Abstract
Helicobacter pylori is well adapted to colonize the epithelial surface of the human gastric mucosa and can cause persistent infections. Its pathogenic effects consist of gastritis, peptic ulcers and increased risk for the development of gastric cancer. In order to infect the gastric mucosa, H. pylori has to survive in the gastric acidic pH. H. pylori has well developed mechanisms to neutralize the effects of acidic pH. The exact role of many bacterial factors as well as environmental factors still remains unsolved and how these factors involve in the acid mediated survival of bacterium is unknown yet. In this review, we have discussed and summarized the various information published in scientific literatures regarding functional and molecular aspects by which the bacterium can combat and survive the adverse effects of stomach acidic pH in order to establish the persistent infections.
Keywords: Acid acclimation, gastric acidic environment, Helicobacter pylori, survival
Introduction
Helicobacter pylori (H. pylori), a Gram-negative helical bacterium, has been estimated to infect at least half of the world’s population. This bacterium was first identified by two Australian scientists Barry J. Marshal and Robin Warren in 1982. British scientist Stewart Goodwin further investigated this bacterium and reported that it was present in patients with chronic gastritis and gastric ulcers, previously not believed to be bacterial cause [1].
H. pylori has been established as the causative agent of human gastric mucosal infection with several gastro-duodenal diseases such as chronic gastritis, gastric ulcer, duodenal ulcer and increased risk for gastric cancer [2]. The International Agency for Research on Cancer (IARC) classified H. pylori as a group I carcinogen in 1994 [3] and based on the epidemiological data the carcinogenic behavior of H. pylori was reconfirmed in 2009 [4]. Despite of the several attempts the clear mechanism of development of cancer is not understood clearly [5, 6]. This bacterium possesses several virulence factors such as outer membrane proteins (OMPs) for adhesion, cag pathogenicity island (PAI), VacA, gGT and DupA; however the pathogenicity of H. pylori depends on its ability to survive in the harsh gastric environment characterized by high acidity [7]. After transmission, the organism is reached and lodged in the stomach; the ecological niche of the bacterium where the pH tends to be very low. Therefore, to establish the infectious process H. pylori must survive in the high acidic environment of stomach.
Ecological niche
Although the exact means of transmission of H. pylori is not elucidated clearly, evidences suggest that the bacterium is often acquired during childhood via oral-oral or faecal-oral routes from family members [8, 9]. After transiting to the stomach it localizes to specific locations; the corpus and antrum simultaneously within the stomach [10, 11]. In the stomach H. pylori is highly adapted to the challenges encountered [8]. Once the persistent infection is established, the bacterium stays within the stomach for lifetime and it becomes the dominant inhabitants of the stomach [12]. In the gastric lumen the pH is around 2.0 and more in the mucus layer. However, H. pylori stays for short duration within the lumen and enters the mucus layer, the habitat of the H. pylori, where the pH ranges from 4.5–6.5; the bacterium is highly capable to neutralize the gastric lumen acidity [13, 14].
Many studies have revealed that H. pylori has several mechanisms to overcome the lethal effects of gastric acidic pH. This review gives the concise overview about the utilization of different factors by which H. pylori maintains the survival capability in acidic environment (Table 1).
Table 1.
Factors involved in the survival of acidic pH.
| Factors involved | Mechanisms | Reference |
|---|---|---|
| Urease dependent mechanisms | ||
| Urease | Breakdown of urea and production of ammonia that neutralizes the acidity. | 17, 42 |
| NixA and NikR | Regulates the availability of nickel molecules for full action of metallo-enzyme, urease. | 27, 28, 30, 31, 33–36 |
| UreI | Mediates entry of urea after exposure of bacterial cell to the high acidic pH. | 38–39 |
| Arginase | Production of endogenous urea that is hydrolyzed by urease. | 41 |
| glutamine synthetase (GS), glutamate dehydrogenase (GDH), glutaminase (Ggt) and asparaginase (AsnB) | Mediates the metabolism and assimilation of NH4+ generated by periplasmic alpha-carbonic anhydrase. | 43 |
| Alpha-carbonic anhydrase | Conversion of CO2 to HCO3− and maintenance of periplasmic pH close to 6.1. | 45 |
| ExbD | Transfers energy to the outer membrane for active transportation of urea and other essential molecules at acidic pH. | 48–50 |
| ArsR and ArsS | Regulation, expression of urease gene cluster and recruitment of urease. | 51, 52 |
| Urease independent mechanism | ||
| Gastric mucus layer | Gel-like viscoelastic property of mucus facilitates movement of bacteria at acidic pH. | 54 |
| Helical bacterial shape | Provides corkscrew like movement that facilitates penetration into mucus layer | 58, 60 |
| Flagella | Acid activates the flagellar protein and movement. | 65, 66 |
| Chemotaxis and chemo-receptor | Chemoreceptor TlpB senses the environmental pH for chemotaxis | 78–80 |
| Recombinational repair proteins; RecA, RecN, RecO, RecR, Hup and PriA | Mediates recombinational repair of DNA damage caused by acid stress. | 83, 91, 94, 98, 101 |
| DNA binding protein, HP0119 | Histone like hypothetical protein HP0119 physically protects the bacteria at acidic pH | 105 |
| AmiE, AmiF and aspartase | Urease independent ammonia production that neutralizes acidic pH | 110–113 |
| AtoS and CrdS | Senses acid pH and regulates expression of acid responsive genes. | 117–120 |
| Fur | Mediates iron uptake and regulates expression of acid responsive genes. | 65, 121, 122, 129 |
Acid stress (Urease dependent) survival of H. pylori
The essential colonization factor urease and its associates
Several bacterial species including normal flora and non-pathogens requires the enzymatic activity of several proteins such as urease and hydrogenase to colonize the acidic niche. Urease, a high molecular weight multi-subunit metallo-enzyme, has been demonstrated as a potent virulence factor for some species like Proteus mirabilis [15], Staphylococcus saprophyticus [16] and H. pylori [17]. H. pylori produces large amount of intracellular (cytoplasmic) urease, around 10% of total bacterial proteins; however, H. pylori also contains urease on the bacterial surface due to the lysis of some organisms [18, 19]. The environmental pH regulates the synthesis of intracellular urease that acts to increase the periplasmic pH and membrane potential allowing synthesis of urease at low pH [20, 21]. In a recent study by Schoep et al., it was found that the surface properties of H. pylori urease complex play an important role for persistence during gastric colonization, directly interacting with host components that underlie H. pylori persistence [18]. Actually, the urease complex is multi-functional and the surface property of this protein is distinct from the urealytic activity. Uberti et al. [22] demonstrated the involvement of H. pylori urease in pro-inflammatory and neutrophil activation activity different from the urealytic activity. Urease, the essential component of colonization, is a nickel containing enzyme and it requires efficient acquisition of nickel from the environment for its activity [23, 24]. One active urease molecule requires 24 nickel ions for full enzymatic action [25]. The nickel molecules found as trace molecules in the blood are taken by uptake proteins of bacteria, FecA3 and FrpB4, anchored in outer membrane [26]. After entering the outer membrane the nickel molecules are transported to the cytoplasm through the protein channel NixA, a monomeric high affinity nickel uptake protein localized in the cytoplasmic (inner) membrane of the bacteria [27, 28]. Since, the activation of urease system requires nickel, therefore, when cytoplasmic nickel availability is insufficient, the urease system cannot be fully activated and this inactivation impairs survival of H. pylori at acidic pH [29]. Another ribbon-helix-helix (RHH) family of DNA binding nickel-responsive regulatory protein (NikR) mediates its repressor function via nickel-dependent binding to a palindromic sequence in the promoter region of the nik operon [30, 31] resulting in the expression of the Nik system only when nickel is insufficient in the cell [32]. Since, H. pylori shows reduced growth at higher concentration of environmental nickel and in the absence of nickel, NikR has been suggested to function as the main nickel-responsive regulatory system for acid responsive induction of urease expression [33–36]. Too little entry of nickel impairs urease activity and acid survival while too much nickel generates reactive oxygen species leading to cell damage; therefore, entry of nickel needs to be well regulated [37]. NikR regulates the nickel responsive genes including ureA, ureB, nixA, frpB4, and fecA3 [26, 36, 34]. NikR binds to the promoter regions of frpB4 and fec3A genes in high concentration of nickel leading to the depression of these genes and decreased nickel uptake [26].
