Bukhari A, Zhao X, Deonarine A, Westerheide SD, Kang DE. Loss of function CHCHD10 mutations in cytoplasmic TDP-43 accumulation and synaptic integrity. Nat Commun. 2017;8:15558. doi: 10.1038/ncomms15558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–95. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 92.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(Suppl):S10–7. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 93.Devi L, Prabhu BM, Galati DF, Avadhani NG, Anandatheerthavarada HK. Accumulation of amyloid precursor protein in the mitochondrial import channels of human Alzheimer’s disease brain is associated with mitochondrial dysfunction. J Neurosci. 2006;26:9057–68. doi: 10.1523/JNEUROSCI.1469-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cenini G, Rub C, Bruderek M, Voos W. Amyloid beta-peptides interfere with mitochondrial preprotein import competence by a coaggregation process. Mol Biol Cell. 2016;27:3257–3272. doi: 10.1091/mbc.E16-05-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mattiazzi M, D’Aurelio M, Gajewski CD, Martushova K, Kiaei M, Beal MF, Manfredi G. Mutated human SOD1 causes dysfunction of oxidative phosphorylation in mitochondria of transgenic mice. J Biol Chem. 2002;277:29626–33. doi: 10.1074/jbc.M203065200. [DOI] [PubMed] [Google Scholar]
- 96.Igoudjil A, Magrane J, Fischer LR, Kim HJ, Hervias I, Dumont M, Cortez C, Glass JD, Starkov AA, Manfredi G. In vivo pathogenic role of mutant SOD1 localized in the mitochondrial intermembrane space. J Neurosci. 2011;31:15826–37. doi: 10.1523/JNEUROSCI.1965-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lasagna-Reeves CA, Castillo-Carranza DL, Sengupta U, Clos AL, Jackson GR, Kayed R. Tau oligomers impair memory and induce synaptic and mitochondrial dysfunction in wild-type mice. Mol Neurodegener. 2011;6:39. doi: 10.1186/1750-1326-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Deng J, Yang M, Chen Y, Chen X, Liu J, Sun S, Cheng H, Li Y, Bigio EH, Mesulam M, Xu Q, Du S, Fushimi K, Zhu L, Wu JY. FUS interacts with HSP60 to promote mitochondrial damage. PLoS Genet. 2015;11:e1005357. doi: 10.1371/journal.pgen.1005357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xu YF, Zhang YJ, Lin WL, Cao X, Stetler C, Dickson DW, Lewis J, Petrucelli L. Expression of mutant TDP-43 induces neuronal dysfunction in transgenic mice. Mol Neurodegener. 2011;6:73. doi: 10.1186/1750-1326-6-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Martin LJ, Pan Y, Price AC, Sterling W, Copeland NG, Jenkins NA, Price DL, Lee MK. Parkinson’s disease alpha-synuclein transgenic mice develop neuronal mitochondrial degeneration and cell death. J Neurosci. 2006;26:41–50. doi: 10.1523/JNEUROSCI.4308-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tabrizi SJ, Workman J, Hart PE, Mangiarini L, Mahal A, Bates G, Cooper JM, Schapira AH. Mitochondrial dysfunction and free radical damage in the Huntington R6/2 transgenic mouse. Ann Neurol. 2000;47:80–6. doi: 10.1002/1531-8249(200001)47:1<80::aid-ana13>3.3.co;2-b. [DOI] [PubMed] [Google Scholar]
- 102.Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011;144:67–78. doi: 10.1016/j.cell.2010.11.050. [DOI] [PubMed] [Google Scholar]
- 103.Ciryam P, Tartaglia GG, Morimoto RI, Dobson CM, Vendruscolo M. Widespread aggregation and neurodegenerative diseases are associated with supersaturated proteins. Cell Rep. 2013;5:781–90. doi: 10.1016/j.celrep.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ciryam P, Kundra R, Morimoto RI, Dobson CM, Vendruscolo M. Supersaturation is a major driving force for protein aggregation in neurodegenerative diseases. Trends Pharmacol Sci. 2015;36:72–7. doi: 10.1016/j.tips.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bose A, Beal MF for the NISC Comparative Sequencing Program. Mitochondrial dysfunction in Parkinson’s disease. J Neurochem. 2016;139(Suppl 1):216–231. doi: 10.1111/jnc.13731. [DOI] [PubMed] [Google Scholar]
