Abstract
Purpose
To explore the cause of functional changes of tumor necrosis factor alpha (TNF-α) in development of gastric cancer through the structural changes of each site in TNF-α before and after mutation.
Methods
Three typical mutant sites (TNF-α-308G/A, 857C/T and 863C/A of TNF-α) were chosen and methods like ab initio modeling was adopted for 3D modeling of TNF-α before and after mutation, besides, the structural changes were also analyzed.
Results
Mutation of TNF-α-308G/A led to the production of multiple helical structures and that of 863C/A caused the production of one helical structure in its adjacent region. Mutation of 857C/T, however, did not cause the change in the basic structure of TNF-α.
Conclusions
Structural changes of TNF-α may have a significant effect on development of gastric cancer.
Keywords: TNF-α, Gastric cancer, Polymorphism, Helical structure
1. Introduction
Gastric cancer is one of common tumors for human, and according to the research of Chen et al. the incidence and mortality rate of gastric cancer is the second only to lung cancer in China (Chen et al., 2016). Helicobacter pylori infection is correlated to gastric cancer. But it’s reported that there’s a large disparity in numbers between infected people and infected people who finally had gastric cancer which might result from bacteria pathogenicity, host genetic susceptibility and environment (Rokkas et al., 2014). When it comes to host genetic susceptibility, relationship between polymorphism and gastric cancer has caught people’s eyes in recent years. And as a tumor necrosis factor, tumor necrosis factor alpha (TNF-α) plays an important role in the onset and development of gastric cancer; besides, the influence of its gene mutation on growth of cancers has been found more and more.
TNF-α is located in the human leukocyte antigen (HLA) III gene region of the human chromosome 6, between HLA-B and HLA-DR. TNF-α is mainly secreted by activated mononuclear macrophage, T cells stimulated by antigens, activated NK cells and mast cells. Moderate number of TNF-α can regulate the body’s immunologic and metabolic functions and is significant in maintaining internal stability and resisting various disease factors (Lili, 2006); while excessive TNF-α will cause multiple pathological changes (Fang, 2009). Some studies (Sugimoto et al., 2007) showed that with helicobacter pylori infection, gastric secretion of TNF-α of patients had elevated apparently, and there’s a positive correlation between the increasing level and inflammation. Moreover, many studies have proved that polymorphism of TNF-α is closely related to gastric cancer. Studies made by Canedo et al. (2008) believed that polymorphism of TNF-α-308 makes stomach low-acid and depauperate after helicobacter pylori infection, which would lead to gastric cancer. Tahara et al. (2011) found that TNF-α-857 T allele is the risk factor of gastric cancer; similarly, TNF-α-863 T allele is correlated to the risk of gastric cancer (Sugimoto et al., 2007).
Nowadays there are many studies on the polymorphism of TNF-α and findings of its relation with gastric cancer are also a lot. Three mutant types (TNF-α-308 G/A, 857C/T and 863C/A) were chosen. By means of bioinformatics, the amino acid sequences, models of secondary and tertiary structures before and after mutation were obtained. Then the reason of its functional changes was mainly explored from the structural changes.
To investigate the change of the amino acid sequences, models of secondary and tertiary structures and the effect of these structural changes on the function of TNF-α in the development and progression of gastric cancer after three mutant types (TNF-α-308G/A, 857C/T and 863C/A) were mutated. Finally, to study the effect of polymorphism of TNF-α gene on gastric cancer, and to lay a theoretical foundation for the study of the effect of gene polymorphism on the occurrence and development of cancer.
2. Materials and methods
Nucleotide sequence of the analyzed TNF-α gene came from Genbank Home and the Accession number was NM_000594.
Manually site-directed mutagenesis of the nucleotide sequence of TNF-α was carried out. Nucleotide A of 308 site was replaced with G (308G/A), nucleotide T was replaced with C (857 C/T) and nucleotide A of 863 site was replaced with C (863C/A). The changed nucleotide sequence was inputted into ORF Finder to be translated, thus obtaining the corresponding open reading frame and amino acid sequence.
