Abstract
Microbial secondary metabolites represent a rich source of valuable compounds with a variety of applications in medicine or agriculture. Effective exploitation of this wealth of chemicals requires the functional expression of the respective biosynthetic genes in amenable heterologous hosts. We have previously established the TREX system which facilitates the transfer, integration and expression of biosynthetic gene clusters in various bacterial hosts. Here, we describe the yTREX system, a new tool adapted for one-step yeast recombinational cloning of gene clusters. We show that with yTREX, Pseudomonas putida secondary metabolite production strains can rapidly be constructed by random targeting of chromosomal promoters by Tn5 transposition. Feasibility of this approach was corroborated by prodigiosin production after yTREX cloning, transfer and expression of the respective biosynthesis genes from Serratia marcescens. Furthermore, the applicability of the system for effective pathway rerouting by gene cluster adaptation was demonstrated using the violacein biosynthesis gene cluster from Chromobacterium violaceum, producing pathway metabolites violacein, deoxyviolacein, prodeoxyviolacein, and deoxychromoviridans. Clones producing both prodigiosin and violaceins could be readily identified among clones obtained after random chromosomal integration by their strong color-phenotype. Finally, the addition of a promoter-less reporter gene enabled facile detection also of phenazine-producing clones after transfer of the respective phenazine-1-carboxylic acid biosynthesis genes from Pseudomonas aeruginosa. All compounds accumulated to substantial titers in the mg range. We thus corroborate here the suitability of P. putida for the biosynthesis of diverse natural products, and demonstrate that the yTREX system effectively enables the rapid generation of secondary metabolite producing bacteria by activation of heterologous gene clusters, applicable for natural compound discovery and combinatorial biosynthesis.
Keywords: Synthetic biology, Yeast recombinational cloning, Tn5 transposition, Heterologous gene cluster expression, Secondary metabolite production, Pseudomonas putida
Abbreviations: yTREX, yeast recombinational cloning-enabled pathway transfer and expression tool; CIS, cluster integration site; pig, prodigiosin biosynthesis genes; vio, violacein biosynthesis genes; phz, phenazine biosynthesis genes; PCA, phenazine-1-carboxylic acid
1. Introduction
Microorganisms exhibit an immense biosynthetic capability for the production of valuable compounds offering versatile bioactivities, applicable in sectors like human medicine or agriculture [1]. A vast multitude of gene sequences has become available, in which more and more gene clusters are identified that encode secondary metabolite biosynthetic pathways [2]. One key technology enabling effective exploration of the encoded chemical wealth is the functional expression in amenable heterologous hosts [3]. Therefore, increasing efforts are put in the development of diverse genetic systems for accessing natural compounds by heterologous expression of biosynthetic genes and gene clusters [4]. Here, the critical determinants for successful heterologous compound production currently represent (i) the efficient gene cluster cloning and (ii) the functional expression of all pathway genes requiring an appropriate host strain which offers a genetic codon usage compatible with the genes to be expressed, can provide metabolic precursors and is tolerant against putative toxicity of heterologous biosynthetic products [5].
Regarding cloning, restriction-independent methods have proven to be a key enabling technology in natural product research [6]. Phage enzyme-dependent recombination in E. coli and in vitro homology-based methods have been developed and successfully applied for gene cluster cloning and engineering [7], [8], [9], [10], and recently, increasing use of yeast-based recombination cloning highlights the value of such approaches [6], [11].
Regarding heterologous expression, the number of sophisticated tools refined for the use in different hosts increases likewise. Here, especially P. putida KT2440 represents one promising host for heterologous secondary metabolite biosynthesis [12], [13]. Valuable tools include diverse vector and promoter systems enabling calibrated gene expression [14], [15]. Furthermore, we have previously established the pathway transfer and expression (TREX) system which allows the straight-forward generation of stable expression strains in different species, employing random chromosomal integration of the heterologous gene cluster in the host by transposition and bidirectional expression of all biosynthetic genes by T7 RNA polymerase [16]. Moreover, we recently applied the tool for random integration of a unidirectional gene cluster into the chromosome of P. putida which resulted in strains exhibiting effective heterologous expression via a chromosomal promoter [17]. Nonetheless, the lack of suitable advanced cloning and expression systems for gene clusters was identified as one drawback hampering the broad utilization of this bacterium [5]. Thus, novel easy to apply tools for the fast activation of heterologous pathways in the host are needed.
