Abstract
Class 2 CRISPR–Cas systems are characterized by effector modules that consist of a single multidomain protein, such as Cas9 or Cpf1. We designed a computational pipeline for the discovery of novel class 2 variants and used it to identify six new CRISPR–Cas subtypes. The diverse properties of these new systems provide potential for the development of versatile tools for genome editing and regulation. In this Analysis article, we present a comprehensive census of class 2 types and class 2 subtypes in complete and draft bacterial and archaeal genomes, outline evolutionary scenarios for the independent origin of different class 2 CRISPR–Cas systems from mobile genetic elements, and propose an amended classification and nomenclature of CRISPR–Cas.
CRISPR–Cas systems provide adaptive immunity in archaea and bacteria1–5. The structural features and mechanisms of CRISPR–Cas are described in detail in several recent reviews3–6. In brief, the CRISPR–Cas response consists of three stages. During the first stage, known as adaptation, the Cas1–Cas2 protein complex (which, in some cases, contains additional subunits) excises a segment of the target DNA (known as the protospacer) and inserts it between the repeats at the 5′ end of a CRISPR array, yielding a new spacer. In the expression and processing stage, a CRISPR array, together with the spacers, is transcribed into a long transcript known as the pre-CRISPR RNA (pre-crRNA) and is processed by a distinct complex of Cas proteins (which, in some cases, involves additional proteins and RNA molecules) into mature small CRISPR RNAs (crRNAs). Finally, during the interference stage, a complex of Cas proteins (typically, a modified processing complex) uses the crRNA as a guide to cleave the target DNA or RNA. Similarly to other defence mechanisms, CRISPR–Cas systems have evolved in the context of an incessant arms race with mobile genetic elements, which has resulted in extreme diversification of Cas protein sequences and in the architecture of the CRISPR–cas loci7–12. Owing to this diversity and the lack of universal cas genes, a comprehensive classification of CRISPR–Cas systems cannot be generated as a single phylogenetic tree, but requires a multifaceted approach that combines the identification of signature genes with phylogenetic trees and the analysis of sequence similarity between partially conserved cas genes, as well as the comparison of the loci organization13, 14. The latest published classification of CRISPR–Cas systems includes two classes that are subdivided into five types and 16 subtypes15. Shortly after this classification, a sixth type and three additional subtypes were identified16.
Class 1 CRISPR–Cas systems, which have multisubunit effector complexes, are most common in bacteria and archaea (including in all hyperthermophiles), comprising ~90% of all identified CRISPR–cas loci15. The remaining ~10% of CRISPR–cas loci belong to class 2 CRISPR–Cas systems (which use a type II, type V or type VI effector protein); these systems are found almost exclusively in bacteria and have not been identified in hyperthermophiles15, 17.
CRISPR–Cas systems are characterized by pronounced functional and evolutionary modularity8. The adaptation module, which is responsible for spacer acquisition shows limited variation among the diverse CRISPR–Cas systems15. By contrast, the CRISPR–Cas effector module, which mediates the maturation of crRNAs, as well as target recognition and cleavage, is more versatile in gene composition and locus architecture; this led to the two classes of CRISPR–Cas system being defined based on the organization of their effector modules15. The effector complexes of class 1 systems consist of 4–7 Cas protein subunits in an uneven stoichiometry, as exemplified by the CRISPR-associated complex for antiviral defence (Cascade) of type I systems18–21, and the Csm–Cmr complexes of type III systems22–25. By contrast, the characteristic feature of class 2 systems is an effector module that consists of a single, multidomain protein. The relatively simple architecture of their effector complexes has made class 2 CRISPR–Cas systems an attractive choice for use in the new generation of genome-editing tools26–29.
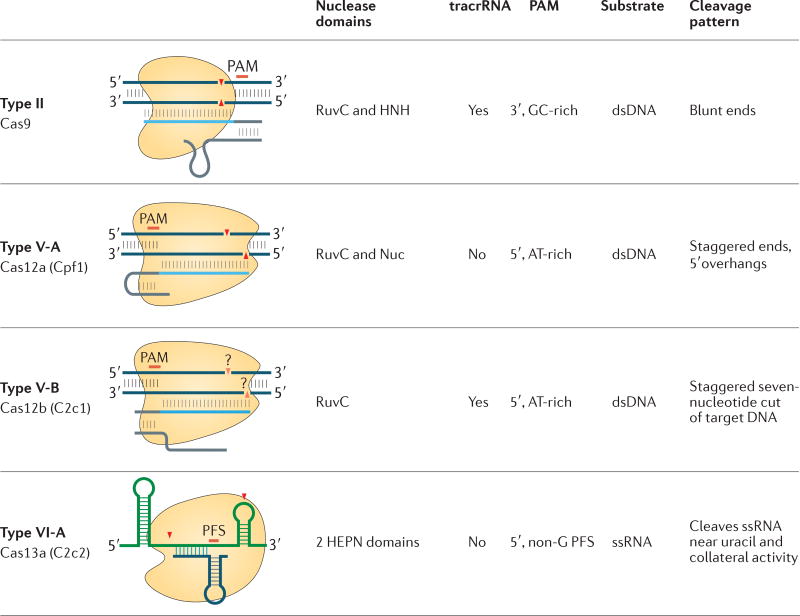
Before the analysis reported here, five (predicted) class 2 effectors had been described — Cas9, Cpf1, C2c1, C2c2 and C2c3 — the most common and best studied of which is the type II effector, Cas9. Cas9 is a crRNA-dependent endonuclease that contains two unrelated nuclease domains, RuvC and HNH, which are responsible for cleavage of the displaced (non-target) and target DNA strands, respectively, in the crRNA–target DNA complex26,29–33. Type II CRISPR–cas loci also encode a trans-acting CRISPR RNA (tracrRNA) that might have evolved from the corresponding CRISPR and that is essential for pre-crRNA processing and target recognition in type II systems26,34–36. The amino acid sequence of Cpf1, which is the prototype type V effector, contains only one readily detectable nuclease domain, RuvC15,37,38. However, structures of Cpf1 in complex with crRNA, or with both crRNA and target DNA, revealed a second nuclease domain with a unique fold that is functionally analogous to the HNH domain of Cas9 (REFS 39,40). An important difference between Cpf1 and Cas9 is that Cpf1 is a single-RNA-guided nuclease that does not require a tracrRNA. Furthermore, the Cpf1 protein itself is responsible for pre-crRNA processing, although the nature of its RNase activity is not characterized41. Cpf1 also differs from Cas9 in its cleavage pattern and in its protospacer-adjacent motif (PAM), which determines which targets are cleaved38. These differences suggest that the discovery of novel class 2 effectors could enhance the application of CRISPR systems to genome engineering. Furthermore, the discovery of two distantly related class 2 effector proteins, Cas9 and Cpf1, suggests that other distinct variants of such systems could exist. Prompted by these findings, we developed a computational pipeline to systematically identify novel class 2 CRISPR–cas loci in genomic and metagenomic sequences. Using Cas1, which is the most highly conserved Cas protein, as a seed, we identified three previously unknown class 2 subtypes, two of which contained effectors that are distantly related to Cpf1 and were included as additional subtypes in type V; the third novel class 2 subtype became the newly classified type VI subtype16. The expression and ability to cause interference of two of these proteins, denoted C2c1 and C2c2, have been experimentally demonstrated16,42.
In this Analysis article, we expand on our previous findings16,42 and describe further analysis that we believe provides a comprehensive census of class 2 effectors in sequenced bacterial and archaeal genomes. This new analysis stems from the observation that many known CRISPR–Cas systems are non-autonomous; that is, they depend on Cas1 and Cas2 proteins that are supplied by other CRISPR–cas loci in the same genome, and, as such, their loci lack cas1 (REF. 15) and will not have been detected in our previous analyses15,16. We extended the search for novel class 2 systems by using the CRISPR array itself as the seed. Consequently, we identified novel, putative class 2 effectors that were missed in the previous analyses15,16 and which belong to at least three new CRISPR–Cas subtypes. We further discuss the evolutionary implications of our findings, including evidence of a crucial role for mobile genetic elements in the independent origin of different types and subtypes of class 2 CRISPR–Cas systems.
Comparative genomics and evolution
The previously developed computational pipeline that is extended in this analysis is shown in BOX 1 and is further explained in Supplementary information S1 (box). Using Cas1 as the seed, two new type V subtypes (effectors that contain a RuvC-like nuclease domain that is distantly related to that of Cas9) and one new type VI subtype (a putative effector that contains two higher eukaryotes and prokaryotes nucleotide-binding domains (HEPN domains)) were identified16. When CRISPR was used as the seed to detect non-autonomous class 2 systems in this analysis, three new subtypes were detected, including an additional heterogeneous subtype of putative type V systems and two subtypes of type VI systems (see Supplementary information S2 (box), part a). We expect that the detected variants almost completely represent the diversity of class 2 CRISPR–Cas systems that are detectable in currently available genomes, given that all large proteins (that is, putative class 2 effectors) that are encoded near a cas1 gene and/or a CRISPR array were analysed in detail in this work.
