Abstract
The evolutionarily conserved Piezo family of proteins, including Piezo1 and Piezo2, encodes the long‐sought‐after mammalian mechanosensitive cation channels that play critical roles in various mechanotransduction processes such as touch, pain, proprioception, vascular development and blood pressure regulation. Mammalian Piezo proteins contain over 2500 amino acids with numerous predicted transmembrane segments, and do not bear sequence homology with any known class of ion channels. Thus, it is imperative, but challenging, to understand how they serve as effective mechanotransducers for converting mechanical force into electrochemical signals. Here, we review the recent major breakthroughs in determining the three‐bladed, propeller‐shaped structure of mouse Piezo1 using the state‐of‐the‐art cryo‐electron microscopy (cryo‐EM) and functionally dissecting out the molecular bases that define its ion permeation and mechanotransduction properties, which provide key insights into clarifying its oligomeric status and pore‐forming region. We also discuss the hypothesis that the complex Piezo proteins can be deduced into discrete mechanotransduction and ion‐conducting pore modules, which coordinate to fulfil their specialized function in mechanical sensing and transduction, ion permeation and selection.
Keywords: Piezo1, Piezo2, mechanotransduction, mechanosensitive ion channels, ion permeation, ion selectivity, mechanogating, topology, cryo‐electron microscopy, membrane protein
Introduction
Mechanosensitive (MS) cation channels represent a principal type of mechanotransducer specialized in effectively converting mechanical force into electrochemical signals, which is critical for various forms of mammalian physiology and pathophysiology involving mechanotransduction, such as touch, proprioception, pain, vascular development and blood pressure regulation (Chalfie, 2009; Ranade et al. 2015). Despite awareness of the functional significance of MS cation channels in mammals, their molecular identities have remained elusive for decades, significantly hampering our understanding of the mechanotransduction processes in vivo and the molecular mechanisms underlying the mechano‐gating and ion permeation properties.
In 2010, Patapoutian and colleagues made the groundbreaking discovery that the Piezo family of genes, including Piezo1 and Piezo2, encodes essential components of mammalian MS cation channels (Coste et al. 2010). Combining microarray analysis and RNAi‐mediated knockdown of a total of 73 candidate genes, they identified a single gene, Piezo1 (also known as Fam38A), that is required for the mechanically activated cationic currents in the mouse Neuro2A neuroblastoma cell line. Further sequence homology analysis led to the identification of the homologous gene Piezo2. Remarkably, heterologous expression of either Piezo1 or Piezo2 in various mammalian cell lines generated MS cationic currents, making them the first family of genes identified in mammals that is both necessary and sufficient for generating MS cationic currents.
Piezo proteins are evolutionarily conserved, but possess unique features in their primary sequences. Mammalian Piezos are large transmembrane proteins that are composed of about 2500–2800 amino acids and are predicted to encompass between 26 and 40 transmembane segments (TMs). Furthermore, they lack apparent sequence homology with any known protein classes including ion channels (Coste et al. 2010). Thus, it was unknown whether Piezo proteins are the pore‐forming subunits of the MS cation channels, or are non‐conducting subunits of yet‐to‐be‐identified ion channels required for proper expression or for modulating channel properties.
In a subsequent study, Patapoutian and colleagues provided several lines of evidence to support the hypothesis that Piezo proteins are the pore‐forming subunits (Coste et al. 2012). First, they found that Piezo‐mediated MA currents display species‐dependent pore properties. For instance, Ruthenium Red (RR) acts as an effective pore blocker of mouse Piezo1/2 (mPiezo1/2) (Coste et al. 2010). In contrast, Drosophila Piezo (dPiezo) is insensitive to RR blockage (Coste et al. 2012). Furthermore, under similar recording conditions, the slope conductances for mPiezo1 and dPiezo are about 30 pS and 3 pS, respectively. These data indicate that Piezo proteins form the ion channel by themselves. Secondly, they successfully purified the glutathione S‐transferase (GST)‐tagged C‐terminal mPiezo1 fusion protein (mPiezo1‐GST) using HEK293T cells as the heterologous expression system. Both native gel and formaldehyde cross‐linking analyses indicate that the purified mPiezo1‐GST proteins form oligomers. Importantly, when the purified proteins were reconstituted into lipid bilayers, they detected spontaneous single‐channel activities, which were blocked by RR. The single channel conductances were ∼120 pS and 60 pS in symmetric 0.5 m and 0.2 m KCl, respectively. The reconstituted proteins also conducted sodium, consistent with Piezo1 proteins being non‐selective cation channels. Taken together, these data strongly suggest that Piezo proteins are the pore‐forming subunits of MS cation channels. However, given that the ion‐conducting pore was not identified in this study, Nilius et al. had argued that it was still premature to claim that Piezos are indeed pore‐forming subunits of the MS cation channels (Nilius & Honore, 2012). As described later, after resolving the cryo‐EM structure of the full‐length mPiezo1 protein and unveiling its ion‐conducting pore (Ge et al. 2015; Zhao et al. 2016), we provided definitive evidence to establish Piezo proteins as the prototypic class of MS cation channels in mammals.