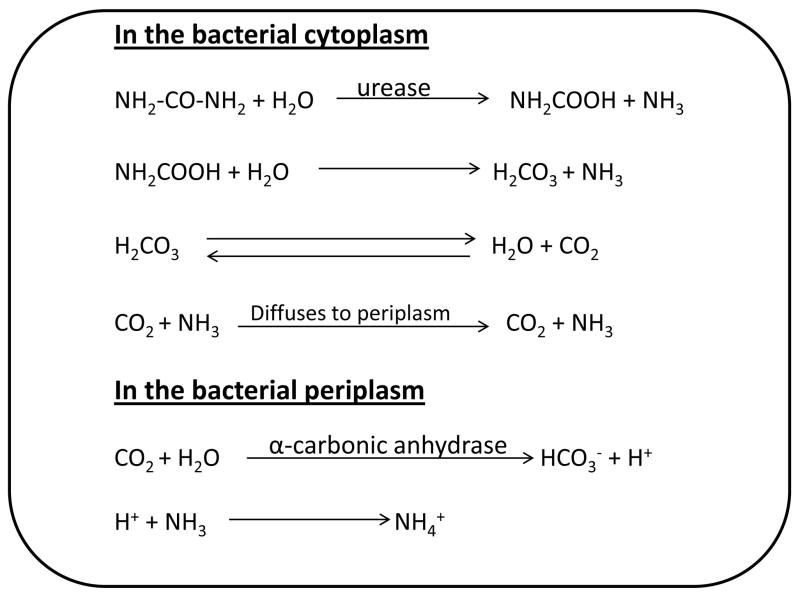
Urea, the substrate for urease enzyme, after entering the outer membrane through porins can passively diffuse across the bacterial inner membrane and upon exposure of H. pylori to high acidity; the influx of urea becomes faster. Active influx of urea is acid dependent property that is mediated by urea specific influx protein, permease (UreI) an inner membrane localized, proton gated urea channel [38, 39]. However, the urea concentration in gastric content is around 1 mM at physiological condition and it is assumed that this concentration of urea is not sufficient to provide the enough ammonia and acid survival benefit especially during the starvation period when the pH is around 1.0 [40]. Therefore, the mechanisms either producing the urea endogenously or promoting the other mechanisms are of most importance for the survival in vitro and during starvation period. Therefore, the urea can also be produced endogenously by arginase in vitro, the highly active enzyme of the urea cycle, encoded by rocF gene. The endogenously produced urea also provides concomitant acid survival activity in vitro. In a study, the rocF knock-out mutants were found to express increased susceptibility to acid treatment in vitro [41]. This observation indicates the importance of the enzyme arginase in the survival of bacterium to acidic pH in vitro and this could be also important for in vivo survival in case there is inadequate level of urea at the site of colonization. Arginine being a substrate of arginase enzyme can protect the H. pylori from the adverse effect of acidity by raising the pH in vitro. Urea (endogenous and exogenous) is hydrolyzed by urease resulting in the production of ammonia (NH3) and carbamate. The carbamate spontaneously decomposed to give ammonia (NH3) and carbonic acid. The carbonic acid is broken down to CO2 and H2O (Figure 1). Both ammonia and CO2 participate in the lowering of pH. Ammonium (NH4+), the protonated form of ammonia neutralizes the stomach acidity, plays an important role to promote favorable environment for H. pylori survival in the stomach [42]. However, despite of the role of acid survival advantage, the ammonium produced must be metabolized or effluxed because its presence within the bacterial cells is counterproductive to the goal of rising pH while maintaining a viable proton motive force. It is unknown how NH4+ rapidly exits across the inner membrane as the H. pylori lacks the homologs to known NH3/NH4+ transport system. However, Miller and Maier [43] in 2014 demonstrated the role of ammonium assimilating enzymes, glutamine synthetase (GS), glutamate dehydrogenase (GDH) and the ammonium evolving periplasmic enzymes glutaminase (Ggt) and asparaginase (AsnB) in metabolism and effluxing of NH4+. The assimilating enzymes GDH and GS convert the NH4+ in to glutamate and glutamine respectively. Glutamine can diffuse to the periplasmic space or can be further converted to aspergine and diffused to periplasmic space. In the periplasmic space glutamine and aspergine are converted to NH4+ catalyzed by Ggt and AsnB respectively and involves in the hydrolysis of urea inside the cell or the expulsion/assimilation of ammonium elaborating the link between urea-derived ammonium and urease-mediated acid resistance. After exit from the bacterial cell, the NH4+ neutralizes the acidity and creates a nearly neutral micro-environment for survival of H. pylori (Figure 2).
Figure 1.
Hydrolysis of urea by urease. Urease breaks down the urea to form carbamate and NH3. Carbamate is further decomposed to produce carbonic acid and NH3. Carbonic acid is broken down spontaneously aqueous to produce H2O and CO2. Therefore, one molecule of urea after hydrolysis produces two molecules of ammonia (NH3) and one molecule of carbon dioxide (CO2). CO2 and NH3 are defused to periplasm where α-carbonic anhydrase catalyzes the breakdown of CO2 to produce HCO3− and H+. The H+ produced combines with NH3 to produce NH4+.
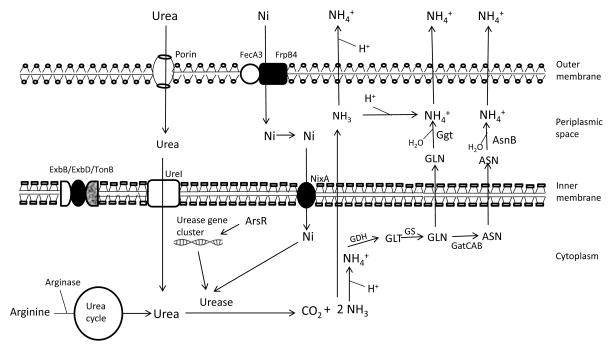
Figure 2.
The metallo-enzyme, urease requires nickel (Ni) for its activity. Ni slowly enters the outer membrane through porin (outer membrane protein) and actively enters through FecA3/FrpB4 proteins anchored in outer membrane and crosses inner membrane through NixA (inner membrane protein). However, in acidic pH more Ni is required for increased activity of urease and for the active transport of essential molecules (Ni), ExbD provides energy to the outer membrane. Urea enters the outer membrane through the porin and inner membrane through UreI (found in inner membrane) which opens in acid exposure of bacteria. Endogenously produced by arginase in urea cycle (during in vitro survival) and exogenously entered urea molecules are hydrolyzed by urease to ammonia (NH3) and carbon dioxide (CO2). ArsR regulates urease gene cluster expression. Ammonia (NH3) produced is diffused out and binds with proton (H+) leading to the neutralization of acidic pH. However, NH3 is also converted to ammonium (NH4+) inside the cell (cytoplasm and periplasm) which is toxic to the bacteria. The cytoplasmic NH4+ is assimilated to glutamate (GLT) by GDH and to glutamine (GLN) by GS. GLN can diffuse to periplasmic space or can be converted to aspergine (ASN) by GatCAB aminoacyl-tRNA amidotransferase (GatCAB) and then can diffuse to periplasmic space. GLN and ASN bind with water molecule and led to the formation of NH4+ and catalyzed by Ggt and AsnB respectively. Finally the NH4+ exits to the bacterial cell and increases the pH that leads to the survival of bacteria at acidic pH.
In summary, the nickel, transportation mediated by NixA and NikR, acts as the cofactor of urease enzyme that catalyzes the hydrolysis of urea, the entry of which is enhanced in acid exposure. The NH4+ produced is transported outside of bacterial cell which creates a neutral microenvironment.
Acid acclimation
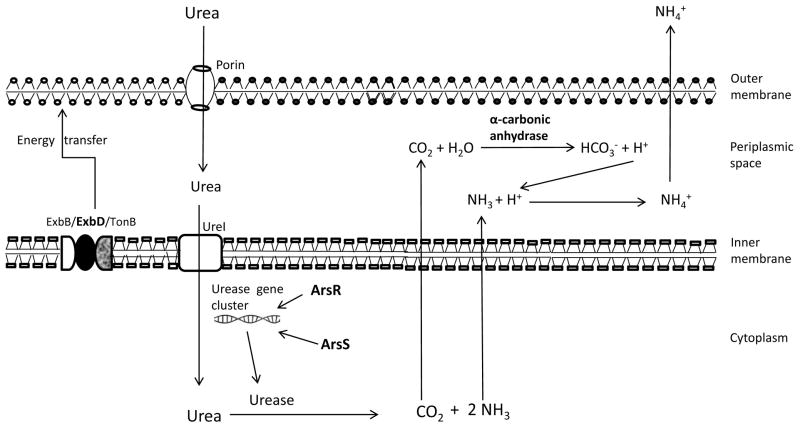
The acidic pH of the mammalian stomach is important for killing the potentially injurious bacteria that transit to the intestine. However, H. pylori can survive the acidic gastric environment by expressing some acid tolerance genes [44]. The CO2 and NH3 produced by the hydrolysis of urea, diffuse to the periplasmic space and provide the two major arms of the acid acclimation. The CO2 in periplasmic space converted in to bicarbonate (HCO3−) and H+ by periplasmic alpha-carbonic anhydrase. The HCO3− provides one arm of the acid acclimation maintaining the periplasmic pH close to 6.1. The protons (H+) produced by alpha-carbonic anhydrase and exogenously entered to the periplasmic space are neutralized by NH3 [45]; another arm of acid acclimation. Hence, both the NH3 and CO2 produced by the intracellular urease activity enables the acid acclimation mechanism in H. pylori and can maintain periplasmic pH at 6.1 in the face of an external pH below 2.5 [39, 45]. However, acid acclimation in H. pylori is a multifactorial mechanism for effective acid neutralization and the H. pylori is able to up-regulate acid acclimation and other group of genes under acidic pH [46]. Marcus et al. demonstrated the role of bacterial inner membrane protein ExbD in acid acclimation mechanism [47]. ExbD, a product of gene HP1340 in strain 26695 is an inner membrane protein and a part of a 3 protein complex (ExbB/ExbD/TonB) [48]. After exposure of bacteria to the acidic pH, it needs more urea and urease production to help combat the lethal effect of acidic pH. The ExbB/ExbD/TonB complex anchored to the inner membrane transfers energy to the outer membrane in the form of conformational changes in TonB for the active transport of essential molecules that are found in trace amounts in environment such as iron and nickel [49, 50]. Recently, the role of ArsR and ArsS was demonstrated to function in acid acclimation. ArsR is involved in regulation of urease gene cluster expression whereas ArsS is involved in regulation of urease gene transcription and recruitment of urease protein to the cytoplasmic membrane to augment the acid acclimation during acute acid exposure. ArsR and ArsS work through phosphorylation-independent and phosphorylation-dependent regulatory mechanisms to impact acid acclimation and allow gastric colonization [51]. It has been also concluded that the arsS mediate acid survival regulated by transcriptional up-regulation of arsS and by an enhanced capacity to sense the pH change in environment [52]. Urease, catalyzing the hydrolysis of urea to carbon dioxide and ammonia, is also critical in the development of diseases caused by H. pylori [53]. In this way H. pylori is capable of maintaining a near neutral periplasmic pH, the cytoplasmic pH experiences relatively small excursions from its optimal pH in the acidity as if it were in a neutral environment (Figure 3). Therefore, taking en mass, the data depict that the breakdown of urea whose entry is enhanced by ExbD and UreI after acid exposure produces CO2 and NH3 that provides the major pathways of acid acclimation for the neutralization of acidity in periplasmic space where pH rises to 6.1.
Figure 3. Acid acclimation.
Exposure of bacterial cells to acidic pH opens the urea channel, UreI and urea enters the cytoplasm to be hydrolyzed by urease to NH3 and CO2. However, the outer membrane proteins (porins) need energy for active transport of essential molecules (urea) which is provided by ExbD located in inner membrane in the form of complex of three proteins (ExbB/ExbD/TonB). ArsR and ArsS regulate the expression of urease gene cluster and increase the urease activity at strong acidic pH. Ammonia (NH3) and carbon dioxide (CO2) diffuse to the periplasmic space. In the periplasmic space, CO2 binds with water molecule to synthesize HCO3− and H+ catalyzed by periplasmic α-carbonic anhydrase. The synthesized HCO3− acts as a strong buffer and can maintain the periplasmic pH to 6.1. Protons (H+), entering to the periplasm and synthesized by α-carbonic anhydrase, are neutralized by NH3 converting to NH4+ and diffuses out of the bacterial cell membrane.
