Abstract
Carotenoids, found in the fruit and different organs of bitter melon (Momordica charantia), have attracted great attention for their potential health benefits in treating several major chronic diseases. Therefore, study related to the identification and quantification of the medically important carotenoid metabolites is highly important for the treatment of various disorderes. The present study involved in the identification and quantification of the various carotenoids present in the different organs of M. charantia and the identification of the genes responsible for the accumulation of the carotenoids with respect to the transcriptome levels were investigated. In this study, using the transcriptome database of bitter melon, a partial-length cDNA clone encoding geranylgeranyl pyrophosphate synthase (McGGPPS2), and several full-length cDNA clones encoding geranylgeranyl pyrophosphate synthase (McGGPPS1), zeta-carotene desaturase (McZDS), lycopene beta-cyclase (McLCYB), lycopene epsilon cyclases (McLCYE1 and McLCYE2), beta-carotene hydroxylase (McCHXB), and zeaxanthin epoxidase (McZEP) were identified in bitter melon. The expression levels of the mRNAs encoding these eight putative biosynthetic enzymes, as well as the accumulation of lycopene, α-carotene, lutein, 13Z-β-carotene, E-β-carotene, 9Z-β-carotene, β-cryptoxanthin, zeaxanthin, antheraxanthin, and violaxanthin were investigated in different organs from M. charantia as well as in the four different stages of its fruit maturation. Transcripts were found to be constitutively expressed at high levels in the leaves where carotenoids were also found at the highest levels. Collectively, these results indicate that the putative McGGPPS2, McZDS, McLCYB, McLCYE1, McLCYE2, and McCHXB enzymes might be key factors in controlling carotenoid content in bitter melon. In conclusion, the over expression of the carotenoid biosynthetic genes from M. charantia crops to increase the yield of these medically important carotenoids.
Keywords: Medically important carotenoids, Momordica charantia, Transcriptome level, HPLC analysis
1. Introduction
Momordica charantia (family Cucurbitaceae), commonly known as bitter gourd or bitter melon, is a popular herb found in Asia, Africa, and the Caribbean. As a medicinal plant, bitter melon is used in the treatment of several diseases or conditions including diabetes, HIV, viral infections, cancer, inflammation, ulcers, and sepsis (Chao et al., 2014, Liaw et al., 2015). Researchers have found that, with respect to its pharmaceutical applications, the important components of bitter melon are the phenolic, flavonoid, triterpene, and carotenoid compounds, including alpha and beta-carotene, lycopene, and zeaxanthin (Liaw et al., 2015).
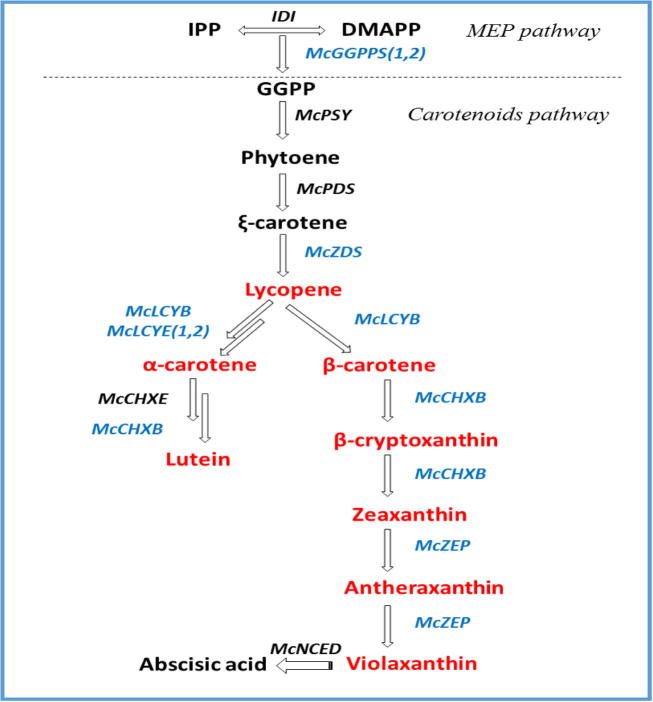
The Carotenoids derivative present in the vegetables and fruits were known for their medical applications especially in controlling the chronic and vascular diseases (Khoo et al., 2011). Till today more than 600 types of Carotenoids are identified from various plant specieses (Khoo et al., 2011). The color of carotenoids also attracts both pollinators and seed dispersal agents to flowers and fruit and also the starting molecules for the synthesis of abscisic acid which is mainly involved in the plant stress regulations. Vitamin A is synthesized from the intermediary molecules of carotenoids such as such as α-carotene and β-carotene (Tuan et al., 2011a, Tuan et al., 2011b). In human vitamin A deficiency causes various visionary diseases such as xerophthalmia, and blindness. Also, consumption of carotenoids help in reducing the risks of cancer, cataract formation and heart related disease in human. In carotenoid biosynthetic pathway, dimethylallyl pyrophosphate (DMAPP) catalysis by geranylgeranyl pyrophosphate synthetase (GGPPS) (Fig. 1) (Tuan et al., 2011a, Tuan et al., 2011b). The genes responsible for the metabolic pathways enzymes were already amplified and characterized in Arabidopsis (Ruiz-Sola and Rodriguez-Concepcion, 2012), Brassica rapa (Li et al., 2015), tomato (Namitha et al., 2011), carrot (Clotault et al., 2008), Momordica cochinchinensis (Hyun et al., 2012), rice (Beyer et al., 2002), maize (Messias et al., 2014), and Scutellaria baicalensis Georgi (Tuan et al., 2015).