The identification and demonstration of Piezo proteins as the bona fide MS cation channels has heralded a new era of research in the mechanotransduction field and tremendous progress has been made towards characterizing their physiological and pathophysiological significance, human diseases, biophysical properties and molecular mechanisms (Ranade et al. 2015; Geng et al. 2017; Gottlieb, 2017). Physiologically, Piezo1 has been proposed to function as a mechanotransducer required for vascular development, remodelling and blood pressure regulation (Li et al. 2014; Ranade et al. 2014; Retailleau et al. 2015; Wang et al. 2016; Rode et al. 2017). Additionally, it plays critical roles in red blood cell volume regulation (Faucherre et al. 2014; Cahalan et al. 2015), cell migration (McHugh et al. 2012), homeostasis of epithelial cell numbers (Eisenhoffer et al. 2012; Gudipaty et al. 2017), high‐strain mechanotransduction of cartilage (Lee et al. 2014; Rocio Servin‐Vences et al. 2017), regulation of urinary osmolarity (Martins et al. 2016), neural stem cell fate determination (Pathak et al. 2014), and neuronal axon guidance (Koser et al. 2016). Piezo2 functions as a major mechanotransduction channel for sensation of gentle touch (Ikeda et al. 2014; Maksimovic et al. 2014; Ranade et al. 2014; Woo et al. 2014), proprioception (Woo et al. 2015), airway stretch and lung inflation (Nonomura et al. 2016). Pathophysiologically, mutations in Piezo1 are associated with dehydrated hereditary stomatocytosis, lymphatic dysplasia, hereditary high phosphatidylcholine haemolytic anaemia and haemochromatosis‐induced diabetes mellitus (Zarychanski et al. 2012; Albuisson et al. 2013, Bae et al. 2013; Szklarczyk et al. 2013; Beneteau et al. 2014, Sandberg et al. 2014, Andolfo et al. 2015; Fotiou et al. 2015). Mutations in Piezo2 are linked to distal arthrogryposis, Gordon syndrome, Marden‐Walker syndrome and scoliosis with peripheral sensory dysfunction (Coste et al. 2013; McMillin et al. 2014; Chesler et al. 2016). Taken together, these studies demonstrate the importance of Piezo‐associated mechanotransduction in physiology and pathophysiology and their potential as validated drug targets for therapeutic development.
Given their importance functionally and as the prototype of mammalian MS cation channels, it is pivotal to understand how Piezo channels function at the molecular level. Here we focus on reviewing the recent progress towards uncovering the structure–function relationship that enables Piezo proteins to function as sophisticated MS cation channels, with a particular emphasis on the recently resolved cryo‐electron microscopy (cryo‐EM) structure of Piezo1 and the structure‐guided characterization of the molecular bases that underlie its ion permeation and mechanotransduction properties (Ge et al. 2015; Zhao et al. 2016). We also discuss the specific scenarios where the application of advanced techniques not only transforms the structure–function characterization of this novel and complex Piezo channel but also leads to effective clarification of paradoxical findings arising along the course of studies in the field.
The three‐bladed, propeller‐like homo‐trimeric architecture of mPiezo1
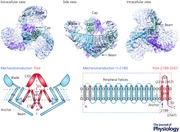
Taking advantage of a recent technical breakthrough, the application of cryo‐EM for determining high‐resolution three‐dimensional (3D) structure of membrane proteins (Cao et al. 2013; Liao et al. 2013, 2014; Paulsen et al. 2015), in combination with our expertise in Piezo protein engineering, expression and purification in mammalian expression systems, we have spearheaded the effort to resolve the 3D structure of Piezo proteins. After testing various constructs and detergents, a combination of the full‐length mouse Piezo1 protein with the C12E10 detergent was selected for structural determination using the state‐of‐the‐art Titan Krios microscope operated at a voltage of 300 kV with a K2 Summit direct electron detector. These collaborative efforts eventually led to the determination of the 3D cryo‐EM structure of the full‐length (2547 amino acids) mPiezo1 at a medium resolution (Ge et al. 2015). Strikingly, mPiezo1 is organized into a unique three‐bladed, propeller‐shaped architecture with distinct domains resembling the typical structural components of a propeller, including a central cap and three distal blades in one side, three featured beams in the opposite side, and the transmembrane (TM) region in between (Abstract Figure). The relatively better density associated with the TM regions allowed us to build a de novo alanine model with 502 amino acids for the apparent TM segments, the beams and the C‐terminal domain (CTD). Together with the 227 amino acids of the C‐terminal extracellular domain (CED) that was crystallized, 719 out of 2547 amino acids of each protomer were built into the density map (Abstract Figure). The lack of information on side‐chains of the structure prevented us from unambiguously assigning a primary sequence to the featured structural domains. Here we first review the distinct structural domains and then discuss the studies that have addressed the oligomeric status, bona fide ion‐conducting pore and mechanotransduction components of the Piezo1 channel.
The central cap
The cap‐forming region was identified through two independent approaches (Ge et al. 2015). The topological model predicts that the residues from 2210 to 2457 between the last two TMs constitute a large extracellular loop, termed CED. When this region was deleted and the resulting deletion‐mutant of mPiezo1Δ2218‐2453 was purified and observed by negative‐staining EM, the central cap was found to be absent in the 2D class averages, suggesting that this region accounts for the cap. Furthermore, the soluble CED fragment (residues from 2218 to 2457) was crystallized. The CED proteins form a trimeric complex both in solution and in the crystal lattice. The crystallographic trimer fits well into the cryo‐EM density map of the cap. Taken together, these results conclusively demonstrate that the CED trimerizes to form the cap. Using live labelling of the FLAG‐tag from HEK293T cells expressing the mPiezo1 with a FLAG‐tag inserted after the residue 2419, CED was experimentally demonstrated to be on the extracellular side. Consequently, the overall topology of the homotrimeric structure was determined. Both the central cap and the three blades are extracellular, while the beams and CTDs are intracellular (Abstract Figure).
The extended and twisted TM wing
The whole TM region displays a three‐winged arrangement, with each extended wing being slightly twisted (Abstract Figure). Unexpectedly, each wing has only 14 apparently resolved TMs, much lower than the predicted number. However, given the low resolution at the peripheral blade region, it remains possible that additional TMs might exist, but were not resolved in the current Piezo1 structure. The two innermost TMs converged at the centre are termed inner helix (IH) and outer helix (OH), respectively, while those TMs located in the extended wings are termed as peripheral helix (PH) (Abstract Figure and Fig. 1). The OH and IH could be tentatively traced to the N‐ and C‐terminus of the CED, respectively, albeit the lack of reliable density connections at the junction points made this assignment ambiguous (Abstract Figure and Fig. 1). Functional characterizations validated that the pore‐lining IH is formed by the last TM, while OH is composed of the second‐to‐the‐last TM (Abstract Figure and Fig. 1) (Zhao et al. 2016).
Figure 1. Structure of the pore module of mPiezo1.