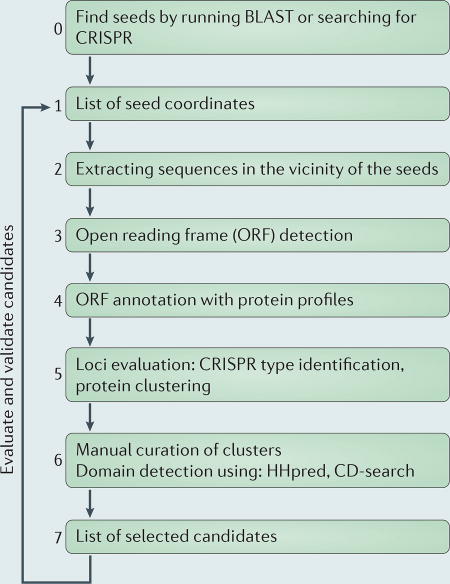
Box 1. The computational pipeline for the discovery of class 2 CRISPR–cas loci.
We have developed a computational pipeline for the systematic detection of class 2 CRISPR–Cas systems (see the figure). The procedure begins with the identification of a ‘seed’ that signifies the likely presence of a CRISPR–cas locus in a given nucleotide sequence (see the figure; the steps in the procedure are numbered in the order in which they occur). In the previously reported analyses15,16, we used Cas1 as the seed, as it is the most common Cas protein in CRISPR–Cas systems and is most highly conserved at the sequence level9. In this article, we update this part of the analysis by searching the current sequence databases (Supplementary information S1 (box)). To ensure the maximum sensitivity of detection, the search was carried out by comparing a Cas1 sequence profile to translated genomic and metagenomic sequences. After the cas1 genes were detected, their respective ‘neighbourhoods’ were examined for the presence of other cas genes by searching with ~400 previously developed profiles for Cas proteins and applying the criteria for the classification of the CRISPR–cas loci15. In a complementary approach, to extend the search to non-autonomous CRISPR–Cas systems, the same procedure was repeated using the CRISPR array as the seed. To ensure that the CRISPR array was detected at a high level of sensitivity, the predictions that were made using the Piler-CR72 and CRISPRfinder73 methods were pooled and taken as the final CRISPR set (see the figure). This procedure yielded 47,174 CRISPR arrays, which is more than twice the number of cas1 genes that were detected, reflecting the fact that many CRISPR–cas loci lack the adaptation module and that numerous ‘orphan’ arrays, some of which seem to be functional, also exist74.
All loci that were assigned to known CRISPR–Cas subtypes through the Cas protein profile search were discarded from the subsequent analysis, given that the search aimed to discover new subtypes. Among the remaining cas1 and CRISPR neighbourhoods, those that encoded large proteins (>500 amino acids) were analysed in detail, given that Cas9 and Cpf1 are large proteins (typically >1000 amino acids) and that their protein structures suggest that this large size is required to accommodate the CRISPR RNA (crRNA)–target DNA complex30,31,40. The sequences of such large proteins were then screened for known protein domains using sensitive profile-based methods, such as HHpred, secondary structure prediction and manual examination of multiple alignments (Supplementary information S1 (box)). Under the premise that class 2 effector proteins contain nuclease domains, even if they are distantly related or unrelated to known families of nucleases, the proteins that contain domains that are deemed irrelevant in the context of the CRISPR–Cas function (for example, membrane transporters or metabolic enzymes) were discarded. The retained proteins either contained readily identifiable, or completely unknown, nuclease domains. The sequences of these proteins were then analysed using the most sensitive methods for domain detection, such as HHpred, with a curated multiple alignment of the respective protein sequences that were used as the query. The use of sensitive methods is essential because proteins that are involved in antiviral defence, and the Cas proteins in particular, typically evolve extremely fast9,75.
Note that the depicted procedure for the discovery of class 2 CRISPR–Cas systems, at least in principle, is expected to be exhaustive, because all loci that contain a gene that encodes a large protein (that is, a putative class 2 effector) in the vicinity of cas1 and/or CRISPR are analysed in detail. The assumption of the structural requirements for a class 2 effector, which underlie the protein size cut-off that is used, and the precision of cas1 and CRISPR detection, are the only limitations of this approach. BLAST, basic local alignment search tool.
Subtypes V-A, V-B and V-C identified using a Cas1 seed: large multidomain effectors
The distinctive feature of type II and type V CRISPR–Cas sequences is the presence of a RuvC-like nuclease domain in their multidomain effector proteins. In the type II effector Cas9, the RuvC-like domain contains an inserted HNH nuclease domain (FIGS 1,2). Other than the RuvC-like domain, the effector proteins of the three type V subtypes do not share any detectable sequence similarity to each other or to Cas9. However, the only available crystal structures of class 2 effectors, specifically those of Cas9 and Cpf1, reveal that they have a common structural framework (see above)39,40. The structures of the putative, large, type V effectors that were discovered using the cas1 seed, namely those of the subtype V-B and subtype V-C, are unsolved, but the subtype V-B effector C2c1 was shown to have robust interference activity16. All of the class V effectors that were identified at this stage share a similar, large size (typically, 1,000–1,300 amino acid residues) and a single common domain, the RuvC-like endonuclease domain, although the sequence similarity between the effector proteins of different subtypes is extremely low. It is likely that all type V effectors adopt similar bilobed structures that hold together the crRNA and target DNA, although the effector proteins of different subtypes do not seem to be directly related.
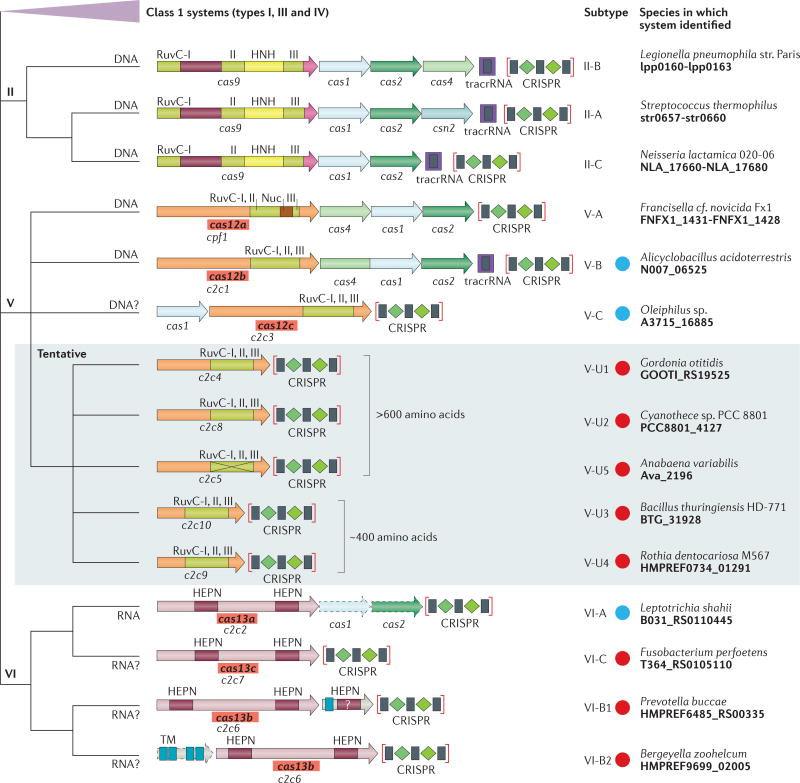
Figure 1. The updated classification scheme for class 2 CRISPR–Cas systems.
The class 1 systems are collapsed; all other systems shown are class 2 systems. New class 2 systems that were discovered using the computational pipeline in this study (see BOX 1) are indicated with blue circles for those that were described previously16 and with red circles for those that are presented here for the first time. For each class 2 system subtype, as well as for the five distinct variants of the provisional V-uncharacterized (V-U) subtype, the locus organization and the domain architecture of the effector and accessory proteins are schematically shown. RuvC-I, RuvC-II and RuvC-III are the three distinct motifs that contribute to the nuclease catalytic centre; numerals in the figure correspond to the respective RuvC motif. The portions of Cas9 proteins that roughly correspond to the recognition lobe and the protospacer-adjacent motif (PAM)-interacting domain are shown by maroon and pink shapes, respectively. The proposed new systematic gene names are shown in bold type in red boxes. Provisional gene names for effector protein candidates are shown below the respective shapes as follows: C2c1–10, class 2 candidate proteins 1–10; for subtype V-A, the previously introduced vernacular cpf1 is indicated. For subtype VI-A, cas1 and cas2 are shown with dashed contours to indicate that only some of these loci include the adaptation module. For the V-U5 variant, the inactivation of the RuvC-like nuclease domain is indicated by a cross. The specific strains of bacteria in which these systems were identified and locus tags for the respective protein-coding genes are also indicated. The abbreviation TM indicates a predicted transmembrane helix. The predicted type of target, namely DNA or RNA, is indicated for each subtype. A question mark next to the target indicates that the activity is only predicted and has not been demonstrated experimentally. The target is not indicated for the type V-U systems because their RNA-guided interference capacity is questionable, which is additionally emphasized by shading. tracrRNA, trans-acting CRISPR RNA.