Fig. 1.
Carotenoid biosynthetic pathway in plants. Red color denotes the carotenoids measured in this study by HPLC analysis and blue color indicates enzymatic activities for which gene expression was monitored via real time-PCR. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In recent years, several genes in the M. charantia carotenoid biosynthesis pathway have been cloned and characterized including phytoene synthase (McPSY), phytoene desaturase (McPDS), carotenoid cleavage dioxygenase 1 (McCCD1), carotenoid cleavage dioxygenase 4 (McCCD4), 9-cis-epoxycarotenoid dioxygenase (McNCED) (Tuan and Park, 2013). However, there are still a number of genes that remain uncharacterized, including geranylgeranyl pyrophosphate synthase (McGGPPS), zeta-carotene desaturase (McZDS), lycopene beta-cyclase (McLCYB), lycopene epsilon cyclase (McLCYE), beta-carotene hydroxylase (McCHXB), and zeaxanthin epoxidase (McZEP) (Tuan and Park, 2013). To date, no comparative studies have been performed examining the genes from M. charantia. Here, we have examined the levels of McGGPPS, McZDS, McLCYE, McLCYB, McCHXB, and McZEP in different organs, as well fruit at different stages of maturation, from M. charantia. This is the first description of these enzymes in M. charantia, and it marks a first step toward possible bioengineering of M. charantia crops to increase the yield of these medically important carotenoids.
2. Materials and methods
2.1. Plant material
Seeds of a Chinese cultivar of bitter melon (Momordica charantia L.) were purchased from Beijing Namo Tech.-Trade Co. Ltd (Beijing, China). After three months, different bitter melon organs including roots, stems, old leaves, young leaves, male flowers, female flowers, and fruit at four different stages of maturation (Table S1) were collected and harvested.
2.2. RNA isolation and cDNA synthesis
RNeasy Plant mini kit (QIAGEN, Valencia, CA, USA) was used for the extraction and purification of the total RNA from different organs of M. charantia (Tuan and Park, 2013). After extraction, 1 μg of high-quality total RNA was used for the preparation of cDNA synthesis. The cDNA was synthesized using the ReverTra Ace-α-kit (Toyobo Co. Ltd., Osaka, Japan).
2.3. Sequence analysis
Using sequence data from the sequencing of complementary DNA (cDNA) libraries obtained from M. charantia seedlings (data not shown). The genes that showed maximum identity and similarity were selected for further study (Tuan et al., 2011a, Tuan et al., 2011b).
2.4. Quantitative real-time polymerase chain reaction (qRT- PCR) analysis
Real-time PCR primers (Table 1) were designed using the Primer 3 website (http://frodo.wi.mit.edu/primer3/) based on the sequences of geranylgeranyl pyrophosphate synthase (McGGPPS1 and McGGPPS2), zeta-carotene desaturase (McZDS), lycopene beta-cyclase (McLCYB), lycopene epsilon cyclase (McLCYE1 and McLCYE2), beta-carotene hydroxylase (McCHXB), zeaxanthin epoxidase (McZEP), and based on the published gene sequences of phytoene synthase (McPSY) (GenBank Accession Number: AY494789), and phytoene desaturase (McPDS) (GenBank Accession Number: AY494790.1). The levels of gene expression were calculated by relative quantification using the M. charantia cyclophilin gene (McCYP) (GenBank Accession Number HQ171897) as reference. Standard amplification procedures such as initial denaturation 95 °C for 5 min, 95 °C for 15 s, template annealing at 65 °C for 15 s, and final extension 72 °C for 20 s, respectively. To the PCR mixture SYBR Green was added for the quantification of the expression level of the individual genes (Tuan et al., 2011a, Tuan et al., 2011b).
Table 1.
Sequences of specific primers used for quantitative real-time PCR.