Here, we describe the yTREX system, a new tool which like TREX enables the transfer, chromosomal integration and expression of gene clusters, but is enhanced by the key feature of fast one-step yeast recombinational cloning. As an application example, we moreover present the rapid generation of P. putida secondary metabolite production strains based on yTREX-mediated random chromosomal integration of biosynthetic genes. Employing the biosynthetic gene clusters of prodigiosin from Serratia marcescens, of violacein from Chromobacterium violaceum, and of phenazines from Pseudomonas aeruginosa, we demonstrate the system's applicability not only for i) the rapid transfer of metabolic pathways to the host, but also for ii) straightforward pathway engineering via targeted gene cluster re-design, and iii) the implementation of reporter systems for indication of biosynthetic gene expression.
2. Materials and methods
2.1. Bacterial and yeast strains
Escherichia coli strains DH5α [18] and S17-1 [19], applied for cloning and conjugation, were cultivated in shake flasks under constant agitation (120 rpm) at 37 °C in LB liquid medium (Carl Roth®, Karlsruhe, Germany: 10 g/L tryptone, 5 g/L yeast extract, 10 g/L sodium chloride) or on LB agar plates (15 g/L Agar-Agar, Kobe I, Carl Roth®, Karlsruhe, Germany). Pseudomonas putida KT2440 [20] was cultivated in shake flasks under constant agitation (120 rpm) at 30 °C in either LB or TB liquid medium (Terrific broth, modified, Carl-Roth Karlsruhe, Germany: 12 g/L Casein, enzymatically digested, 24 g/L yeast extract, 9.4 g/L dipotassium phosphate, 2.2 g/L monopotassium phosphate, 4 mL/L glycerol) or on LB agar plates. Antibiotics were added to the culture medium to the following final concentrations [μg/mL]: E. coli: kanamycin 50, tetracycline 10; P. putida: tetracycline 50, irgasan 25.
Saccharomyces cerevisiae strain VL6-48 [21], [22] (ATCC® MYA-3666, LGC Standards GmbH, Wesel, Germany) was cultivated in shake flasks under constant shaking (120 rpm) at 30 °C in YPD (yeast peptone dextrose) liquid medium (10 g/L yeast extract, 20 g/L peptone, 20 g/L glucose) [23] or on YPD agar plates (20 g/L Agar-Agar). Plasmid-bearing VL6-48 were cultivated in SD (synthetic dextrose) minimal medium [23] without uracil (SD-Ura), composed of 6.7 g/L Yeast-Nitrogen-Base (without Amino Acids) (Carl Roth®, Karlsruhe, Germany), 1.926 g/L Kaiser Uracil drop-out mixture [24] (Formedia™, Norfolk, United Kingdom), 20 g/L glucose, and for SD-Ura agar plates 20 g/L agar.
2.2. Standard molecular genetic methods
Standard recombinant DNA techniques were performed essentially as previously described [25]. Plasmid DNA was amplified using E. coli DH5α and isolated with innuPREP Plasmid Mini Kit (Analytik Jena AG, Jena, Germany). Genomic DNA was isolated with DNeasy Blood & Tissue Kit (Quiagen® GmbH, Hilden, Germany). Restriction endonuclease enzymes and phosphatase FastAP (ThermoFisher Scientific GmbH, Walkham, USA) were applied according to the manufacturer's instructions. DNA fragments were purified using innuPREP DOUBLEpure Kit (Analytik Jena AG, Jena, Germany). Commercial services were engaged for DNA synthesis of primer oligonucleotides and yTREX cassettes, and for DNA sequencing (Eurofins Genomics GmbH, Ebersberg, Germany).
2.3. Yeast recombinational cloning
DNA fragment generation: DNA fragments with homology arms ranging from 28 bp to 52 bp were generated by PCR or restriction hydrolysis and, if necessary, purified by agarose gel electrophoresis and spin column purification. Vector fragments were furthermore treated with FastAP. All DNA fragments were combined to a total volume of 20 μL (5 μL yTREX vector, 15 μL insert solution; amounts of all DNA parts were adjusted to 0.1–1 μg) and co-transformed into S. cerevisiae for assembly by yeast recombinational cloning.