Since the reason of functional changes was explored mainly from structural changes, what this study had focused was the local modeling of mutant sites. There were many kinds of tools for protein modeling, including Swiss-model and Phyre and so on. And Swiss-model, the most widely used method, maintains regular data updates to ensure the integrity and accuracy of the collected data. However, Swiss-model adopted the method of homology modeling. If the inputted sequence was too short, Swiss-model cannot find effective homologous template, causing the failure of modeling. If the inputted whole sequence was too long, however, the amino acids of mutant sites were easy to be neglected. Then the structural change before and after mutation cannot be effectively detected. Therefore, Swiss-model was not a feasible method (Rong et al., 2006). Phyre established the 3D model of proteins by the method of threading, which was also easy to neglect the mutant sites. So Phyre2 was used for modeling of more than 200 amino acids of TNF-α and found the structural changes of proteins cannot be detected. Through a comprehensive analysis, software QUARK and I-TASSER invented by Zhang Lab in Michigan University were chosen as modeling tools. QUARK was a method of calculation, analyzing models of foldable and tertiary structures of proteins, which was based on ab initio prediction method. With only the amino acid sequence, it aimed to establish exact 3D models. QUARK model was established by synthesizing the small fragments (the length of 1–20 amino acids) by copy convert of Monte-Carlo simulation, which was completed based on knowledge of the force field on atomic energy level. Since there was no universal template information, QUARK was applicable in protein modeling without homologous templates and can exactly predict the 3D models of short sequences consisting of less than 100 amino acids. I-TASSER server was an online platform for the prediction of protein structures and functions. Its 3D model was established through multi-threading method of LOMETS and iterative template fragments. I-TASSER synthesized the two methods of homologous template modeling and threading. Its advantage lies in that there is relatively low request of the similarity than Swiss-model when seeking the homologous templates. It can not only get the models of proteins, but also give the confidence values as well as showing the first five models with the highest confidence values (Yang et al., 2014). In addition, it can provide the first ten protein structures with high similarity and binding sites as well as the prediction of products, which was very convenient for users to consult. I-TASSER was the prediction server of protein structures which ranked first in free modeling of recent experiments of CASP7, CASP8, CASP9 and CASP10.
Two kinds of tools, QUARK and I-TASSER, were used to establish 3D models of each site of TNF-α before and after mutation. Then MATLAB was used to draw paths of α carbon atom in each mutant site in order to analyze the change in skeletons before and after mutation.
3. Results
Nucleotide sequences before and after mutation were inputted into open reading frame analyzing software ORF Finder. After analysis and translation, they were compared with the original sequences. The results showed TNF-α contained an open reading frame consisting of 701 nucleotides, which was translated into a sequence consisting of 233 amino acids. The 238G/A mutation did not contribute to the change in amino acid sequence while 308G/A mutation led to the change of the 45th encoding amino acid from threonine T to alanine A, the 857 C/T mutation caused the 228th encoding amino acid to change from phenylalanine F to leucine L and 863C/A mutation caused the 230th encoding amino acid to change from isoleucine I to leucine L.
The whole sequences of amino acid (233 amino acids) before and after mutation were inputted into I-TASSER for modeling. The results were shown in the following figures.
From Fig. 1, Fig. 2, Fig. 3, Fig. 4, it can be seen there was one α helical structure in the region where the 308th mutant site was located, and after the mutation there were 4 α helical structures in that region. The mutant site 857 was located in the foldable region of TNF-α before mutation, after mutation, the structure did not change obviously. The mutant site 863 was located in the margin of foldable region of TNF-α before mutation, after mutation, α helical structure appeared in the adjacent region of the mutant site. The whole sequence was used to establish the model. To further study the local change in the structure, QUARK need to be adopted for a second-time modeling.
Fig. 1.