| Primers name | Forward primer sequences (5′–3′) | Reverse primer sequences (5′–3′) | Size (bp) |
|---|---|---|---|
| McGGPPS1 | GGCACCAATTCGATGTTCTT | GATCTCGTCGGGGTACTGAA | 154 |
| McGGPPS2 | GCCGTGTGTGGAGGACTAAT | GCTTCCCCTTCTTCGTTCTT | 154 |
| McPSY | GCTTCATCGTTGGTTGTCTCTCT | TGCTCCATTTCTGCCTCTTACTC | 154 |
| McPDS | TTTGCTTGGATTACCCTAGACCA | TGCACCAGCGATCACTACTTTTA | 128 |
| McZDS | TCTTGGCTTTATTCCCATCG | AGTTGCTCCTTCCATGCTGT | 194 |
| McLCYB | CGGGAGGGTTAATAGGAAGC | GGACAAGGCATCGAGAGAAG | 194 |
| McLCYE1 | AAGCGTTTTGAAGCAAGGAA | AGTGCAAGCCCAAAGAGAAA | 141 |
| McLCYE2 | CTTGTAGCCTGCGAACATGA | TCCTACCTCGACCTCCACAC | 147 |
| McCHXB | ACGATGTTTTCGCCATTACC | GGACCCACAGGGAATCTTTT | 178 |
| McZEP | GAGCGTGCTGTGCATTAGAA | ACAACTGGGTTCTCCCACTG | 193 |
| McCYP | GGCAAACCCTAAAGTTTTCTTCG | GATGAGCCCTTGTAATGAAGTGG | 174 |
2.5. Extraction and high performance liquid chromatography (HPLC) analysis of carotenoids from M. charantia
The extraction method used for carotenoid analysis in bitter melon was similar to that described by Tuan et al., 2011a, Tuan et al., 2011b . Gradient elution system was used for the complete separation of the individual carotenoid components. For the mobile solvent preparation 10 mM ammonium acetate was dissolved in (92% of methanol and 8% of water (Solvent A) and 100% methyl tert-butyl ether (MTBE) (Solvent B). Initially the column was eluted with 83% A and 17% B for 23 min, after that 70% A and 30% B for 29 min, 59% A and 41% B for 35 min, 30% A and 70% B for 40 min, 30% A and 70% B for 44 min, 83% A and 17% B for 55 min respectively (Howe and Tanumihardjo, 2006)
3. Results
3.1. Sequence analyses of carotenoid biosynthetic genes from M. charantia
In our study, complementary DNA (cDNA) libraries from M. charantia seedlings were sequenced using the Illumina Next Seq500 platform. Out of 68,073,862 total reads, approximately 88,703 unigenes were identified (data not shown). Among the unigenes from the M. charantia database, several full-length cDNAs encoding McGGPPS1, McZDS, McLCYB, McLCYE1, McLCYE2, McCHXB, and McZEP, and a partial-length cDNA clone encodingMcGGPPS2 were identified (Table 2). Their homology was confirmed by BLAST program, and the cDNAs were designated as McGGPPS1 (335 amino acids, aa), McGGPPS2 (261 aa), McZDS (579 aa), McLCYB (504 aa), McLCYE1 (249 aa), McLCYE2 (308 aa), McCHXB (311 aa), McZEP (450 aa).
Table 2.
Comparison of carotenoid genes of M. charantia with the most orthologous genes.
| Genes of M. charantia | Length (amino acid) | Orthologous genes | Accession no. | Identity (%) |
|---|---|---|---|---|
| McGGPPS1 | 335 | Cucumis melo | XP_008447910.1 | 88 |
| Cucumis sativus | XP_004144868.1 | 87 | ||
| Populus euphratica | XP_011046617.1 | 80 | ||
| McGGPPS2 | 261 | Cucumis sativus | XP_004144868.1 | 82 |
| Morus notabilis | XP_010102651.1 | 82 | ||
| Cucumis melo | XP_008447910.1 | 81 | ||