Physiological acid adaptation (Urease independent survival) of H. pylori
Characteristic role of mucus and helical shape of bacteria
After transit to the stomach the survival in acidic pH is important for H. pylori cells during the early stages of gastric infection before colonization to the gastric mucus that provides a protective layer against the acid content of the stomach. H. pylori crosses the gastric mucus layer and starts colonizing the gastric mucosa by adhering to the gastric epithelium where necessary nutrients and protection from the host immune system is obtained [54]. The urea hydrolyzing property exhibited by urease enzyme increases the pH in mucus microenvironment and modifies it to less gel-like, facilitating the movement of bacterium [55]. The viscoelasticity property of mucus layer is high at strongly acidic pH whereas decreases as the gastric pH increases and becomes gel like above pH 4.0. The inter-chain disulphide bonds of mucus glycoproteins (mucins) are reduced by thioredoxin system found in H. pylori that alters the mucus micro-structure that provides favorable environment and motility through the viscoelastic mucus gel layer [56, 57]. The characteristic helical shape of bacteria also determines to cope and adapt to different environments [58]. The rapid movement of bacterium within less acidic mucus layer is facilitated by its helical shape, allowing the bacterium to escape extremely low pH [59]. The alterations in the cross linking of cell determining layer, peptidoglycan found in periplasmic space of cell wall, has been shown to modify the bacterial shape. In an in vitro study, the cell shape mutants were found unable to efficiently colonize when compared with helical shaped bacteria despite possessing motility similar to the wild type [60]. The cell shape role in determination of bacterial cell movement was also elaborated in a recent study where 7–21% reduction in the speed was found in the helical shape determination mutants with straight rod morphology when compared with the helical shape bacteria in a viscous environment [54]. In another study it has been demonstrated that both the helical shape and motility of bacterium is necessary to penetrate the mucus layer in corkscrew like movement in order to colonize the gastric epithelium [61]. Therefore, the mucus protects the bacteria from acid stress and the helical shape of bacteria is of importance which provides the cork-screw like active motility to swim towards the higher pH environment.
Flagella mediating the movement
H. pylori normally possesses about 3 μm long two to six sheathed flagella at one pole (i.e. lophotrichous) that enables the bacteria to move in their ecological niche [62]. The flagella are composed of three structural elements like those of enteric bacteria: a basal body, hook and filament. The filament consists of the two flagellin protein subunits, more abundant FlaA lies in the outer region and minor subunit FlaB localizes to the base of the flagellum and both proteins are necessary for full motility [63, 64]. In a study by Merrell et al., it was found that acid exposure can activate flagellar proteins that leads to enhanced changes in motility of H. pylori, as larger percentage of acid exposed bacterial cells displayed motility and moved at significantly higher speeds [65]. It has been also demonstrated by Yoshiyama et al. that flagellar motor is powered by a proton motive force at low pH and H. pylori can swim faster at low pH [66]. The number of flagella also seems to play an important role for speed of bacterial cell during movement. In a recent study, the cells having 4 flagella showed 19% increase in speed when compared with cells having 3 flagella in viscous environment [54]. Thus orally up taken H. pylori can move quickly towards the epithelial surface by chemo-attractive substances and promptly evade the acidic periphery of the mucous layer. The role of flagella in the attachment to the host cells and in the survival has been evaluated in several bacterial species [67–69]. However, in H. pylori, it was shown that the flagellin (flaA and/or flaB) and flagellar regulator gene (flbA) mutants adhered well to the gastric cells [70]. In another recent study, Kao et al. using the experimental model of H. pylori J99 strain depicted that those flagella regulator gene (csrA or rpoN) mutant strains showed decreased bacterial adhesion to AGS cells [71]. Together these data suggest that although the flagella are not involved directly in the attachment, the regulators of flagella related genes do involve in the expression of adhesins in H. pylori.
Chemotactic activity
Some chemicals that are continuously secreted from the gastric epithelial cells to the lumen such as urea [72], sodium carbonate [73] and potassium carbonate [74] serve as potent attractants for H. pylori towards higher pH. Despite these data, little is known about the chemotactic behavior of H. pylori at various pH of the stomach, ranging from highly acidic to pH 4.5–6.5, which is the ecological niche of this bacterium. In a recent study conducted by Tadjrobehkar and Abdollahi about the chemotactic behavior of H. pylori showed that the maximum chemotactic activity occurred at pH 5.5 to 6.5, but no chemotaxis was found in solution with pH 3 and they concluded that chemotactic responses of H. pylori were severely affected by pH condition [75]. In H. pylori, smooth movement in the presence of chemical attractants and increased stopping behavior in the presence of repellent is facilitated by presence of four chemoreceptors, TlpA, TlpB, TlpC and TlpD for sensing the chemicals found in environment and tuft of flagella found at one pole mediate the movement in response to chemo-attractants [76, 77]. Chemoreceptors are trans-membrane proteins of which the TlpB consisting of two domains, periplasmic and cytoplasmic, is the most important for chemotactic behavior of bacterium that belongs to the class of methyl-accepting chemotaxis proteins (MCPs) [78, 79]. Croxen et al. in a study found that tlpB knockout mutants were defective for pH taxis and concluded that the receptor TlpB is essential for sensing pH in H. pylori [80]. The periplasmic domain mediating the chemical sensing property accounts for about one-third of the total amino acid found in TlpB that binds to urea and confers its pH sensing function [79]. Therefore, by chemotactic mechanism H. pylori senses the environmental pH and drives itself towards the nearly neutral pH that minimizes the bacterial exposure to acidic environment of stomach to survive and colonize for persistent infections.
Recombinational repair of DNA damage
DNA repair is a fundamental process in all free-living as well as pathogenic bacteria and it acts as one of the defense mechanisms that allow the pathogenic bacteria to survive in their hosts. Double strand breaks (DSBs) occur as a result of a variety of physical or chemical insults that modify the DNA. Damage at a single-strand site, if not repaired immediately, leads to a DSB. More commonly, if a replication fork meets damaged bases that cannot be replicated, the fork can collapse, leading to a DSB. The physiological condition found in stomach frequently cause acid stress in H. pylori leading to DNA damage. However, H. pylori harbors the transformation mediated recombination repair mechanism that can repair the DNA damage caused by stress conditions [81, 82]. Bacteria can repair damaged DNA by using homologous recombination which involves three steps, presynaptic, synaptic and postsynaptic steps. In presynaptic step, the production of single-stranded DNA coated with RecA protein occurs. Synaptic step involves strand exchange and joining of DNA molecule promoted by RecA protein. In postsynaptic step priming of new DNA synthesis occurs. Recombinational DNA repair requires a large number of proteins among which RecA is critical in DNA recombination and repair [83]. In the well-studied bacterial model Escherichia coli (E. coli), a 3 enzyme complex; the RecBCD mediating the helicase and nuclease activities initiates the recombinational DNA repair of double-stranded DNA (dsDNA) [84]. The RecBCD binds to the damaged duplex DNA end and unwinds it. After unwinding, the both nascent DNA strands are degraded until the recognition of specific sequence (Chi-sequence), the DNA ends are resected to form a 3′-ssDNA overhang that terminates at Chi sequences. Chi (χ)-sequence (Chi = crossover hotspot instigator) is an 8-nuleotide sequence (5′GCTGGTGG-3′), hotspot for homologous recombination [85]. However, in H. pylori the homologous enzymes complex AddAB performs the helicase and nuclease activity [86]. A recent study suggests that, Chi-sequence after binding within the AddAB enzyme complex sequesters the 3′-terminated strand and prevents from the action of AddA nuclease domain, thereby resulting in nuclease activity attenuation beyond Chi-sequence [87, 88]. The sequestration of 3′-terminated strand and continued unwinding and degradation of the 5′-terminated strand by the AddB nuclease domain results in the production of a long 3′-ssDNA overhang. The 3′-ssDNA overhang acts as the substrate for the loading of RecA protein for strand exchange [89]. Loading of the RecA protein to the Chi-containing ssDNA overhang leads to the formation of nucleoprotein complex and promoting the search for homologous DNA sequence and DNA strand invasion with the homologous donor duplex. The donor DNA acts as a template for DNA synthesis resulting in the formation of two Holliday junctions that resolves to yield intact duplex products [90]. Thus, the role of RecA is very important for the initial step of the recombinational DNA repair. Similarly, the role of RecN, another protein involved in DNA recombination process was evaluated under low pH condition, as the recN knockout mutants were found highly susceptible to the low pH conditions [91]. Although the mechanism of the RecN protein in the recombinational repair in H. pylori is unknown it has been shown in E. coli that RecA dependent recruitment of RecN to the damaged dsDNA serving a scaffolding function facilitates a critical role for searching of homology DNA by RecA. Failure to the recruitment of RecN to the damaged strand causes the accumulation of fragmented chromosomes [92].