The search for homologues of the type II and type V effectors showed that the RuvC-like nuclease domains are related to TnpB proteins, an extremely abundant but poorly characterized family of nucleases that are encoded by many autonomous (that is, those that encode an active transposase, denoted TnpA, and mediate their own transposition) and even more numerous non-autonomous (that is, those that consist solely of the tnpB gene and rely on transposases from other elements for their transposition) bacterial and archaeal transposons43–45 (FIG. 3a). In addition to the RuvC-like nuclease domain, TnpB proteins contain a predicted, positively charged, long α-helix that seems to be the counterpart to the bridge helix, which is a common feature of Cas9 and Cpf1 (FIG. 2). Thus, similar to the class 2 effectors, the TnpB proteins are predicted to bind to RNA. Moreover, it has been reported that a TnpB protein from the haloarchaeon Halobacterium salinarum binds to short overlapping sense transcripts of its own gene46. Biochemical and biological characterization of TnpB should shed light on the evolution of the functions of class 2 CRISPR–Cas effectors.
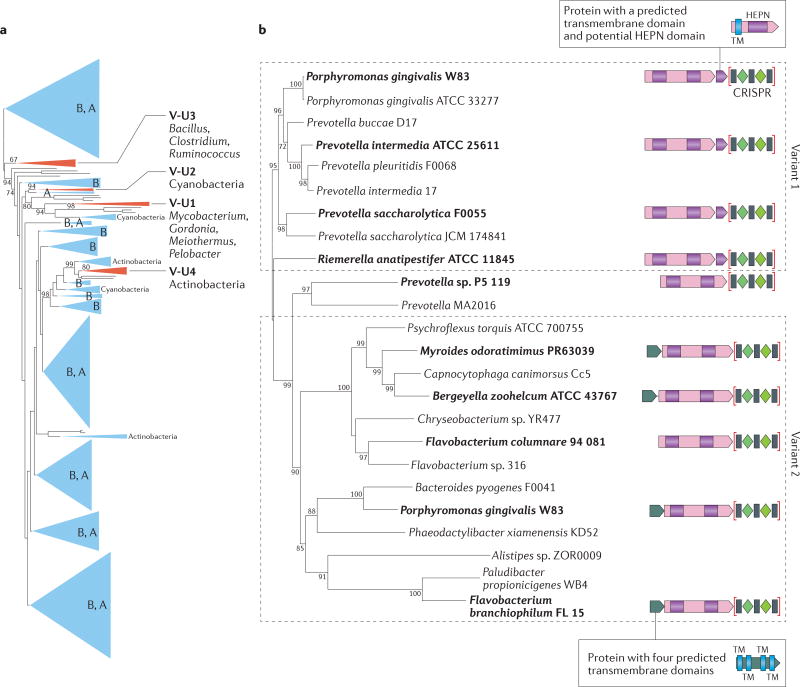
Figure 3. Phylogenies of the type V and type VI-B effectors.
a | A maximum-likelihood phylogenetic tree of TnpB nucleases, including the putative type V-uncharacterized (V-U) effectors that have a predicted active RuvC domain (Supplementary information S1 (box)). The major subtrees of transposon-encoded TnpB proteins are collapsed and indicated by triangles; some of these large groups include tnpB genes that are adjacent to CRISPR arrays, but these do not show evolutionary stability and thus cannot be identified as effectors. The four distinct evolutionarily stable groups of CRISPR-associated TnpB assigned to subtype V-U are shown by red triangles. Altogether, the tree includes 1,770 unique TnpB sequences, 403 of which are TnpB proteins that are encoded next to TnpA (autonomous transposons); 168 of these tnpB genes are adjacent to CRISPR arrays, and of these, 49 are assigned to four variants of subtype V-U (none of these belongs to autonomous transposons). In the subtrees that include the subtype V-U variants, bootstrap values (percentages) are shown for those subtrees that include the distinct V-U variants. For each type V-U variant, the bacterial taxa that harbour the majority of the respective loci are indicated. Dominant bacterial or archaeal lineages, if there are any, are indicated in the triangles. For the complete tree and accession numbers of all sequences, see Supplementary information S2 (box), part c and part h. b | Phylogenetic tree of the subtype VI-B Cas13b effector proteins. The tree was constructed as in part a, and the bootstrap values that are larger than 70% are indicated. The organization of typical cas13b loci for selected representatives (specifically those that are shown in bold) is schematically shown on the right. Variant 1 and variant 2 correspond to the two major branches of the tree and differ with respect to the domain architectures of the second smaller protein encoded in the locus; the domain architectures of these putative accessory proteins are shown above (for variant 1) and below (for variant 2) the respective loci schematics. The CRISPR arrays are shown schematically in brackets. TM indicates a predicted transmembrane domain, shown by blue boxes. Higher eukaryotes and prokaryotes nucleotide-binding (HEPN) domains are shown as maroon boxes. A, diverse archaea; B, diverse bacteria.
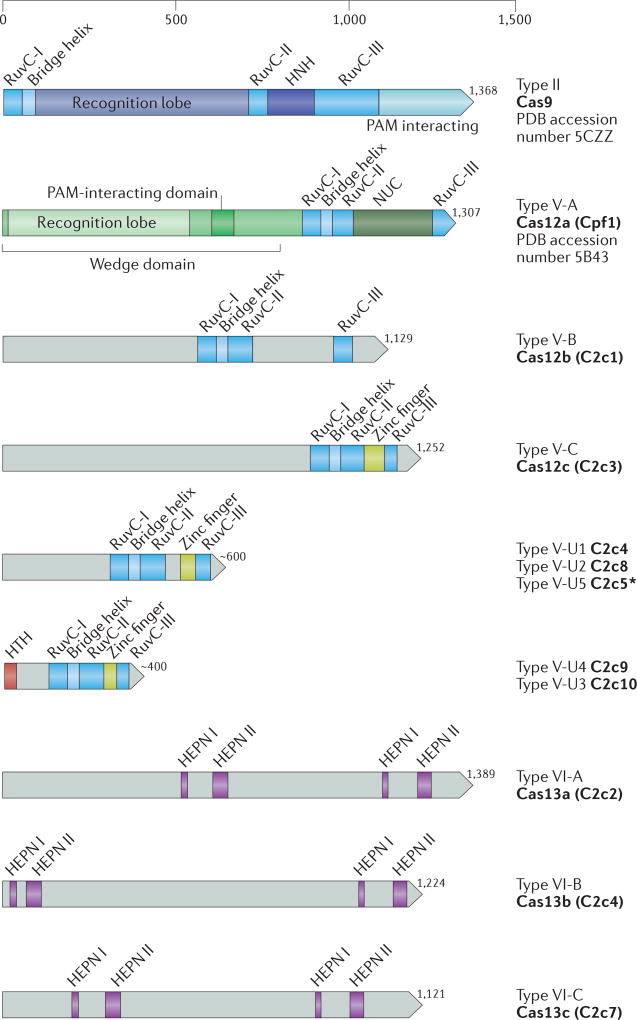
Figure 2. The domain architecture of class 2 CRISPR effector proteins.
For the type II and subtype V-A effectors, the crystal structures (indicated here by their RCSB Protein Data Bank (PDB) accession numbers (5CZZ and 5B43, respectively)) are available and the corresponding domain architectures are shown in detail. For the remainder of the proteins, the grey areas indicate structurally and functionally uncharacterized portions. RuvC-I, RuvC-II and RuvC-III, as well as higher eukaryotes and prokaryotes nucleotide-binding I (HEPN I) and HEPN II, denote the catalytic motifs of the respective nuclease domains of the CRISPR effectors. The bridge helix corresponds to an arginine-rich region that follows the RuvC-I motif. Other domains shown in the figure are denoted as follows: PAM interacting, protospacer-adjacent motif (PAM)-interacting domain; HNH, HNH family endonuclease domain, zinc finger domain with a CXXC.. CXXC motif (dots represent the variable distance between the two pairs of cysteines); HTH, putative DNA-binding helix–turn–helix domain; NUC, nuclease domain. The proteins and domains are shown approximately to scale. For each protein, the corresponding number of amino acids is indicated, and a ruler is shown on top of the figure to guide the eye. For the functionally characterized full-length effectors, the proposed new nomenclature (Cas12 and Cas13) is indicated, whereas for the uncharacterized putative effectors of type V-uncharacterized (V-U), only the provisional names are indicated. When, and if, functional evidence of a bona fide CRISPR response is reported for these effectors, they should be referred to as Cas12 proteins with the corresponding specifying letters. The putative V-U1, V-U2 and V-U5 effectors are larger than the typical TnpB proteins, whereas the V-U3 and V-U4 effectors are in the characteristic size range of TnpB. The asterisk at C2c5 indicates that this putative effector protein contains replacements of the catalytic residues of the RuvC-like nuclease domain and lacks the zinc finger.
The closest relatives and possible ancestors of Cas9 were identified on the basis of readily detectable sequence similarity and on the presence of the HNH insert in the RuvC-like nuclease domain of a distinct family of TnpB proteins that was denoted IscB (insertion sequences Cas9-like protein B)17,45. It is difficult to confidently trace a direct connection between type V effector proteins and a particular group of TnpB proteins, because type V effector proteins show less similarity to TnpB proteins than Cas9 shows to IscB proteins. Nevertheless, the effectors of the three subtypes of type V systems are similar to different TnpB families, which suggests independent origins of the effectors of different type V subtypes from the pool of tnpB genes16.