| McZDS | 579 | Cucurbita moschata | AEK86566.1 | 90 |
| Cucumis melo | XP_008462722.1 | 89 | ||
| Cucumis sativus | XP_004142522.1 | 89 | ||
| McLCYB | 504 | Citrullus lanatus | ABM90917.1 | 96 |
| Cucumis sativus | XP_004150761.1 | 95 | ||
| Cucurbita moschata | AEN94902.1 | 91 | ||
| Cucumis melo | XP_008457615.1 | 95 | ||
| McLCYE1 | 249 | Cucumis sativus | XP_004141172.1 | 95 |
| Cucurbita moschata | AEN94903.1 | 92 | ||
| Cucumis melo | XP_008459489.1 | 94 | ||
| McLCYE2 | 308 | Cucumis melo | XP_008459489.1 | 87 |
| Cucumis sativus | XP_004141172.1 | 84 | ||
| Cucumis melo var. makuwa | AID51466.1 | 87 | ||
| McCHXB | 311 | Cucurbita moschata | AEK86567.1 | 87 |
| Cucumis sativus | XP_004140758.1 | 88 | ||
| Cucumis melo | XP_008439286.1 | 88 | ||
| McZEP | 450 | Cucumis sativus | XP_004148358.2 | 88 |
| Morus notabilis | XP_010104831.1 | 72 | ||
| Vitis vinifera | XP_002265622.3 | 74 | ||
3.2. Gene expression of carotenoid biosynthesis genes in different M. charantia organs
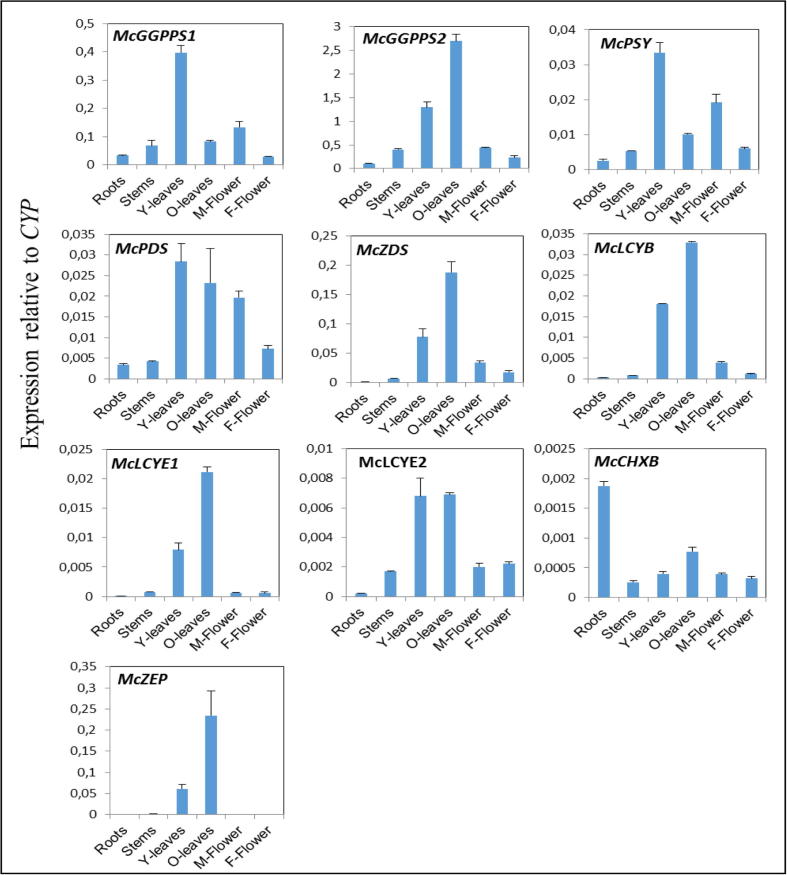
The expression levels of the studied genes in roots, stems, young leaves, old leaves, male flowers, female flowers, and four fruit stages from M. charantia using real-time PCR was presented in Fig. 2, Fig. 3. Among the carotenoid biosynthetic genes of M. charantia, considerable levels of almost all the genes were detected in leaves, whereas only McCHXB was expressed at high levels in the roots. Considerable levels of McGGPPS1 and McGGPPS2 were detected in leaves (Fig. 2), whereas only small levels were found in other organs. However, expression levels of these two GGPPSs in leaves varied depending on the age of the leaf; McGGPPS1 had high expression levels in young leaves, whereas McGGPPS2 had high expression levels in old leaves. The transcription patterns of McZDS, McLCYB, McLCYE1, and McZEP were all essentially similar, with high expression observed in old leaves, intermediate levels in young leaves, and low levels in male flowers, female flowers, stems, and roots. Considerable levels of McLCYE2 were detected in leaves, lower levels in stems and flowers, and trace levels in the roots.
Fig. 2.
Expression levels of genes in carotenoid biosynthesis pathway in different organs of M. charantia.
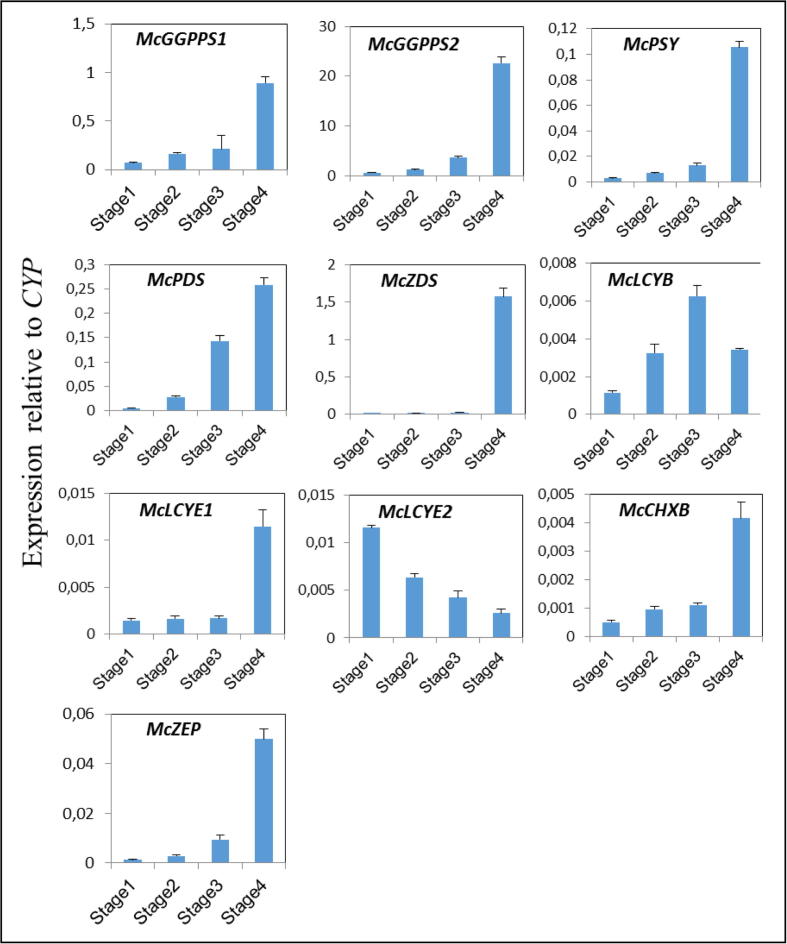
Fig. 3.
Expression levels of genes in carotenoid biosynthesis pathway during fruit maturation of M. charantia fruit. CYP, M. charantia cyclophilin.
With the exception of McLCYB and McLCYE2, the M. charantia expression patterns of McGGPPS1, McGGPPS2, McPSY, McPDS, McZDS, McLCYE1, McCHXB, and McZEP were similar during the four stages of fruit maturation (Fig. 3). Specifically, expression barely changed from stage 1 to stage 3.In contrast, McLCYE2 decreased from stage 1 to stage 4, whereas expression of McLCYB increased from stage 1 to stage 3, but then decreased at stage 4.
