Abstract
An ancestral configuration is one of the combinatorially distinct sets of gene lineages that, for a given gene tree, can reach a given node of a specified species tree. Ancestral configurations have appeared in recursive algebraic computations of the conditional probability that a gene tree topology is produced under the multispecies coalescent model for a given species tree. For matching gene trees and species trees, we study the number of ancestral configurations, considered up to an equivalence relation introduced by Wu (2012) to reduce the complexity of the recursive probability computation. We examine the largest number of non-equivalent ancestral configurations possible for a given tree size n. Whereas the smallest number of non-equivalent ancestral configurations increases polynomially with n, we show that the largest number increases with kn, where k is a constant that satisfies . Under a uniform distribution on the set of binary labeled trees with a given size n, the mean number of non-equivalent ancestral configurations grows exponentially with n. The results refine an earlier analysis of the number of ancestral configurations considered without applying the equivalence relation, showing that use of the equivalence relation does not alter the exponential nature of the increase with tree size.
1 Introduction
Under the multispecies coalescent model for the evolution of gene trees conditional on species trees, symmetries and identities among gene tree probabilities and algebraic perspectives for examining the probability computations have contributed to advances in understanding the properties of evolutionary descent in closely related species (Allman et al., 2011). Calculations of the probabilities of gene tree topologies can proceed by one of two computational approaches: nonrecursive (Degnan and Salter, 2005) or recursive (Wu, 2012). Both methods involve combinatorial and probabilistic components, in which probabilities are evaluated for each element of a set of objects that can be defined purely in mathematical terms. Computational complexity is affected both by the size of the underlying set of objects and by the complexity of the probability calculation.
In the recursive approach, the relevant combinatorial set consists of ancestral configurations, each of which represents a set of gene lineages that can be extant at a given node of the species tree (Wu, 2012). We have previously studied the set of ancestral configurations possible for a given gene tree and matching species tree, showing that the largest number of ancestral configurations across labeled tree topologies of a fixed tree size n increases exponentially with n (Disanto and Rosenberg, 2017).
To lower the computation time of the recursive evaluation of gene tree probabilities, Wu (2012) introduced an equivalence relation that, taking into account symmetries in tree shapes, reduces the set of ancestral configurations to a potentially much smaller set of non-equivalent ancestral configurations. The computation of gene tree probabilities can then make use of intermediate steps calculated for the elements of this smaller set, rather than for the full set of ancestral configurations.
Here, for gene trees and species trees with a matching labeled topology t, we study the number of non-equivalent ancestral configurations that can appear at the nodes of a species tree t. We determine the number of non-equivalent ancestral configurations when t belongs to special families of trees characterized by balanced and unbalanced patterns. We study the largest number of non-equivalent ancestral configurations possible for a given tree size n, showing that this number grows exponentially with kn, where k is a constant that satisfies . Although tree families exist for which the number of non-equivalent ancestral configurations grows polynomially in n (Wu, 2012), we show that under a uniform distribution on the set of labeled trees of size n, the mean number of non-equivalent ancestral configurations of a random labeled tree shape also grows exponentially in n. Finally, we compare our results on the number of non-equivalent ancestral configurations with corresponding results for the full set of ancestral configurations (Disanto and Rosenberg, 2017). Although by definition, the non-equivalent ancestral configurations are no more numerous than ancestral configurations that do not take into account the equivalence relation—and indeed, are intended to be less numerous—the base k for the maximal number of non-equivalent ancestral configurations kn across trees of size n is bounded below by a constant only slightly smaller than the corresponding base for the maximal number of ancestral configurations.
2 Preliminaries
We study the number of non-equivalent ancestral configurations of rooted binary labeled trees. We start by giving definitions and preliminary results. In Section 2.1, we recall some properties of rooted binary labeled trees. In Section 2.2, we discuss properties of the exponential growth of sequences of non-negative numbers. Following Wu (2012), Section 2.3 defines ancestral configurations for a gene tree and a species trees with a matching labeled topology t. In Section 2.4, we recall related enumerative results of Disanto and Rosenberg (2017).
2.1 Labeled topologies
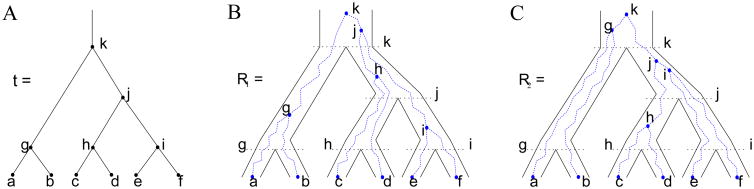
A labeled topology t of size |t| = n is a bifurcating rooted tree with n labeled leaves, also termed “taxa” (Fig. 1A). We sometimes refer to labeled topologies simply as “trees.” We define a total order a ≺ b ≺ c ≺ … for the set {a, b, c, …} of labels of the leaves of a tree, proceeding alphabetically. That is, without loss of generality, we assume that a tree of size n has its taxa labeled using the first n symbols that appear in the order ≺.
Figure 1.
A matching gene tree and species tree with labeled topology t. (A) A tree t of size 6 isomorphic to the gene tree and species tree in (B) and (C). Tree t is uniquely determined by the labeling of its leaves and by its unlabeled shape. It is convenient to assign arbitrary labels to the internal nodes of t as well. We use letters g, h, i, j, k in this case. Each lineage (edge) of t is identified by the lowest node it intersects; for example, lineages h and i descend from lineage j. (B) A possible realization R1 of a gene tree (dotted lines) in a species tree (solid lines). The gene tree and the species tree have a matching topology that follows (A). At species tree node j, the ancestral configuration is {c, d, i}. At node k, the configuration is {g, h, i}. (C) A non-equivalent realization R2 of the gene tree in (A) in the matching species tree. At species tree nodes j and k, the configurations are {h, e, f} and {a, b, j}, respectively.
We represent labeled topologies in Newick notation (Felsenstein, 2004), in which t = (t1, t2) is the tree obtained by appending trees t1 and t2 to a common root node. For example, ((a, b), ((c, d), (e, f))) gives the Newick notation for the tree depicted in Fig. 1A. We term non-leaf nodes of a tree “internal” nodes. By “subtree” of a tree t, we mean a node of t together with all its descendants; a “root subtree” of t is a subtree—one of two possible—immediately descended from the root of t.
For two trees t1, t2, we say that t1 is isomorphic to t2 and write t1 ≅ t2 when, after their leaf labels are removed, t1 and t2 have the same unlabeled topology. Moreover, given trees t1 and t2 with |t1| ≥ |t2|, we say that a subtree t of t1 is equal to t2 up to “rescaling” labels when, respecting the order ≺, we can replace the labels of t to obtain t2. For instance, the largest root subtree ((c, d), (e, f)) of the tree depicted in Fig. 1A is equal to ((a, b), (c, d)) up to rescaling, as we can replace the labels c → a, d → b, e → c, f → d. Note that alphabetical order is preserved in this replacement.
We denote by Tn the set of trees of size n, and by the set of all trees of any size. The number of trees of size n ≥ 2 is given by
| (1) |
(Felsenstein, 1978), which assuming n ≥ 1 can be rewritten
| (2) |
We will have occasion to employ a uniform probability distribution over the set of trees of fixed size. In this distribution, each tree of size n has probability 1/|Tn|.
2.2 Exponential growth of a sequence
As in Flajolet and Sedgewick (2009), we say that a sequence of positive numbers an is of exponential order k or, equivalently, has exponential growth kn, when
This relation holds when an = kns(n), where s is a subexponential factor, so that limsupn→∞[s(n)1/n] = 1. According to these definitions, a sequence an grows exponentially in n if its exponential order strictly exceeds 1.