Subtype V-U identified using a CRISPR seed: small putative effectors
The search for CRISPR–cas loci that lack the adaptation module (that is, loci that were identified with a CRISPR seed but not with a cas1 seed; see BOX 1) yielded several additional variants of putative type V systems (FIGS 1,2) that might help to explain how CRISPR–Cas effectors evolved from TnpB. The putative effector proteins of these loci, which we have provisionally assigned to subtype V-U (where the ‘U’ stands for ‘uncharacterized’; see below), share two features that distinguish them from type II and type V effectors that are found at CRISPR–cas loci that contain Cas1 (FIG. 2). First, these proteins are much smaller than class 2 effectors that contain Cas1, comprising between ~500 amino acids (only slightly larger than the typical size of TnpB) and ~700 amino acids (between the size of TnpB and the typical size of the bona fide class 2 effectors). Second, these putative effectors show a higher level of similarity to TnpB proteins than the larger type I and type V effectors (see Supplementary information S3 (figure)). In particular, three groups of TnpB homologues, which are included here in subtype V-U (denoted V-U1, V-U2 and VU-5), showed evolutionary stability in terms of sequence conservation, consistent association with CRISPR arrays and presence in distinct groups of bacteria (FIGS 1,2; see below). A more detailed examination showed that, within each of these groups, in closely related bacterial genomes the respective loci were genuinely orthologous, as indicated by the gene synteny conservation.
In view of the identification of these smaller CRISPR-associated TnpB homologues, we ran the pipeline (BOX 1) with the requirement for the minimal length of the protein adjacent to the CRISPR array removed, and examined the results for the presence of additional TnpB homologues. Numerous CRISPR-associated TnpB homologues were detected in the size range that is typical of the transposon-encoded TnpB, that is, ~400 amino acids (Supplementary information S2 (box), part a). Most of these loci were not evolutionarily conserved and were thus of questionable functional relevance. However, we additionally detected two distinct groups of such smaller CRISPR-associated TnpB (V-U3 and V-U4) with characteristics that are similar to those of the three subtype V-U groups that have intermediately sized CRISPR-associated TnpB (FIGS 1,2; Supplementary information S4 (figure)).
Notably, the genes for the putative effectors of subtype V-U showed signs of purifying selection on protein sequences (as indicated by the low values of the non-synonymous to synonymous nucleotide substitutions, dN/dS), which was found to be particularly strong for the subtype V-U3 group (Supplementary information S2 (box), part b, and Supplementary information S4 (figure)). Taken together, these observations imply that the respective TnpB homologues have CRISPR-dependent functions and, in our view, justify the designation of the respective loci as subtype V-U.
For the larger bona fide type V effectors, low sequence conservation precluded reliable phylogenetic analysis, whereas a robust tree could be constructed for the smaller CRISPR-associated homologues, together with the typical transposon-encoded TnpB (see Supplementary information S1 (box) and Supplementary information S2 (box), part c). The topology of this tree indicated that four of the five distinct variants of subtype V-U (hereafter referred to as subtypes V-U1, V-U2, V-U3, V-U4 and V-U5) originated from different TnpB families (FIG. 3a), which is in agreement with the hypothesis of the independent evolution of different class 2 subtype effectors from transposon-encoded nucleases. The fifth variant (subtype V-U5), which is found in various cyanobacteria, consists of diverged TnpB homologues that have several mutations in the catalytic motifs of their RuvC-like domain and was accordingly not included in the phylogeny here. Of the five stable variants, subtype V-U1 is found in diverse bacteria, whereas the remaining subtypes are largely limited in their spread to particular bacterial taxa (FIG. 3a; Supplementary information S2 (box), part d). We further extended this evolutionary analysis to all putative type V effectors by building a cluster dendrogram based on the distances that were derived from profile-to-profile comparisons of the respective protein sequences (Supplementary information S1 (box)). The results suggest that the effectors of each of the identified subtypes, as well as the five distinct variants in subtype V-U, originated independently from different TnpB families (Supplementary information S5 (figure)).
The subtype V-U TnpB-like proteins are too small to adopt a bilobed structure of sufficient size to accommodate the crRNA–target DNA complex, as the typical class 2 effectors do, and, therefore, are unlikely to function in that capacity without additional partners. Furthermore, the subtype V-U loci lack any additional cas genes (FIG. 1), which, together with the above structural considerations, calls for caution in predicting that they have fully fledged CRISPR activity. Nevertheless, the evolutionarily stable association of at least five distinct subtype V-U variants with CRISPR arrays implies that at least some of these proteins do carry out CRISPR-dependent biological functions. Such functions might involve a typical CRISPR response that is aided by Cas proteins from other loci and/or by additional non-Cas proteins. Remarkably, the CRISPR arrays that are associated with group V-U3, which is mostly found in bacilli and clostridia, contain several spacers that match the genomic sequences of bacteriophages that infect these bacteria (Supplementary information S2 (box), part e). Furthermore, the sets of spacers in each subtype V-U group were completely different, even between closely related bacterial genomes (Supplementary information S2 (box), part e), which implies active spacer turnover. The diversity of the spacers and the presence of the phage-specific spacers in group V-U3 imply that at least some subtype V-U variants are functional CRISPR–Cas systems that are engaged in anti-phage adaptive immunity. Many of the complete genomes that contain group V-U3 and group V-U4 loci lack any additional CRISPR–Cas systems (Supplementary information S2 (box), part f), which makes it puzzling as to how these systems acquire their spacers. Alternatively, some of the subtype V-U systems might have distinct regulatory roles that do not require the formation of a ternary complex with the crRNA and the DNA target; indeed, several non-defence functions of CRISPR–Cas have been described47. This possibility is particularly plausible for the V-U5 variant, which seems to encompass a catalytically inactive TnpB homologue (FIG. 2, denoted C2c5*; Supplementary information S3 (box)). Furthermore, in genomes that contain the group V-U2 and group V-U5 loci, along with other CRISPR– Cas systems, the CRISPR sequences that are associated with the former loci are unique (Supplementary information S2 (box), part f), which suggests that these subtype V-U systems have distinct functions.
Subtypes VI-B and VI-C identified using a CRISPR seed: RNA-targeting CRISPR–Cas
The signature of type VI systems is the presence of an effector protein that contains two HEPN domains (FIGS 1,2). HEPN domains are common in various defence systems, the experimentally characterized of which, such as the toxins of numerous prokaryotic toxin–antitoxin systems or eukaryotic RNase L, all have RNase activity48,49. Therefore, the first putative type VI effector, denoted C2c2, was predicted to function as an RNA-guided RNase16. Subsequently, this prediction was experimentally validated, and the type VI effectors were shown to protect against the RNA bacteriophage MS2 (REF. 42). In addition, a novel feature of C2c2 is that, once primed with the cognate target RNA, the effector becomes a promiscuous RNase that has a toxic, growth-inhibitory effect on bacteria. These findings demonstrate a coupling between adaptive immunity and programmed cell death (or dormancy induction) that was previously predicted through comparative genomic analysis50 and mathematical modelling51. More recently, the C2c2 protein was shown to mediate not only interference but also the processing of pre-crRNA52.
The search for CRISPR–cas loci using the CRISPR seed identified two additional large putative effectors that contained two HEPN domains and which we assigned to subtype VI-B and subtype VI-C, respectively (accordingly, the C2c2-encoding loci became subtype VI-A). This classification of the type VI systems into separate subtypes is justified by the extremely low sequence similarity between the three groups of effectors, which is practically limited to the catalytic motif of the HEPN domain, the different positions of the HEPN domains with the large protein sequences, and the additional features of the locus architecture in the case of subtype VI-B (FIGS 1,2; Supplementary information S2 (box), part d). Specifically, the two distinct variants of subtype VI-B (variants VI-B1 and VI-B2) both encode additional proteins that contain predicted transmembrane domains; VI-B1 encodes four of these and VI-B2 encodes one (FIG. 3b; Supplementary information S2 (box), part d). Phylogenetic analysis of the effector proteins suggests that the VI-B1 and VI-B2 variants diverged during evolution in accordance with the distinct architectures of the associated predicted membrane proteins (FIG. 3b; Supplementary information S2 (box), part d). VI-B1 systems that contain several transmembrane domains might localize to membranes and thus could include membrane-associated RNA-targeting systems, which would be a novel feature in the biology of CRISPR– Cas. Furthermore, the single-transmembrane protein of variant VI-B2 encompasses an additional HEPN domain, which is the third one in the type VI system (FIG. 3b; Supplementary information S2 (box), part d, and Supplementary information S6 (figure)).
Given that all of the putative type VI effectors that have been discovered so far are similar in size to the active class 2 effectors of subtype VI-A48, even the loci that lack cas1 are likely to be functional CRISPR–Cas systems that rely on adaptation modules from other loci in the same genome. Moreover, given that RNA viruses only represent a minor part of the prokaryotic virome53, type VI systems might primarily elicit toxin activity in response to the active transcription of foreign DNA. This mechanism might not be limited to type VI systems, given the presence of HEPN domains in poorly characterized Cas proteins in many other CRISPR–Cas systems. Indeed, the RNase activity of the HEPN-containing Csm6 and Csx1 proteins in type III systems has been demonstrated54,55, although their functions in the CRISPR response remain to be studied.