3.3. HPLC analyses of carotenoid in different M. charantia organs
Carotenoids in the roots, stems, young leaves, old leaves, male flowers, female flowers, and the four stages of fruit maturation, were determined by HPLC analysis of tissue extracts (Table 3, Table 4). Carotenoid accumulation was lowest in the roots, the only abundant carotenoid in roots being lutein (4.81 μg/gdry weight (DW)); however, this accumulation was significantly lower than that observed in other organs of bitter melon. These high levels of lutein may explain why low levels of its precursor, α- carotene, were found in M. charantia. α-carotene accumulation was highest in old leaves (41.06 μg/g DW), whereas only small amounts were found in young leaves, stems, male flowers, and female flowers (18.77 μg/g DW, 7.29 μg/g DW, 5.75 μg/g DW, and 5.34 μg/g DW, respectively); there were no detectable levels in roots. β-carotene, including E-β-carotene, 9z-β-carotene, and 13z-β-carotene, synthesized high amounts in M. charantia. Especially the the content of E-β-carotene was higher in old leaves and young leaves (287.03 μg/g DW and 220.12 μg/g DW, respectively), comparable contents were documented the stems, male flowers, and female flowers (51.11 μg/g DW, 65.13 μg/g DW and 56.64 μg/g DW, respectively), and not detected in roots. The accumulation of the isomers 9z-β-carotene and 13z-β-carotenewassimilar, being abundant in old leaves (49.16 μg/g DW and 45.62 μg/g DW, respectively), and young leaves (41.54 μg/g DW and 33.22 μg/g DW, respectively), with low accumulation in male flowers (13.9 μg/g DW and 16 μg/g DW, respectively), female flowers (9.38 μg/g DW and 8.26 μg/g DW, respectively), and stems (8.10 μg/g DW and 8.6 μg/g DW, respectively), and undetectable in roots. M. charantia also contained a small amount of violaxanthin and antheraxanthin in the male flowers (48.91 μg/g DW and 32.29 μg/g DW, respectively) whereas only trace amounts of these carotenoids were found in other organs.
Table 3.
Carotenoid content in different organs of M. charantia (µg/g dry weight).
| Roots | Stems | Y-leaves | O-leaves | M-Flower | F-Flower | |
|---|---|---|---|---|---|---|
| Lycopene | ND | ND | ND | ND | ND | ND |
| α-carotene | ND | 7.29 ± 0.57 | 18.77 ± 2.13 | 41.06 ± 1.03 | 5.75 ± 1.18 | 5.34 ± 0.44 |
| Lutein | 4.81 ± 0.83 | 93.64 ± 7.69 | 314.83 ± 48.78 | 412.33 ± 59.89 | 57.33 ± 2.70 | 86.12 ± 3.83 |
| 13Z-β-carotene | ND | 8.60 ± 1.23 | 33.22 ± 6.62 | 45.62 ± 6.27 | 16.00 ± 1.27 | 8.26 ± 1.13 |
| E-β-carotene | ND | 51.11 ± 6.80 | 220.12 ± 14.50 | 287.03 ± 21.81 | 65.13 ± 4.92 | 56.64 ± 6.71 |
| 9Z-β-carotene | ND | 8.10 ± 0.22 | 41.54 ± 0.64 | 49.16 ± 0.89 | 13.90 ± 2.97 | 9.38 ± 1.23 |
| β-cryptoxanthin | ND | 8.07 ± 0.05 | 10.97 ± 1.11 | 10.87 ± 0.13 | 15.26 ± 1.09 | 9.12 ± 0.33 |
| Zeaxanthin | ND | 11.78 ± 1.17 | 28.15 ± 2.67 | 15.38 ± 1.17 | 13.42 ± 0.57 | 9.63 ± 0.35 |
| Antheraxanthin | ND | 1.70 ± 0.20 | 12.08 ± 3.39 | 4.79 ± 0.63 | 32.29 ± 1.06 | 4.95 ± 0.24 |
| Violaxanthin | ND | ND | 17.32 ± 1.20 | 21.02 ± 1.96 | 48.91 ± 1.27 | 12.37 ± 0.34 |
| Total carotenoids | 4.81 ± 0.83 | 190.29 ± 17.92 | 697.01 ± 81.04 | 887.26 ± 93.77 | 267.98 ± 17.04 | 201.82 ± 14.60 |
Table 4.
Carotenoid content in different fruit of M. charantia (µg/g dry weight).