Marsin et al. [93] identified a novel RecO orthologue by using bioinformatics analysis and suggested the presence of RecRO pathway in the dsDNA damage repair. Subsequently, Wang et al. [94] demonstrated that the RecRO pathway is not responsible for dsDNA damage repair caused by mitomycin C (MMC) in H. pylori; however, the proteins RecR and RecO were found responsible for damage repair caused by acid stress. The recR and recO knockout mutants were unable to survive at pH 3.0 whereas wild-type strains survived well in the study [94]. In E. coli, in addition to the RecBCD pathway; the RecFOR (or RecF) pathway can also repair double strand breaks [95]. In RecF pathway, RecJ, a recombinational exonuclease binds to the 5′-ends of the damaged DNA and starts cleaving the strand in upstream direction creating a 3′-overhang. Single-strand binding protein (SSBP) binds to the 3′-overhang [96] and inhibits the binding of RecA protein to the 3′-overhang. The RecO protein facilitates the displacement of SSBP from 3′-overhang and promoting the binding of RecA protein [97]. The RecR, another key component involved in the recombinational damage repair, mediates the loading of RecA on the 3′-overhang [97]. Recently, Wang et al. elucidated the role of hup, encoding a histone like protein by suspending wild-type and the hup knockout mutants at different acidic pH (pH 7.0, pH 5.0 and pH 3.0). They found that suspending at pH 5.0 for 1 hour killed 90% of the hup mutants but caused no significant effect on survival of the wild-type strains while 40% of the wild-type H. pylori survived at pH 3.0 for 1 hour, whereas more than 98% of the hup mutants were killed by the same treatment. Thus, the H. pylori Hup protein has a critical role in the protection from acid stress as the Hup protein has an ability to physically protect DNA from stress damage [98]. The exact mechanism of how the Hup protein involves in the recombinational repair of damaged DNA in H. pylori is unknown. However, in E. coli the homologous histone like protein HU regulates the expression of genes involved in anaerobic respiration, acid stress and response to DNA damage [99, 100]. Therefore, it has been suggested that Hup protein physically protects the DNA or it may cause expression of genes that involve in the stress resistance. Further study is demanded to understand; how Hup protein interacts with other recombinational protein in DNA repair mechanism. Another DNA replication priming protein, PriA, of H. pylori is a component of the priming system which primes the DNA synthesis, has recently been shown to play an essential role in the acid damaged DNA repair and colonization to the epithelial cells. The priA knockout mutants were unable to survive in acidic pH and level of infection was much less when compared with the wild-type strains [101]. The mechanism of this protein in recombinational DNA repair due to acid stress in H. pylori is also unknown. However, in well-studied bacterial model E. coli, the PriA is a key DNA replication protein that involves in the major processes of DNA replication, recombination, damaged DNA repair and restart initiation of the stalked replication fork at damaged DNA sites [102–104].
In summary, the data above mentioned indicate that the homologous recombination is an important mechanism which comes in action after double-stranded DNA damage caused by acid and oxidative stress and involves in the damage repair. The RecA mediated DNA repair involving homology search in addition involves several other recombinational proteins such as RecN, RecO, RecR, Hup, and PriA in the acid stress DNA damage repair.
DNA binding protein
HP0119 (HP number comes from protein number of strain 26695) is a histone like hypothetical protein (HLP) found in H. pylori. Its role in DNA binding and acid stress tolerance was investigated by Wang and Maier [105]. In their study, the treatment at pH 5.0 and 3.0 for one hour killed 90% and 95% of the hp0119 knockout mutants respectively but same treatment to wild-type H. pylori did not affect the survival at pH 5.0 and about 40% cells were killed at pH 3.0. Thus, the hp0119 mutants were susceptible to acid stress while wild-type strains were less susceptible [105]. This study indicates the role of HP0119 protein in protection of the bacterium at low pH. The mechanism of HP0119 in protection from acid stress is unknown and remains to be elucidated in H. pylori. However, multiple functions of HP0119 have been proposed in acid stress survival. It may possess an ability to physically protect DNA from oxidative and acid stress like Mycobacterium tuberculosis histone like protein Lsr2 that protects DNA from stress generated by reactive oxygen intermediates [106]. HP0119 also involves in increased expression of the enzymes that aids the survival of H. pylori [105]. It may also act like Salmonella enterica histone like protein HU that controls three regulons that coordinate virulence, maintains survival in stress and maintains cellular physiology [107]. Therefore, the DNA binding protein HP0119 may protect the bacteria from acid stress in several ways although the exact underlying mechanism is not known.
Urease independent ammonia production
H. pylori infection is potentiated by the production of ammonia which is of great importance for nitrogen source and for neutralization of gastric acidity. Urease, which is found in abundant amount in H. pylori is essential for the production of ammonia from the urea and its survival in the acidic condition [108]. It has been suggested that the large amount of ammonia produced by H. pylori can cause tissue damage during colonization and long term ammonia administration induces gastric mucosal atrophy in rat [109]. Short chain aliphatic amides can be used by some bacteria as source of nitrogen for the growth as they possess the ability to hydrolyze the amides to ammonia and the corresponding organic acid, using an aliphatic amidase. For ammonia production, in addition to the urease; H. pylori possess two aliphatic amidases and aspartase. AmiE, a classical amidase [110] and AmiF, a new type of formamidase, which was detected when the analysis of the complete genome was sequenced [111]. Apart from these amidases there was also increased expression of aspA, an aspartase encoding gene in vivo [112]. In another study it was illustrated that these aliphatic amidases are found in Helicobacter species capable of colonizing the stomach epithelia [113].
Histidine kinase protein
H. pylori encounters a range of acidic conditions within the human stomach and the survival in the acidic pH is mediated by regulation of bacterial gene expression in response to pH. Detection and response to environmental signals requires two component signal transduction system (TCSTS) comprised of a sensor histidine kinase and a response regulator [114, 115]. Three histidine kinase proteins ArsS, AtoS (FlgS, FleS) and CrdS [116–118] also designated as HP0164/HP0165, HP0244 and HP1364 or JHP0151, JHP0229 and JHP1282 are encoded by H. pylori genome, respectively (JHP number come from protein number of strain J99). The histidine kinase CrdS is a strain specific which is required to grow at pH 5.0 in strain J99 [41]. In a study, individual histidine kinase gene (hp0165, hp0244 and hp1364) mutant strains were defective to colonize the mouse stomach [119]. It was also reported that the up-regulation of transcription of three genes (hp0119, hp1432 and ureA) in acidic pH was HP0165 dependent as expression was found in wild-type H. pylori but not in isogenic hp0165 mutants suggesting the role of histidine kinase HP0165 as acid sensor [117]. Recently, Loh and Cover also reported the role of histidine kinases HP0165 and HP1364 in the expression of other several genes, including those that encode urease, amidase (HP0294) and formamidase (HP1238) for acid resistance in H. pylori [120]. All these enzymes are involved in ammonia generation, are likely to play an important role in acid resistance [35, 121, 122]. Marcus et al. [123] demonstrated that during acute acid exposure ArsS was involved in the recruitment of urease protein to the inner membrane to augment acid acclimation in addition to its role as urease gene transcription as described in acid acclimation (Figure 3).
In summary, the histidine kinase proteins regulating the expression of several other genes maintain the survival advantage of H. pylori in acid stress.
Ferric uptake regulator (Fur)
H. pylori is equipped with a repertoire of response regulator gene able to regulate gene expression in response to environmental change as with many other organisms [124]. The regulatory proteins involved in urease, amidase and formamidase regulation are encoded by two genes in H. pylori; the HP1027 (fur) encoding Fur regulator, which mediates iron responsive regulation of amidases [122] and HP1338 (nikR) encoding NikR regulator; which mediates nickel-responsive urease expression [34, 125]. Fur is a best known regulatory protein for regulation of a complicated system that controls the iron uptake and storage in bacterial cell [126, 127]. This regulatory system is accomplished as Fur binds to the promoter region known as Fur box and regulates the expression of Fur-regulatory genes [128]. Fur also regulates gene expression in response to low pH, although, the exact mechanism is not well understood [121, 122, 129]. However, the essential role of Fur in acidic pH adaptation may be partially linked to regulation of amiE and amiF, the amidases that degrades the amides to produce ammonia [122]. In addition, the critical role of Fur under acid stress may be likely linked to the NikR, which directly regulates the trace elements like nickel availability, can also regulate the iron availability in the cell and Fur expression in the acid response [33, 34]. Indeed, the adaptation to the acid stress in H. pylori is a complex process that is mediated by more than one regulatory protein. In a study conducted by Merrell et al. [65] in an experimental model of H. pylori 26695, out of 93 genes, 41 were found to be regulated in a Fur dependent manner and 23 genes were identified in both acid and Fur dependent. Bury-Mone et al. [33] also identified 101 genes that showed altered expression during the growth of H. pylori in acidic pH. Of these 101 genes, 36 genes were shown to be regulated by NikR and/or Fur for pH dependent changes in gene expression.
Fur plays an important role in acid resistance of H. pylori independent of urease [121, 127] and independent of its role in iron acquisition [121]. Gancz et al. [129] using a Mongolian gerbil model demonstrated that although a fur mutant was able to colonize the gastric mucosa if large infectious dose is given, it does inefficiently at early stage of colonization. The oral ID50 for the fur mutant strain was also 100 times higher than that of the wild-type strain. The in vivo colonization defect of fur mutant strains can be likely explained by the fact that Fur modulates expression and changes in transcription in response to acid stress of such a large number of genes. Nevertheless, the role of Fur is more important at early stage of colonization. During initial steps the bacteria are challenged to adapt to new environment that requires changes in gene expression to ensure rapid accumulation to the gastric acidic niche. It was evident that the slight decrease in colonization of the fur mutant at early time points suggests that Fur is essential for the initial stages of colonization [129]. In another in vivo experiment by Miles et al. [130] using Mongolian gerbil models confirmed the role of Fur during the earliest stages of infection establishments where gerbils were first infected with the fur knockout mutants and subsequently super-infected with the wild-type strains at 1, 3, 7, 14 and 28 days after the initial infection. The fur mutants were efficiently displaced with the wild-type strains during first week of infection, but this ability progressively diminished overtime. In converse experiment, the fur mutants were unable to displace the wild-type strains at any time point tested. When co-infected with fur mutants and wild-type strains and monitored the colonization, a 10-fold and 100-fold colonization defect was found at day 1 and 3 respectively [130]. Thus, the Fur plays an important role in the regulation and expression of some genes in response to the acid stress which provides survival advantage in acid environment. Fur also involves in the colonization of H. pylori during the initial stage of colonization.