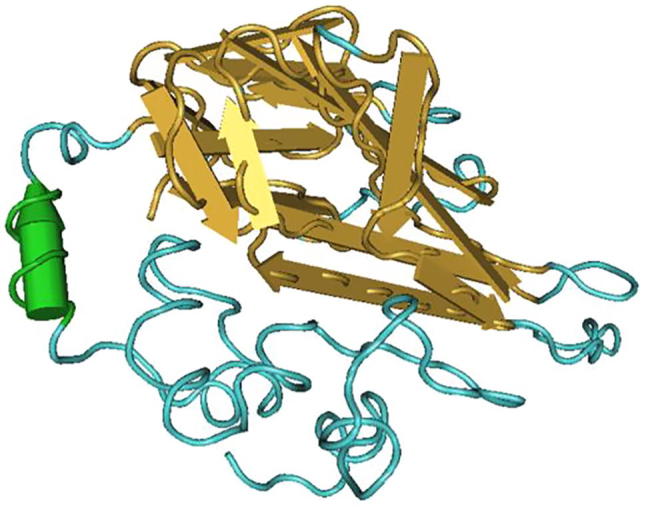
Structure of TNF-α before mutation. Note: green column structure is α helix, yellow flat arrow is folding structure, blue linearity is ordinary peptide chain; there’s only one α helix before the mutation.
Fig. 2.
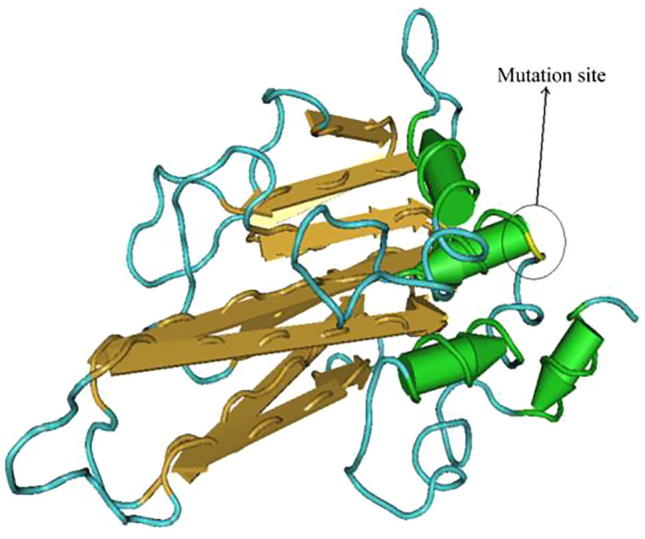
Structure of TNF-α-308G/A after mutation. Note: green column structure is α helix, yellow flat arrow is folding structure, blue linearity is ordinary peptide chain; the marked position in the figure is the mutation site of the 308th site and after the mutation there are 4 α helixes.
Fig. 3.
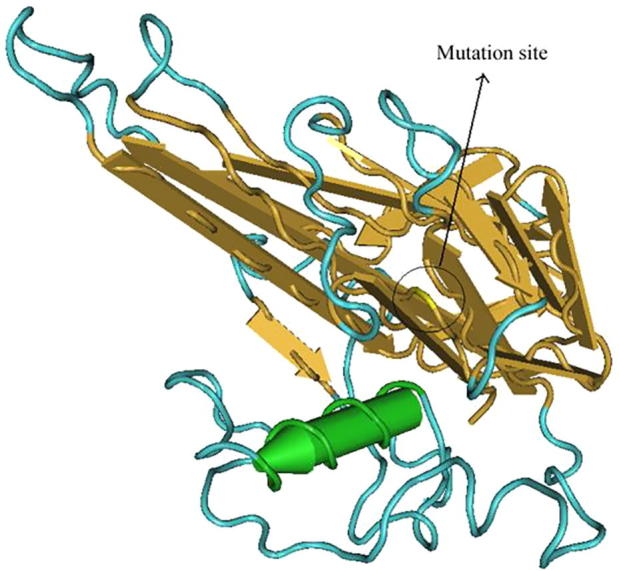
Structure of TNF-α-857C/T after mutation. Note: green column structure is α helix, yellow flat arrow is folding structure, blue linearity is ordinary peptide chain; the marked position in the figure is the mutation site of the 857th site whose structural changes is not significant.