The exponential order of a sequence describes its asymptotic growth. It follows from the definition that if (an) has exponential order ka and (bn) has exponential order kb > ka, then an/bn converges to 0 exponentially fast as (ka/kb)n for n → ∞. When two sequences (an) and (bn) have the same exponential order, we write an ⋈ bn. If an ⋈ bn and limn→∞(an/bn) = 1, we write an ∼ bn.
2.3 Ancestral configurations
This section defines the set of ancestral configurations of a gene tree G in a species tree S. In our setting, exactly one gene lineage is selected from each species. We assume a matching labeled topology t for G and S.
Consider a realization R of a gene tree G in a species tree S, with G = S = t (Fig. 1). Equivalently, R is one of the possible evolutionary scenarios for gene tree G on species tree S. Given a node κ of t, we denote by C(κ, R) the set of gene lineages, i.e. edges of G, that are present in S at the point right before node κ looking backward in time. Following Wu (2012), we call the set C(κ, R) the ancestral configuration of G at node κ of S.
For the tree t in Fig. 1A, if we consider the realization R1 of the gene tree G = t in the species tree S = t depicted in Fig. 1B, then we see that C(k, R1) = {g, h, i} is the ancestral configuration of the gene tree at node k of the species tree. The gene lineages g, h, and i are those present in the species tree at the point right before the root node k. Similarly, the ancestral configuration of the gene tree at node j of the species tree is given by the set of gene lineages C(j, R1) = {c, d, i}. In Fig. 1C, a different realization R2 of the same gene tree is described. The ancestral configuration at the root k of the species tree is in this case C(k, R2) = {a, b, j}, whereas the ancestral configuration at node j is C(j, R2) = {h, e, f}.
We denote the set of all possible realizations of the gene tree G = t in the species tree S = t by ℜ(G, S). By considering all elements R ∈ ℜ(G, S), for a given node κ of t we define the set of all possible ancestral configurations at node κ,
| (3) |
and the number of such configurations,
| (4) |
In particular, c(κ) counts the number of ways the gene lineages of G can reach the point right below node κ in S, when all possible realizations of G in S are taken into account. For example, if we set t as in Fig. 1A, then we have C(g) = {{a, b}} and C(j) = {{c, d, e, f}, {h, e, f}, {c, d, i}, {h, i}}. At the root node k, the set of all possible ancestral configurations is
Note that two different realizations R1, R2 ∈ ℜ(G, S) can generate the same ancestral configuration C(κ, R1) = C(κ, R2) at an internal node κ.
Following Disanto and Rosenberg (2017), for each internal node κ, our definition of ancestral configuration excludes the case {κ} ∈ C(κ). This choice accords with the fact that each configuration at node κ is considered at the point right below node κ in the species tree, with no time for the gene lineages from the left and right subtrees of κ to coalesce together. With the exception that we say that a leaf or 1-taxon tree has 0 ancestral configurations, our definition is identical to that of Wu (2012), which assigns these cases 1 ancestral configuration.
Under our assumption of a matching gene tree and species tree G = S = t, the set C(κ) defined in (3) and its cardinality c(κ) (4) depend only on node κ and tree t. When we refer to an element of C(κ), we use the term configuration at node κ of t. When κ is the root node, we use the term root configuration to describe an element of C(κ). Also, considering the union of all the sets C(κ) of configurations across all internal nodes κ of t, we can count the total number of configurations.
2.4 The number of configurations
We recall some of the results of Disanto and Rosenberg (2017) on the number of configurations possessed by a tree. These results are used to measure the decrease in the number of configurations when, as in Wu (2012), an equivalence relation is introduced in Section 3 to merge topologically equivalent configurations.
-
If A, B are two sets of sets, define A ⊗ B = {a ∪ b : a ∈ A, b ∈ B}. For a given tree t with |t| > 1, the set C(r) of configurations at the root r of t satisfies the following decomposition
where rℓ and rr respectively denote the left and right children of r.
-
For a given tree t with |t| > 1, the number c(r) of possible configurations at the root node r of t can be recursively computed as
(5) where we set c(r) = 0 when |t| = 1. At each node κ of t, the number of configurations c(κ) is bounded as c(κ) ≤ c(r). Thus, the total number of configurations c = Σκc(κ) satisfies c(r) ≤ c ≤ (2|t| − 1)c(r). In particular, the quantities c and c(r) are equal up to a factor that is at most polynomial in |t|, and they have the same exponential order when measured across families of trees of increasing size.
Denote by Mn(r) and Mn, respectively, the largest number of root configurations and the largest total number of configurations that a tree of size n can have. The exponential growth of the sequences Mn(r) and Mn is , where k0 is a constant, k0 ≈ 1.5028.
A completely balanced tree of size n = 2h has root configurations. A caterpillar tree of size n has n − 1 root configurations.
For a tree of given size n leaves selected uniformly at random, the mean number of root configurations c(r) and the mean total number of configurations c have exponential growth 𝔼n[c(r)] ⋈ 𝔼n[c] ⋈ (4/3)n with n.
3 Equivalent and non-equivalent configurations
Wu (2012) introduced an equivalence relation over the set of configurations at a given node of a species tree, using this equivalence relation to evaluate the probability of a gene tree topology by performing computations over the sets of non-equivalent configurations of the gene tree at species tree nodes (e.g. eq. (7) of Wu (2012)). Following the definition of Wu (2012), in this section, we introduce the notion of equivalent configurations for gene trees and species trees with matching topology t. Under certain assumptions on t, in Section 3.3, we provide a recursion analogous to the one in (5) for counting non-equivalent configurations at the root of t.
3.1 An equivalence relation
We begin with some notation. If κ is a node of a tree t, denote by tκ the subtree of t generated by κ (i.e., κ and all nodes below it). If X is a set of nodes of a subtree tκ, the restriction tκ(X) of tκ to X is the tree shape obtained by removing from tκ all nodes that remain strictly below the nodes belonging to X. For instance, if tj is the subtree generated by node j in the tree t in Fig. 1A and X = {h, e, f}, then tj(X) is obtained by removing nodes c and d from tj, and thus is the caterpillar tree shape of size 3. Similarly, if X = {a, b, h, i}, then tk(X) is the balanced tree shape of size 4.
The definition of equivalent configurations given by Wu (2012) reduces to the following one when gene trees and species trees are matching. Given a tree t and a node κ, two configurations γ1, γ2 at node κ, γ1, γ2 ∈ C(κ), are equivalent at κ—with the equivalence denoted by γ1 ∼κ γ2—when the tree shape tκ(γ1) is isomorphic to the tree shape tκ(γ2). For instance, in Fig. 1A, we have {h, e, f} ∼j {c, d, i} and {a, b, j} ∼k {g, h, i}. The set of non-equivalent configurations at a given node κ is denoted by C*(κ), and its cardinality is c*(κ) = |C*(κ)|
The notion of equivalent configurations groups together at a given node configurations for which exactly the same topological constraints apply in ordering the coalescent events of their gene lineages. In other words, gene lineages of equivalent configurations at a node κ of a species tree have completely topologically equivalent transitions when they move from node κ backward in time (upward in the species tree).
For instance, consider the tree in Fig. 1A, where the configurations {a, b, j} and {g, h, i} at node k satisfy {a, b, j} ∼k {g, h, i}. Consider the mapping ϕ(a) = h, ϕ(b) = i, ϕ(j) = g, ϕ(g) = j, ϕ(k) = k. The transition in Fig. 1C that along the root branch of the species tree transforms the set of gene lineages {a, b, j} into the single lineage k corresponds topologically to the transition in Fig. 1B that transforms {g, h, i} into k. Indeed, the two trees tk({a, b, j}) with nodes {a, b, j, g, k}, and tk({g, h, i}) with nodes {g, h, i, j, k} are isomorphic through ϕ.