Role of other virulence factors in acid tolerance
Cytotoxin-associated gene A (cagA)
Many virulence factors have been described in association with different clinical outcomes of H. pylori infections. CagA is an onco-protein and one of the most widely studied virulence factors [131]. Many studies since last one decade have shown that after secretion, the CagA is injected into the target epithelial cells via type-IV secretory system (T4SS) encoded by the cag PAI [132]. After entering to the host cells, phosphorylation of CagA protein undergoes at specific tyrosine residues present in Glu-Pro-Ile-Tyr-Ala (EPIYA) motifs found at the C-terminus of the protein and led to the CagA mediated pathogenicity [133, 134]. However, this virulence factor despite of the role in pathogenicity also involves in the acid susceptibility although the mechanism is not known yet (Table 2). Karita and Blaser first reported that the cagA-positive strains pre-exposed at pH 6.0 for 48 hours were more susceptible to re-exposure at pH 3.0 than the cagA-negative strains [135]. Subsequently, we also found that the H. pylori strains with 5 EPIYA repeats were more susceptible to pH 3.0 than strains having 4 or less EPIYA repeats using Western type strains [136]. Currently, sequences of the EPIYA repeat regions have been annotated according to the segments (20–50 amino acids) flanking the EPIYA motifs (i.e., segments EPIYA-A, B, C or D; C is specific for Western type strains and D is specific for East Asian strains) [137]. Therefore, our previous study indicated that the Western type strains carrying the 2 or less EPIYA-C motif are more acid tolerable in comparison to strains carrying 3 EPIYA-C motif and this phenomena could be the advantageous for survival in gastric acidic environment for strains possessing ABC and ABCC type CagA strains. Kalaf et al. recently reported that strains containing 2 or more EPIYA-C caused infection in patients with higher mean age rather than early age patients [138]. The in vivo data support our previous data that the elder patients are supposed to produce less amount of gastric acid than early age group. In summary, the CagA negative strains are more acid resistant than positive ones and the Western strains with less number of EPIYA-C motifs are more acid resistant than strains with more number of EPIYA-C motifs.
Table 2.
Virulence factors involved in acid tolerance
| Virulence factors | Role of virulence factors | References |
|---|---|---|
| cagA |
cagA-negative strains were more resistant to low pH than cagA-positive strains. Strains with 2 or less EPIYA-C repeats were more acid resistant |
135 136, 138 |
| dupA | dupA-positive strains possess more acid resistant capability than dupA-negative strains | 139–141 |
Role of duodenal ulcer promoting gene (dupA)
Duodenal ulcer promoting gene (dupA) is located in the plasticity region of the H. pylori genome. We first described the role of the gene; one gene that encompassed both jhp0917 and jhp0918 called dupA was associated with a specific clinical outcome [139]. The prevalence of this gene was significantly higher in strains from duodenal ulcer patients than those from gastritis or gastric cancer patients. We also reported the role of dupA in acid survival although the mechanism is unknown yet. We pre-exposed the dupA deleted mutants and wild-type strains at pH 6.0 for 24 hours and subsequently re-exposed to pH 3.0 for 20 minutes and found that the dupA deleted strains were more susceptible at pH 3.0 than wild-type strains [140]. Further the role of dupA in acid shock tolerance was also confirmed by other studies. In a study, Imagawa et al. [140] isolated dupA positive strains from patients with significantly higher gastric acid secretion and dupA negative strains from patients with low gastric acid secretion. In another study by Abadi et al. [141], 12 dupA positive and 20 dupA negative strains from gastritis patients were subjected to a pH range from 3.0 to 7.0. All of the dupA positive strains were able to grow well at pH 4.0 and 33.3% strains did grow at pH 3.0. In contrast, only 40% and 10% dupA negative strains were able to grow at pH 4.0 and 3.0 respectively. The reports of these studies indicate the importance of dupA in the survival of organism in gastric acidic pH.
Conclusions
Given the above information currently compiled suggest that the H. pylori is well habituated for its ecological niche of gastric acidity at pH below 3.0 to cause the persistent infections. The adaptation mechanism of bacterium to the adverse effects of acidic pH is a complex mechanism involving several factors such as bacterial factors (proteins, enzymes, shape and flagella) and environmental factors (urea, mucus and acid). However, the exact mechanism of several factors involved in the acid toleration still remains unsolved and it demands extensive study to evaluate the exact role in the acid survival.
Acknowledgments
This work was supported in part by grants from the National Institutes of Health (DK62813) and the Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan (24406015, 24659200, 25293104, 26640114 and 15H02657) (YY). It was also supported by the Japan Society for the Promotion of Science (JSPS) Institutional Program for Young Researcher Overseas Visits (YY), the Strategic Funds for the Promotion of Science and Technology from Japan Science and Technology Agency (JST) (YY). SA is a PhD student supported by The Japanese Government (Monbukagakusho: MEXT) Scholarship Program
Footnotes
disclosure: The authors declare that they have no any conflict of interests.
References
- 1.Blaser MJ. Who are we? Indigenous microbes and the ecology of human diseases. EMBO Reports. 2006;7(10):956–60. doi: 10.1038/sj.embor.7400812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruggiero P. Helicobacter pylori and inflammation. Curr Pharm Des. 2010;16:4225–4236. doi: 10.2174/138161210794519075. [DOI] [PubMed] [Google Scholar]
- 3.IARC Working-Group; World Health Organization, International Agency for Research on Cancer, editor. Schistosomes, Liver Flukes and Helicobacter pylori. Lyon, France: 1994. [Google Scholar]
- 4.IARC Working Group. Helicobacter pylori Eradication as a Strategy for Preventing Gastric Cancer. Lyon, France: International Agency for Research on Cancer; 2014. (IARC Working Group Reports, No. 8) [Google Scholar]
- 5.Hardbower DM, de Sablet T, Chaturvedi R, Wilson KT. Chronic inflammation and oxidative stress: the smoking gun for Helicobacter pylori-induced gastric cancer? Gut Microbes. 2013;4:475, e481. doi: 10.4161/gmic.25583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rutherford JC. The Emerging role of urease as a general microbial virulence factor. Plos Pathog. 2014;10(5):e1004062. doi: 10.1371/journal.ppat.1004062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olczak AA, Olson JW, Maier RJ. Oxidative-stress resistance mutants of Helicobacter pylori. J Bacteriol. 2002;184(12):3186–93. doi: 10.1128/JB.184.12.3186-3193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown LM. Helicobacter pylori: epidemiology and routes of transmission. Epidemiol Rev. 2000;22:283–97. doi: 10.1093/oxfordjournals.epirev.a018040. [DOI] [PubMed] [Google Scholar]
- 9.Goh KL, Chan WK, Shiota S, Yamaoka Y. Epidemiology of Helicobacter pylori infection and public health implications. Helicobacter. 2011;16(Suppl 1):1–9. doi: 10.1111/j.1523-5378.2011.00874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fock KM, Graham DY, Malfertheiner P. Helicobacter pylori research: historical insights and future directions. Nat Rev Gastroenterol Hepatol. 2013;10:495–500. doi: 10.1038/nrgastro.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rolig AS, Shanks J, Carter JE, Ottemann KM. Helicobacter pylori requires TlpD-driven chemotaxis to proliferate in the antrum. Infect Immun. 2012;80:3713–20. doi: 10.1128/IAI.00407-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–70. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schade C, Flemstrom G, Holm L. Hydrogen ion concentration in the mucus layer on top of acid-stimulated and -inhibited rat gastric mucosa. Gastroenterology. 1994;107:180–88. doi: 10.1016/0016-5085(94)90075-2. [DOI] [PubMed] [Google Scholar]
- 14.Schreiber S, Bucker R, Groll C, Azevedo-Vethacke M, Garten D, Scheid P, Friedrich S, Gatermann S, Josenhans C, Suerbaum S. Rapid loss of motility of Helicobacter pylori in the gastric lumen in vivo. Infect Immun. 2005;73:1584–9. doi: 10.1128/IAI.73.3.1584-1589.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones BD, Lockatell CV, Johnson DE, Warren JW, Mobley HLT. Construction of a urease-negative mutant of Proteus mirabilis: analysis of virulence in a mouse model of ascending urinary tract infection. Infect Immun. 1990;58:1120–1123. doi: 10.1128/iai.58.4.1120-1123.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gatermann S, Marre R. Cloning and expression of Staphylococcus saprophyticus urease gene sequences in Staphylococcus carnosus and contribution of the enzyme to virulence. Infect Immun. 1989;57:2998–3002. doi: 10.1128/iai.57.10.2998-3002.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eaton KA, Brooks CL, Morgan DR, Krakowka S. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect Immun. 1991;59:2470–75. doi: 10.1128/iai.59.7.2470-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schoep TD, Fulurija A, Good F, Lu W, Himbeck RP, Schwan C, Choi SS, Berg DE, Mittl PR, Benghezal M, Marshall BJ. Surface properties of Helicobacter pylori urease complex are essential for persistence. PLoS One. 2010;5(11):e15042. doi: 10.1371/journal.pone.0015042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bode G, Malfertheiner P, Nilius M, Lehnhardt G, Ditschuneit H. Ultrastructural localisation of urease in outer membrane and periplasm of Campylobacter pylori. J Clin Pathol. 1989;42:778–779. doi: 10.1136/jcp.42.7.778-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott DR, Weeks D, Hong C, Postius S, Melchers K, Sachs G. The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterol. 1998;114:58–70. doi: 10.1016/s0016-5085(98)70633-x. [DOI] [PubMed] [Google Scholar]