Yeast transformation: S. cerevisiae cells were transformed with DNA fragments using a protocol based on the high-efficiency yeast transformation LiAc/SS carrier DNA/PEG method developed by Gietz and Schiestl in the most recent version [26], with the following adaptions: Prior to transformation, cells were cultivated in YPD liquid medium (Gietz step 1/3). Instead of water, 1 mL 100 mM LiAc was used to wash yeast cells and for aliquotation (Gietz step 7). Heatshock was performed for 30 min at 42 °C followed by a regeneration phase of 30 min at 30 °C under constant shaking (120 rpm) (Gietz step 9). Subsequently, transformation mixtures were plated on SD-Ura medium for selection and incubated for 2 days at 30 °C (Gietz step 12) [26].
Plasmid isolation: For plasmid isolation from yeast cells, colonies were transferred from plates to 1 mL SD-Ura medium and incubated in FlowerPlates® (m2p-labs GmbH, Baesweiler, Germany) at 30 °C under constant shaking (1400 rpm) over night. Cells were harvested by centrifugation and solved in resuspension buffer from innuPREP Plasmid Mini Kit, supplemented with 12.5 U of Zymolyase (Zymo Research Europe GmbH, Freiburg, Germany), incubated for 2 h at 37 °C, pelleted again and subjected to plasmid isolation using innuPREP Plasmid Mini Kit. Plasmid DNA was finally introduced into E. coli DH5α for amplification, isolation and analysis for correct assembly.
2.4. Generation of the yTREX vector
The vector yCP50-poly [27] (ATCC® 87555™, LGC Standards GmbH, Wesel, Germany) was employed as a starting point to create the shuttle vector backbone, which was modified by deletion of the β-lactamase gene (AmpR) together with the MCS, and integration of a kanamycin resistance providing gene (KmR) in exchange. Therefore, the vector yCP50-poly was hydrolyzed using PvuI and EheI. The kanamycin resistance gene aphII from plasmid pK18 was obtained as 1055 bp amplicon by PCR, with oligos AD9+AD10 which provided appropriate complementary overhangs to vector sequences. Both DNA fragments were assembled via yeast recombinational cloning, creating vector yCP50-poly-KmR. The new L-yTREX and R-yTREX cassettes described in this study were obtained as one synthetic DNA fragment from eurofins Genomics, bearing appropriate cloning overhangs for introduction into the vector. After hydrolysis of yCP50-poly-KmR using SpeI, the yTREX cassettes were inserted via yeast recombinational cloning, resulting in the yTREX vector. Correct integration of the aphII gene and both yTREX cassettes, respectively, was verified by restriction analysis and sequencing using the oligonucleotides AD39, AD40, AD41, and AD42.
2.5. Assembly of yTREX constructs
For the generation of any yTREX construct, the yTREX vector was linearized by hydrolyzation at the so-called CIS (cluster integration site) between the CIS1 and CIS2 sequences using homing endonuclease I-SceI prior to assembly cloning, exposing the partial I-SceI recognition sequence as well as the CIS sequences at either end of the linear DNA fragment. The CIS1 and CIS2 sequences were used for homologous recombination to integrate inserts without reconstitution of the I-SceI site. For our first application, they were designed to match the sequences up- and downstream of prodigiosin biosynthetic genes in plasmid pPIG [16].
The prodigiosin pig gene cluster from Serratia marcescens W838 was obtained as one 23.7 kb fragment of vector pPIG by restriction digestion with ScaI and XbaI. In this fragment, the sequences up- and downstream of pigA and pigN, respectively, were compatible with the ends of the linearized yTREX vector, promoting the integration by yeast recombinational cloning of a 21.1 kb fragment containing pigA-pigN for construction of vector yTREX-pig.