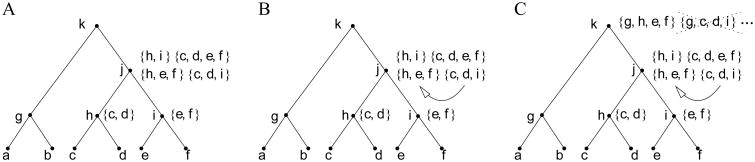
As described in Fig. 2, for a given tree t, the effective computation of non-equivalent configurations can be performed recursively as in the algorithm STELLS (Wu, 2012) by scanning t from bottom to top with a postorder traversal. At each visited node κ, we first compute the set
Figure 2.
Merging of equivalent configurations at node κ = j. (A) At node j, the set C̃(j) = {{h, i}, {h, e, f}, {c, d, i}, {c, d, e, f}} of configurations is computed from the non-equivalent configurations at the child nodes h and i by using (6). (B) Two equivalent configurations appear in C̃(j), namely {h, e, f} ∼j {c, d, i}. Configuration {c, d, i} is merged into {h, e, f} (or vice versa). (C) The configurations in C*(j) = {{h, e, f}, {h, i}, {c, d, e, f}} are used to determine configurations at node k. In particular, {g, h, e, f} ∈ C̃(k) and {g, c, d, i} ∉ C̃(k), as {c, d, i} has been merged into {h, e, f}. Configuration {g, c, d, i}, which is not present in C̃(k), is represented by the equivalent configuration {g, h, e, f} ∼k {g, c, d, i}. Similarly, {a, b, c, d, i} ∉ C̃(k), and it is represented by {a, b, h, e, f} ∼k {a, b, c, d, i}.
| (6) |
from the sets of non-equivalent configurations of the two child nodes κℓ, κr (Fig. 2A with κ = j). Next, we merge all the equivalent configurations present in C̃(κ) into a single representative, one for each class of equivalence of the relation ∼κ, to determine the set C*(κ) of non-equivalent configurations at κ (Fig. 2B). Only the configurations in C*(κ) are used to determine configurations at the parent node of κ (Fig. 2C). Note that from (6), the cardinality of the set C̃(κ) ⊇ C*(κ) satisfies
| (7) |
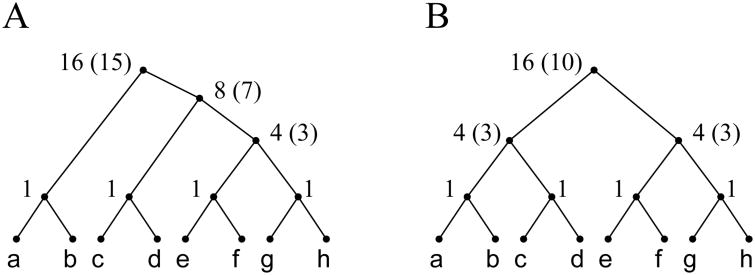
Following this procedure in Fig. 3, we report the quantities |C̃(κ)| and c*(κ) at each internal node κ of two trees of size 8. When |C̃(κ)| > c*(κ), the latter value is given in parentheses. The same trees are considered in the enumerations provided in Table A1 (Fig. 3A) and Table 1 (Fig. 3B) by Wu (2012).
Figure 3.
Computing the number of non-equivalent configurations in two trees of size 8. By using (7), at each internal node κ, |C̃(κ)| is computed from the number of non-equivalent configurations at the nodes descending from κ. When |C̃(κ)| > c*(κ), c*(κ) appears in parentheses. (A) A tree considered in Table A1 by Wu (2012). Adding |t| = 8 to the value Σκ |C̃(κ)| = 32 to take into account the fact that Wu (2012) counts a configuration for each leaf whereas our definition does not do so, we produce entry 40 of the table of Wu (2012). (B) The completely balanced tree of size 8 considered in Table 1 by Wu (2012). Adding |t| = 8 to Σκ |C̃(κ)| = 28, we produce entry 36. The numbers c*(κ) satisfy recursion (10).
In the next sections, we study the number c*(κ) = |C*(κ)| of pairwise non-equivalent configurations at a given node κ of a fixed or random tree t ∈ Tn selected uniformly as well as the total number of non-equivalent configurations c* = Σκ c*(κ) in t. To measure the strength of the equivalence relation ∼κ, we focus on c*(r), the number of non-equivalent configurations at the root κ = r of t, comparing our results with those in Section 2.4.
When there is no need to distinguish between the number of non-equivalent root configurations and the total number of non-equivalent configurations, we simply write “number of non-equivalent configurations”. It is then understood that a statement applies to both root and total non-equivalent configurations. Similarly, “number of configurations” stands for both “number of root configurations” and “total number of configurations.”
3.2 Non-equivalent root configurations in small trees
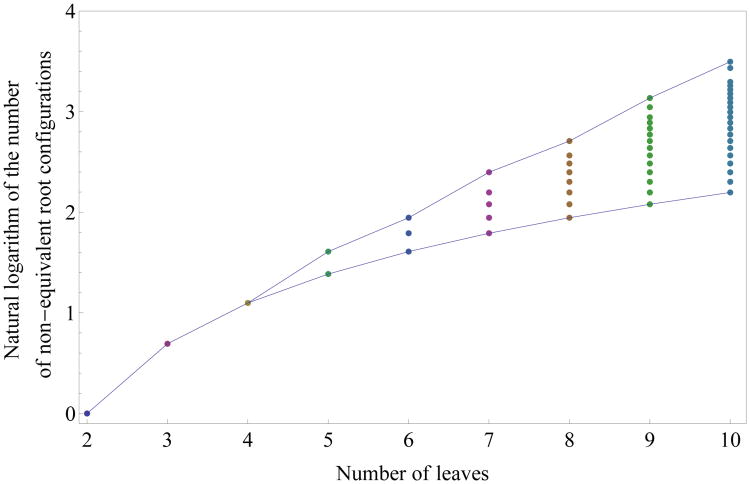
For small values of n, it is possible to exhaustively compute the number of non-equivalent root configurations c*(r) for representative labelings of each of the unlabeled topologies of size n. In Fig. 4, each dot corresponds to the logarithm of the number of non-equivalent root configurations for a certain tree shape of size determined by its x-coordinate. The points associated with the largest values of c*(r) are connected by the top line, whose growth appears to be linear in n. Indeed, as we show in Section 4, tree families exist for which the growth of the number of non-equivalent root configurations is exponential in the tree size.
Figure 4.
Natural logarithm of the number of non-equivalent root configurations for all possible tree shapes of size 2 ≤ n ≤ 10. The value for n = 1, log(0), is omitted. Points corresponding to the largest and smallest numbers of root configurations for each n are connected by the top and bottom lines, respectively.
The tree shapes whose labeled topologies possess the largest number of non-equivalent root configurations among trees of fixed size n ≤ 20 appear in Fig. 5. For 12 ≤ n ≤ 20, each shape in the sequence is produced by connecting the tree with three taxa and the tree of size n − 3 already in the sequence to a shared root. This pattern is used in Section 4.3 to determine a lower bound for the exponential growth of the sequence describing the largest number of non-equivalent root configurations among trees at fixed n.
Figure 5.
Tree shapes of size 5 ≤ n ≤ 20 with the largest number of non-equivalent root configurations. For n = 4, both unlabeled topologies have c*(r) = 3. For 12 ≤ n ≤ 20, the tree with the largest value of c*(r) is obtained by appending a caterpillar of size 3 and the tree of size n – 3 with the largest value of c*(r) to a common root node. From n = 2 to n = 20, the largest values of c*(r) follow the sequence 1, 2, 3, 5, 7, 11, 15, 23, 33, 47, 69, 99, 141, 207, 297, 423, 621, 891, 1269.