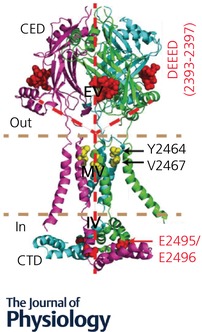
The structurally revealed pore module with functionally characterized residues highlighted.
Coste et al. have carried out extensive efforts to experimentally determine the TM topological model (Coste et al. 2015). They generated a total of 48 constructs with insertion of Myc‐tag into predicted loops, then used live‐labelling to locate those extracellularly facing tag positions. These studies led to the identification of nine extracellular loops. Together with bioinformatics analysis and identification of intracellular phosphorylation sites, they have proposed possible topological models of 38‐, 18‐ or 10‐TMs. Thus, the TM topology remains elusive and clearly requires better structural resolution for validation.
Anchor and subunit swapping
Interestingly, a unique hairpin structure, termed anchor domain, exists at the interface of two adjacent subunits, which contains several helices and penetrates into the inner leaflet of the membrane, with a long helix roughly parallel to the membrane (Ge et al. 2015). This anchor domain connects OH to the PHs (Abstract Figure). Given that the OH is formed by the second‐to‐last TM (around residues 2101–2213), anchor is composed of residues from ∼2100 to 2190 where the most evolutionally conserved sequence motif PF(X2)E(X6)W (2129–2140) among Piezo homologues (Prole & Taylor, 2013) and the disease causing mutation T2142 (T2127 of human Piezo1) have been identified (Andolfo et al. 2013), indicating its structural and functional importance. Intriguingly, the anchor domain results in swapping the OH‐CED‐IH‐containing region of one subunit into the neighbouring subunit in a clockwise direction (viewed from the cap), leading to a clear separation of the three subunits on the 3D structure (Abstract Figure). This domain swapping mechanism might be critical for stabilizing the trimeric structure.
The long intracellular beam
The Piezo1 structure displays three apparent beam‐like structures exposed at the intracellular surface (Abstract Figure). The beam is about 90 nm in length and positioned in a 30 degree angle relative to the membrane plane. It appears to originate peripherally at the intracellular side of the PH7–8 pair, and ends near the central axis of the channel complex, where it forms extensive interactions with the anchor and CTD. The composition of the beam remains unidentified. Nevertheless, the unique organization suggests that the three beams may not only support the TM region, but also transmit conformational changes from peripheral TMs and blades to the central region where the ion‐conduction pore is located.
The flexible blade
The local resolution map shows that the three blade‐resembling structures of Piezo1 have much lower resolution than other regions, including the cap, TMs, beam and CTD. Comparison of different classes of the structures from symmetry‐free 3D classification revealed that the blades are highly mobile (Ge et al. 2015). The large conformational heterogeneity in the blades could be the main factor prevented us from obtaining high‐resolution structure. The composition of the blade remains unassigned. A single blade has a volume comparable to the cap region, which is made up of about 700 residues. Given that the PHs are organized in a curved fashion (Abstract Figure), it remains possible that some of the predicted N‐terminal TMs might reside in the blade, which would make an unusually curved TM wing.
Piezo1 forms a homo‐trimer
The Piezo1 structure clearly shows a trimeric oligomerization state (Abstract Figure) (Ge et al. 2015). In the previous study (Coste et al. 2012), the purified mPiezo1‐GST fusion proteins were subjected to stoichiometry analysis using both native gel and cross‐linking. The calculated molecular weight of the mPiezo1‐GST fusion protein is ∼318 kDa. When assayed with native gel, a major Piezo1‐specific band at the position near the 1236 kDa‐marker and a very weak band near the 1048 kDa‐marker were detected. Similar results were obtained by Ge et al using the mPiezo1‐GST protein sample (Ge et al. 2015). When assayed with formaldehyde cross‐linking experiments, four Piezo1‐specific bands were detected, albeit the topmost band was always weak. These data suggest that mPiezo1‐GST fusion proteins form oligomers, likely to be tetramers. However, when examined by negative staining EM, the mPiezo1‐GST fusion protein was apparently found to exist as dimer of trimers organized in a back‐to‐back manner (Ge et al. 2015). By contrast, the GST‐tag‐cleaved mPiezo1 proteins displayed individual trimers. On the basis of the Piezo1 structure, it is now clear that the three C‐terminal ends of the trimeric complex, where GST is fused in the mPiezo1‐GST protein, are located in the centre at the intracellular side (Abstract Figure), allowing three pairs of GST to dimerize and consequently form stable dimer of trimers of the mPiezo1‐GST fusion proteins. It is unclear what drives the co‐incident migration of the 1.8 MDa mPiezo1‐GST dimer of trimers near the 1.2 MDa marker. The large native size of the fusion protein together with its numerous TMs might be the cause. Together with the finding that CED alone forms a trimer, the structural and biochemical data are reconciled to demonstrate that purified mPiezo1 forms a homo‐trimer instead of tetramer.
The bona fide ion‐conducting pore module (residues 2189–2547)
As pore‐forming subunits, Piezo proteins should encode ion‐conducting pores. Indeed, the mPiezo1 structure shows a continuous central channel along the Z‐axis including, an extracellular vestibule (EV) within the cap, a transmembrane vestibule (MV) enclosed by the three IHs, and an intracellular vestibule (IV) formed by the CTD (Fig. 1) (Ge et al. 2015). Thus, this central region, including the IH‐OH pairs, CED and CTD, comprises the putative pore‐module of Piezo1 channels (Abstract Figure and Fig. 1). This pore module displays a remarkable architectural similarity to trimeric acid‐sensing ion channel 1 (ASIC1) and ATP‐gated P2X4 receptors that essentially contain two TMs and a large extracellular domain in each promoter (Gonzales et al. 2009; Kawate et al. 2009).
The pore module has been tentatively assigned into the most distal C‐terminal region (residues from 2189 to 2547) of mPiezo1 that contains the last two putative TMs, CED and CTD (Abstract Figure) (Ge et al. 2015). However, the lack of information about side‐chains and reliable density connections between CEDs, which alone have atomic‐resolution structures, and the flanking OH‐IH TM pairs introduces important uncertainties about the assignment of a primary sequence into the pore region (Fig. 1) (Ge et al. 2015).