- 106.Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, Albanese A, Nussbaum R, Gonzalez-Maldonado R, Deller T, Salvi S, Cortelli P, Gilks WP, Latchman DS, Harvey RJ, Dallapiccola B, Auburger G, Wood NW. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–60. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 107.Unal Gulsuner H, Gulsuner S, Mercan FN, Onat OE, Walsh T, Shahin H, Lee MK, Dogu O, Kansu T, Topaloglu H, Elibol B, Akbostanci C, King MC, Ozcelik T, Tekinay AB. Mitochondrial serine protease HTRA2 p.G399S in a kindred with essential tremor and Parkinson disease. Proc Natl Acad Sci USA. 2014;111:18285–90. doi: 10.1073/pnas.1419581111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nicklas WJ, Vyas I, Heikkila RE. Inhibition of NADH-linked oxidation in brain mitochondria by 1-methyl-4-phenyl-pyridine, a metabolite of the neurotoxin, 1-methyl-4-phenyl-1,2,5,6-tetrahydropyridine. Life Sci. 1985;36:2503–8. doi: 10.1016/0024-3205(85)90146-8. [DOI] [PubMed] [Google Scholar]
- 109.Olanow CW, Brundin P. Parkinson’s disease and alpha synuclein: is Parkinson’s disease a prion-like disorder? Mov Disord. 2013;28:31–40. doi: 10.1002/mds.25373. [DOI] [PubMed] [Google Scholar]
- 110.Gispert S, Ricciardi F, Kurz A, Azizov M, Hoepken HH, Becker D, Voos W, Leuner K, Muller WE, Kudin AP, Kunz WS, Zimmermann A, Roeper J, Wenzel D, Jendrach M, Garcia-Arencibia M, Fernandez-Ruiz J, Huber L, Rohrer H, Barrera M, Reichert AS, Rub U, Chen A, Nussbaum RL, Auburger G. Parkinson phenotype in aged PINK1-deficient mice is accompanied by progressive mitochondrial dysfunction in absence of neurodegeneration. PLoS One. 2009;4:e5777. doi: 10.1371/journal.pone.0005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mann VM, Cooper JM, Daniel SE, Srai K, Jenner P, Marsden CD, Schapira AH. Complex I, iron, and ferritin in Parkinson’s disease substantia nigra. Ann Neurol. 1994;36:876–81. doi: 10.1002/ana.410360612. [DOI] [PubMed] [Google Scholar]
- 112.Power JH, Barnes OL, Chegini F. Lewy bodies and the mechanisms of neuronal cell death in Parkinson’s disease and dementia with Lewy bodies. Brain Pathol. 2017;27:3–12. doi: 10.1111/bpa.12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hashimoto M, Takeda A, Hsu LJ, Takenouchi T, Masliah E. Role of cytochrome c as a stimulator of alpha-synuclein aggregation in Lewy body disease. J Biol Chem. 1999;274:28849–52. doi: 10.1074/jbc.274.41.28849. [DOI] [PubMed] [Google Scholar]
- 114.Gandhi S, Muqit MM, Stanyer L, Healy DG, Abou-Sleiman PM, Hargreaves I, Heales S, Ganguly M, Parsons L, Lees AJ, Latchman DS, Holton JL, Wood NW, Revesz T. PINK1 protein in normal human brain and Parkinson’s disease. Brain. 2006;129:1720–31. doi: 10.1093/brain/awl114. [DOI] [PubMed] [Google Scholar]
- 115.Leverenz JB, Umar I, Wang Q, Montine TJ, McMillan PJ, Tsuang DW, Jin J, Pan C, Shin J, Zhu D, Zhang J. Proteomic identification of novel proteins in cortical lewy bodies. Brain Pathol. 2007;17:139–45. doi: 10.1111/j.1750-3639.2007.00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Onyango IG, Dennis J, Khan SM. Mitochondrial dysfunction in alzheimer’s disease and the rationale for bioenergetics based therapies. Aging Dis. 2016;7:201–14. doi: 10.14336/AD.2015.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hirai K, Aliev G, Nunomura A, Fujioka H, Russell RL, Atwood CS, Johnson AB, Kress Y, Vinters HV, Tabaton M, Shimohama S, Cash AD, Siedlak SL, Harris PL, Jones PK, Petersen RB, Perry G, Smith MA. Mitochondrial abnormalities in Alzheimer’s disease. J Neurosci. 2001;21:3017–23. doi: 10.1523/JNEUROSCI.21-09-03017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Parker WD, Jr, Parks J, Filley CM, Kleinschmidt-DeMasters BK. Electron transport chain defects in Alzheimer’s disease brain. Neurology. 1994;44:1090–6. doi: 10.1212/wnl.44.6.1090. [DOI] [PubMed] [Google Scholar]