The violacein vio gene cluster from Chromobacterium violaceum ATCC 12472 was obtained as a PCR product using vector pAra-vio [28] as template, with appropriate homology arms matching the CIS sequences added in primer sequences for integration into the yTREX vector. Different variants of the vio gene cluster were generated by assembly of PCR products from specific vio genes, harboring suitable homology arms for reconstitution of engineered operons and integration at CIS1 and CIS2 of the yTREX vector, added in the primers. Therefore, the 7.4 kb full length operon vioABCDE, as well as 5.7 kb vioABC, 4.4 kb vioAB, and 0.6 kb vioE were PCR amplified and employed to assemble vectors yTREX-vio, yTREX-vioABCE and yTREX-vioABE.
The phenazine-1-carboxylic (PCA) acid biosynthesis encoding phz genes were obtained as a 6.4 kb PCR product using the plasmid pUC18-pyo containing the Pseudomonas aeruginosa PAO1 phzMA1B1C1D1E1F1G1S genes (S. Thies, unpublished) as template, using primers with homology arms to the yTREX vector CIS1 sequence on the 5′-end and to another PCR product containing the lacZ gene on the 3′-end. The 3.1 kb lacZ PCR product was generated using vector pRcExpII2-YF1-FixJ-PFixK2lacZ (A. Loeschcke, unpublished) as template, adding homologous sequences to the phz fragment at the 5′-end and to the CIS2 sequence at the 3′-end of lacZ. Thereby, one-step assembly of the phz genes together with lacZ as a unidirectional synthetic operon into the yTREX vector by yeast recombinational cloning was enabled, producing vector yTREX-phzA-G-lacZ.
All used oligonucleotides are listed in Table S1, all generated PCR products are summarized in Table S2. Correct insertion of the gene clusters in final yTREX constructs was verified by restriction analysis and sequencing using the oligonucleotides AD90 and AD93. All plasmids used in this study are listed in Table S3, respective plasmid maps are shown in Fig. S1.
2.6. Generation of yTREX production strains
To generate P. putida production strains, gene cluster-carrying yTREX constructs were transformed into E. coli S17-1 and further transferred to P. putida KT2440 via conjugation as previously described [16]. Since yTREX constructs do not replicate in P. putida, positive selection for strains in which Tn5 transposition of the recombinant yTREX transposon including the respective gene clusters occurred, could be conducted by using LB medium supplemented with tetracycline. In addition, 25 μg/mL irgasan were added to prevent E. coli growth. Among exconjugants, production strains were identified visually as colored colonies on agar plates. Prodigiosin producers showed a red colored phenotype. Violacein and deoxyviolacein or prodeoxyviolacein/deoxychromoviridans producers exhibited a violet or green color, respectively. In case of phenazine-1-carboxylic acid, selection medium after conjugation was additionally supplemented with 0.3 mM X-Gal (stock solution: 50 mM in DMF), and expression strains were identified by blue color due to β-galactosidase activity.
2.7. Production and analysis of prodigiosin
Precultures of P. putida strains producing prodigiosin were grown in TB medium and used to inoculate production cultures starting with a cell density of OD650 = 0.05 in FlowerPlates® with 800 μL TB medium per well. Cultures were incubated at 30 °C under constant shaking at 1400 rpm in a ThermoMixer® C (Eppendorf AG, Hamburg, Germany) or in a TiMix MTP-shaker with TH Incubationhood (EB GmbH, Hechingen, Germany). After 24 h of cultivation, cell material of 100 μL culture was harvested by centrifugation. Extraction was performed with 1 mL acidified ethanol (4% of 1 M HCl), extracts were cleared by centrifugation, and prodigiosin content was determined spectrophotometrically based on molar extinction coefficient (ε535 [M−1cm−1] = 139,800) in acidified ethanol as described before [17]. HPLC-PDA analysis of prodigiosin containing extracts was performed on an LC-10Ai series (Shimadzu Deutschland GmbH, Duisburg, Germany) equipped with SPD-M10Avp photodiode array detector (PDA) and installed with LabSolutions program. Samples (10 μL injection volume) were injected onto an Accucore™ C18 HPLC column (100 × 2.1 mm i.d., 2.6 μm particle size, 80 Å pores; ThermoFisher Scientific GmbH, Walkham, USA), equipped with a guard column filled with the same material. The column oven temperature was maintained at 30 °C. Solvent flow rate was 1 mL/min. The mobile phase consisted of water (A) and acetonitrile (B), both containing 0.1% formic acid. Starting conditions of 11.5 min analysis runs were 5% B for 1.5 min, followed by a gradient over 5.5 min to 98% B. This proportion was maintained for 2 min, before returning to 5% B within 0.5 min which was maintained for re-equilibration for 2 min. Chromatograms were recorded at 535 nm. The PDA detector was used to scan wavelengths from 425 to 625 nm. Prodigiosin signals (min 6.6/λmax 534) were identified based on spectral properties in comparison to published data [17].