| Stage 1 | Stage 2 | Stage 3 | Stage 4 | |
|---|---|---|---|---|
| Lycopene | ND | ND | ND | 12.44 ± 3.20 |
| α-carotene | 12.67 ± 1.26 | 4.54 ± 0.39 | 3.19 ± 0.36 | 2.48 ± 0.18 |
| Lutein | 41.16 ± 3.21 | 22.66 ± 1.26 | 17.43 ± 0.61 | 21.89 ± 1.33 |
| 13Z-β-carotene | 4.44 ± 0.70 | 1.79 ± 0.16 | 1.93 ± 0.32 | 5.89 ± 0.64 |
| E-β-carotene | 26.13 ± 2.59 | 12.25 ± 1.00 | 10.84 ± 0.89 | 28.08 ± 1.64 |
| 9Z-β-carotene | 3.84 ± 0.25 | 2.03 ± 0.15 | 2.23 ± 0.36 | 2.83 ± 0.09 |
| β-cryptoxanthin | ND | ND | ND | 69.05 ± 15.76 |
| Zeaxanthin | ND | ND | 9.82 ± 0.17 | 45.91 ± 3.81 |
| Antheraxanthin | ND | ND | ND | 7.07 ± 1.54 |
| Violaxanthin | ND | ND | ND | ND |
| Total carotenoids | 88.24 ± 8.00 | 43.28 ± 2.97 | 45.44 ± 2.71 | 195.63 ± 28.20 |
In the fruit of M. charantia, the noted amount of the carotenoids was varied between covering stage 1 to stage 3 to orange (stage 4) (Table 4). For example, zeaxanthin was highest in stage 4 fruit (45.91 g/gDW), followed by stage 3 fruit (9.82 μg/gDW), but undetectable in stage 1 and stage 2 fruit. Lycopene, β-cryptoxanthin, and antheraxanthin only accumulated at the final stage of fruit maturation (stage 4) (being 12.4 μg/gDW, 69.05 μg/gDW, and 7.07 g/gDW, respectively).The accumulation of lutein decreased from stage 1 (41.16 g/gDW) to stage 3 (17.43 μg/gDW), and then increased again at stage 4 (21.89 μg/gDW). The α-carotene content decreased from stage 1 (12.67 μg/gDW) to stage 4 (2.48 μg/gDW). The β-carotene content, including 13Z-β-carotene, e-β-carotene, 9Z-β-carotene, varied during fruit maturation from stage 1 to stage 4. 13Z-β-carotene and 9Z-β-carotene contents decreased from stage 1 (4.44 μg/gDW, and 3.84 μg/gDW, respectively) to stage 2 (1.79 μg/gDW, and 2.03 g/gDW, respectively), and then further increased at stage 3 (1.93 μg/gDW, and 2.23 g/gDW, respectively) and stage 4 (5.89 μg/gDW, and 2.83 μg/gDW, respectively). In contrast E-β-carotene content decreased from stage 1 (26.13 g/gDW) to stage 3 (10.84 g/gDW), and then increased at stage 4 (28.08 μg/g DW).
4. Discussion
In our study, two genes encoding geranylgeranyl pyrophosphate synthase (McGGPPS1 and McGGPPS2) were identified. However, the expression level of McGGPPS2 was higher than McGGPPS1 in all different M. charantia organs. Moreover, the gene expression level of McGGPPS2 correlated with the accumulation of total carotenoids being highest in old leaves, followed by young leaves, male flowers, female flowers, stems, and lowest in roots. This result suggests that McGGPPS2 is involved in the production of carotenoids in M. charantia. This result was similar to what has previously been described in a range of other plants. For example, a statistical analysis of the correlation between carotenoid content and candidate gene transcript levels in a maize germ plasam collection revealed that the expression of only one of the three plastidial GGPPS enzymes (GGPPS1) positively correlated with endosperm carotenoid content (Vallabhaneni and Wurtzel, 2009). Similarly, the Arabidopsis genome contains a family of 12 genes encoding putative GGPPS isoforms (Lange and Ghassemian, 2003); however, only one Arabidopsis gene, encoding GGPPS1, has been suggested to be involved in the production of carotenoids (Meier et al., 2011).
The first committed step in plant carotenoid biosynthesis is the synthesis of phytoene from GGPP; this reaction is catalyzed by phytoene synthase (PSY). The number of PSY genes differs in different plants; Arabidopsis (Ruiz-Sola and Rodriguez-Concepcion, 2012), and Tartary buckwheat (Tuan et al., 2013) contain one PSY gene, tobacco (Busch et al., 2002) and tomato (Giorio et al., 2008) contain two PSY genes, and maize (Li et al., 2008), rice (Welsch et al., 2008), and cassava (Arango et al., 2010) contain three PSY genes. Some PSY isoforms are involved in the biosynthesis of carotenoids in other tissues. For example, tomato PSY1 participates in the production of carotenoids in fruit, the seed endosperm (maize PSY1), or the root (maize and rice PSY3). In our study, a single M. charantia PSY gene (McPSY) was expressed in virtually all tissues, and positively correlated with McGGPPS1 expression levels showing high expression levels in young leaves and stage 4 fruit compared to other organs (Fig. 2, Fig. 3).
The α-carotene content was highest in old leaves, followed by young leaves and lowest in roots, and this correlated well with McLCYE1 expression across the different M. charantia organs. Similarly the expression level of McLCYB correlated well with β-carotene content. Accordingly, we propose that McLCYE1 and McLCYB are involved in the production of carotenoids in M. charantia.
During the maturation of M. charantia fruit, of the contents of zeaxanthin and β-cryptoxanthin, was higher. When the color of the fruit changed from green (stage 1-3) to orange (stage 4), the concomitant increase in zeaxanthin and β-cryptoxanthin content is likely brought about because of increased McCHXB expression. However, in other organs, such as the roots, which have high McCHXB expression, the levels of β-cryptoxanthin and zeaxanthin content did not correspond with McCHXB expression. Based on this, we proposed that McCHXB causes accumulation of β-cryptoxanthin and zeaxanthin in fruit. We also suggested that there is another isoform of CHXB in M. charantia, which has direct influence on the synthesis of β-cryptoxanthin and zeaxanthin in the different organs. It is also evidenced that the two isoforms of the CHXB gene was specific for chromoplasts in flowers and/or the fruit (Galpaz et al., 2006).