The assignment apparently contradicts a previous study showing that human Piezo1 lacking the entire C‐terminus starting from residue 2218 (the corresponding residue 2234 in mPiezo1) retains stretch‐activated channel activities (Bae et al. 2013), indicating that this region does not encode the ion‐conducting pore. However, neither mPiezo1‐2234 nor hPiezo1‐2218 gave rise to MA whole‐cell currents (Bae et al. 2013; Zhao et al. 2016). Furthermore, a portion of either the mutant‐ or control‐transfected cells (also observed in a previous study: Bae et al. 2011) showed stretch‐activated currents only at high negative pressures, which were substantially different from wild‐type Piezo1‐mediated currents and were insensitive to RR blocking (Zhao et al. 2016). These data indicate that this type of potentially endogenous current or artifact is unlikely to be mediated by the bona fide pore of Piezo1 channels.
Using chimeras and alanine scanning of negatively charged residues (the C‐terminal region after residue 2234 was excluded from screening based on the study by Bae et al. as described above; Bae et al. 2013), Coste et al. have identified E2133 as a residue affecting unitary conductance and ion selectivity of Piezo channels (Coste et al. 2015). Based on topology prediction, the E2133‐residing region is located in the extracellular side. As identification of residues that affect pore properties has been generally considered as the gold standard for indication of pore location, these results could suggest that the E2133‐residing region is the pore‐forming region in the absence of structural information (Coste et al. 2015). However, on the basis of the mPiezo1 structure, E2133 is located in the anchor domain at the intracellular side rather than in the structurally indicated pore module (Ge et al. 2015). Thus, this residue most likely affects the essential pore properties via an allosteric effect.
Given the ambiguity of the structural assignment and inconsistency with reported structure‐function evidence, we carried out systematic structure‐guided studies and obtained the following experimental evidence to support the structural assignment of the pore‐forming region of the Piezo1 channel (Zhao et al. 2016). First, replacing residues 2189–2547 of mPiezo1 with the corresponding region of dPiezo resulted in a chimeric channel that was insensitive to RR blockage and had dPiezo‐like single‐channel properties. These data suggest that residues 2189–2547 of mPiezo1 determine the fundamental pore properties, including unitary conductance and pore blockage. Second, using the substituted‐cysteine accessibility method (SCAM) that has been widely used for probing channel pore positions (Akabas et al. 1992; Li et al. 2011), among eight cysteine mutants of the consecutive residues (2461–2468) that likely span two turns of alpha helix in the extracellular side of the last TM, Y2464C and V2467C were found to be covalently modified by the thiol‐reactive reagent 2‐sulfonatoethyl methanethiosulfonate sodium salt (MTSES) (Fig. 1). These data suggest that the last TM forms the pore‐lining IH of the Piezo1 channel. Third, deleting the CED or mutating a large patch of negatively charged residues (DEEED at the position from 2393 to 2397) at the fenestration sites drastically reduced the MA currents (Fig. 1). Furthermore, the DEEED (2393–2397)‐AAAAA mutant had significantly higher permeability to chloride (P Cl/P Na = ∼27%) than mPiezo1 (P Cl/P Na = ∼8%), but normal Ca2+ permeability over monovalent Cs+. We propose that Piezo1 may employ a negative electrostatic potential for selecting cation over anion entry to the pore and ensuring efficient ion conduction. Additionally, the CED was found to determine the distinct pore properties between mPiezo1 and dPiezo. Last, the acidic residues, E2495 and E2496, located in the intracellular CTD, were identified as playing critical roles in discriminating divalent Ca2+ from monovalent Cs+, as well as determining unitary conductance and RR sensitivity. Mutating the corresponding residues, E2769 and E2770, in mouse Piezo2 to alanine also resulted in altered ion selectivity and loss of RR blockade, indicating conserved roles of these residues.
Taken together, these structural and functional characterizations provide compelling evidence that the last‐two‐TM‐containing region of residues 2189–2547 encodes the channel pore, and also indicate that trimeric Piezo1 channels possess a central ion‐conducting pore instead of multiple pores formed by individual subunits, as proposed by a previous study (Bae et al. 2013). Thus, Piezo proteins are bona fide pore‐forming subunits of MS cation channels, establishing them as the prototypic class of mammalian MS cation channels.
Peripheral propeller‐resembling structures as mechanotransduction modules?
Piezo channels can respond to various forms of mechanical stimulation including poking, suction and flow‐induced shear stress (Coste et al. 2010; Ranade et al. 2014). The tension of half‐maximal activation (T 50) of Piezo1 is estimated to be ∼2.7 mN m−1, which is lower than that for MscS and MscL (T 50 = ∼5 mN m−1 and 10 mN m−1, respectively) (Lewis & Grandl, 2015). By characterizing Piezo1 activity in membrane blebs that are largely free of cytoskeleton, Cox et al. have found that Piezo1 can be activated by membrane tension, leading them to propose that Piezo1 senses force directly transmitted through the bilayer (Cox et al. 2016). Syeda et al. have provided direct evidence that reconstituted Piezo1 proteins can respond to a change of membrane tension in the droplet interface lipid bilayers (DIBs) system (Syeda et al. 2016). Collectively, these data demonstrate that Piezo1 proteins are intrinsically mechanosensitive, and thus should contain designated mechanotransduction components.
The three peripheral propeller‐resembling structures of mPiezo1, which contain the featured blades, PH wing, anchors, and beams (Abstract Figure), are absent in other trimeric ion channels, thus defining the unique structural architecture of Piezo1.
The striking similarity of the mPiezo1 structure to a three‐bladed propeller leads us to propose that Piezo channels might mechanistically function in a similar manner to the way a propeller senses and transduces force, which is to employ the peripheral structures formed by the large N‐terminal region of residues 1–2190 as designated mechanotransduction modules to gate the central ion‐conducting pore module (Abstract Figure) (Zhao et al. 2016).