2.8. Production and analysis of violaceins
P. putida strains producing violacein and pathway-related metabolites were cultivated and harvested like prodigiosin producing strains. Cells were extracted with 1 mL ethanol and the extract was cleared by centrifugation. The content of violacein with byproduct deoxyviolacein was estimated spectrophotometrically using the molar extinction coefficient of violacein (ε575 [M−1cm−1] = 25,400) in ethanol [29]. In strains expressing engineered pathway variants that synthesized mainly deoxyviolacein or prodeoxyviolacein together with deoxychromoviridans in mixtures with more compounds, product accumulation was evaluated as specific absorption at 575 nm, and 610 nm, respectively, and was in addition roughly estimated using the molar extinction coefficient of deoxyviolacein (ε575 [M−1cm−1] = 15,700) [29] as an approximation. HPLC-PDA analysis of bacterial extracts (10 μL injection volume for violacein, 35 μL for deoxyviolacein or prodeoxyviolacein and deoxychromoviridans) was performed based on published methods [30], [31] with the same instrument and method as described for prodigiosin. Chromatograms were recorded at 600 nm. The PDA detector was used to scan wavelengths from 315 to 715 nm for spectral analysis. Compounds were identified based on their relative retention times and spectral properties (violacein, min 5.9/λmax 374, 571; deoxyviolacein, min 6.3/λmax 372, 562; prodeoxyviolacein, min 5.6/λmax 418, 610; deoxychromoviridans, min 6.9/λmax 354, 479, 643) in comparison to published data [30], [31].
2.9. Phenazine production and analysis
Cultures of P. putida strains carrying phz genes were inoculated from LB medium-grown precultures to a starting cell density of OD600 = 0.05 in Round Well Plates (m2p-labs GmbH, Baesweiler, Germany) with 1 mL TB medium per well. Cultures were incubated at 30 °C under constant shaking (900 rpm). After 24 h of cultivation, cells were removed by centrifugation. The entire culture supernatant was acidified with 100 μL of 6 M HCl, and extracted twice with 500 μL ethyl acetate by vortexing for 15 s. Ethyl acetate fractions were combined and evaporated (Concentrator 5301, Eppendorf AG, Hamburg, Germany), at 45 °C for 1 h. For discrimination between non-producing and producing strains, the residue was solved in 150 μL water/acetonitrile (95:5), both containing 0.1% formic acid, under shaking at 50 °C for 1 h and subjected to HPLC analysis (10 μL injection volume). Since residual insoluble yellow material was observed to remain adhered to the plastic material, quantitative analysis was performed using 1 mL ethanol to re-dissolve the residue after ethyl acetate evaporation. HPLC analysis of appropriately diluted extracts (10 μl injection volume) was performed with the same instrument as described above. Here, a C30-reverse-phase HPLC column (250 × 4.6 mm i.d., 5 μm particle size, YMC-Europe GmbH, Dinslaken, Germany), equipped with a guard column filled with the same material (20 mm × 4.0 mm i.d.), was used for analysis at 30 °C column oven temperature and 1 mL/min solvent flow rate. The mobile phase consisted of water (A) and acetonitrile (B), both containing 0.1% formic acid. Starting conditions of 30 min analysis runs were 5% B for 2.5 min, followed by a gradient to 98% B in 16.5 min. This proportion was maintained for 5 min, before returning to 5% B within 1 min which was maintained for re-equilibration for 5 min. Chromatograms were recorded at 366 nm. The PDA detector was used to scan wavelengths from 225 to 450 nm. PCA signals (min 18.1/λmax 249, 370) were identified by comparison to an authentic reference (Apollo Scientific Ltd, Cheshire UK) which was further employed for quantitative calibration in a dilution series from 0.008 to 0.25 mg/mL, enabling estimation of PCA production titers.