For values of n ≤ 20, the tree shape that minimizes the number of non-equivalent root configurations is the caterpillar topology. The number of non-equivalent root configurations in the caterpillar of size n is n − 1 (Wu, 2012). The bottom line in Fig. 4, which connects points corresponding to the smallest number of non-equivalent root configurations for a tree with n taxa, grows with log(n − 1).
These observations show that tree topology can have a considerable impact on the number of non-equivalent configurations possible at a given tree size. Indeed, Section 4 investigates the effect of symmetries in a tree on its number of non-equivalent configurations. In Section 5, we show that although tree families (e.g. caterpillars) exist for which the growth of the number of non-equivalent configurations is polynomial in the tree size n, the expected number of non-equivalent configurations in a labeled topology selected uniformly at random in Tn grows exponentially in n.
3.3 A recursion for the number of non-equivalent root configurations
In this section, we provide a recursive procedure for computing the number of non-equivalent root configurations in trees satisfying certain topological constraints. We later use this recursion to study the number of non-equivalent root configurations for several families of trees.
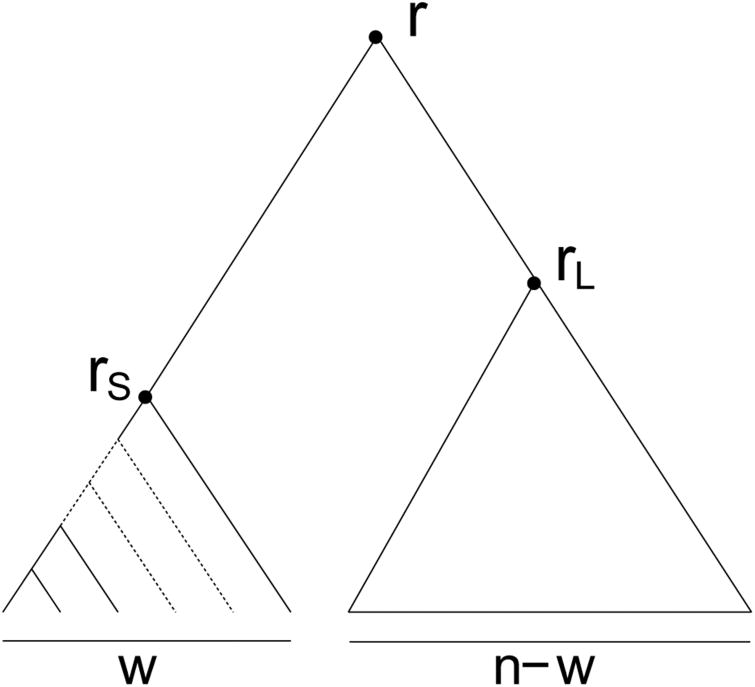
Let r be the root of a tree t. We denote by rS and rL the nodes descending from r that generate the smaller, trS, and the larger, trL, root subtrees of t (we will soon see that if the root subtrees of t have equal size, then we can choose either labeling). As depicted in Fig. 6, suppose subtree trS can be displayed inside subtree trL by a configuration at node rL; that is, assume there is a configuration γ at node rL such that
Figure 6.
A tree t in which the smaller root subtree trS can be displayed as trS ≅ trL (γ) in the larger root subtree trL through a configuration γ at node rL. The configuration γ is determined by the black squares.
| (8) |
Note that it immediately follows that when (8) is satisfied, if trS and trL have the same size, then they must have the same unlabeled shape, and it does not matter which is assigned the label trS and which is assigned trL. It is trivial that (8) is satisfied when trS ≅ trL, by the configuration γ that simply consists of all leaves of trL.
When condition (8) is satisfied, as shown in Appendix 1, the number of non-equivalent configurations c*(r) at the root r of a tree t with |t| > 1 can be directly computed from the corresponding numbers at the children rS and rL:
| (9) |
where c*(r) = 0 if |t| = 1. Note that if the smaller root subtree has size |trS| = 1, then condition (8) is technically not satisfied, as each configuration at node rL has at least 2 elements (unless |t| = 2). However, in this case as well, with |trS| = 1 and c*(rS) = 0, formula (9) holds, yielding c*(r) = 1 + c*(rL).
4 Non-equivalent configurations for special tree families
In this section, we study the number of non-equivalent configurations for special families of trees. We consider completely unbalanced caterpillar trees in Section 4.1 and completely balanced trees in Section 4.2. The number of non-equivalent configurations in the caterpillar family has been investigated by Wu (2012). For the completely balanced family, we show that the number of non-equivalent configurations grows exponentially in the tree size, though in a manner slower than the exponential growth of the number of configurations (see point (iv) in Section 2.4). By considering a particular family of unbalanced trees, in Section 4.3, we bound the exponential growth of the sequence of the largest number of non-equivalent root configurations for a given tree size n.
4.1 Completely unbalanced trees
Consider the family of caterpillar trees. Recursive application of (9) shows that, as was already observed by Wu (2012), the number of non-equivalent root configurations in the caterpillar with n taxa is n − 1. In particular, for caterpillar trees, trS has only one leaf, and c*(r) = 1 + c*(rL). For a caterpillar tree of size n, subtree rL is simply a caterpillar tree of size n − 1. Noting that c*(r) = 1 for a two-taxon caterpillar tree, we can iterate to obtain c*(r) = n − 1 for an n-taxon caterpillar tree. Considering all internal nodes of an n-taxon caterpillar, each of which has one fewer non-equivalent configuration than the number of leaves it subtends, the total number of non-equivalent configurations in the caterpillar of size n is .
We have thus found a family of trees for which the growth of the number of non-equivalent configurations is polynomial in the tree size. This result suggests that 𝔼n[c*(r)]—the expected number of non-equivalent root configurations in a random tree selected uniformly among those of size n—could, in theory, grow as a subexponential function of n. We study the growth of this expectation in Section 5, showing that 𝔼n[c*(r)] in fact grows exponentially in n.
4.2 Completely balanced trees
Now consider the family of completely balanced trees b0, b1, b2, …, where bh is the completely balanced tree of size n = 2h (Fig. 3B). Each tree bh satisfies condition (8), as trS ≅ trL. Because of this equivalence of unlabeled shapes, c*(rS) = c*(rL). Therefore, denoting by γh the number of non-equivalent root configurations in bh, from (9) we have the recursion
| (10) |
where γ0 = 0. Setting xh = (γh + 1)/2, this recursion can be written
| (11) |
with x0 = 1/2. The sequence (xh) can be studied as in Appendix 2. A constant exists for which
| (12) |
The constant can be approximated using the recursive definition of xh, summing terms in a series
| (13) |
Switching back to γh, we obtain
where n = 2h = |bh|.
The following proposition summarizes our result.
Proposition 1 Consider the family of completely balanced trees (bh), with n = 2h = |bh|. Its sequence of the number of non-equivalent root configurations, c*(r), grows asymptotically as , where (13). In particular, c*(r) and the sequence of the total number of non-equivalent configurations, c*, both have exponential growth .
Proof. It remains to show that for tree family (bh), the exponential growth of the total number of non-equivalent configurations equals the exponential growth of the number of non-equivalent root configurations. Because the sequence γh (10) is increasing, in the completely balanced tree bh, the maximum number of non-equivalent configurations across all internal nodes is reached at the root of the tree, equaling c*(r). The total number of nodes (including the leaves) in bh is 2n − 1. We therefore have the inequality c*(r) ≤ c* ≤ (2n − 1)c*(r). In particular, the quantities c* and c*(r) are equal up to a factor that is at most polynomial in the size n. It follows that the exponential growth of c* equals the exponential growth of c*(r).