To obtain a proof of concept that the peripheral region can indeed function as an integrated an mechanotransduction module, we took advantage of the structural similarity of the Piezo1 pore module and ASIC1 channels and engineered a series of chimeric mutants by fusing the N‐terminal regions 1–2170, 1–2181, or 1–2190 of Piezo1 with ASIC1 (with a GGGGG linker in between) (Zhao et al. 2016, 2017). Intriguingly, we found that only the chimeric construct Piezo1(1–2190)‐ASIC1 gave rise to a portion of the transfected HEK293T cells with MA currents, which show distinct pharmacological and channel properties from Piezo1‐mediated currents (Zhao et al. 2016). For instance, Piezo1(1–2190)‐ASIC‐mediated MA currents were partially blocked by amiloride at 100 μm concentration which effectively blocked ASIC1 but not Piezo1. Furthermore, a portion of the Piezo1(1–2190)‐ASIC1‐transfected cells also responded to acid. Given that Piezo1 channels are instead inhibited by acid (Bae et al. 2015), these data suggest that the mechanical force‐ or acid‐induced current in Piezo1(1–2190)‐ASIC1‐transfected cells is mediated by the ASIC1 channel pore of the hybrid protein. By contrast, other chimeric constructs failed to give rise to MA currents, indicating the specificity of the Piezo1(1–2190)‐ASIC chimera in mediating MA currents. Based on these studies, we suggested that the N‐terminal region of 1–2190 in principle is sufficient to confer mechanosensitivity to trimeric channel pores. In line with this hypothesis, Moroni has followed the same strategy to show that fusing the region of 1–2190 of mPiezo1 with the pore region of Piezo2 (Y2472–N2822) conferred stretch sensitivity to the Piezo2 channel pore, as Piezo2 appears to specifically respond to poking but not stretch (Moroni et al. 2017).
The interpretation of the Piezo1(1–2190)‐ASIC1 chimera implication, however, has been complicated by a recent study by Dubin et al., driven by the application of the CRISPR/Cas9 technology to generate Piezo1‐KO cell lines in which the endogenous Piezo1 gene was disrupted (Dubin et al. 2017). They replicated the results that a portion of HEK293T cells transfected with the Piezo1 (1–2190)‐ASIC1 chimera showed significantly enhanced MA currents. However, Dubin et al. found the construct to be ineffective in generating MA currents when tested in the Piezo1‐KO HEK293T cells. Based on this study, they have suggested that the MA currents observed in the chimera‐transfected wild‐type HEK293T cells are due to endogenous Piezo1. However, given that HEK293T cells express nearly no detectable level of endogenous Piezo1 and that the chimera‐mediated currents are clearly different from Piezo1‐dependent currents as described above (Zhao et al. 2016), it is not clear why only overexpression of Piezo1(1–2190)‐ASIC1, but not other chimeras differing slightly in amino acid composition, could consistently generate MA currents in HEK293T cells (Zhao et al. 2016, 2017).
Nevertheless, together with the structural features of the Piezo1 channel (Ge et al. 2015), the concept that the N‐terminal propeller‐resembling structure might serve as an intrinsic mechanotransduction module for the Piezo1 channel is worthy of further exploration. On the one hand, more chimeras can be generated and tested for identifying more robust mechanosensitive chimeric channels, even in the Piezo1‐KO cells. On the other hand, it is critical to identify specific regions/residues that constitute the featured blade and beam domains in the N‐terminal region, and test their functional relevance in mechanotransduction.
A proposed working model for the mechanosensitive Piezo channels
Based on the revelation of the three‐bladed, propeller‐shaped structure and identification of the ion‐conducting pore, we propose that Piezo proteins consist of discrete pore module and mechanotransduction modules, which are coordinated for ion conduction, mechanical force sensing and transduction (Ge et al. 2015; Zhao et al. 2016). Although the concept that the N‐terminal region might form an independent mechanotransduction module requires further experimental validation, we speculate that the mechanotransduction module possesses all the needed structural components to fulfil their mechanical sensing and transduction roles. For instance, the peripheral blades, which are highly mobile, could be involved in mechanotransduction. The twisted and tilted PH wings could be involved in sensing membrane tension by changing its exposure area in the membrane. The featured beams may transmit conformational changes from peripheral blades and TMs to the central region where the ion‐conduction pore is located, which might enable a lever‐like mechanotransduction mechanism for long‐distance mechanogating. The anchor, connecting the peripheral structural modules to the central pore module, may play a critical role in functionally coupling the two modules. Although the extracellular cap and intracellular CTD constitute key parts of the pore module, they might also be involved in mechanotransduction. For instance, the CTD, structurally positioned in between the beam and anchor, might serve as a key relaying‐component for ultimately gating the pore by the peripheral mechanotransduction module.
Conclusion and perspective
Piezo proteins play essential roles in mechanotransduction in mammalian physiology and pathophysiology. The cryo‐EM structure of Piezo1 shows that it forms a three‐bladed, propeller‐shaped trimeric complex. Examination of both the structure and function of the ion‐conducting pore provides definitive proof that Piezo1 forms the bona fide MS cation channel. The proposal that the complicated Piezo proteins can be deduced into the discrete ion‐conducting pore module formed by the C‐terminal region of ∼350 residues and the mechanotransduction module formed by the large N‐terminal region of ∼2200 residues might represent a simplified, but testable hypothesis for exploring the speculative roles of the distinct domains and the functional interaction of the two modules. Future efforts towards fully understanding the structure‐function relationship of this novel and distinct type of MS channel should also be oriented towards obtaining Piezo structures in high resolution and in different conformational states, and making in‐depth structure‐guided functional characterizations and dynamic analysis.
Additional information
Competing interests
The authors declare no conflict of interest.
Author contributions
Y. W. and B.X. discussed and wrote the manuscript.
Funding
This work was supported by the grants 2016YFA0500402, 31630090, 31422027, 2015CB910102, 31371118 from either the Ministry of Science and Technology or the National Natural Science Foundation of China.