Census of class 2 CRISPR–cas loci
The design of our CRISPR–Cas discovery pipeline implies that the analysis described in this article has identified nearly all variants of class 2 systems present in the bacterial and archaeal genomes that are currently available (BOX 1). Given that the current databases include only a small proportion of the entire inferred microbial diversity of the biosphere56–59, the discovery of new CRISPR–Cas subtypes, or even of novel CRISPR–Cas types, is likely. However, such novel variants are expected to be either extremely rare or limited in their spread to specific groups of microorganisms that are, at present, poorly sampled.
Comprehensive census of class 2 CRISPR–cas loci in bacteria and archaea
We were interested in a comprehensive census of class 2 types and subtypes in the current set of complete bacterial and archaeal genomes. To this end, we constructed sequence profiles for the effectors of all identified class 2 subtypes (two separate profiles were used for the variants V-U1, V-U2 and V-U5; the V-U3 and V-U4 variants were not included in the census because, in database searches, they cannot be readily distinguished from transposon-encoded TnpB) and compared these with the proteins that are encoded in the 4,961 completely sequenced prokaryotic genomes and 43,599 partial prokaryotic genomes that are available from the National Center for Biotechnology Information (NCBI) database (Supplementary information S1 (box)). This procedure should detect almost all instances of each effector, including highly diverged variants. The neighbourhoods of the respective genes were then examined for the presence of CRISPR arrays and additional cas genes, as described previously15.
The most remarkable observation is the substantial dominance of type II, which is represented in about 8% of bacterial genomes, among the class 2 systems (TABLE 1). Both type V and type VI are more than an order of magnitude less abundant, which is in agreement with the expectation that the CRISPR–Cas types and subtypes that remain to be discovered are rare variants15. An intriguing question is whether the type II CRISPR–Cas system provides a substantial fitness advantage, perhaps being more efficient in defence and/or incurring a lower cost than other class 2 variants. Most of the class 2 subtypes are represented in taxonomically diverse bacteria, and, furthermore, for type II and subtype V-A, the effector tree topologies differ from the topology of the species tree17,38. These observations indicate that horizontal gene transfer might be a key process in the evolution of CRISPR–Cas. However, it is notable that the relatively abundant subtype VI-B seems to be restricted to the phylum Bacteroidetes, which perhaps reflects a unique aspect of the biology of these bacteria. Similarly, the V-U5 variant, which contains an inactivated TnpB homologue, is limited to cyanobacteria (see above), and could be involved in a distinct cyanobacterial regulatory pathway. As has been previously noted13,15, and is emphasized by this expansion of the diversity of class 2 systems, apart from the identification of subtype V-A in mesophilic archaea in two instances, class 2 systems are unique to bacteria. The exclusion of class 2 systems from archaea, particularly from hyperthermophiles in which class 1 systems are ubiquitous, implies that there is a major functional distinction between the two classes of CRISPR–Cas system, the nature of which remains enigmatic.
Table 1.
A comprehensive census of class 2 CRISPR–Cas systems in bacterial and archaeal genomes
| Subtype | |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| II | V-A | V-B | V-U* | VI-A | VI-B | VI-C | |
| Effector‡ | Cas9 | Cas12a (Cpf1) | Cas12b (C2c1) | C2c4, C2c5; five distinct subgroups (V-U 1–5) | Cas13a (C2c2) | Cas13b (C2c6) | Cas13c (C2c7) |
| Number of loci in bacterial and archaeal genomes |
|
70 | 18 | 92 | 30 | 94 | 6 |
| Representation | Diverse bacteria | Diverse bacteria and two archaea | Diverse bacteria | Diverse bacteria | Diverse bacteria | Bacteroidetes | Fusobacteria and Clostridia |
| Other cas genes | 85% cas1 and cas2; 55% csn2; 3% cas4 | 70% cas1 and cas2; 55% cas4 | 65% cas1, cas2 and cas4 | None | 25% cas1 and cas2 | None | None |
| Percent of loci that contain CRISPR array | 65% | 68% | 60% | ~50% | 73% | 90% | 83% |
The subtype V-uncharacterized (V-U) loci were originally identified on the basis of the adjacency of tnpB genes to CRISPR arrays and the evolutionary conservation of this association. Then, this putative subtype of class 2 CRISPR–Cas systems was expanded by searching for homologues of the respective effector proteins, irrespective of their adjacency to CRISPR arrays. Hence, only about half of the V-U loci include CRISPR.
Both the proposed systematic Cas names and the provisional vernacular names are used for the effectors, with the exception of type II effectors, which have only systematic names, and type V-U effectors, to which a systematic name has so far not been assigned.
Origins of class 2 CRISPR–Cas systems
In an extension of the previous hypothesis on the independent origins of the effectors in different types and subtypes of class 2 CRISPR–Cas systems, we use the findings on incomplete type V loci to propose a more specific evolutionary scenario (FIG. 4). As discussed above, at least five distinct variants within subtype V-U show a substantial degree of evolutionary stability and consistent association with CRISPR arrays, and typically contain TnpB homologues that are intermediate in size between the compact transposon-encoded TnpB proteins and the large class 2 effectors (FIGS 2,3b). These groups of TnpB homologues might represent intermediate stages in independent pathways to the emergence of new CRISPR–Cas variants. The other CRISPR–tnpB associations are not evolutionarily conserved and are likely to result from more or less random insertions of tnpB genes next to CRISPR arrays; some of these loci could represent the earliest stages of the evolution of CRISPR–Cas systems.
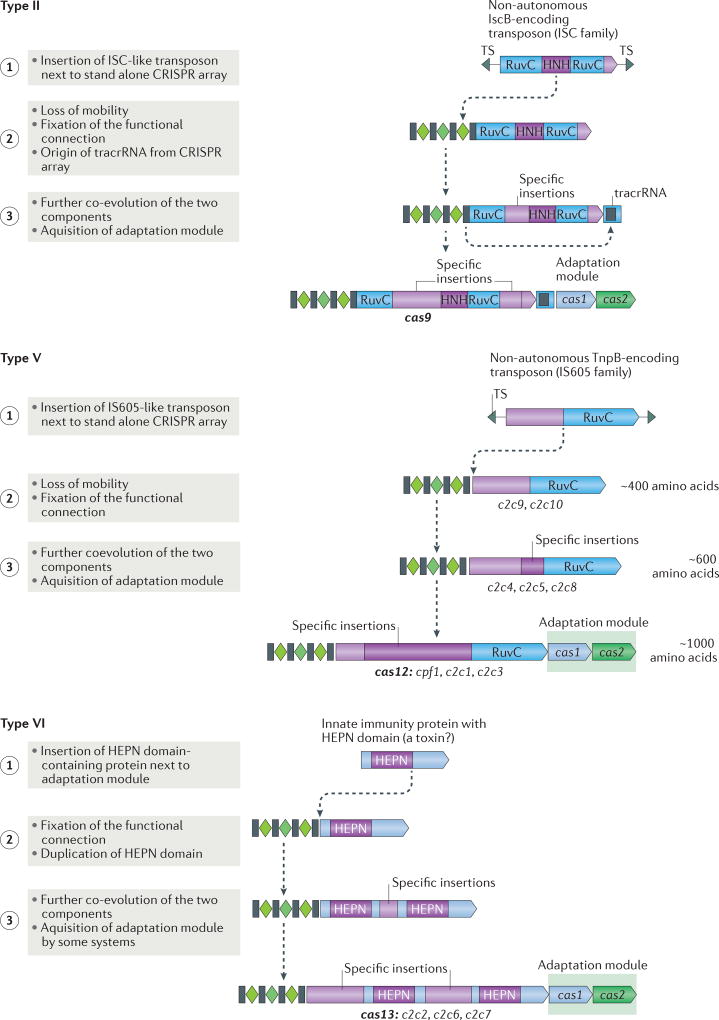
Figure 4. Possible routes of evolution for class 2 CRISPR–Cas systems.
The figure depicts the three-step pathway of the evolutionary ‘maturation’ of type II, type V and type VI CRISPR–Cas systems. The systematic and/or provisional gene names are indicated below the respective ‘mature’ effector protein schematics and the proposed intermediate forms of type V systems. The first step involves the random insertion of a TnpB-encoding or insertion sequences Cas9-like protein B (IscB)-encoding transposon or a higher eukaryotes and prokaryotes nucleotide-binding (HEPN) domain RNase-encoding gene next to a CRISPR cassette for type II, type V and type VI systems, respectively. During the second step, the functional connection between this protein and the CRISPR array is established and co-evolution begins, in particular, in the form of the accumulation of specific insertions that facilitate CRISPR RNA (crRNA) binding. For type V systems, the intermediate forms that correspond to the first and second step are identified as different type V-uncharacterized (V-U) variants. Additional components of the system could have originated during the second step, such as trans-acting CRISPR RNA (tracrRNA) in the case of type II systems. During the third step, further insertions lead to increased specificity of crRNA and target binding, and enable interactions with accessory proteins, such as Csn2 for type II-A and a protein with predicted transmembrane (TM) domains for type VI-B. The adaptation module is only inserted into some of the class 2 CRISPR–cas loci during the third step. TS, target site.