The expression of McZDS was highest in stage 4 fruit and corresponded with lycopene accumulation. Similarly, the α-carotene content decreased from stage 1 to stage 4 and corresponded with the level of McLCYE2 expression in fruit. Based on this, we suggested that McZDS and McLCYE2cause the accumulation of lycopene and α-carotene, respectively, in the fruit of M. charantia. In this study, we have also reported the levels of McZEP gene expression in different organs and, unlike for several of the previously discussed genes, we found that carotenoid content did not correlate with the levels of McZEP gene expression (Clotault et al., 2008).
In our study, carotenoids were generally found to be at high levels in organs that are exposed to direct light (e.g. leaves, flowers, and stems). This finding is similar to previous studies conducted in M. charantia (Tuan et al., 2011a, Tuan et al., 2011b), Allium sativum (Tuan et al., 2012), and Chinese cabbage (Fraser et al., 2000) which also found that the levels of carotenoids were very high in the leaves, but very low in the organs not exposed to light (e.g. roots).
5. Conclusion
Summary, eight genes related to the carotenoid biosynthetic pathway in bitter melon including McGGPPS1, McGGPPS2, McZDS, McLCYB, McLCYE1, McLCYE2, McCHXB, and McZEP were identified and their expression levels and carotenoid accumulation were high in leaves and comparatively lower in roots. Alternatively, the obtained results have guided to propose that the putative McGGPPS2, McZDS, McLCYB, McLCYE1, McLCYE2 and McCHXB might be the key factors for controlling carotenoid content in M. charantia. Collectively, the obtained results were evidenced and were marked as a first step toward possible bioengineering of medically important M. charantia crops to increase the yield of these medically important carotenoids. In the future, the production rate of carotenoids might be increased by the bioengineering tools.
Acknowledgments
This Research supported by grants from the Research Program of KAERI, Republic of Korea The authors extend their sincere appreciation to the International Scientific Partnership Program ISPP at King Saud University for funding this research work through ISPP#0021.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.sjbs.2017.11.039.
Contributor Information
Naif Abdullah Al-Dhabi, Email: naldhabi@ksu.edu.sa.
Sang Un Park, Email: supark@cnu.ac.kr.
Appendix A. Supplementary material
References
- Arango J., Wust F., Beyer P., Welsch R. Characterization of phytoene synthases from cassava and their involvement in abiotic stress-mediated responses. Planta. 2010;232:1251–1262. doi: 10.1007/s00425-010-1250-6. [DOI] [PubMed] [Google Scholar]
- Beyer P., Al-Babili S., Ye X.D., Lucca P., Schaub P., Welsch R., Potrykus I. Golden rice: introducing the beta-carotene biosynthesis pathway into rice endosperm by genetic engineering to defeat vitamin a deficiency. J. Nutr. 2002;132:506s–510s. doi: 10.1093/jn/132.3.506S. [DOI] [PubMed] [Google Scholar]
- Busch M., Seuter A., Hain R. Functional analysis of the early steps of carotenoid biosynthesis in tobacco. Plant Physiol. 2002;128:439–453. doi: 10.1104/pp.010573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao C.Y., Sung P.J., Wang W.H., Kuo Y.H. Anti-inflammatory effect of Momordica charantia in sepsis mice. Molecules. 2014;19:12777–12788. doi: 10.3390/molecules190812777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clotault J., Peltier D., Berruyer R., Thomas M., Briard M., Geoffriau E. Expression of carotenoid biosynthesis genes during carrot root development. J. Exp. Bot. 2008;59:3563–3573. doi: 10.1093/jxb/ern210. [DOI] [PubMed] [Google Scholar]
- Fraser P.D., Pinto M.E., Holloway D.E., Bramley P.M. Technical advance: application of high-performance liquid chromatography with photodiode array detection to the metabolic profiling of plant isoprenoids. Plant J. 2000;24:551–558. doi: 10.1046/j.1365-313x.2000.00896.x. [DOI] [PubMed] [Google Scholar]
- Galpaz N., Ronen G., Khalfa Z., Zamir D., Hirschberg J. A chromoplast-specific carotenoid biosynthesis pathway is revealed by cloning of the tomato white-flower locus. Plant Cell. 2006;18:1947–1960. doi: 10.1105/tpc.105.039966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorio G., Stigliani A.L., D'Ambrosio C. Phytoene synthase genes in tomato (Solanum lycopersicum L.) - new data on the structures, the deduced amino acid sequences and the expression patterns. FEBS J. 2008;275:527–535. doi: 10.1111/j.1742-4658.2007.06219.x. [DOI] [PubMed] [Google Scholar]
- Howe J.A., Tanumihardjo S.A. Evaluation of analytical methods for carotenoid extraction from biofortified maize (Zea mays sp.) J. Agric. Food Chem. 2006;54:7992–7997. doi: 10.1021/jf062256f. [DOI] [PubMed] [Google Scholar]
- Hyun T.K., Rim Y., Jang H.J., Kim C.H., Park J., Kumar R., Lee S., Kim B.C., Bhak J., Nguyen-Quoc B. De novo transcriptome sequencing of momordica cochinchinensis to identify genes involved in the carotenoid biosynthesis. Plant Mol. Biol. 2012;79:413–427. doi: 10.1007/s11103-012-9919-9. [DOI] [PubMed] [Google Scholar]