- 119.Reddy PH. Abnormal tau, mitochondrial dysfunction, impaired axonal transport of mitochondria, and synaptic deprivation in Alzheimer’s disease. Brain Res. 2011;1415:136–48. doi: 10.1016/j.brainres.2011.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hoglinger GU, Lannuzel A, Khondiker ME, Michel PP, Duyckaerts C, Feger J, Champy P, Prigent A, Medja F, Lombes A, Oertel WH, Ruberg M, Hirsch EC. The mitochondrial complex I inhibitor rotenone triggers a cerebral tauopathy. J Neurochem. 2005;95:930–9. doi: 10.1111/j.1471-4159.2005.03493.x. [DOI] [PubMed] [Google Scholar]
- 121.Escobar-Khondiker M, Hollerhage M, Muriel MP, Champy P, Bach A, Depienne C, Respondek G, Yamada ES, Lannuzel A, Yagi T, Hirsch EC, Oertel WH, Jacob R, Michel PP, Ruberg M, Hoglinger GU. Annonacin, a natural mitochondrial complex I inhibitor, causes tau pathology in cultured neurons. J Neurosci. 2007;27:7827–37. doi: 10.1523/JNEUROSCI.1644-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Roses AD, Lutz MW, Amrine-Madsen H, Saunders AM, Crenshaw DG, Sundseth SS, Huentelman MJ, Welsh-Bohmer KA, Reiman EM. A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer’s disease. Pharmacogenomics J. 2009;10:375–84. doi: 10.1038/tpj.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ciryam P, Kundra R, Freer R, Morimoto RI, Dobson CM, Vendruscolo M. A transcriptional signature of Alzheimer’s disease is associated with a metastable subproteome at risk for aggregation. Proc Natl Acad Sci USA. 2016;113:4753–8. doi: 10.1073/pnas.1516604113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sergeant N, Wattez A, Galvan-valencia M, Ghestem A, David JP, Lemoine J, Sautiere PE, Dachary J, Mazat JP, Michalski JC, Velours J, Mena-Lopez R, Delacourte A. Association of ATP synthase alpha-chain with neurofibrillary degeneration in Alzheimer’s disease. Neuroscience. 2003;117:293–303. doi: 10.1016/s0306-4522(02)00747-9. [DOI] [PubMed] [Google Scholar]
- 125.Wang Q, Woltjer RL, Cimino PJ, Pan C, Montine KS, Zhang J, Montine TJ. Proteomic analysis of neurofibrillary tangles in Alzheimer disease identifies GAPDH as a detergent-insoluble paired helical filament tau binding protein. FASEB J. 2005;19:869–71. doi: 10.1096/fj.04-3210fje. [DOI] [PubMed] [Google Scholar]
- 126.Shi P, Gal J, Kwinter DM, Liu X, Zhu H. Mitochondrial dysfunction in amyotrophic lateral sclerosis. Biochim Biophys Acta. 2010;1802:45–51. doi: 10.1016/j.bbadis.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Blokhuis AM, Groen EJ, Koppers M, van den Berg LH, Pasterkamp RJ. Protein aggregation in amyotrophic lateral sclerosis. Acta Neuropathol. 2013;125:777–94. doi: 10.1007/s00401-013-1125-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, Jiang H, Hirano M, Rampersaud E, Jansen GH, Donkervoort S, Bigio EH, Brooks BR, Ajroud K, Sufit RL, Haines JL, Mugnaini E, Pericak-Vance MA, Siddique T. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–5. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Szczesny B, Hazra TK, Papaconstantinou J, Mitra S, Boldogh I. Age-dependent deficiency in import of mitochondrial DNA glycosylases required for repair of oxidatively damaged bases. Proc Natl Acad Sci USA. 2003;100:10670–5. doi: 10.1073/pnas.1932854100. [DOI] [PMC free article] [PubMed] [Google Scholar]