Biographies
Yubo Wang received her PhD from the Academy of Military Medical Sciences in China, and is currently a postdoctoral fellow in Dr. Bailong Xiao's laboratory.

Bailong Xiao is an Associate Professor and Principal Investigator. He pursued his PhD study with Dr Wayne Chen from the University of Calgary in Canada, and investigated the regulation of the cardiac ryanodine receptor by protein interaction and phosphorylation. Then he did his postdoctoral training with Dr Ardem Patapoutian at the Scripps Research Institute in the United States, and contributed to the identification and characterization of several classes of ion channels that serve as receptors for noxious chemicals, temperature and mechanical force, respectively, including the long‐sought‐after mammalian mechanosensitive cation channels – Piezo channels. After setting up his own independent laboratory in Tsinghua University, one of his major research interests has been to understand how the Piezo channels serve as effective mechanotransducers for converting mechanical force into electrochemical signals.
Edited by: Ole Petersen & David Beech
This review was presented at the symposium ‘Piezo channel mechanisms in health and disease’, which took place at the IUPS 38th World Congress, Rio de Janeiro, Brazil, 1–5 August 2017.
References
- Akabas MH, Stauffer DA, Xu M & Karlin A (1992). Acetylcholine receptor channel structure probed in cysteine‐substitution mutants. Science 258, 307–310. [DOI] [PubMed] [Google Scholar]
- Albuisson J, Murthy SE, Bandell M, Coste B, Louis‐Dit‐Picard H, Mathur J, Feneant‐Thibault M, Tertian G, de Jaureguiberry JP, Syfuss PY, Cahalan S, Garcon L, Toutain F, Simon Rohrlich P, Delaunay J, Picard V, Jeunemaitre X & Patapoutian A (2013). Dehydrated hereditary stomatocytosis linked to gain‐of‐function mutations in mechanically activated PIEZO1 ion channels. Nat Commun 4, 1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfo I, Alper SL, De Franceschi L, Auriemma C, Russo R, De Falco L, Vallefuoco F, Esposito MR, Vandorpe DH, Shmukler BE, Narayan R, Montanaro D, D'Armiento M, Vetro A, Limongelli I, Zuffardi O, Glader BE, Schrier SL, Brugnara C, Stewart GW, Delaunay J & Iolascon A (2013). Multiple clinical forms of dehydrated hereditary stomatocytosis arise from mutations in PIEZO1. Blood 121, 3925–3935, S3921–3912. [DOI] [PubMed] [Google Scholar]
- Andolfo I, Russo R, Manna F, Shmukler BE, Gambale A, Vitiello G, De Rosa G, Brugnara C, Alper SL, Snyder LM & Iolascon A (2015). Novel Gardos channel mutations linked to dehydrated hereditary stomatocytosis (xerocytosis). Am J Hematol 90, 921–926. [DOI] [PubMed] [Google Scholar]
- Bae C, Gnanasambandam R, Nicolai C, Sachs F & Gottlieb PA (2013). Xerocytosis is caused by mutations that alter the kinetics of the mechanosensitive channel PIEZO1. Proc Natl Acad Sci USA 110, E1162–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae C, Sachs F & Gottlieb PA (2011). The mechanosensitive ion channel Piezo1 is inhibited by the peptide GsMTx4. Biochemistry 50, 6295–6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae C, Sachs F & Gottlieb PA (2015). Protonation of the human PIEZO1 ion channel stabilizes inactivation. J Biol Chem 290, 5167–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beneteau C, Thierry G, Blesson S, Le Vaillant C, Picard V, Bene MC, Eveillard M & Le Caignec C (2014). Recurrent mutation in the PIEZO1 gene in two families of hereditary xerocytosis with fetal hydrops. Clin Genet 85, 293–295. [DOI] [PubMed] [Google Scholar]
- Cahalan SM, Lukacs V, Ranade SS, Chien S, Bandell M & Patapoutian A (2015). Piezo1 links mechanical forces to red blood cell volume. Elife 4, e07370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao E, Liao M, Cheng Y & Julius D (2013). TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 504, 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M (2009). Neurosensory mechanotransduction. Nat Rev Mol Cell Biol 10, 44–52. [DOI] [PubMed] [Google Scholar]
- Chesler AT, Szczot M, Bharucha‐Goebel D, Ceko M, Donkervoort S, Laubacher C, Hayes LH, Alter K, Zampieri C, Stanley C, Innes AM, Mah JK, Grosmann CM, Bradley N, Nguyen D, Foley AR, Le Pichon CE & Bonnemann CG (2016). The role of PIEZO2 in human mechanosensation. N Engl J Med 375, 1355–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste B, Houge G, Murray MF, Stitziel N, Bandell M, Giovanni MA, Philippakis A, Hoischen A, Riemer G, Steen U, Steen VM, Mathur J, Cox J, Lebo M, Rehm H, Weiss ST, Wood JN, Maas RL, Sunyaev SR & Patapoutian A (2013). Gain‐of‐function mutations in the mechanically activated ion channel PIEZO2 cause a subtype of distal arthrogryposis. Proc Natl Acad Sci USA 110, 4667–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste B, Mathur J, Schmidt M, Earley TJ, Ranade S, Petrus MJ, Dubin AE & Patapoutian A (2010). Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 330, 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste B, Murthy SE, Mathur J, Schmidt M, Mechioukhi Y, Delmas P & Patapoutian A (2015). Piezo1 ion channel pore properties are dictated by C‐terminal region. Nat Commun 6, 7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste B, Xiao B, Santos JS, Syeda R, Grandl J, Spencer KS, Kim SE, Schmidt M, Mathur J, Dubin AE, Montal M & Patapoutian A (2012). Piezo proteins are pore‐forming subunits of mechanically activated channels. Nature 483, 176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox CD, Bae C, Ziegler L, Hartley S, Nikolova‐Krstevski V, Rohde PR, Ng CA, Sachs F, Gottlieb PA & Martinac B (2016). Removal of the mechanoprotective influence of the cytoskeleton reveals PIEZO1 is gated by bilayer tension. Nat Commun 7, 10366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin AE, Murthy S, Lewis AH, Brosse L, Cahalan SM, Grandl J, Coste B & Patapoutian A (2017). Endogenous Piezo1 can confound mechanically activated channel identification and characterization. Neuron 94, 266–270, e263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhoffer GT, Loftus PD, Yoshigi M, Otsuna H, Chien CB, Morcos PA & Rosenblatt J (2012). Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature 484, 546–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucherre A, Kissa K, Nargeot J, Mangoni ME & Jopling C (2014). Piezo1 plays a role in erythrocyte volume homeostasis. Haematologica 99, 70–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotiou E, Martin‐Almedina S, Simpson MA, Lin S, Gordon K, Brice G, Atton G, Jeffery I, Rees DC, Mignot C, Vogt J, Homfray T, Snyder MP, Rockson SG, Jeffery S, Mortimer PS, Mansour S & Ostergaard P (2015). Novel mutations in PIEZO1 cause an autosomal recessive generalized lymphatic dysplasia with non‐immune hydrops fetalis. Nat Commun 6, 8085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J, Li W, Zhao Q, Li N, Chen M, Zhi P, Li R, Gao N, Xiao B & Yang M (2015). Architecture of the mammalian mechanosensitive Piezo1 channel. Nature 527, 64–69. [DOI] [PubMed] [Google Scholar]
- Geng J, Zhao Q, Zhang T & Xiao B (2017). In touch with the mechanosensitive Piezo channels: structure, ion permeation, and mechanotransduction. Curr Top Membr 79, 159–195. [DOI] [PubMed] [Google Scholar]
- Gonzales EB, Kawate T & Gouaux E (2009). Pore architecture and ion sites in acid‐sensing ion channels and P2X receptors. Nature 460, 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb PA (2017). A tour de force: the discovery, properties, and function of piezo channels. Curr Top Membr 79, 1–36. [DOI] [PubMed] [Google Scholar]
- Gudipaty SA, Lindblom J, Loftus PD, Redd MJ, Edes K, Davey CF, Krishnegowda V & Rosenblatt J (2017). Mechanical stretch triggers rapid epithelial cell division through Piezo1. Nature 543, 118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda R, Cha M, Ling J, Jia Z, Coyle D & Gu JG (2014). Merkel cells transduce and encode tactile stimuli to drive Aβ‐afferent impulses. Cell 157, 664–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawate T, Michel JC, Birdsong WT & Gouaux E (2009). Crystal structure of the ATP‐gated P2X4 ion channel in the closed state. Nature 460, 592–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koser DE, Thompson AJ, Foster SK, Dwivedy A, Pillai EK, Sheridan GK, Svoboda H, Viana M, Costa LD, Guck J, Holt CE & Franze K (2016). Mechanosensing is critical for axon growth in the developing brain. Nat Neurosci 19, 1592–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Leddy HA, Chen Y, Lee SH, Zelenski NA, McNulty AL, Wu J, Beicker KN, Coles J, Zauscher S, Grandl J, Sachs F, Guilak F & Liedtke WB (2014). Synergy between Piezo1 and Piezo2 channels confers high‐strain mechanosensitivity to articular cartilage. Proc Natl Acad Sci USA 111, E5114–E5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AH & Grandl J (2015). Mechanical sensitivity of Piezo1 ion channels can be tuned by cellular membrane tension. Elife 4 e12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Hou B, Tumova S, Muraki K, Bruns A, Ludlow MJ, Sedo A, Hyman AJ, McKeown L, Young RS, Yuldasheva NY, Majeed Y, Wilson LA, Rode B, Bailey MA, Kim HR, Fu Z, Carter DA, Bilton J, Imrie H, Ajuh P, Dear TN, Cubbon RM, Kearney MT, Prasad RK, Evans PC, Ainscough JF & Beech DJ (2014). Piezo1 integration of vascular architecture with physiological force. Nature 515, 279–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Yang Y & Canessa CM (2011). Outlines of the pore in open and closed conformations describe the gating mechanism of ASIC1. Nat Commun 2, 399. [DOI] [PubMed] [Google Scholar]
- Liao M, Cao E, Julius D & Cheng Y (2013). Structure of the TRPV1 ion channel determined by electron cryo‐microscopy. Nature 504, 107–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M, Cao E, Julius D & Cheng Y (2014). Single particle electron cryo‐microscopy of a mammalian ion channel. Curr Opin Struct Biol 27, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh BJ, Murdoch A, Haslett C & Sethi T (2012). Loss of the integrin‐activating transmembrane protein Fam38A (Piezo1) promotes a switch to a reduced integrin‐dependent mode of cell migration. PLoS One 7, e40346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillin MJ, Beck AE, Chong JX, Shively KM, Buckingham KJ, Gildersleeve HI, Aracena MI, Aylsworth AS, Bitoun P, Carey JC, Clericuzio CL, Crow YJ, Curry CJ, Devriendt K, Everman DB, Fryer A, Gibson K, Giovannucci Uzielli ML, Graham JM Jr, Hall JG, Hecht JT, Heidenreich RA, Hurst JA, Irani S, Krapels IP, Leroy JG, Mowat D, Plant GT, Robertson SP, Schorry EK, Scott RH, Seaver LH, Sherr E, Splitt M, Stewart H, Stumpel C, Temel SG, Weaver DD, Whiteford M, Williams MS, Tabor HK, Smith JD, Shendure J, Nickerson DA, University of Washington Center for Mendelian Genomics & Bamshad MJ (2014). Mutations in PIEZO2 cause gordon syndrome, Marden‐Walker syndrome, and distal arthrogryposis Type 5. Am J Hum Genet 94, 734–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimovic S, Nakatani M, Baba Y, Nelson AM, Marshall KL, Wellnitz SA, Firozi P, Woo SH, Ranade S, Patapoutian A & Lumpkin EA (2014). Epidermal Merkel cells are mechanosensory cells that tune mammalian touch receptors. Nature 509, 617–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins JR, Penton D, Peyronnet R, Arhatte M, Moro C, Picard N, Kurt B, Patel A, Honore E & Demolombe S (2016). Piezo1‐dependent regulation of urinary osmolarity. Pflugers Arch 468, 1197–1206. [DOI] [PubMed] [Google Scholar]