- 21.Stingl K, Altendorf K, Bakker EP. Acid survival of Helicobacter pylori: how does urease activity trigger cytoplasmic pH homeostasis? Trends Microbiol. 2002;10:70–4. doi: 10.1016/s0966-842x(01)02287-9. [DOI] [PubMed] [Google Scholar]
- 22.Uberti AF, Olivera-Severo D, Wassermann GE, Scopel-Guerra A, Moraes JA, Barcellos-de-Souza P, Barja-Fidalgo C, Carlini CR. Pro-inflammatory properties and neutrophil activation by Helicobacter pylori urease. Toxicon. 2013;69:240–49. doi: 10.1016/j.toxicon.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 23.Eaton KA, Gilbert JV, Joyce EA, Wanken AE, Thevenot T, Baker P, Plaut A, Wright A. In vivo complementation of ureB restores the ability of Helicobacter pylori to colonize. Infect Immun. 2000;70:771–78. doi: 10.1128/iai.70.2.771-778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olson JW, Maier RJ. Molecular hydrogen as an energy source for Helicobacter pylori. Science. 2002;298:1788–90. doi: 10.1126/science.1077123. [DOI] [PubMed] [Google Scholar]
- 25.Ha NC, Oh ST, Sung JY, Cha KA, Lee MH, Oh BH. Supramolecular assembly and acid resistance of Helicobacter pylori urease. Nature Structural Biol. 2001;8(6):505–9. doi: 10.1038/88563. [DOI] [PubMed] [Google Scholar]
- 26.Ernst FD, Stoof J, Horrevoets WM, Kuipers EJ, Kusters JG, van Vliet AH. NikR mediates nickel responsive transcriptional repression of the Helicobacter pylori outer membrane proteins FecA3 (HP1400) and FrpB4 (HP1512) Infect Immun. 2006;74(12):6821–28. doi: 10.1128/IAI.01196-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fulkerson JF, Jr, Mobley HL. Membrane topology of the NixA nickel transporter of Helicobacter pylori: two nickel transport-specific motifs within trans-membrane helices II and III. J Bacteriol. 2000;182:1722–30. doi: 10.1128/jb.182.6.1722-1730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolfram L, Bauerfeind P. Conserved low-affinity nickel-binding amino acids are essential for the function of the nickel permease NixA of Helicobacter pylori. J Bacteriol. 2002;184:1438–43. doi: 10.1128/JB.184.5.1438-1443.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Vliet AHM, Ernst FD, Kusters JG. NikR-mediated regulation of Helicobacter pylori acid adaptation. Trends Microbiol. 2004;12:489–94. doi: 10.1016/j.tim.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Chivers PT, Sauer RT. NikR is a ribbon-helix-helix DNA-binding protein. Protein Sci. 1999;8:2494–2500. doi: 10.1110/ps.8.11.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chivers PT, Sauer RT. Regulation of high affinity nickel uptake in bacteria. Ni2+-dependent interaction of NikR with wild-type and mutant operator sites. J Biol Chem. 2000;275:19735–741. doi: 10.1074/jbc.M002232200. [DOI] [PubMed] [Google Scholar]
- 32.De-Pina K, Desjardin V, Mandrand-Berthelot MA, Giordano G, Wu LF. Isolation and characterization of the nikR gene encoding a nickel-responsive regulator in Escherichia coli. J Bacteriol. 1999;181:670–74. doi: 10.1128/jb.181.2.670-674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bury-Mone S, Thiberge JM, Contreras M, Maitournam A, Labigne A, De-Reuse H. Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol Microbiol. 2004;53:623–38. doi: 10.1111/j.1365-2958.2004.04137.x. [DOI] [PubMed] [Google Scholar]
- 34.Contreras M, Thiberge JM, Mandrand-Berthelot MA, Labigne A. Characterization of the roles of NikR, a nickel-responsive pleiotropic autoregulator of Helicobacter pylori. Mol Microbiol. 2003;49:947–63. doi: 10.1046/j.1365-2958.2003.03621.x. [DOI] [PubMed] [Google Scholar]
- 35.Van-Vliet AHM, Kuipers EJ, Stoof J, Poppelaars SW, Kusters JG. Acid-responsive gene induction of ammonia-producing enzymes in Helicobacter pylori is mediated via a metal-responsive repressor cascade. Infect Immun. 2004;72:766–73. doi: 10.1128/IAI.72.2.766-773.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Vliet AHM, Poppelaars SW, Davies BJ, Stoof J, Bereswill S, Kist M, Penn CW, Kuipers EJ, Kusters JG. NikR mediates nickel-responsive transcriptional induction of urease expression in Helicobacter pylori. Infect Immun. 2002;70(6):2846–52. doi: 10.1128/IAI.70.6.2846-2852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mulrooney SB, Hausinger RP. Nickel uptake and utilization by microorganisms. FEMS Microbiol Rev. 2003;27(2–3):239–61. doi: 10.1016/S0168-6445(03)00042-1. [DOI] [PubMed] [Google Scholar]
- 38.Bury-Moné S, Skouloubris S, Labigne A, De-Reuse H. The Helicobacter pylori UreI protein: role in adaptation to acidity and identification of residues essential for its activity and for acid activation. Mol Microbiol. 2001;42:1021–34. doi: 10.1046/j.1365-2958.2001.02689.x. [DOI] [PubMed] [Google Scholar]
- 39.Weeks DL, Eskandari S, Scott DR, Sachs G. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science. 2000;287:482–85. doi: 10.1126/science.287.5452.482. [DOI] [PubMed] [Google Scholar]
- 40.Wen Y, Marcus EA, Matrubutham U, Gleeson MA, Scott DR, Sachs G. Acid-adaptive genes of Helicobacter pylori. Infect Immun. 2003;71:5921–39. doi: 10.1128/IAI.71.10.5921-5939.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mcgee DJ, Radcliff FJ, Mendz GL, Ferrero RL, Mobley HLT. Helicobacter pylori rocF is required for arginase activity and acid protection in vitro but is not essential for colonization of mice or for urease activity. J Bacteriol. 1999;181(23):7314–22. doi: 10.1128/jb.181.23.7314-7322.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Athmann C, Zeng N, Kang T, Marcus EA, Scott DR, Rektorschek M, Buhmann A, Melchers K, Sachs G. Local pH elevation mediated by the intrabacterial urease of Helicobacter pylori cocultured with gastric cells. J Clin Invest. 2000;106:339–47. doi: 10.1172/JCI9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller EF, Maier RJ. Ammonium metabolism enzymes aid Helicobacter pylori acid resistance. J Bacteriol. 2014;196(17):3074–81. doi: 10.1128/JB.01423-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Young GM, Amid D, Miller VL. A bifunctional urease enhances survival of pathogenic Yersinia enterocolitica and Morganella morganii at low pH. J Bacteriol. 1996;178:6487–95. doi: 10.1128/jb.178.22.6487-6495.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marcus EA, Moshfegh AP, Sachs G, Scott DR. The periplasmic alpha-carbonic anhydrase activity of Helicobacter pylori is essential for acid acclimation. J Bacteriol. 2005;187:729–38. doi: 10.1128/JB.187.2.729-738.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ang S, Lee CZ, Peck K, Sindici M, Matrubutham U, Gleeson MA, Wang JT. Acid induced gene expression in Helicobacter pylori: study in genomic scale by microarray. Infect Immun. 2001;69:1679–86. doi: 10.1128/IAI.69.3.1679-1686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marcus EA, Sachs G, Scott DR. The role of ExbD in periplasmic pH homeostasis in Helicobacter pylori. Helicobacter. 2013;18(5):363–72. doi: 10.1111/hel.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388(6642):539–47. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 49.Ollis AA, Manning M, Held KG, Postle K. Cytoplasmic membrane proton motive force energizes periplasmic interactions between ExbD and TonB. Molec Microbiol. 2009;73(3):466–81. doi: 10.1111/j.1365-2958.2009.06785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Postle K, Larsen RA. TonB-dependent energy transduction between outer and cytoplasmic membranes. Biometals. 2007;20(3–4):453–65. doi: 10.1007/s10534-006-9071-6. [DOI] [PubMed] [Google Scholar]
- 51.Marcus EA, Sachs G, Wen Y, Scott DR. Phosphorylation-dependent and phosphorylation-independent regulation of Helicobacter pylori acid acclimation by the ArsRS two-component system. Helicobacter. 2016;21(1):69–81. doi: 10.1111/hel.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krishna U, Romero-Gallo J, Suarez G, Azah A, Krezel AM, Varga MG, Forsyth MH, Peek RM., Jr Genetic evolution of a Helicobacter pylori acid-sensing histidine kinase and gastric disease. J Infect Dis. 2016;214:644–48. doi: 10.1093/infdis/jiw189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gobert AP, McGee DJ, Akhtar M, Mendz GL, Newton JC, Cheng Y, Mobley HLT, Wilson KT. Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: a strategy for bacterial survival. Proc Natl Acad Sci USA. 2001;98:13844–49. doi: 10.1073/pnas.241443798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martínez LE, Hardcastle JM, Wang J, Pincus Z, Tsang J, Hoover TR, Bansil R, Salama NR. Helicobacter pylori strains vary cell shape and flagellum number to maintain robust motility in viscous environments. Mol Microbiol. 2016;99(1):88–110. doi: 10.1111/mmi.13218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Celli JP, Turner BS, Afdhal NH, Keates S, Ghiran I, Kelly CP, Ewoldt RH, McKinley GH, So P, Erramilli S, Bansil R. Helicobacter pylori moves through mucus by reducing mucin viscoelasticity. Proc Natl Acad Sci USA. 2009;106:14321–26. doi: 10.1073/pnas.0903438106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Windle HJ, Fox A, NíEidhin D, Kelleher D. The thioredoxin system of Helicobacter pylori. J Biol Chem. 2000;275:5081–89. doi: 10.1074/jbc.275.7.5081. [DOI] [PubMed] [Google Scholar]
- 57.Celli JP, Turner BS, Afdhal NH, Ewoldt RH, McKinley GH, Bansil R, Erramilli S. Rheology of gastric mucin exhibits a pH-dependent sol-gel transition. Biomacromolecules. 2007;8:1580–86. doi: 10.1021/bm0609691. [DOI] [PubMed] [Google Scholar]
- 58.Young KD. Bacterial morphology: why have different shapes? Curr Opin Microbiol. 2007;10:596–600. doi: 10.1016/j.mib.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee A, Fox J, Hazell S. Pathogenicity of Helicobacter pylori: a perspective. Infect Immun. 1993;61:1601–10. doi: 10.1128/iai.61.5.1601-1610.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sycuro LK, Pincus Z, Gutierrez KD, Biboy J, Stern CA, Vollmer W, Salama NR. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori’s helical shape and stomach colonization. Cell. 2010;141:822–33. doi: 10.1016/j.cell.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sycuro LK, Wyckoff TJ, Biboy J, Born P, Pincus Z, Vollmer W, Salama NR. Multiple peptidoglycan modification networks modulate Helicobacter pylori’s cell shape, motility, and colonization potential. PLoS Pathog. 2012;8:e1002603. doi: 10.1371/journal.ppat.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suerbaum S. The complex flagella of gastric Helicobacter species. Trends Microbiol. 1995;3:168–70. doi: 10.1016/s0966-842x(00)88913-1. [DOI] [PubMed] [Google Scholar]