3. Results
3.1. yTREX design
The previously constructed TREX system allows for the transfer and expression of gene clusters in bacteria [16], [17], [32]: After labeling a gene cluster of interest with the TREX cassettes on a plasmid, this construct can be transferred to a Gram-negative bacterial host of choice where integration into the host chromosome is implemented by Tn5 transposition. Expression of the gene cluster can be realized by using T7 RNA polymerase promoters [16] or by a chromosomal promoter [17]. This system served as the starting point to develop the new yTREX system.
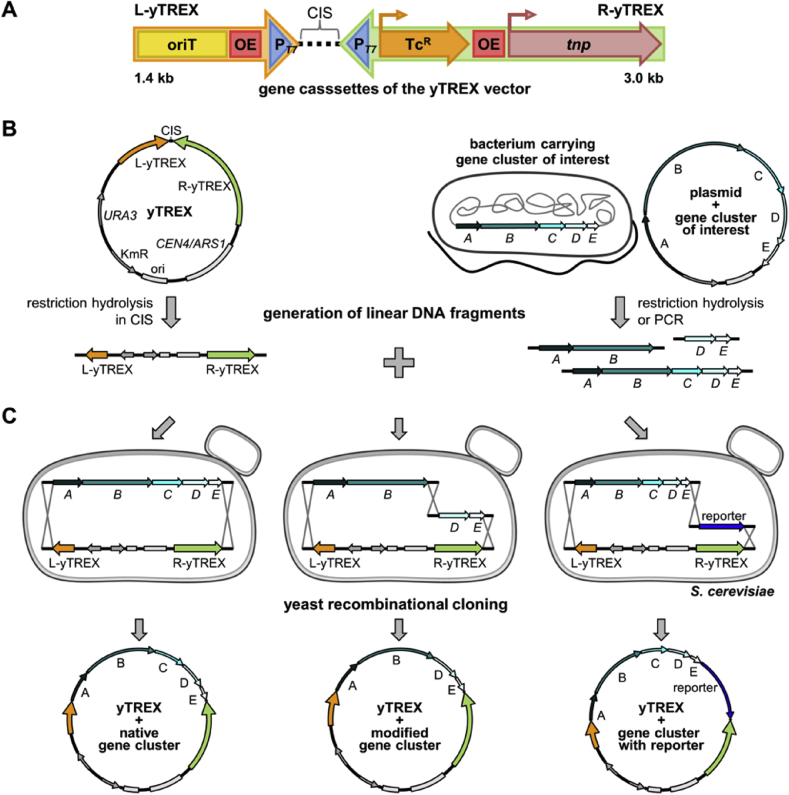
Since we identified the cloning step via conventional restriction-/ligation-based methods as a time consuming bottleneck of the procedure, hampering the straightforward application of the system, we exploited here the benefits of yeast homologous recombination. To this end, we designed an adapted version of the TREX system, based on an E. coli-yeast shuttle vector which carries adapted TREX cassettes, designated as yTREX cassettes, which convergently enclose a site for targeted integration of a gene cluster of interest (Fig. 1A). The functional elements of this construct shall allow the application procedure following the four steps of labeling, transfer, integration and expression (Fig. S2):
Fig. 1.
Schematic representation of yTREX cassettes and cloning options. (A) Composition of yTREX cassettes. The yTREX system consists of two gene cassettes, the L-yTREX (orange) and R-yTREX (green) cassette. TcR, tetracycline resistance gene; oriT, origin of transfer; OE, outside end of transposon Tn5; PT7, T7 bacteriophage promoter; tnp, Tn5 transposase gene. (B) Generation of linear DNA fragments for yeast recombinational yTREX cloning. The yTREX vector, which harbors the yTREX cassettes and elements for replication and selection in both E. coli and S. cerevisiae (CEN4/ARS1, S. cerevisiae origin of replication; URA3, pyrimidine ribonucleotide biosynthetic gene; ori, E. coli origin of replication; KmR, kanamycin resistance gene), can be linearized for yTREX construct assembly by I-SceI endonuclease restriction in the cluster integration site, or short CIS. A gene cluster of interest can be obtained from genomic or plasmid DNA via endonuclease restriction hydrolysis or PCR. (C) Yeast recombinational yTREX cloning options. Design of DNA fragments with appropriate homologous overhangs, which can be added by PCR primers, and transformation into yeast enables recombinational cloning of a gene cluster in its native form (left), as an engineered variant, e.g. with gene deletions (middle) or with additionally included elements like transcription reporter genes such as β-galactosidase encoding lacZ (right). Cloned yTREX vectors can be readily employed for transfer and integration of biosynthetic genes in a bacterial expression host.