Comparing the constant with the value of k0 ≈ 1.5028 that describes the exponential growth of the number of configurations for the completely balanced family of trees (Disanto and Rosenberg, 2017), the proposition shows that in this family, the sequence of the number of non-equivalent configurations grows exponentially slower than the sequence of the number of configurations. However, the growth is still exponential in the tree size, and it is not true that non-equivalent configurations always grow polynomially—as they do for caterpillar trees.
4.3 Bounds for the largest number of non-equivalent configurations for a given tree size
We now seek to bound the value of , the largest number of non-equivalent root configurations among trees of size n.
Proposition 2 Let k0 ≈ 1.5028 be the exponential order of the sequence (Mn(r)) describing the largest number of root configurations in trees of size n (point (iii) of Section 2.4). Then , where .
Proof. For the upper bound, because non-equivalent configurations are no more numerous than configurations, , and the upper bound follows.
For the lower bound, it suffices to exhibit a tree family in which the number of non-equivalent root configurations has exponential order . For n ≥ 9, we define the family of unlabeled topologies (un) by taking un as the tree shape of size n depicted in Fig. 5 if n ∈ {9, 10, 11} and un = (un−3, c3)—where c3 is the caterpillar with 3 taxa—when n ≥ 12. Note that for n ≥ 12, the tree t = un satisfies condition (8) with trS = c3 (Fig. 6).
Let γn be the number of non-equivalent root configurations in un. For n ≥ 12, (9) yields the recursion
| (14) |
with γ9 = 23, γ10 = 33, and γ11 = 47. We set xn = [2(n − 3⌊n/3⌋)2 + 8(n − 3⌊n/3⌋) + 23]/27 to produce a function that cycles through the values 23/27, 33/27, and 47/27 as n is incremented. From (14), we have
| (15) |
when n ≥ 9. In particular, using (15), we see that (γn) has exponential growth as desired.
The recursive definition un = (un−3, c3) of the tree family (un) matches the pattern found by exhaustive computation for the unlabeled topologies of trees of size 12 ≤ n ≤ 20 with the largest number of non-equivalent root configurations (Fig. 5). Applying the floor function to the expression in (15), we obtain
| (16) |
This formula, which equals (15) for n ≥ 9, computes the correct values of from Figure 5 for 2 ≤ n ≤ 20. Based on this result, it is a plausible conjecture that (16) gives the exact value for the maximum number of non-equivalent root configurations at a given n ≥ 2.
Note that the constant bounds from below the exponential order of the sequence of the largest total number of non-equivalent configurations among trees of given size, as total non-equivalent configurations are at least as numerous as non-equivalent root configurations. Further, because k0 is the exponential order of the sequence Mn of the largest total number of configurations in trees of fixed size (see point (iii) of Section 2.4), k0 bounds from above the exponential order of the sequence .
Because , another consequence of Propositions 1 and 2 is that sequences and grow exponentially faster than the sequence of the number of non-equivalent configurations in the family of completely balanced trees. This property illustrates a remarkable effect of merging equivalent configurations. From points (iii) and (iv) of Section 2.4, the number of configurations for completely balanced trees follows the sequence of the largest number of configurations for trees of size n. When equivalent configurations are merged together, however, other tree families, such as the unbalanced family (un), possess a number of non-equivalent configurations that grows faster than the corresponding number for completely balanced trees.
5 Mean number of non-equivalent root configurations
We denote by 𝔼n[c*(r)] the expected number of non-equivalent root configurations in a random tree of size n drawn under a uniform distribution. This section shows that 𝔼n[c*(r)] grows as an exponential function of n. We first present a lower bound for 𝔼n[c*(r)]. Next, we show that this lower bound is itself bounded below by a quantity that increases exponentially with n.
For the first step, we bound the expectation 𝔼n[c*(r)] by considering a certain set in which each tree satisfies formula (9). For n ≥ 2, define the quantity x = x(n) as the solution of 2x−2 + x = n − 1, and consider the function w′(n) given by w′(2) = 1 and for n ≥ 3,
| (17) |
In Appendix 3, it is shown that w′(n) satisfies w′(n) ≤ n/2, and that w′(n) = n/2 holds only when n = 2, 4, or 6. For 2 ≤ n ≤ 10, the values of (n, w′(n)) are (2, 1), (3, 1), (4, 2), (5, 2), (6, 3), (7, 3), (8, 3), (9, 4), and (10, 4).
The growth of w′(n) is logarithmic. Indeed, for increasing values of n, the ratio x/n becomes small, so that x − 2 = log2[n(1 − (x + 1)/n)] ≈ (log2n) − (x + 1)/(n log 2), where the Taylor approximation log(1 − u) ≈ −u for u near 0 is used. We then obtain x(n) ≈ [n log(4n) − 1]/(n log 2 + 1) ∼ (log n)/(log 2).
For a given n ≥ 2 and a given w ∈ [1, w′(n)], we denote by Tn,w the set of trees of size n such that trS, the smaller root subtree, is a caterpillar of size w, and trL, the larger root subtree, has an unconstrained labeled topology of size n − w (Fig. 7). For a given n ≥ 2, we define the set of trees
Figure 7.
Schematic representation of the unlabeled topology of a tree in set Tn,w. The smaller root subtree, trS, is a caterpillar of size w ∈ [1, w′(n)]. The larger trL, has an unconstrained labeled topology of size n – w. The largest possible value of w, or w′(n), is small enough for trS to be displayed in trL, as in (8). Note that Tn,w1 ⋂ Tn,w2 = ø if w1 ≠ w2.
Four properties can be demonstrated for trees in Tn,w. (i) If w ≥ 2, then each tree t ∈ Tn,w satisfies (8) (Appendix 4), and thus, the number of non-equivalent root configurations in t satisfies (9). Furthermore, note that as was observed in Section 3.3, if t ∈ Tn,1, we have c*(rS) = 0, and (9) holds even though (8) does not.
(ii) For any fixed n ≥ 2 and w ∈ [1, w′(n)], with w ≠ n/2, the probability of observing a given tree t̄ ∈ Tn−w as the rescaled larger root subtree of a tree t ∈ Tn,w selected uniformly at random is, as shown in Appendix 5,
| (18) |
(iii) Because γw = w!/(2 − δw,1) is the number of caterpillar trees of size w ≥ 1 given a set of w labels, the probability pn,w = ℙ[t ∈ Tn,w] for a random tree of size n drawn under a uniform distribution to be in Tn,w can be computed as pn,w = |Tn,w|/|Tn|, or
| (19) |
Here, counts the number of ways of choosing the w taxa for the caterpillar subtree, and we have used (1) to expand |Tn|, |Tw|, and |Tn−w|.
(iv) If w1 ≠ w2, then the sets Tn,w1 and Tn,w2 are disjoint, with Tn,w1 ⋂ Tn,w2 = ∅. Indeed, if t ∈ Tn,w1 ⋂ Tn,w2, then we would have w1 + w2 = n, as t must have a caterpillar of size w1 and a caterpillar of size w2 as root subtrees. However, w1 + w2 cannot equal n, as either w1 < w2 ≤ n/2 or w2 < w1 ≤ n/2.
For a tree t of size n ≥ 2 selected uniformly at random, the mean number 𝔼n[c*(r)] of non-equivalent root configurations can be written by conditioning on , that is,
| (20) |
Here, the probability has been calculated as the sum because is a disjoint union.
The expression in (20) can be replaced by
| (21) |
because for a random tree t ∈ Tn,w selected under a uniform distribution, (9) applies with c*(rS) = w − 1 and c*(rL) = 𝔼n−w[c*(r)]. In particular, c*(rS) = w − 1, as a caterpillar of size w has w − 1 non-equivalent root configurations (Section 4.2), and c*(rL) = En−w[c*(r)], as the larger root subtree trL of a random t ∈ Tn,w selected uniformly has a uniform distribution over Tn−w if w ≠ n/2 (18). If w = n/2—which can happen only for n = 2, 4, or 6—(21) holds because , , and , while 𝔼1[c*(r)] = 0, 𝔼2[c*(r)] = 1, and 𝔼3[c*(r)] = 2.