All subtype V-U loci lack adaptation modules, which suggests that the first stage of the evolution of new class 2 CRISPR–Cas systems involves the random insertion of a TnpB-encoding element next to an orphan CRISPR array (FIG. 4). At the next stage of evolution, the association between CRISPR and a TnpB derivative would become fixed in the microbial population, conceivably owing to the emergence of a novel function, the exact nature of which remains to be understood. This would be accompanied by an increase in the size of the protein through internal duplications and/or the insertion of additional domains (FIG. 5). The final stages include further growth of the effector protein, resulting in the typical bilobed structure, and, in some cases, its association with an adaptation module through recombination with a different CRISPR–cas locus (FIG. 4). Compatible with this scenario, the Cas1 proteins of different subtypes of type II and of type V are homologous to different subtypes of type I16. The fact that no subtype V-U loci contain cas1 and cas2 genes, whereas many of the loci that encode typical large effector proteins do, strongly suggests that the adaptation modules came last.
Figure 5. Functional diversity of the experimentally characterized class 2 CRISPR–Cas systems.
For each type of the class 2 CRISPR–Cas systems (and two subtypes in the case of type V), a schematic of the complex between the effector protein, the target, crRNA and, in the case of type II and type V-B systems, trans-acting CRISPR RNA (tracrRNA), is shown. The position of the protospacer adjacent motif (PAM) or the protospacer flanking site (PFS) is indicated by a red bar. The small red triangles show the position of the cut, or cuts, in the target DNA or RNA molecule. dsDNA, double-stranded DNA; ssRNA, single-stranded RNA.
The above scenario might be challenged in regard to the directionality of evolution: the possibility could be considered that the transposon-encoded TnpB protein actually evolved from class 2 effectors. However, the scenario in which transposon-encoded TnpB is the ancestral form (FIG. 4) seems much more likely. First, TnpB-encoding transposons (autonomous and non-autonomous, including some that have lost mobility) are far more abundant across a broad range of bacteria and archaea than class 2 CRISPR–Cas systems, which are relatively rare and limited in their spread to a subset of bacterial phyla (see above; TABLE 1; Supplementary information S2 (box), part d). Second, and perhaps more important, the class 2 effectors are much larger and more complex than TnpB proteins, which makes them unlikely ancestral forms. Third, the TnpB proteins are encoded in transposons, which, through their mobility, are well suited to move into the vicinity of CRISPR arrays; by contrast, CRISPR–Cas systems lack active mobility mechanisms. Finally, the observations that are reported here on the phylogeny of TnpB, in which the CRISPR-associated variants are lodged among the transposon-encoded proteins (FIG. 3a), imply the ancestral status of TnpB.
Hypothetically, a similar scenario could apply to the type VI systems (FIG. 4). A comprehensive database search for HEPN domain-containing proteins that are encoded in the vicinity of CRISPR arrays failed to identify any evolutionarily stable configurations that might have been analogous to subtype V-U, whereas it detected numerous members of the HEPN-containing Cas protein families, Csm6 and Csx1 (Supplementary information S2 (box), part g). Thus, it seems possible that, during evolution, type VI systems recruited one of the HEPN-containing Cas proteins, which was followed by duplication of the HEPN domain and further expansion of the protein to the typical size of a class 2 effector (FIG. 4). However, the possibility that type VI effectors directly originate from HEPN-containing toxins cannot be ruled out; further screening of new genomes and metagenomes for likely ancestors of the two HEPN domain proteins should establish the origin of type VI effectors.
Amended classification and proposed nomenclature
The systematic search for novel class 2 CRISPR–cas loci described here led to a major expansion of the known diversity of these systems. Instead of the two types and four subtypes that were included in the latest classification15, there are now three types and at least 10 subtypes (FIG. 1). Some uncertainty remains, owing to the lack of functional data on subtype V-U, but it seems likely that evolutionarily stable and apparently functional variants that are currently grouped into this provisional subtype, particularly V-U3, will eventually be ‘upgraded’ to subtypes in their own right. The functional characterization of V-U variants will provide a more precise classification, although it is likely that many V-U loci do not encode typical active CRISPR–Cas systems. Given the comprehensive nature of the search described here (BOX 1), we expect that the new variants will be extremely rare or restricted in their spread to particular groups of bacteria and archaea that are not adequately represented in current sequence databases.
We believe that the expansion of the CRISPR–Cas classification calls for a corresponding change to the nomenclature, in which at least the experimentally characterized effectors and their homologues are given new names that correspond to numbered Cas proteins (FIG. 2; TABLE 1). Thus, the type V effectors would become Cas12a, Cas12b and Cas12c, and those of type VI would become Cas13a, Cas13b and Cas13c (numerical continuity with Cas9 is not possible because Cas10 and Cas11 are already used for other proteins)15. We currently refrain from renaming the putative subtype V-U effectors until functional evidence of a bona fide CRISPR response for these effectors is reported, at which time we propose that they are referred to as Cas12 proteins.
Applications in genome engineering
Most applications of CRISPR systems have focused on the programmable DNA-targeting activity of Cas9. The cleavage activity of Cas9 can be harnessed for genome editing, including gene knockout and precise editing through homology-directed repair. Catalytically inactive (‘dead’) variants of Cas9 have been used for transcriptional control60, epigenetic modulation61 and imaging62–64. All of these advances notwithstanding, Cas9 has its limitations, due to the potential for off-target effects, challenges that are associated with delivery and the difficulty of targeting RNA rather than DNA. Thus, alternative tools for CRISPR-mediated editing are in high demand.
Although functional characterization of the class 2 subtypes is far from complete, even at this stage, remarkable functional diversity is apparent. The manifestations of this diversity include different targets (dsDNA for type II and type V, but RNA for type VI); the requirement for tracrRNA (type II and subtype V-B, but not subtype V-A or type VI, require this), the sequence of the PAM and the type of cut that is introduced into the target nucleic acid (FIG. 5). This functional diversity is a major incentive for further characterization of different class 2 systems, as it creates opportunities for the enhancement and expansion of the capabilities of the genome editing toolbox for research, biotechnology and medicine65. The use of Cas12a (better known as Cpf1) from the type V-A family of effectors has already yielded simpler, single RNA-guided and more specific enzymes than Cas9 for genome-editing applications38,41,66–70, as well as offering an alternative PAM that would facilitate genome editing in AT-rich genomes, such as the genome of Plasmodium falciparum.
The continued exploration of CRISPR effector diversity, such as the recently characterized type VI-A effector Cas13a (previously known as C2c2)42, also opens the door for the development of new RNA-guided RNA-targeting technologies that enable the perturbation, modulation, modification and monitoring of specific RNA transcripts in cells. The development of an efficient programmable RNA-binding protein (for example, of a ‘dead’ Cas13a that has mutated HEPN domains) could rapidly advance our existing understanding of RNA biology. Such a tool would enable the sensing of different cellular states, the manipulation of translation, and tracking of RNA levels and localization in live cells. Although Cas9 has been modified to provide some RNA-targeting capabilities71, this system requires the delivery of chemically modified DNA, which limits its use for many applications, including genome-wide screening or virus delivery. Upon binding to a complementary RNA target, Cas13a engages both specific and nonspecific RNase activities, and induces growth inhibition in Escherichia coli71. This feature complicates the use of Cas13a for specific RNA knockdown, but potentially could be harnessed for other applications, such as the selective ablation of cell types based on expression profiles. It remains to be investigated whether the nonspecific RNase activity of Cas13a can be inactivated independently of its target-specific activity and whether other type VI effectors, such as Cas13b, have similar properties. Further mining of CRISPR–Cas systems, and, more broadly, of the diversity of bacterial and archaeal defence systems and of mobile genetic elements, is expected to enable new applications in biotechnology. In particular, programmable integrases or transposases that have yet to be discovered would be powerful tools for targeted genomic integration and rearrangement.
Concluding remarks
The genomic analysis that is presented here expands the diversity of class 2 CRISPR–Cas systems. In particular, the inclusion of non-autonomous CRISPR–Cas systems that lack the adaptation module, combined with the search of expanded genomic and metagenomics databases, led to the discovery of three new subtypes which, together with our previous analysis, increases the number of class 2 subtypes from 4 to 10. Furthermore, at present, one of the new subtypes, V-U, is a collection of diverse variants, some of which are expected to become new subtypes once they have been functionally characterized. It seems especially notable that the newly discovered class 2 systems all fall into the two previously defined subclasses: those that cleave the non-target strand of the target dsDNA using a RuvC-like nuclease and those that attack RNA targets using a two HEPN domain RNase. The apparent repeated emergence of these CRISPR–Cas variants might reflect strict demands for protein structure to accommodate the crRNA and the target molecule, to which only a few protein folds are conducive.
The new class 2 variants show some unprecedented functional features; for example, subtype V-A does not require a tracrRNA, whereas other variants, such as subtype VI-A (and probably all type VI systems), exclusively target RNA and seem to induce a toxic response in bacterial cells. Subtype V-U is expected to show even more unusual properties. This functional diversity provides the potential for the development of new, versatile genome-editing and regulation tools. We provide indications that different class 2 types and subtypes independently originate from mobile elements that encode diverse TnpB proteins (type II and type V) and from HEPN domain-containing proteins (type VI) that ultimately originate from mRNA-cleaving toxins. The remarkable diversity notwithstanding, we believe that the computational pipeline that is applied here provides for a nearly exhaustive identification of class 2 systems. Additional variants that remain to be found will be either extremely rare or confined to bacterial phyla that are currently unknown or poorly sampled. However, as shown by the example of type VI, despite the rarity and/or narrow spread of such variants, their biological features could be of major interest and potential value for new applications.