Labeling: First, a gene cluster of interest is labeled with the yTREX cassettes, i.e. the cluster is cloned into the cluster integration site, or short CIS, of the yTREX vector, located in between the two yTREX cassettes (Fig. 1B). To enable the use of yeast recombinational cloning to accomplish this step, the yTREX vector backbone is based on E. coli-yeast shuttle vector yCP50-poly [27] in which the ampicillin resistance gene and MCS were replaced with a kanamycin resistance gene. The yTREX vector can be linearized at the CIS by restriction hydrolysis with endonuclease I-SceI for gene cluster incorporation. A gene cluster of interest can be obtained from a given source (e.g. plasmid or chromosomal DNA) via restriction or PCR. To facilitate immediate use of the yTREX construct for an initial proof-of-concept application, the CIS sequence was designed to match the sequences upstream and downstream of prodigiosin biosynthetic genes in plasmid pPIG [16] which can be isolated via restriction hydrolysis. Notably, effective recombination can be achieved with homologous regions of only about 30 bp length [33], so they can also easily be integrated into oligonucleotides used for PCR priming. An often exploited capability of yeast-based DNA assembly is the concerted recombination of several DNA fragments in one step, which enables to reassemble large clusters from several amplicons or to reorganize clustered genes [34]. By choice of amplicon and addition of appropriate overhangs, a gene cluster can thus be cloned in its native form or easily manipulated for diverse purposes (Fig. 1C). Options include the reorganization of transcriptional units within gene clusters to construct unidirectional arrangements or the targeted deletion of individual pathway genes. Moreover, additional features such as reporter genes can easily be added to a gene cluster of interest for indication of full length gene expression.
After assembly of DNA fragments, a uracil biosynthetic gene (URA3) included in the vector backbone facilitates selection of yeast clones carrying a yTREX construct in a strain with the respective auxotrophy. A kanamycin resistance gene (KmR) likewise allows selection of respective bacterial clones for amplification in E. coli. After cloning, the yTREX construct (Fig. S2A) carrying a gene cluster of interest enables conducting the following typical steps of the TREX application, i.e. transfer, integration and expression [16], [17].
Transfer: The construct is introduced into the host of choice, e.g. via chemical transformation or electroporation. To additionally enable conjugational transfer to a broad range of bacterial hosts irrespective of the plasmid size [35], the L-yTREX contains an origin of transfer (oriT) (Fig. S2B).
Integration: The gene cluster of interest is subsequently integrated into the bacterial host chromosome by random transposition, implemented by Tn5 transposase (tnp) [36] and respective OE-sequences in the yTREX cassettes. A narrow host range E. coli pMB1 origin of replication in the yTREX vector backbone and a tetracycline resistance gene in the R-yTREX cassette (TcR) enable direct selection for clones carrying the gene cluster and TREX elements after a transposition event. In this step, not only the oriT, but, in contrast to the initial setup [16], also the transposase gene is lost as they are not included in the transposon thus ruling out any chance of re-transposition after integration (Fig. S2C).
Expression: As reported for the TREX system, two strategies can be applied for expression. T7 RNA polymerase can be employed for gene expression from two T7 promoters (PT7) included in the yTREX cassettes to enable bidirectional expression of complex gene clusters composed of transcriptional units in different orientations [16]. This may potentially also be achieved by random-transposition mediated integration in proximity to two host promoters which are convergently oriented toward the integration locus. Alternatively, in the case of a gene cluster consisting of unidirectionally oriented genes, the random integration can evidentially result in gene expression from a chromosomal promoter [17] (Fig. S2D).