Using (21) and ignoring the second term in (20) yields the inequality
This inequality can be iterated if n − w ≥ 2 by applying the same procedure to 𝔼n−w[c*(r)]. It follows that for each n ≥ 1, the integer en defined recursively for n ≥ 2 by
| (22) |
where e1 = 0, bounds from below the expectation 𝔼n[c*(r)]. The first values of en and 𝔼n[c*(r)] are reported in Table 1. The values of en match the values of 𝔼n[c*(r)] for n ≤ 7, that is, as long as and the second term in (20) is 0. We also have the following result.
Table 1.
The sequences en, 𝔼n[c*(r)] and 𝔼n[c(r)] for small values of n.
| n | en | 𝔼n[c*(r)] | 𝔼n[c(r)] | |||
|---|---|---|---|---|---|---|
|
| ||||||
| 1 | 0 | 0 | 0 | |||
| 2 | 1 | 1 | 1 | |||
| 3 | 2 | 2 | 2 | |||
| 4 | 3 | 3 |
|
|||
| 5 |
|
|
|
|||
| 6 |
|
|
|
|||
| 7 |
|
|
|
|||
| 8 |
|
|
|
|||
| 9 |
|
|
|
|||
| 10 |
|
|
|
|||
| 11 |
|
|
|
|||
| 12 |
|
|
|
|||
| 13 |
|
|
|
|||
| 14 |
|
|
|
|||
| 15 |
|
|
|
|||
| 16 |
|
|
|
|||
| 17 |
|
|
|
|||
| 18 |
|
|
|
|||
| 19 |
|
|
|
|||
| 20 |
|
|
|
|||
Values of en were computed by using (22). Values of 𝔼n[c*(r)] were computed by generating all possible unlabeled topologies of size n and then using STELLS (Wu, 2012) to obtain the number of non-equivalent root configurations for each unlabeled topology t. The probability of t under a uniform distribution over labeled topologies of size n was obtained by noting that its number of labelings L(t) follows the recursion in Eq. 5.1 of Harding (1971); nonrecursively, the number of labelings is n!/2s(t), where s(t) is the number of internal nodes of t, including cherries and possibly the root, whose two descendant subtrees are isomorphic (this result is obtained by taking the quotient of the results of Theorems 3.5 and 3.3 of Rosenberg (2006)). To compute , we ran STELLS on tree (t, •) in which the two root subtrees were t and the one-taxon tree •. According to (7), the number of root configurations computed by STELLS is , from which the desired is obtained. Values of 𝔼n[c(r)] were computed by the method of Disanto and Rosenberg (2017, Fig. 7).
Proposition 3 The expected number 𝔼n[c*(r)] of non-equivalent root configurations in a random tree of size n ≥ 1 selected under a uniform distribution can be bounded
| (23) |
where en is defined in (22) and 𝔼n[c(r)] is the expected number of root configurations. Furthermore, the sequence 𝔼n[c*(r)] grows exponentially in n, with exponential order at most 4/3.
Proof. The upper bound follows from the fact that for any tree, c*(r) ≤ c(r), and by point (v) in Section 2.4, 𝔼n[c(r)] has exponential order 4/3. All that remains is to show that 𝔼n[c*(r)] grows exponentially in n. To achieve this goal, we prove that the exponential order of the lower bound sequence en strictly exceeds one.
Truncating the sum (22) after the first four terms, for n ≥ 9, we have
| (24) |
The last step follows because according to (19), when n ≥ 9, pn,1 = n/(2n−3), pn,2 = n(n−1)/[2(2n−3)(2n−5)], pn,4 = n(n − 1)(n − 2)(n − 3)/[2(2n − 3)(2n − 5)(2n − 7)(2n − 9)], and
Define the sequence an by an = en for 1 ≤ n ≤ 8, and an = pn,1an−1 + 2pn,2an−2 + 3pn,3an−3 + 4pn,4an−4 for n ≥ 9. From (24), we have, for each n ≥ 1,
| (25) |
When n ≥ 9 and 1 ≤ w ≤ 4, because w ≠ n/2 and δn,w/2 = 0, the probability pn,w in (19) can be written
The recursion for an then becomes
| (26) |
Setting qn = an(2n − 3)!!/n!, we obtain from (26)
| (27) |
Recursion (27) is homogeneous and linear with constant coefficients, and therefore (Sedgewick and Flajolet, 1996, Theorems 3.3 and 4.1), the exponential order of the sequence qn is the inverse of the unique positive solution z0 of the characteristic equation 1 = z + z2 + 3z3/2 + 2z4.
Solving the equation numerically, we find qn ⋈ (1/z0)n, where z0 ≈ 0.4845. In particular, the exponential order 1/z0 of the sequence qn strictly exceeds 2. Using (2) to rewrite (2n − 3)!!, and observing by Stirling's formula that , it follows that sequence an = qnn!/(2n − 3)!! has exponential growth
Therefore, the exponential order of the sequence an is 1/(2z0) ≈ 1.0320 > 1. By inequality (25), the sequence en grows exponentially in n.
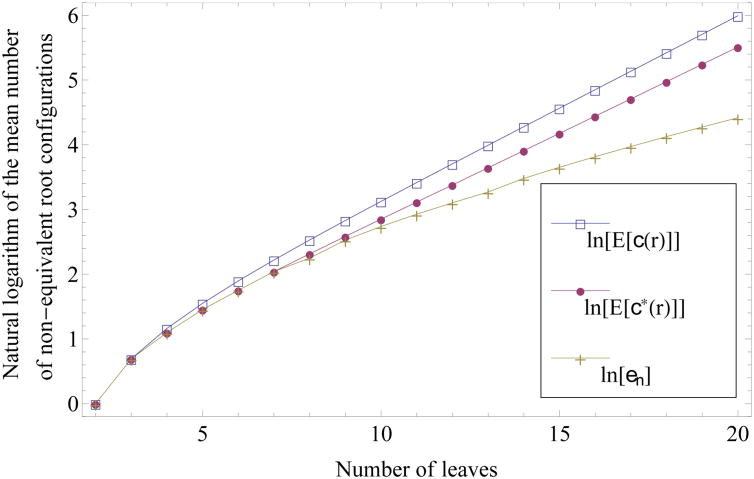
For n ≤ 20, the exact values of en, 𝔼n[c*(r)], and 𝔼n[c(r)] are reported in Table 1 and plotted in Fig. 8. The figure illustrates that the numerical values of log 𝔼n[c*(r)], though initially coincident with the values of log en, are already closer to the values of log 𝔼n[c(r)] by n = 20. This observation suggests that in bounding 𝔼n[c*(r)] from below to demonstrate its exponential growth, the steps we have taken have led to a bound that is quite loose; the exponential growth of 𝔼n[c*(r)] is likely to have a comparable magnitude to that of 𝔼n[c(r)], or 4/3.
Figure 8.
Natural logarithm of the mean number 𝔼n[c*(r)] of non-equivalent root configurations for labeled topologies of size 2 ≤ n ≤ 20. The value for n = 1, log(0), is omitted. The natural logarithms of the bounds en and 𝔼n[c(r)] (23) determine the lower and upper lines. Exact values for the three quantities are reported in Table 1.
6 Discussion
For labeled gene tree topologies t that match the labeled species tree topology, we have extended the enumerative study of ancestral configurations, considering non-equivalent configurations specified by an equivalence relation that groups ancestral configurations according to symmetries in t. We have focused on the exponential growth in the tree size |t| = n of the number of non-equivalent configurations present at the root of t.