- 63.Suerbaum S, Josenbaus C, Labigne A. Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flagellin genes and construction of Helicobacter pylori flaA- and flaB-negative mutants by electroporation-mediated allelic exchange. J Bacteriol. 1993;175:3278–88. doi: 10.1128/jb.175.11.3278-3288.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Josenhans C, Labigne A, Suerbaum S. Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J Bacteriol. 1995;177:3010–20. doi: 10.1128/jb.177.11.3010-3020.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Merrell DS, Goodrich ML, Otto G, Tompkins LS, Falkow S. pH-regulated gene expression of the gastric pathogen Helicobacter pylori. Infect Immun. 2003;71(6):3529–39. doi: 10.1128/IAI.71.6.3529-3539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoshiyama H, Nakamura H, Kimoto M, Okita K, Nakazawa T. Chemotaxis and motility of Helicobacter pylori in a viscous environment. J Gastroenterol. 1999;34(Suppl XI):18–23. [PubMed] [Google Scholar]
- 67.Troge A, Scheppach W, Schroeder BO, Rund SA, Heuner K, Wehkamp J, Stange EF, Oelschlaeger TA. More than a marine propeller--the flagellum of the probiotic Escherichia coli strain Nissle 1917 is the major adhesin mediating binding to human mucus. Int J Med Microbiol. 2012;302:304–314. doi: 10.1016/j.ijmm.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 68.Bucior I, Pielage JF, Engel JN. Pseudomonas aeruginosa pili and flagella mediate distinct binding and signaling events at the apical and basolateral surface of airway epithelium. PLoS Pathog. 2012;8:e1002616. doi: 10.1371/journal.ppat.1002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Olsen JE, Hoegh-Andersen KH, Casadesus J, Rosenkranzt J, Chadfield MS, Thomsen LE. The role of flagella and chemotaxis genes in host pathogen interaction of the host adapted Salmonella enterica serovar Dublin compared to the broad host range serovar S. Typhimurium. BMC Microbiol. 2013;13:67. doi: 10.1186/1471-2180-13-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Clyne M, Ocroinin T, Suerbaum S, Josenhans C, Drumm B. Adherence of isogenic flagellum-negative mutants of Helicobacter pylori and Helicobacter mustelae to human and ferret gastric epithelial cells. Infect Immun. 2000;68:4335, e9. doi: 10.1128/iai.68.7.4335-4339.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kao CY, Sheu BS, Wu JJ. CsrA regulates Helicobacter pylori J99 motility and adhesion by controlling flagella formation. Helicobacter. 2014;19:443, e54. doi: 10.1111/hel.12148. [DOI] [PubMed] [Google Scholar]
- 72.Nakamura H, Yoshiyama H, Takeuchi H, et al. Urease plays an important role in the chemotactic motility of Helicobacter pylori in a viscous environment. Infect Immun. 1998;66:4832–37. doi: 10.1128/iai.66.10.4832-4837.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ottemann KM, Lowenthal AC. Helicobacter pylori uses motility for initial colonization and attain robust infection. Infect Immun. 2002;70:1984–90. doi: 10.1128/IAI.70.4.1984-1990.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Parsonnet J, Hansen S, Rodruguez L, Gelb AB, Warnke RA, Jellum E, Orentreich N, Vogelman JH, Friedman GD. Helicobacter pylori infection and gastric lymphoma. N Engl J Med. 1994;330:1267–71. doi: 10.1056/NEJM199405053301803. [DOI] [PubMed] [Google Scholar]
- 75.Tadjrobehkar O, Abdollahi H. Survival and chemotactic behavior of Helicobacter pylori at different media pH. IJMS. 2004;29(2):81–4. [Google Scholar]
- 76.Lowenthal AC, Simon C, Fair AS, Mehmood K, Terry K, Anastasia S, Ottemann KM. A fixed-time diffusion analysis method determines that the three cheV genes of Helicobacter pylori differentially affect motility. Microbiol. 2009;155:1181–91. doi: 10.1099/mic.0.021857-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rader BA, Wreden C, Hicks KG, Sweeney EG, Ottemann KM, Guillemin K. Helicobacter pylori perceives the quorum-sensing molecule AI-2 as a chemorepellent via the chemoreceptor TlpB. Microbiol. 2011;157:2445–55. doi: 10.1099/mic.0.049353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sweeney EG, Henderson JN, Goers J, Wreden C, Hicks KG, Foster JK, Parthasarathy R, Remington SJ, Guillemin K. Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB. Structure (Cell press) 2012;20:1177–88. doi: 10.1016/j.str.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Croxen MA, Sisson G, Melano R, Hoffman PS. The Helicobacter pylori chemotaxis receptor TlpB (HP0103) is required for pH taxis and for colonization of the gastric mucosa. J Bacteriol. 2006;188(7):2656–65. doi: 10.1128/JB.188.7.2656-2665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.O’Rourke EJ, Chevalier C, Pinto AV, Thiberge JM, Ielpi L, Labigne A, Radicella JP. Pathogen DNA as target for host generated oxidative stress: role for repair of bacterial DNA damage in Helicobacter pylori colonization. Proc Natl Acad Sci USA. 2003;100:2789–94. doi: 10.1073/pnas.0337641100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang G, Alamuri P, Maier RJ. The diverse antioxidant systems of Helicobacter pylori. Mol Microbiol. 2006;61:847–60. doi: 10.1111/j.1365-2958.2006.05302.x. [DOI] [PubMed] [Google Scholar]
- 83.Fischer W, Haas R. The RecA protein of Helicobacter pylori requires a post-translational modification for full activity. J Bacteriol. 2004;186:777–84. doi: 10.1128/JB.186.3.777-784.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Spies M, Kowalczykowski SC. Homologous recombination by RecBCD and RecF pathways. In: Higgins NP, editor. The Bacterial Chromosome. Washington, D.C: ASM Press; 2005. pp. 389–403. [Google Scholar]
- 85.Lam ST, Stahl MM, McMilin KD, Stahl FW. Rec-mediated recombinational hot spot activity in bacteriophage lambda. II. A mutation which causes hot spot activity. Genetics. 1974;77:425–433. doi: 10.1093/genetics/77.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Amundsen SK, Fero J, Hansen LM, Cromie GA, Solnick JV, Smith GR, Salama NR. Helicobacter pylori AddAB helicase-nuclease and RecA promote recombination-related DNA repair and survival during stomach colonization. Molecular Microbiol. 2008;69(4):994–1007. doi: 10.1111/j.1365-2958.2008.06336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Saikrishnan K, Yeeles JT, Gilhooly NS, Krajewski WW, Dillingham MS, Wigley DB. Insights into Chi recognition from the structure of an AddAB-type helicase-nuclease complex. EMBO J. 2012;31:1568–78. doi: 10.1038/emboj.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chedin F, Handa N, Dillingham MS, Kowalczykowski SC. The AddAB helicase/nuclease forms a stable complex with its cognate chi sequence during translocation. J Biol Chem. 2006;281:18610–617. doi: 10.1074/jbc.M600882200. [DOI] [PubMed] [Google Scholar]
- 89.Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–65. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 91.Wang G, Maier RJ. Critical role of RecN in recombinational DNA repair and survival of Helicobacter pylori. Infect Immun. 2008;76(1):153–60. doi: 10.1128/IAI.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Keyamura K, Sakaguchi C, Kubota Y, Niki H, Hishida T. RecA recruits structural maintenance of chromosomes (SMC)-like RecN to DNA double-strand breaks. J Biol Chem. 2013;288(41):29229–237. doi: 10.1074/jbc.M113.485474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Marsin S, Mathieu A, Kortulewski T, Guerois R, Radicella JP. Unveiling novel RecO distant orthologues involved in homologous recombination. PLoS Genet. 2008;4(8):e1000146. doi: 10.1371/journal.pgen.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang G, Lo LF, Maier RJ. The RecRO pathway of DNA recombinational repair in Helicobacter pylori and its role in bacterial survival in the host. DNA Repair (Amst) 2011;10(4):373–79. doi: 10.1016/j.dnarep.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Morimatsu K, Kowalczykowski SC. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Molecular Cell. 2003;1(5):1337–47. doi: 10.1016/s1097-2765(03)00188-6. [DOI] [PubMed] [Google Scholar]
- 96.Handa N, Morimatsu K, Lovett ST, Kowalczykowski SC. Reconstitution of initial steps of dsDNA break repair by the RecF pathway of E. coli. Genes & Development. 2009;23(10):1234–45. doi: 10.1101/gad.1780709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Inoue H, Honda M, Ikawa S, Shibata T, Mikawa T. The process of displacing the single-stranded DNA-binding protein from single-stranded DNA by RecO and RecR proteins. Nucleic Acids Research. 2008;36(1):94–109. doi: 10.1093/nar/gkm1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang G, Lo LF, Maier RJ. A histone-like protein of Helicobacter pylori protects DNA from stress damage and aids host colonization. DNA Repair (Amst) 2012;11(9):733–40. doi: 10.1016/j.dnarep.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Oberto J, et al. The HU regulon is composed of genes responding to anaerobiosis, acid stress, high osmolarity and SOS induction. PLoS One. 2009;4(2):e4367. doi: 10.1371/journal.pone.0004367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bi H, et al. HU participates in expression of a specific set of genes required for growth and survival at acidic pH in Escherichia coli. Current Microbiol. 2009;58(5):443–48. doi: 10.1007/s00284-008-9340-4. [DOI] [PubMed] [Google Scholar]
- 101.Singh A, Blaskovic D, Joo J, Yang Z, Jackson SH, Coleman WG, Yan M. Investigating the role of Helicobacter pylori PriA protein. Helicobacter. 2016 doi: 10.1111/hel.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Heller RC, Marians KJ. Replication fork reactivation downstream of the blocked nascent leading strand. Nature. 2006;439:557–62. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- 103.Kogoma T, Cadwell GW, Barnard KG, Asai T. The DNA replication priming protein, PriA, is required for homologous recombination and double-strand break repair. J Bacteriol. 1996;178:1258–64. doi: 10.1128/jb.178.5.1258-1264.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jones JM, Nakai H. Escherichia coli PriA helicase: fork binding orients the helicase to unwind the lagging strand side of arrested replication forks. J Mol Biol. 2001;312:935–47. doi: 10.1006/jmbi.2001.4930. [DOI] [PubMed] [Google Scholar]