Fig. 4.
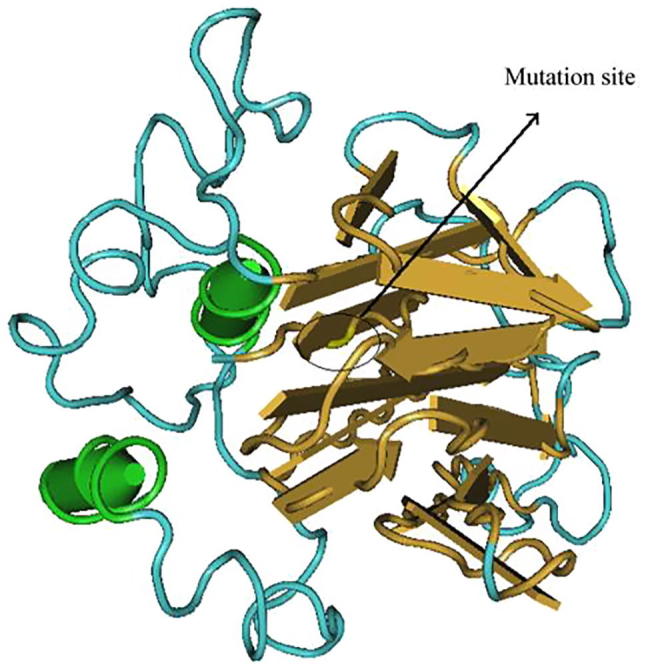
Structure of TNF-α-863C/A after mutation. Note: green column structure is α helix, yellow flat arrow is folding structure, blue linearity is ordinary peptide chain; the marked position in the figure is the mutation site of the 863th site and after the mutation there are two α helixes.
To ensure the accuracy of the model, only no more than 100 amino acids sequences can be inputted into QUARK. 15–30 genes before and after mutant sites in amino acid sequences were chosen to compose a short sequence consisting of 30–70 amino acids, which was inputted into QUARK for modeling. The results of I-TASSER modeling showed that the 308 mutation site caused a wide range change in helical structures. 30 amino acids before and after mutant site respectively were used to compose a sequence consisting of 71 amino acids for modeling. The results were shown in Fig. 5.
Fig. 5.
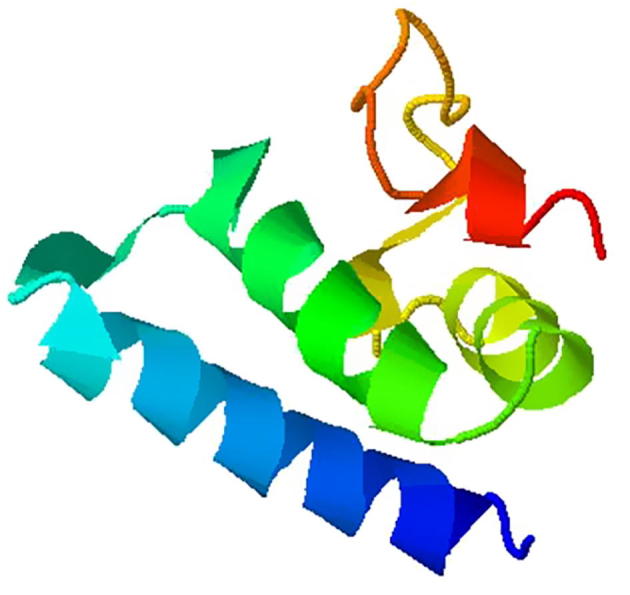
Structure of TNF-α-308G/A after mutation. Note: α helix and folding structure are included in the figure.
In terms of mutant site 857 and mutant site 863, given that they were located in the end of a sequence, several amino acids were selected to compose a short sequence of 35 amino acids. Then it was inputted into QUARK for modeling. The results were shown in Fig. 6, Fig. 7. It can be seen that the foldable structures did not change before and after mutation, the folding angles, however, became large.
Fig. 6.