We have shown that when t satisfies certain constraints, its number of non-equivalent root configurations can be recursively computed from corresponding quantities for its root subtrees. The recursion (9), which shares three of its five terms with an analogous recursion for root configurations (Disanto and Rosenberg, 2017, Proposition 1), enables the study of the number of non-equivalent root configurations for special tree families. For the family of completely balanced trees, the number of non-equivalent root configurations and the total number of non-equivalent configurations grow exponentially with order in n (Proposition 1). Comparing this constant with the exponential orders of the numbers of root configurations and total configurations in the family, both of which equal k0 ≈ 1.5028 (Disanto and Rosenberg, 2017), we see that for the completely balanced trees, the number of configurations grows exponentially faster than the number of non-equivalent configurations. Their symmetric structure collapses the set of configurations into fewer non-equivalent configurations.
A different recursively defined tree family (un), however, has asymptotically more non-equivalent configurations than the balanced trees, its number of root configurations growing with exponential order (Proposition 2). This value is close to the upper bound of k0 ≈ 1.5028 on the exponential order of the maximal number of configurations across all labeled topologies of size n (Disanto and Rosenberg, 2017, Corollary 1). Although the unlabeled shapes that give rise to the largest numbers of non-equivalent root configurations (Figure 5) and root configurations (Disanto and Rosenberg, 2017, Figure 3) are not in general the same, the maximal numbers of non-equivalent configurations and configurations have comparable exponential order.
As was found by Wu (2012), the growth of the number of non-equivalent configurations for some tree families (e.g. caterpillars) can be polynomial in n. Assuming a uniform distribution over the labeled topologies with size n, however, we have shown that the expected number of non-equivalent configurations for a random labeled topology of size n grows exponentially (Proposition 3). The exponential order of this growth is bounded below by 1/(2z0) ≈ 1.0320; numerical exploration suggests that it is closer to the upper bound of 4/3 that describes the exponential order of the mean number of configurations (Disanto and Rosenberg, 2017, Proposition 5).
We focused on the situation in which the gene tree and species tree have a matching topology. In the non-matching case, in parallel to a similar result for configurations (Disanto and Rosenberg, 2017), it is possible that the number of non-equivalent root configurations and the total number of non-equivalent configurations exceed the corresponding values for matching gene trees and species trees. This claim can be verified in a simple example. Let χn = ((… ((a1, a2), a3), …), an) be a caterpillar species tree, and label the unique internal node with k descendants by bk for 2 ≤ k ≤ n. For a matching caterpillar gene tree, all configurations are non-equivalent, the number of non-equivalent configurations at node bk is c*(bk) = k − 1, the number of root configurations is c*(bn) = n − 1, and the total number of configurations is .
Continuing with χn as the species tree topology, consider a gene tree topology
with n ≥ 6. The gene trees (ξn) represent a caterpillar family (Disanto and Rosenberg, 2016) with seed tree (((a1, a2), a3), (a4, a5)). We label the node of ξn ancestral to a1 and a2 by d2, the node ancestral to a1, a2, and a3 by d3, the node ancestral to a4 and a5 by , and the unique node ancestral to k taxa, 5 ≤ k ≤ n, by dk. Following Wu (2012), the definition of equivalent configurations in the non-matching case generalizes the definition in Section 3.1. Consider a gene tree G, a species tree S, a node κ of S, and two configurations γ1, γ2 at node κ—two possible sets of gene lineages that could be present in S at κ under different realizations of G in S. Let κ′ be the most recent common ancestor of the lineages of G collected in the set γ1, and note that κ′ is also the most recent common ancestor of the lineages collected in γ2. Following the terminology of Section 3.1, we say that γ1, γ2 are equivalent at κ when the unlabeled tree shape Gκ′(γ1) is isomorphic to the unlabeled tree shape Gκ′(γ2). We denote by C*(κ) and c*(κ) the set of non-equivalent configurations at κ and its cardinality, respectively.
Proceeding sequentially through the internal nodes of χn, the non-equivalent configurations are C*(b2) = {{a1, a2}}, C*(b3) = {{a1, a2, a3}, {d2, a3}}, C*(b4) = {{a1, a2, a3, a4}, {d2, a3, a4}, {d3, a4}}, and C*(b5) = {{a1, a2, a3, a4, a5}, {d2, a3, a4, a5}, {d3, a4, a5}}, with c*(b2) = 1, c*(b3) = 2, c*(b4) = 3, and c*(b5) = 3. At node b6 of χn, the non-equivalent configurations are C*(b6) = {{a1, a2, a3, a4, a5, a6}, {d2, a3, a4, a5, a6}, {d3, a4, a5, a6}, {a1, a2, a3, , a6}, {d3, , a6}, {d5, a6}}, and configuration {d2, a3, , a6} is not included owing to equivalence with {d3, a4, a5, a6}.
For 7 ≤ k ≤ n, C*(bk) is obtained by augmenting configuration {dk−1, ak} to the set of all configurations formed by adding taxon ak to the non-equivalent configurations in C*(bk−1); none of the resulting configurations are equivalent, and c*(bk) = c*(bk−1) + 1. The number of non-equivalent root configurations of ξn for n ≥ 6 is c*(bn) = n, and the number of total configurations is . Because n > n − 1 and n(n + 1)/2 − 6 > n(n − 1)/2 for n ≥ 7, non-equivalent root configurations and total non-equivalent configurations are more numerous for the non-matching ξn than for the matching caterpillar.
Our enumerative results on ancestral configurations can help to compare the cost of procedures for calculating gene tree probabilities recursively using ancestral configurations (Wu, 2012) to those that proceed nonrecursively using a different data structure, the “coalescent histories” (Degnan and Salter, 2005; Rosenberg, 2007; Than et al., 2007; Rosenberg and Degnan, 2010; Rosenberg, 2013; Disanto and Rosenberg, 2015, 2016). In this context, it is noteworthy that the trees un, which have many non-equivalent root configurations, have a similar recursive structure to the lodgepole trees, which have large numbers of coalescent histories (Disanto and Rosenberg, 2015).
Note that unlike for root configurations, we did not prove a general result describing the unlabeled shapes of trees that give rise to the most non-equivalent root configurations, merely evaluating the number of non-equivalent root configurations for trees un and noting by exhaustive computation that this value is near the maximum for small trees. We also did not produce a general relationship between non-equivalent root configurations and total non-equivalent configurations. For the family of completely balanced trees, the number of non-equivalent root configurations and the total number of non-equivalent configurations have the same exponential growth, as the maximal number of non-equivalent configurations across all internal nodes of a balanced tree is reached at its root (Proposition 1). However, we did not provide a generalization that such a maximum is applicable for arbitrary trees. Because it is the non-equivalent configurations that are employed by Wu (2012) in gene tree probability computations, their further exploration will be important for understanding the relative computational complexity of gene tree probability computations with different species trees.
Acknowledgments
We thank Elizabeth Allman, James Degnan, and John Rhodes for discussions, and two reviewers for comments. Support was provided by National Institutes of Health grant R01 GM117590 and by a 2014 Rita Levi Montalcini grant to FD from the Ministero dell'Istruzione, dell'Universitá e della Ricerca.
Appendix 1
Proof of (9)
Let C*(rS) = {γS,1, …, γS,q} with c*(rS) = q, and let C*(rL) = {γL,1, …, γL,Q}, with c*(rL) = Q. Because condition (8) is satisfied, the entire tree trS can be displayed in trL, each configuration γS,i ∈ C*(rS) has exactly one corresponding configuration γL,i ∈ C*(rL) such that trS(γS,i) ≅ trL(γL,i), and Q ≥ q.