- Moroni M, Servin‐Vences MR, Fleischer R & Lewin GR (2017). Voltage‐gating of mechanosensitive PIEZO channels. bioRxiv, https://doi.org/10.1101/156489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilius B & Honore E (2012). Sensing pressure with ion channels. Trends Neurosci 35, 477–486. [DOI] [PubMed] [Google Scholar]
- Nonomura K, Woo SH, Chang RB, Gillich A, Qiu Z, Francisco AG, Ranade SS, Liberles SD & Patapoutian A (2016). Piezo2 senses airway stretch and mediates lung inflation‐induced apnoea. Nature 541, 176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak MM, Nourse JL, Tran T, Hwe J, Arulmoli J, Le DT, Bernardis E, Flanagan LA & Tombola F (2014). Stretch‐activated ion channel Piezo1 directs lineage choice in human neural stem cells. Proc Natl Acad Sci USA 111, 16148–16153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen CE, Armache JP, Gao Y, Cheng Y & Julius D (2015). Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature 520, 511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prole DL & Taylor CW (2013). Identification and analysis of putative homologues of mechanosensitive channels in pathogenic protozoa. PLoS One 8, e66068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranade SS, Qiu Z, Woo SH, Hur SS, Murthy SE, Cahalan SM, Xu J, Mathur J, Bandell M, Coste B, Li YS, Chien S & Patapoutian A (2014). Piezo1, a mechanically activated ion channel, is required for vascular development in mice. Proc Natl Acad Sci USA 111, 10347–10352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranade SS, Syeda R & Patapoutian A (2015). Mechanically activated ion channels. Neuron 87, 1162–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranade SS, Woo SH, Dubin AE, Moshourab RA, Wetzel C, Petrus M, Mathur J, Begay V, Coste B, Mainquist J, Wilson AJ, Francisco AG, Reddy K, Qiu Z, Wood JN, Lewin GR & Patapoutian A (2014). Piezo2 is the major transducer of mechanical forces for touch sensation in mice. Nature 516, 121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retailleau K, Duprat F, Arhatte M, Ranade SS, Peyronnet R, Martins JR, Jodar M, Moro C, Offermanns S, Feng Y, Demolombe S, Patel A & Honore E (2015). Piezo1 in smooth muscle cells is involved in hypertension‐dependent arterial remodeling. Cell Rep 13, 1161–1171. [DOI] [PubMed] [Google Scholar]
- Rocio Servin‐Vences M, Moroni M, Lewin GR & Poole K (2017). Direct measurement of TRPV4 and PIEZO1 activity reveals multiple mechanotransduction pathways in chondrocytes. Elife 6 e21074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rode B, Shi J, Endesh N, Drinkhill MJ, Webster PJ, Lotteau SJ, Bailey MA, Yuldasheva NY, Ludlow MJ, Cubbon RM, Li J, Futers TS, Morley L, Gaunt HJ, Marszalek K, Viswambharan H, Cuthbertson K, Baxter PD, Foster R, Sukumar P, Weightman A, Calaghan SC, Wheatcroft SB, Kearney MT & Beech DJ (2017). Piezo1 channels sense whole body physical activity to reset cardiovascular homeostasis and enhance performance. Nat Commun 8, 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg MB, Nybo M, Birgens H & Frederiksen H (2014). Hereditary xerocytosis and familial haemolysis due to mutation in the PIEZO1 gene: a simple diagnostic approach. Int J Lab Hematol 36, e62–65. [DOI] [PubMed] [Google Scholar]
- Syeda R, Florendo MN, Cox CD, Kefauver JM, Santos JS, Martinac B & Patapoutian A (2016). Piezo1 channels are inherently mechanosensitive. Cell Rep 17, 1739–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk R, Wanschers BF, Nijtmans LG, Rodenburg RJ, Zschocke J, Dikow N, van den Brand MA, Hendriks‐Franssen MG, Gilissen C, Veltman JA, Nooteboom M, Koopman WJ, Willems PH, Smeitink JA, Huynen MA & van den Heuvel LP (2013). A mutation in the FAM36A gene, the human ortholog of COX20, impairs cytochrome c oxidase assembly and is associated with ataxia and muscle hypotonia. Hum Mol Genet 22, 656–667. [DOI] [PubMed] [Google Scholar]
- Wang S, Chennupati R, Kaur H, Iring A, Wettschureck N & Offermanns S (2016). Endothelial cation channel PIEZO1 controls blood pressure by mediating flow‐induced ATP release. J Clin Invest 126, 4527–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo SH, Lukacs V, de Nooij JC, Zaytseva D, Criddle CR, Francisco A, Jessell TM, Wilkinson KA & Patapoutian A (2015). Piezo2 is the principal mechanotransduction channel for proprioception. Nat Neurosci 18, 1756–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo SH, Ranade S, Weyer AD, Dubin AE, Baba Y, Qiu Z, Petrus M, Miyamoto T, Reddy K, Lumpkin EA, Stucky CL & Patapoutian A (2014). Piezo2 is required for Merkel‐cell mechanotransduction. Nature. 509, 622–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarychanski R, Schulz VP, Houston BL, Maksimova Y, Houston DS, Smith B, Rinehart J & Gallagher PG (2012). Mutations in the mechanotransduction protein PIEZO1 are associated with hereditary xerocytosis. Blood 120, 1908–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Wu K, Chi S, Geng J & Xiao B (2017). Heterologous expression of the Piezo1‐ASIC1 chimera induces mechanosensitive currents with properties distinct from Piezo1. Neuron 94, 274–277. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Wu K, Geng J, Chi S, Wang Y, Zhi P, Zhang M & Xiao B (2016). Ion permeation and mechanotransduction mechanisms of mechanosensitive Piezo channels. Neuron 89, 1248–1263. [DOI] [PubMed] [Google Scholar]