Structures before and after the mutation of TNF-α-857C/T. Note: after the mutation the folding structure hasn’t changed significantly but showing a larger folding angle.
Fig. 7.
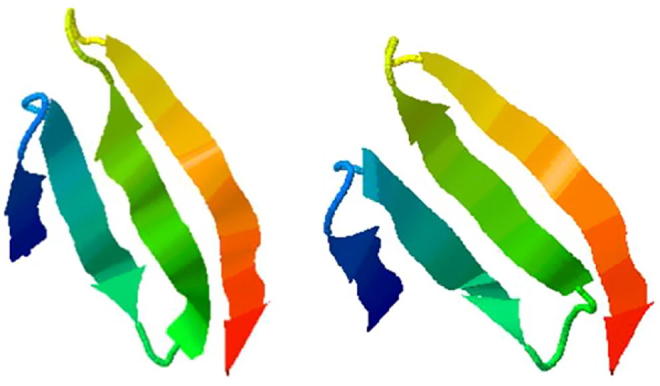
Structures before and after the mutation of TNF-α-863C/A mutation site. Note: the structural change isn’t obvious but the folding angle gets larger.
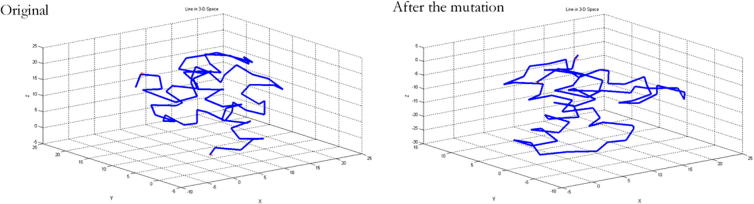
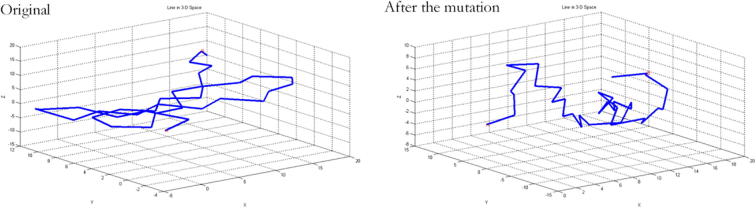
Then, MATLAB was used to draw the paths of carbon atoms in each model. See Fig. 8, Fig. 9, Fig. 10. From the figures, significant changes can be found in these structures.
Fig. 8.
Skeleton carbon atom paths before and after mutation of TNF-α-308G/A mutation site. Note: there’re significant changes before and after the mutation.
Fig. 9.
Skeleton carbon atom paths before and after mutation of TNF-α-857C/T mutation site. Note: the paths become scattered compared with those before the mutation, and the changes is apparently.
Fig. 10.
Skeleton carbon atom paths before and after mutation of TNF-α-863C/A mutation site. Note: the path changes of TNF-α-863C/A and TNF-α-857C/T before and after the mutation are similar, namely, paths are concentrated before the mutation and become scattered after the mutation, and the path of carbon atoms changed remarkably.
4. Discussions
Host genetic factor is crucial to incidence of multiple cancers (Kamangar et al., 2006), such as genetic variation of inflammatory cytokines and anti-inflammatory cytokines have effects on immune response of body to carcinogen, resulting in cancer. And inflammatory cytokines, especially TNF-α and its receptors’ polymorphism are closely linked to the occurrence of gastric cancer (Gorouhi et al., 2008). Related studies showed that there’s single nucleotide polymorphism (SNP) in TNF-α gene promoter methylation, and genotypes in different sites can cause changes in TNF-α transcriptional levels and affect TNF-α secretion (Juszczynski et al., 2002, Pujhari et al., 2012). Current studies found that there are multiple SNPs in TNF-α promoter methylation region, such as −238G/A, −308G/A, −857 C/T, −863 C/A and −193 G/A, etc. (Essadik et al., 2015). And some of them are correlated to gastric cancer but due to different objects, the outcomes vary from study to study. Xu et al. (2016) did research on TNF-α polymorphism of 47 gastric cancer families and 47 non-gastric cancer families and found that TNF-α-308 GA and AA gene have higher frequency in gastric cancer families than non-gastric cancer families, which indicated that TNF-α-308 polymorphism was closely correlated to gastric cancer among Chinese population. Meta analysis by Yang et al. (2014) showed that TNF-α-308 G/A is correlated to occurrence of gastric cancer among Caucasian ethnicities but not among east Asians and other races. While in the study by de Oliveira et al. (2015), it was reported that it is not TNF-α-308 G/A but TNF-α-857 C/T that is correlated to occurrence of gastric cancer. Additionally, research of Sugimoto et al. (2007) indicated that TNF-α-863C/A is also linked to gastric cancer risk.