From (6), we obtain
which can be further decomposed as
| (28) |
| (29) |
| (30) |
| (31) |
| (32) |
We merge equivalent configurations to obtain C*(r) from C̃(r). From (29), we remove those in {γS,1, …, γS,q} ⊗ {{rL}}, as they are equivalent to those in {{rS}} ⊗ {γL,1, …, γL,q}. Thus, we take only q among the 2q configurations in (29). Moreover, due to the equivalence γS,i⋃γL,j ∼r γS,j ⋃ γL,i, we take only those configurations of the form γS,i ⋃ γL,j with i ≤ j among those in {γS,1, …, γS,q} ⊗ {γL,1, …, γL,q}. Thus, among the q2 configurations in (31)—those with 1 ≤ i, j ≤ q—we take only q(q + 1)/2 non-equivalent ones. No equivalences are possible among configurations in (28), (30), and (32), and all are retained in C*(r). From (28)-(32), we then have
Replacing q by c*(rS) and Q by c*(rL) gives (9).
Appendix 2
Proof of (12)
The proof follows the approach of Aho and Sloane (1973, Section 3) for solving certain recurrences. From (11), we have . Taking the logarithm yh = log xh yields yh+1 = 2yh + αh, where . Following Aho and Sloane (1973), yh has solution
| (33) |
Converting back to xh = exp(yh), from (33) we have
where the last step uses the fact that x0 = 1/2.
We then have
When h → ∞, the sum converges to zero because it can be bounded , where, because xh → ∞ as h → ∞, αh → 0 as h → ∞. It follows that converges to 1, producing (12).
Appendix 3
Properties of w′(n)
We prove that for each n ≥ 2, w′(n) ≤ n/2, with equality only for n = 2, 4, or 6. The result is verified by direct computation of w′(n) for 2 ≤ n ≤ 7. For n ≥ 8, by definition, w′(n) = ⌊x⌋, where x satisfies 2x–2 + x = n – 1. Seeking a contradiction, suppose ⌊x⌋ = w′(n) ≥ n/2. Because x ≥ ⌊x⌋, we would have x ≥ n/2, and therefore n – 1 = 2x–2 + x ≥ 2n/2–2 + n/2 ≥ 2(n/2 – 2) + n/2 = 3n/2 – 4, noting that 2u ≥ 2u for u ≥ 2. The inequality n – 1 ≥ 3n/2 – 4 cannot hold if n ≥ 8. Therefore, when n ≥ 8, we must have w′(n) < n/2.
Appendix 4
Proof that trees in Tn,w satisfy (8) for w ≥ 2
We first prove that given any w ≥ 2, a caterpillar tree t1 of size |t1| = w can be displayed in any tree t2 of size |t2| ≥ 2w–2 + 1 through a root configuration γ of t2, that is, t1 ≅ t2(γ). The proof is by induction on w.
For w = 2, we have |t2| ≥ 2 and the result follows by taking the root configuration γ determined by the left and right descendants of the root in t2. For the inductive step, because |t2| ≥ 2w–2 + 1, the larger root subtree of t2 has size at least ⌈|t2|/2⌉ ≥ ⌈2w–3 + 1/2⌉ = 2w–3 + 1. By the inductive hypothesis, the larger root subtree of t2 can display a caterpillar of size w – 1 through a root configuration γ′. Taking the root configuration γ of t2 obtained as γ = γ′ ⋃ {ρ}, where ρ is the root of the smaller root subtree of t2, we have t1 ≅ t2(γ) as desired.
Now suppose we are given a tree t ∈ Tn,w, with 2 ≤ w ≤ w′(n). The smaller root subtree trS of t is by definition a caterpillar of size w ≥ 2, and the larger root subtree trL has size |trL| = n – w. By definition, w ≤ w′(n) = ⌊x⌋ ≤ x, where x = n – 2x–2 – 1, and therefore, w ≤ n – 2w–2 – 1. In particular, |trL| = n – w ≥ 2w–2 + 1. From what we have shown above, a root configuration γ of trL exists such that trS ≅ trL(γ).
Appendix 5
Proof of (18)
Recall that for each tree t ∈ Tn,w, the smaller root subtree trS is a caterpillar of size w ∈ [1,w′] and the larger root subtree trL has size n – w. Because we assume w < n/2, trS and trL have different sizes and different unlabeled topologies. Given a tree t̄ ∈ Tn–w, the number of trees in Tn,w such that trL = t̄ (after rescaling labels for the taxa) is , where γw is the number of caterpillar labeled topologies of size w. Dividing by yields the probability ℙ[trL = t̄|t ∈ Tn,w] = 1/|Tn–w| as desired.
References
- Aho AV, Sloane NJA. Some doubly exponential sequences. Fibonacci Q. 1973;11:429–437. [Google Scholar]
- Allman ES, Degnan JH, Rhodes JA. Identifying the rooted species tree from the distribution of unrooted gene trees under the coalescent. J Math Biol. 2011;62:833–862. doi: 10.1007/s00285-010-0355-7. [DOI] [PubMed] [Google Scholar]
- Degnan JH, Salter LA. Gene tree distributions under the coalescent process. Evolution. 2005;59:24–37. [PubMed] [Google Scholar]
- Disanto F, Rosenberg NA. Coalescent histories for lodgepole species trees. J Comput Biol. 2015;22:918–929. doi: 10.1089/cmb.2015.0015. [DOI] [PubMed] [Google Scholar]
- Disanto F, Rosenberg NA. Asymptotic properties of the number of matching coalescent histories for caterpillar-like families of species trees. IEEE/ACM Trans Comput Biol Bioinf. 2016;13:913–925. doi: 10.1109/TCBB.2015.2485217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disanto F, Rosenberg NA. Enumeration of ancestral configurations for matching gene trees and species trees. J Comput Biol. 2017 doi: 10.1089/cmb.2016.0159. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. The number of evolutionary trees. Syst Zool. 1978;27:27–33. [Google Scholar]
- Felsenstein J. Inferring Phylogenies. Sunderland, MA: Sinauer; 2004. [Google Scholar]
- Flajolet P, Sedgewick R. Analytic Combinatorics. Cambridge: Cambridge University Press; 2009. [Google Scholar]
- Harding EF. The probabilities of rooted tree-shapes generated by random bifurcation. Adv Appl Prob. 1971;3:44–77. [Google Scholar]
- Rosenberg NA. The mean and variance of the numbers of r-pronged nodes and r-caterpillars in Yule-generated genealogical trees. Ann Comb. 2006;10:129–146. [Google Scholar]
- Rosenberg NA. Counting coalescent histories. J Comput Biol. 2007;14:360–377. doi: 10.1089/cmb.2006.0109. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA. Coalescent histories for caterpillar-like families. IEEE/ACM Trans Comp Biol Bioinf. 2013;10:1253–1262. doi: 10.1109/tcbb.2013.123. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Degnan JH. Coalescent histories for discordant gene trees and species trees. Theor Pop Biol. 2010;77:145–151. doi: 10.1016/j.tpb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Sedgewick R, Flajolet P. An Introduction to the Analysis of Algorithms. Boston: Addison-Wesley; 1996. [Google Scholar]
- Than C, Ruths D, Innan H, Nakhleh L. Confounding factors in HGT detection: statistical error, coalescent effects, and multiple solutions. J Comput Biol. 2007;14:517–535. doi: 10.1089/cmb.2007.A010. [DOI] [PubMed] [Google Scholar]
- Wu Y. Coalescent-based species tree inference from gene tree topologies under incomplete lineage sorting by maximum likelihood. Evolution. 2012;66:763–775. doi: 10.1111/j.1558-5646.2011.01476.x. [DOI] [PubMed] [Google Scholar]