- 105.Wang G, Maier RJ. A novel DNA-binding protein plays an important role in Helicobacter pylori stress tolerance and survival in the host. J Bacteriol. 2015;197:973–82. doi: 10.1128/JB.02489-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Colangeli R, Haq A, Arcus VL, Summers E, Magliozzo RS, McBride A, Mitra AK, Radjainia M, Khajo A, Jacobs WR, Jr, Salgamea P, Alland D. The multi-functional histone-like protein Lsr2 protects mycobacteria against reactive oxygen intermediates. Proc Natl Acad Sci USA. 2009;106:4414–18. doi: 10.1073/pnas.0810126106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mangan MW, Lucchini S, Fitzgerald TOCS, Hinton JC, Dorman CJ. Nucleoid-associated protein HU controls three regulons that coordinate virulence, response to stress and general physiology in Salmonella enterica serovar Typhimurium. Microbiol. 2011;157:1075–87. doi: 10.1099/mic.0.046359-0. [DOI] [PubMed] [Google Scholar]
- 108.Clyne M, Labigne A, Drumm B. Helicobacter pylori requires an acidic environment to survive in the presence of urea. Infect Immun. 1995;63:1669–73. doi: 10.1128/iai.63.5.1669-1673.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tsujii M, Kawano S, Tsuji T, Ito K, Sasaki Y, Hayashi N, Fusamoto H, Kamada T. Cell kinetics of mucosal atrophy in rat stomach induced by long-term administration of ammonia. Gastroenterol. 1993;104:796–801. doi: 10.1016/0016-5085(93)91015-a. [DOI] [PubMed] [Google Scholar]
- 110.Skouloubris S, Labigne A, De Reuse H. Identification and characterization of an aliphatic amidase in Helicobacter pylori. Mol Microbiol. 1997;25:989–98. doi: 10.1111/j.1365-2958.1997.mmi536.x. [DOI] [PubMed] [Google Scholar]
- 111.Skouloubris S, Labigne A, De Reuse H. The AmiE aliphatic amidase and AmiF formamidase of Helicobacter pylori: natural evolution of two enzyme paralogues. Mol Microbiol. 2001;40(3):596–09. doi: 10.1046/j.1365-2958.2001.02400.x. [DOI] [PubMed] [Google Scholar]
- 112.Scott DR, Marcus EA, Wen Y, Oh J, Sachs G. Gene expression in vivo shows that Helicobacter pylori colonizes an acidic niche on the gastric surface. Proc Natl Acad Sci USA. 2007;104(17):7235–40. doi: 10.1073/pnas.0702300104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bury-Mone S, Skouloubris S, Dauga C, Thiberge JM, Dailidiene D, Berg DE, Labigne A, Reuse HD. Presence of active aliphatic amidases in Helicobacter species able to colonize the stomach. Infect Immun. 2003;71(10):5613–22. doi: 10.1128/IAI.71.10.5613-5622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hoch JA. Two-component and phosphorelay signal transduction. Curr Opin Microbiol. 2000;3:165–70. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 115.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–15. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 116.Niehus E, Gressmann H, Ye F, Schlapbach R, Dehio M, Dehio C, Stack A, Meyer TF, Suerbaum S, Josenhans C. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol Microbiol. 2004;52:947–61. doi: 10.1111/j.1365-2958.2004.04006.x. [DOI] [PubMed] [Google Scholar]
- 117.Pflock M, Dietz P, Schar J, Beier D. Genetic evidence for histidine kinase HP165 being an acid sensor of Helicobacter pylori. FEMS Microbiol Lett. 2004;234:51–61. doi: 10.1016/j.femsle.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 118.Waidner B, Melchers K, Stahler FN, Kist M, Bereswill S. The Helicobacter pylori CrdRS two-component regulation system (HP1364/HP1365) is required for copper-mediated induction of the copper resistance determinant CrdA. J Bacteriol. 2005;187:4683–88. doi: 10.1128/JB.187.13.4683-4688.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Panthel K, Dietz P, Haas R, Beier D. Two-component systems of Helicobacter pylori contribute to virulence in a mouse infection model. Infect Immun. 2003;71:5381–85. doi: 10.1128/IAI.71.9.5381-5385.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Loh JT, Cover TL. Requirement of histidine kinases HP0165 and HP1364 for acid resistance in Helicobacter pylori. Infect Immun. 2006;74(5):3052–59. doi: 10.1128/IAI.74.5.3052-3059.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bijlsma JJ, Waidner B, van Vliet AHM, Hughes NJ, Hag S, Bereswill S, Kelly DJ, Vandenbroucke-Grauls CMJE, Kist M, Kusters JG. The Helicobacter pylori homologue of the ferric uptake regulator (Fur) is involved in acid resistance. Infect Immun. 2002;70(2):606–11. doi: 10.1128/iai.70.2.606-611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.van Vliet AH, Stoof J, Poppelaars SW, Bereswill S, Homuth G, Kist M, Kuipers EJ, Kusters JG. Differential regulation of amidase and formamidase-mediated ammonia production by the Helicobacter pylori fur repressor. J Biol Chem. 2003;278:9052–57. doi: 10.1074/jbc.M207542200. [DOI] [PubMed] [Google Scholar]
- 123.Marcus EA, Sachs G, Wen Y, Feng J, Scott DR. Role of the Helicobacter pylori sensor kinase ArsS in protein trafficking and acid acclimation. J Bacteriol. 2012;194(20):5545–51. doi: 10.1128/JB.01263-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Baltrus DA, Amieva MR, Covacci A, Lowe TM, Merrell DS, Ottemann KM, Stein M, Salama NR, Guillemin K. The complete genome sequence of Helicobacter pylori strain G27. J Bacteriol. 2009;191(1):447–48. doi: 10.1128/JB.01416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.van Vliet AHM, Poppelaars SW, Davies BJ, Stoof J, Bereswill S, Kist M, Kuipers EJ, Penn CW, Kusters JG. NikR mediates nickel-responsive transcriptional induction of urease expression in Helicobacter pylori. Infect Immun. 2002;70:2846–52. doi: 10.1128/IAI.70.6.2846-2852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Carpenter BM, Whitmire JM, Merrell DS. This is not your mother’s repressor: the complex role of fur in pathogenesis. Infect Immun. 2009;77(7):2590–01. doi: 10.1128/IAI.00116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.van Vliet AH, Stoof J, Vlasblom R, Wainwright SA, Hughes NJ, Kelly DJ, Bereswill S, Bijlsma JJ, Hoogenboenzem T, Vandenbroucke-Grauls CM, Kist M, Kujpers EJ, Kusters JG. The role of the ferric uptake regulator (Fur) in regulation of Helicobacter pylori iron uptake. Helicobacter. 2002;7(4):237–44. doi: 10.1046/j.1523-5378.2002.00088.x. [DOI] [PubMed] [Google Scholar]
- 128.Delany I, Spohn G, Rappuoli R, Scarlato V. The Fur repressor controls transcription of iron-activated and -repressed genes in Helicobacter pylori. Mol Microbiol. 2001;42:1297–1309. doi: 10.1046/j.1365-2958.2001.02696.x. [DOI] [PubMed] [Google Scholar]
- 129.Gancz H, Censini S, Merrell DS. Iron and pH homeostasis intersect at the level of Fur regulation in the gastric pathogen Helicobacter pylori. Infect Immun. 2006;74(1):602–14. doi: 10.1128/IAI.74.1.602-614.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Miles S, Piazuelo MB, Semino-Mora C, Washington MK, Dubois A, Peek RM, Jr, Correa P, Merrell DS. Detailed in vivo analysis of the role of Helicobacter pylori Fur in colonization and disease. Infect Immun. 2010;78(7):3073–82. doi: 10.1128/IAI.00190-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Oldani A, Cormont M, Hofman V, Chiozzi V, Oregioni O, Canonici A, Sciullo A, Sommi P, Fabbri A, Ricci V, Boquet P. Helicobacter pylori counteracts the apoptotic action of its VacA toxin by injecting the CagA protein into gastric epithelial cells. PLoS Pathog. 2009;5(10):e1000603. doi: 10.1371/journal.ppat.1000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tegtmeyer N, Wessler S, Backert S. Role of the Cag-pathogenicity island encoded type IV secretion system in Helicobacter pylori pathogenesis. FEBS J. 2011;278:1190–02. doi: 10.1111/j.1742-4658.2011.08035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hatakeyama M. Helicobacter pylori CagA-a potential bacterial oncoprotein that functionally mimics the mammalian Gab family of adaptor proteins. Microbes Infect. 2003;5:143–50. doi: 10.1016/s1286-4579(02)00085-0. [DOI] [PubMed] [Google Scholar]
- 134.Higashi H, Tsutsumi R, Fujita A, Yamazaki S, Asaka M, Azuma T, Hatakeyama M. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc Natl Acad Sci USA. 2002;99:14428–33. doi: 10.1073/pnas.222375399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Karita M, Blaser MJ. Acid-tolerance response in Helicobacter pylori and differences between cagA+ and cagA− strains. J Infect Dis. 1998;178:213–19. doi: 10.1086/515606. [DOI] [PubMed] [Google Scholar]
- 136.Yamaoka Y, El–Zimaity HMT, Gutierrez O, Figura N, Kim JK, Kodama T, Kashima K, Graham DY. Relationship between the cagA 3′ repeat region of Helicobacter pylori, gastric histology, and susceptibility to low pH. Gastroenterol. 1999;117:342–49. doi: 10.1053/gast.1999.0029900342. [DOI] [PubMed] [Google Scholar]
- 137.Yamaoka Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat Rev Gastroenterol Hepatol. 2010;7(11):629–41. doi: 10.1038/nrgastro.2010.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kalaf EA, Al-Khafaii ZM, Yassen NY, Al-Abbudi FA, Sadwen SN. Study of the cytoxin-associated gene A (cagA gene) in Helicobacter pylori using gastric biopsies of Iraqi patients. Saudi J Gastroenterol. 2013;19(2):69–74. doi: 10.4103/1319-3767.108474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lu H, Hsu PI, Graham DY, Yamaoka Y. Duodenal ulcer promoting gene of Helicobacter pylori. Gastroenterol. 2005;128(4):833–48. doi: 10.1053/j.gastro.2005.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Imagawa S, Ito M, Yoshihara M, Eguchi H, Tanaka S, Chayama K. Helicobacter pylori dupA and gastric acid secretion are negatively associated with gastric cancer development. J Med Microbiol. 2010;59:1484–89. doi: 10.1099/jmm.0.021816-0. [DOI] [PubMed] [Google Scholar]
- 141.Abadi ATB, Taghvaei T, Wolfram L, Kusters JG. Infection with Helicobacter pylori strains lacking dupA is associated with an increased risk of gastric ulcer and gastric cancer development. J Med Microbiol. 2012;61:23–30. doi: 10.1099/jmm.0.027052-0. [DOI] [PubMed] [Google Scholar]