The results in this paper show that mutation of site TNF-α-308 has led to great change in space structure, producing many helical structures. Some studies (Zhong, 2007) showed helix α was more stable than other helixes. Protein structural domains with many helical structures generally appear to be rather compact and stable, not liable to variation. So, the active sites usually locate between helical regions. If the helical structures of proteins were stretched, they were prone to turning into β foldable structures (Fan, 1996). If the foldable structures were squeezed, they may turn into helical structures. Mutation of site 308 was similar to the above description. Currently, among SNP sites, TNF-α-308 has been studied frequently on its relations to gastric cancer, and a majority of researches have proved their relationship. Similarly, in this study TNF-α-308 changed apparently, and structural change directly affected functional change, but the specific acting mechanism still needs deep researches. Given that bioinformatics means was used for prediction in the study, we can also get TNF-α protein by biologic experiments and then normal analytic method of protein structure was used to get its real structure which finally was compared with the predicted protein structure. In addition, through collecting plenty of clinical samples and data we discussed specific influences of different genotypes on patients with gastric cancer for further studies on the polymorphism of TNF-α and for huge breakthroughs. This study only employed software to imitate the mutation of TNF-α, based on bioinformatics, however, due to the limitation of the software and the complexity of protein structure, the predicted results in the study still needs to be further proved through X ray diffraction analysis, 3D reconstruction technology, nuclear magnetic resonance technology and other methods.
In conclusion, through three typical mutant types (308G/A, 857C/T and 863C/A) of TNF- alpha, gastric cancer related gene, for protein structure analysis, this study reveals the potential mechanism of gene mutation leading to the change of the function of TNF- in the development and progression of gastric cancer, laying a theoretical foundation for the study of the effect of gene polymorphism on the occurrence and development of cancer.
Acknowledgments
Ethical Committee
All experimental processes are within the standards of the ethical committee.
Footnotes
Peer review under responsibility of King Saud University.
References
- Canedo P., Durães C., Pereira F., Regalo G., Lunet N., Barros H., Carneiro F., Seruca R., Rocha J., Machado J.C. Tumor necrosis factor alpha extended haplotypes and risk of gastric carcinoma. Cancer Epidem. Biomar. 2008;17(9):2416–2420. doi: 10.1158/1055-9965.EPI-08-0413. [DOI] [PubMed] [Google Scholar]
- Chen W., Zheng R., Baade P.D., Zhang S., Zeng H. Cancer statistics in China, 2015. Ca-Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- de Oliveira J.G., Rossi A.F., Nizato D.M., Cadamuro A.C., Jorge Y.C. Influence of functional polymorphisms in TNF-α, IL-8, and IL-10 cytokine genes on mRNA expression levels and risk of gastric cancer. Tumour Biol. 2015;36(12):9159–9170. doi: 10.1007/s13277-015-3593-x. [DOI] [PubMed] [Google Scholar]