Note added in proof
While this article was in press, several new findings pertaining to the novel class 2 CRISPR–Cas systems described here were published. The structure of Cas12b (C2c1) in complex with crRNA, tracrRNA and the DNA template was solved independently in two laboratories, and the enzymatic mechanism of this effector protein and the structure of its cut were elucidated76,77. Cas12b cleaves both target and non-target strands using its RuvC-like nuclease domain, which successively accommodates each strand to produce a seven-nucleotide staggered cut with 5′ overhangs. In addition, the tertiary structure of Cas13a (C2c2) was determined; this confirmed that Cas13a has two HEPN domains that form the site that cleaves targets and identified a distinct catalytic site that is responsible for cleaving pre-crRNA78. The RNA-guided RNA-targeting VI-B1 and VI-B2 systems have also been characterized79 and it was demonstrated that, similarly to Cas13a, Cas13b proteins have collateral RNase activity that is activated by target recognition and, furthermore, that they are differentially regulated by accessory proteins that are encoded within the VI-B1 and VI-B2 loci. Finally, two new CRISPR variants that belong to type V were discovered in the genomes of uncultivated bacteria80.
Supplementary Material
Acknowledgments
S.S., K.S.M., Y.I.W. and E.V.K. are supported by intramural funds of the US Department of Health and Human Services (to the US National Library of Medicine). K.S. is supported by a US National Institutes of Health (NIH) grant (GM10407), a Russian Science Foundation grant (14-14-00988), and by the Skolkovo Institute of Science and Technology (Skoltech). F.Z. is supported by the NIH through the US National Institute of Mental Health (NIMH; grants 5DP1-MH100706 and 1R01-MH110049); the US National Science Foundation (NSF); the New York Stem Cell Foundation; the Allen Distinguished Investigator Program; through The Paul G. Allen Frontiers Group; the Simons and Vallee Foundations; the Howard Hughes Medical Institute; the Skoltech–MIT Next Generation Program; James and Patricia Poitras and the Poitras Center for Affective Disorders; R. Metcalfe; and D. Cheng. F.Z. is a New York Stem Cell Foundation–Robertson Investigator.
Glossary
- CRISPR–Cas
An adaptive immune system in archaea and bacteria that functions by inserting fragments of foreign genomes into CRISPR arrays and using the transcripts of the resulting spacers as guide RNAs to detect and inactivate the cognate genetic elements.
- Adaptation
The first phase of the CRISPR immune response, during which a piece of foreign DNA is inserted into a CRISPR array to become a spacer that is subsequently used as the template to produce the CRISPR RNA (crRNA).
- CRISPR RNAs
(crRNAs). Small RNA molecules that consist of the RNA complement of a spacer and parts of the two adjacent repeats. crRNAs are produced by processing of the transcript of the entire CRISPR array (pre-crRNA); processing is mediated either by Cas proteins only (class 1, type V-A and type VI-A systems) or by an external RNase, such as bacterial RNase III, in conjunction with Cas proteins.
- Interference
The final phase of the CRISPR immune response, during which the target DNA (or less commonly, RNA) is recognized by a CRISPR effector through the bound CRISPR RNA (crRNA) and cleaved by the effector nuclease or nucleases.
- Effector
A complex of Cas proteins (in class 1 systems), or a single, large protein (in class 2 systems), that is involved in target recognition and inactivation, and, in most cases, in the processing of pre-CRISPR RNA (pre-crRNA).
- Class 2 CRISPR–Cas systems
One of the two major divisions of CRISPR–Cas that is characterized by effector modules that consist of a single, large protein with endonuclease activity.
- Trans-acting CRISPR RNA
(tracrRNA). An accessory RNA molecule that is partially complementary to CRISPR and is involved in pre-crRNA processing in type II and type V-B CRISPR–Cas interference.
- Higher eukaryotes and prokaryotes nucleotide-binding domains
(HEPN domains). An early name that was given when the functions of the domains were not well characterized. An expansive superfamily of domains with RNase activity that are involved in various defence functions, in particular, type VI and some class 1 CRISPR–Cas interference.
- TnpB proteins
A poorly characterized superfamily of transposon-encoded proteins that contain RuvC-like nuclease domains. TnpB proteins are the apparent evolutionary ancestors of type II and type V CRISPR–Cas effectors.
Footnotes
Competing interests statement
The authors declare competing interests: see Web version for details.
DATABASES
RCSB Protein Data Bank: http://www.rcsb.org/pdb/home/home.do 5CZZ | 5B43
See online article: S1 (box) | S2 (box) | S3 (figure) | S4 (figure) | S5 (figure) | S6 (figure)
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 3.Barrangou R. CRISPR–Cas systems and RNA-guided interference. Wiley Interdiscip. Rev RNA. 2013;4:267–278. doi: 10.1002/wrna.1159. [DOI] [PubMed] [Google Scholar]
- 4.Marraffini LA. CRISPR–Cas immunity in prokaryotes. Nature. 2015;526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- 5.Mohanraju P, et al. Diverse evolutionary roots and mechanistic variations of the CRISPR–Cas systems. Science. 2016;353:aad5147. doi: 10.1126/science.aad5147. [DOI] [PubMed] [Google Scholar]
- 6.Van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR–Cas systems. Nat. Rev. Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR–Cas systems. Biol. Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makarova KS, Wolf YI, Koonin EV. The basic building blocks and evolution of CRISPR–Cas systems. Biochem. Soc. Trans. 2013;41:1392–1400. doi: 10.1042/BST20130038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takeuchi N, Wolf YI, Makarova KS, Koonin EV. Nature and intensity of selection pressure on CRISPR-associated genes. J. Bacteriol. 2012;194:1216–1225. doi: 10.1128/JB.06521-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bondy-Denomy J, Davidson AR. To acquire or resist: the complex biological effects of CRISPR–Cas systems. Trends Microbiol. 2014;22:218–225. doi: 10.1016/j.tim.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Bondy-Denomy J, et al. Multiple mechanisms for CRISPR–Cas inhibition by anti-CRISPR proteins. Nature. 2015;526:136–139. doi: 10.1038/nature15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Houte S, et al. The diversity-generating benefits of a prokaryotic adaptive immune system. Nature. 2016;532:385–388. doi: 10.1038/nature17436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makarova KS, et al. Evolution and classification of the CRISPR–Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makarova KS, Koonin EV. Annotation and classification of CRISPR–Cas systems. Methods Mol. Biol. 2015;1311:47–75. doi: 10.1007/978-1-4939-2687-9_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makarova KS, et al. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. This paper presents the latest classification of the CRISPR–Cas systems, prior to the application of the pipeline described here, along with computational approaches for the identification and quantitative comparison of CRISPR–cas loci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shmakov S, et al. Discovery and functional characterization of diverse class 2 CRISPR–Cas systems. Mol. Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. This paper presents the first instalment of the computational pipeline that is described in this article, using Cas1 as the seed, and experimental validation of the activity of subtype V-B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chylinski K, Makarova KS, Charpentier E, Koonin EV. Classification and evolution of type II CRISPR–Cas systems. Nucleic Acids Res. 2014;42:6091–6105. doi: 10.1093/nar/gku241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jore MM, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 20.Beloglazova N, et al. CRISPR RNA binding and DNA target recognition by purified Cascade complexes from. Escherichia coli. Nucleic Acids Res. 2015;43:530–543. doi: 10.1093/nar/gku1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson RN, et al. Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from. Escherichia coli. Science. 2014;345:1473–1479. doi: 10.1126/science.1256328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rouillon C, et al. Structure of the CRISPR interference complex CSM reveals key similarities with cascade. Mol. Cell. 2013;52:124–134. doi: 10.1016/j.molcel.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Staals RH, et al. RNA targeting by the type III A CRISPR–Cas Csm complex of Thermus thermophilus. Mol. Cell. 2014;56:518–530. doi: 10.1016/j.molcel.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osawa T, Inanaga H, Sato C, Numata T. Crystal structure of the CRISPR–Cas RNA silencing Cmr complex bound to a target analog. Mol. Cell. 2015;58:418–430. doi: 10.1016/j.molcel.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 25.Taylor DW, et al. Structural biology. Structures of the CRISPR–Cmr complex reveal mode of RNA target positioning. Science. 2015;348:581–585. doi: 10.1126/science.aaa4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat. Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl Acad. Sci. USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimasu H, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. This paper reports the first structure of Cas9, which provides insight into the interaction of class 2 effectors with crRNA and target DNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishimasu H, et al. Crystal structure of Staphylococcus aureus Cas9. Cell. 2015;162:1113–1126. doi: 10.1016/j.cell.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sternberg SH, LaFrance B, Kaplan M, Doudna JA. Conformational control of DNA target cleavage by CRISPR–Cas9. Nature. 2015;527:110–113. doi: 10.1038/nature15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sapranauskas R, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chylinski K, Le Rhun A, Charpentier E. The tracrRNA and Cas9 families of type II CRISPR–Cas immunity systems. RNA Biol. 2013;10:726–737. doi: 10.4161/rna.24321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Briner AE, et al. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol. Cell. 2014;56:333–339. doi: 10.1016/j.molcel.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 37.Schunder E, Rydzewski K, Grunow R, Heuner K. First indication for a functional CRISPR/Cas system in Francisella tularensis. Int. J. Med. Microbiol. 2013;303:51–60. doi: 10.1016/j.ijmm.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 38.Zetsche B, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR–Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. This work demonstrates the interference activity of Cpf1 and shows that Cpf1 is a single RNA-guided endonuclease that does not require tracrRNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong D, et al. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature. 2016;532:522–526. doi: 10.1038/nature17944. [DOI] [PubMed] [Google Scholar]