- Khoo H.E., Prasad K.N., Kong K.W., Jiang Y.M., Ismail A. Carotenoids and their isomers: color pigments in fruits and vegetables. Molecules. 2011;16:1710–1738. doi: 10.3390/molecules16021710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange B.M., Ghassemian M. Genome organization in arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol. Biol. 2003;51:925–948. doi: 10.1023/a:1023005504702. [DOI] [PubMed] [Google Scholar]
- Li F., Vallabhaneni R., Yu J., Rocheford T., Wurtzel E.T. The maize phytoene synthase gene family: overlapping roles for carotenogenesis in endosperm, photomorphogenesis, and thermal stress tolerance. Plant Physiol. 2008;147:1334–1346. doi: 10.1104/pp.108.122119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Zhang S., Zhang S., Li F., Zhang H., Cheng F., Wu J., Wang X., Sun R. Carotenoid biosynthetic genes in Brassica rapa: comparative genomic analysis, phylogenetic analysis, and expression profiling. BMC Genomics. 2015;16:492. doi: 10.1186/s12864-015-1655-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw C.C., Huang H.C., Hsiao P.C., Zhang L.J., Lin Z.H., Hwang S.Y., Hsu F.L., Kuo Y.H. 5 beta,19-epoxycucurbitane triterpenoids from Momordica charantia and their anti-inflammatory and cytotoxic activity. Planta Med. 2015;81:62–70. doi: 10.1055/s-0034-1383307. [DOI] [PubMed] [Google Scholar]
- Meier S., Tzfadia O., Vallabhaneni R., Gehring C., Wurtzel E.T. A transcriptional analysis of carotenoid, chlorophyll and plastidial isoprenoid biosynthesis genes during development and osmotic stress responses in arabidopsis thaliana. BMC Syst. Biol. 2011;5:77. doi: 10.1186/1752-0509-5-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messias D.S.R., Galli V., dos Anjos e Silva S.D., Rombaldi C.V. Carotenoid biosynthetic and catabolic pathways: gene expression and carotenoid content in grains of maize landraces. Nutrients. 2014;6:546–563. doi: 10.3390/nu6020546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namitha K.K., Archana S.N., Negi P.S. Expression of carotenoid biosynthetic pathway genes and changes in carotenoids during ripening in tomato (Lycopersicon esculentum) Food Funct. 2011;2:168–173. doi: 10.1039/c0fo00169d. [DOI] [PubMed] [Google Scholar]
- Ruiz-Sola M.A., Rodriguez-Concepcion M. Carotenoid biosynthesis in arabidopsis: a colorful pathway. Arabidopsis Book. 2012;10:e0158. doi: 10.1199/tab.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan P.A., Kim J.K., Kim H.H., Lee S.Y., Park N.I., Park S.U. Carotenoid accumulation and characterization of cdnas encoding phytoene synthase and phytoene desaturase in garlic (Allium sativum) J. Agric. Food Chem. 2011;59:5412–5417. doi: 10.1021/jf2009827. [DOI] [PubMed] [Google Scholar]
- Tuan P.A., Kim J.K., Lee J., Park W.T., Kwon do Y., Kim Y.B., Kim H.H., Kim H.R., Park S.U. Analysis of carotenoid accumulation and expression of carotenoid biosynthesis genes in different organs of chinese cabbage (Brassica rapa subsp. Pekinensis) EXCLI J. 2012;11:508–516. [PMC free article] [PubMed] [Google Scholar]
- Tuan P.A., Kim J.K., Park N.I., Lee S.Y., Park S.U. Carotenoid content and expression of phytoene synthase and phytoene desaturase genes in bitter melon (Momordica charantia) Food Chem. 2011;126:1686–1692. doi: 10.1016/j.foodchem.2010.12.058. [DOI] [PubMed] [Google Scholar]
- Tuan P.A., Kim Y.B., Kim J.K., Arasu M.V., Al-Dhabi N.A., Park S.U. Molecular characterization of carotenoid biosynthetic genes and carotenoid accumulation in Scutellaria baicalensis Georgi. EXCLI J. 2015;14:146–157. doi: 10.17179/excli2014-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan P.A., Park S.U. Molecular cloning and characterization of cdnas encoding carotenoid cleavage dioxygenase in bitter melon (Momordica charantia) J. Plant Physiol. 2013;170:115–120. doi: 10.1016/j.jplph.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Tuan P.A., Thwe A.A., Kim Y.B., Kim J.K., Kim S.J., Lee S., Chung S.O., Park S.U. Effects of white, blue, and red light-emitting diodes on carotenoid biosynthetic gene expression levels and carotenoid accumulation in sprouts of tartary buckwheat (Fagopyrum tataricum gaertn.) J. Agric. Food Chem. 2013;61:12356–12361. doi: 10.1021/jf4039937. [DOI] [PubMed] [Google Scholar]
- Vallabhaneni R., Wurtzel E.T. Timing and biosynthetic potential for carotenoid accumulation in genetically diverse germplasm of maize. Plant Physiol. 2009;150:562–572. doi: 10.1104/pp.109.137042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch R., Wust F., Bar C., Al-Babili S., Beyer P. A third phytoene synthase is devoted to abiotic stress-induced abscisic acid formation in rice and defines functional diversification of phytoene synthase genes. Plant Physiol. 2008;147:367–380. doi: 10.1104/pp.108.117028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.