- Essadik A., Jouhadi H., Rhouda T., Nadifiyine S., Kettani A. Polymorphisms of tumor necrosis factor alpha in moroccan patients with gastric pathology: new single-nucleotide polymorphisms in TNF-α(-193) (G/A) Mediat. Inflamm. 2015;2015:143941. doi: 10.1155/2015/143941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y.Z. Helical structure and function of proteins. Biol. Bull. 1996;31(1) [Google Scholar]
- Fang Y. The prediction and functional research of human antibody against tumor necrosis factor-α. Acad. Mil. Med. Sci. 2009 [Google Scholar]
- Gorouhi F., Islami F., Bahrami H., Kamangar F. Tumour-necrosis factor-A polymorphisms and gastric cancer risk: a meta-analysis. Br. J. Cancer. 2008;98(8):1443–1451. doi: 10.1038/sj.bjc.6604277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszczynski P., Woszczek G., Borowiec M., Kowalski M., Robak T. Comparison study for genotyping of a single-nucleotide polymorphism in the tumor necrosis factor promoter gene. Diagn. Mol. Pathol.: Am. J. Surg. Pathol., Part B. 2002;11(4):228–233. doi: 10.1097/00019606-200212000-00006. [DOI] [PubMed] [Google Scholar]
- Kamangar F., Cheng C.C., Rabkin C.S. Interleukin-1B polymorphisms and gastric cancer risk–a meta-analysis. Cancer Epidem. Biomar. 2006;15(10):1920–1928. doi: 10.1158/1055-9965.EPI-06-0267. [DOI] [PubMed] [Google Scholar]
- Lili D. Effect of RNA interference on the secretion of tumor necrosis factor-α in mice spleen lymphocytes. J. China Med. Univ. 2006;35(5):449–466. [Google Scholar]
- Pujhari S.K., Ratho R.K., Prabhakar S., Mishra B., Modi M. TNF-alpha promoter polymorphism: a factor contributing to the different immunological and clinical phenotypes in Japanese encephalitis. BMC Infect. Dis. 2012;12(1):1–6. doi: 10.1186/1471-2334-12-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokkas T., Sechopoulos P., Pistiolas D., Kothonas F., Margantinis G. Population differences concerning TNF-α gene polymorphisms in gastric carcinogenesis based on meta-analysis. Ann. Gastroenterol. 2014;27(2):139–148. [PMC free article] [PubMed] [Google Scholar]
- Rong Z., Min C., Chunxian Y., Liao Zhihua. Three-dimensional structural modeling of protein based on SWISS-MODEL. Chem. Life. 2006;26(1):54–56. [Google Scholar]
- Sugimoto M., Furuta T., Shirai N., Nakamura A., Fang X. Different effects of polymorphisms of tumor necrosis factor-alpha and interleukin-1 beta on development of peptic ulcer and gastric cancer. J. Gastroen. Hepatol. 2007;22(1):51–59. doi: 10.1111/j.1440-1746.2006.04442.x. [DOI] [PubMed] [Google Scholar]
- Tahara T., Shibata T., Nakamura M., Yamashita H., Yoshioka D. Effect of IL-1β and TNF-α polymorphisms on the prognosis and survival of gastric cancer patients. Clin. Exp. Med. 2011;11(4):211–217. doi: 10.1007/s10238-010-0129-y. [DOI] [PubMed] [Google Scholar]
- Xu Y., Cao X., Jiang J., Chen Y., Wang K. TNF-α-308/-238 polymorphisms are associated with gastric cancer: a case-control family study in China. Clin. Res. Hepatol. Gastroen. 2016;S2210-7401(16):30091–30092. doi: 10.1016/j.clinre.2016.05.014. [DOI] [PubMed] [Google Scholar]
- Yang J.P., Hyun M.H., Yoon J.M., Park M.J., Kim D. Association between TNF-α-308 G/A gene polymorphism and gastric cancer risk: a systematic review and meta-analysis. Cytokine. 2014;70(2):104–114. doi: 10.1016/j.cyto.2014.07.005. [DOI] [PubMed] [Google Scholar]
- Yang J., Yan R., Roy A., Xu D., Poisson J. The I-TASSER Suite: protein structure and function prediction. Nat. Meth. 2014;12(1):7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S.L. Proteins and helixes. J. Mod. Med. Health. 2007;21(7):804–805. [Google Scholar]