- 40.Yamano T, et al. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell. 2016;165:949–962. doi: 10.1016/j.cell.2016.04.003. Together with reference 39, this paper presents the structure of Cpf1 in complex with crRNA and target DNA, demonstrating that, despite similar overall shapes, the domain architectures of Cpf1 and Cas9 differ substantially. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fonfara I, Richter H, Bratovic M, Le Rhun A, Charpentier E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016;532:517–521. doi: 10.1038/nature17945. This work demonstrates that Cpf1 is responsible not only for interference but also for pre-crRNA processing. [DOI] [PubMed] [Google Scholar]
- 42.Abudayyeh OO, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. This paper describes the first CRISPR–Cas system that exclusively cleaves RNA, demonstrates the switch from specific to non-specific RNA cleavage following target recognition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pasternak C, et al. ISDra2 transposition in Deinococcus radiodurans is downregulated by TnpB. Mol. Microbiol. 2013;88:443–455. doi: 10.1111/mmi.12194. [DOI] [PubMed] [Google Scholar]
- 44.Bao W, Jurka J. Homologues of bacterial TnpB_IS605 are widespread in diverse eukaryotic transposable elements. Mob. DNA. 2013;4:12. doi: 10.1186/1759-8753-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kapitonov VV, Makarova KS, Koonin EV. ISC, a novel group of bacterial and archaeal DNA transposons that encode Cas9 homologs. J. Bacteriol. 2015;198:797–807. doi: 10.1128/JB.00783-15. In this work, the direct evolutionary ancestors of Cas9 are identified. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomes-Filho JV, et al. Sense overlapping transcripts in IS1341 type transposase genes are functional non-coding RNAs in archaea. RNA Biol. 2015;12:490–500. doi: 10.1080/15476286.2015.1019998. This work demonstrates that TnpB proteins bind to RNA, which is compatible with their role as ancestors of class 2 CRISPR–Cas effectors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Westra ER, Buckling A, Fineran PC. CRISPR–Cas systems: beyond adaptive immunity. Nat. Rev. Microbiol. 2014;12:317–326. doi: 10.1038/nrmicro3241. [DOI] [PubMed] [Google Scholar]
- 48.Anantharaman V, Makarova KS, Burroughs AM, Koonin EV, Aravind L. Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol. Direct. 2013;8:15. doi: 10.1186/1745-6150-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Makarova KS, Anantharaman V, Grishin NV, Koonin EV, Aravind L. CARF and WYL domains: ligand-binding regulators of prokaryotic defense systems. Front. Genet. 2014;5:102. doi: 10.3389/fgene.2014.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Makarova KS, Anantharaman V, Aravind L, Koonin EV. Live virus-free or die: coupling of antivirus immunity and programmed suicide or dormancy in prokaryotes. Biol. Direct. 2012;7:40. doi: 10.1186/1745-6150-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iranzo J, Lobkovsky AE, Wolf YI, Koonin EV. Immunity, suicide or both? Ecological determinants for the combined evolution of anti-pathogen defense systems. BMC Evol. Biol. 2015;15:43. doi: 10.1186/s12862-015-0324-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.East-Seletsky A, et al. Two distinct RNase activities of CRISPR C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538:270–273. doi: 10.1038/nature19802. This paper describes experiments that show that, similar to Cpf1, C2c2, the subtype VI-A effector, catalyses pre-crRNA processing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koonin EV, Dolja VV, Krupovic M. Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology. 2015;479–480:2–25. doi: 10.1016/j.virol.2015.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheppard NF, Glover CV, Terns RM, Terns MP. The CRISPR-associated Csx1 protein of Pyrococcus furiosus is an adenosine-specific endoribonuclease. RNA. 2016;22:216–224. doi: 10.1261/rna.039842.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Niewoehner O, Jinek M. Structural basis for the endoribonuclease activity of the type III-A CRISPR-associated protein Csm6. RNA. 2016;22:318–329. doi: 10.1261/rna.054098.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Curtis TP, Sloan WT, Scannell JW. Estimating prokaryotic diversity and its limits. Proc. Natl Acad. Sci. USA. 2002;99:10494–10499. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Curtis TP, et al. What is the extent of prokaryotic diversity? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006;361:2023–2037. doi: 10.1098/rstb.2006.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fraser C, Alm EJ, Polz MF, Spratt BG, Hanage WP. The bacterial species challenge: making sense of genetic and ecological diversity. Science. 2009;323:741–746. doi: 10.1126/science.1159388. [DOI] [PubMed] [Google Scholar]
- 59.Quince C, Curtis TP, Sloan WT. The rational exploration of microbial diversity. ISME J. 2008;2:997–1006. doi: 10.1038/ismej.2008.69. [DOI] [PubMed] [Google Scholar]
- 60.Chavez A, et al. Comparison of Cas9 activators in multiple species. Nat. Methods. 2016;13:563–567. doi: 10.1038/nmeth.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thakore PI, Black JB, Hilton IB, Gersbach CA. Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat. Methods. 2016;13:127–137. doi: 10.1038/nmeth.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen B, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Knight SC, et al. Dynamics of CRISPR–Cas9 genome interrogation in living cells. Science. 2015;350:823–826. doi: 10.1126/science.aac6572. [DOI] [PubMed] [Google Scholar]
- 64.Nelles DA, et al. Programmable RNA tracking in live cells with CRISPR/Cas9. Cell. 2016;165:488–496. doi: 10.1016/j.cell.2016.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR–Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kleinstiver BP, et al. Genome-wide specificities of CRISPR–Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 2016;34:869–874. doi: 10.1038/nbt.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim Y, et al. Generation of knockout mice by Cpf1-mediated gene targeting. Nat. Biotechnol. 2016;34:808–810. doi: 10.1038/nbt.3614. [DOI] [PubMed] [Google Scholar]
- 68.Hur JK, et al. Targeted mutagenesis in mice by electroporation of Cpf1 ribonucleoproteins. Nat. Biotechnol. 2016;34:807–808. doi: 10.1038/nbt.3596. [DOI] [PubMed] [Google Scholar]
- 69.Kim D, et al. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 2016;34:863–868. doi: 10.1038/nbt.3609. [DOI] [PubMed] [Google Scholar]
- 70.Li SY, Zhao GP, Wang J. C-Brick: a new standard for assembly of biological parts using Cpf1. ACS Synth. Biol. 2016 doi: 10.1021/acssynbio.6b00114. http://dx.doi.org/10.1021/acssynbio.6b00114. [DOI] [PubMed]
- 71.O’Connell MR, et al. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516:263–266. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Edgar RC. PILER-CR: fast and accurate identification of CRISPR repeats. BMC Bioinformatics. 2007;8:18. doi: 10.1186/1471-2105-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Almendros C, Guzman NM, Garcia-Martinez J, Mojica FJ. Anti-cas spacers in orphan CRISPR4 arrays prevent uptake of active CRISPR–Cas I-F systems. Nat. Microbiol. 2016;1:16081. doi: 10.1038/nmicrobiol.2016.81. [DOI] [PubMed] [Google Scholar]
- 75.Makarova KS, Wolf YI, Koonin EV. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res. 2013;41:4360–4377. doi: 10.1093/nar/gkt157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu L, et al. C2c1–sgRNA complex structure reveals RNA-guided DNA cleavage mechanism. Mol. Cell. 2016 doi: 10.1016/j.molcel.2016.11.040. http://dx.doi.org/10.1016/j.molcel.2016.11.040. [DOI] [PubMed]
- 77.Yang H, Gao P, Rajashankar KR, Patel DJ. PAM-dependent target DNA recognition and cleavage by C2c1 CRISPR–Cas endonuclease. Cell. 2016;167:1814–1828.e12. doi: 10.1016/j.cell.2016.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu L, et al. Two distant catalytic sites are responsible for C2c2 RNase activities. Cell. 2017;168:121–134.e12. doi: 10.1016/j.cell.2016.12.031. [DOI] [PubMed] [Google Scholar]
- 79.Smargon AA, et al. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol. Cell. 2017 doi: 10.1016/j.molcel.2016.12.023. http://dx.doi.org/10.1016/j.molcel.2016.12.023. [DOI] [PMC free article] [PubMed]
- 80.Burstein D, et al. New CRISPR–Cas systems from uncultivated microbes. Nature. 2016 doi: 10.1038/nature21059. http://dx.doi.org/10.1038/nature21059. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.