3.2. yTREX application strategy for metabolite production in P. putida
To demonstrate yTREX applicability, we used P. putida KT2440 as host for the expression of three known biosynthetic gene clusters. To assemble yTREX constructs via yeast recombinational cloning, gene clusters or fragments thereof, equipped with appropriate homologous sequences, were co-transformed into S. cerevisiae VL6-48 cells with the yTREX vector, previously linearized at the CIS with restriction endonuclease I-SceI. In all cases, yTREX constructs carrying unidirectional gene clusters were subsequently introduced into the host, aiming for expression by random transposition behind a sufficiently strong native promoter.
3.3. Proof of yTREX functionality by prodigiosin production
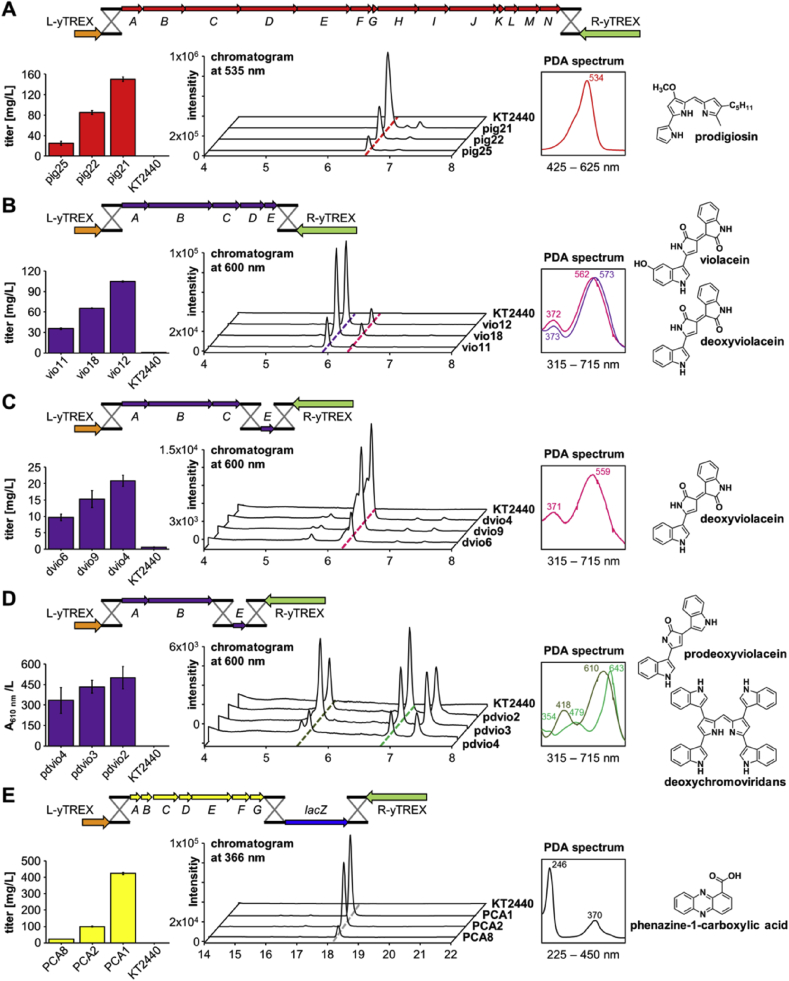
In order to first proof yTREX functionality we employed the native 21 kb prodigiosin (pig) gene cluster from Serratia marcescens which consists of 14 genes encoding the bifurcated tripyrrole biosynthetic pathway [37], [38] (Fig. S3), and was previously successfully expressed in the host using the initial TREX system [16], [17]. The prodiginine family has aroused interest, e.g. for their antibiotic and anticancer bioactivities [39], [40]. To assemble vector yTREX-pig, the pig gene cluster was obtained from plasmid pPIG [16] by restriction hydrolysis and transformed together with the linear yTREX vector into S. cerevisiae for recombinational cloning (Fig. 2A). Due to homology between the regions upstream of pigA and downstream of pigN, respectively, and the yTREX CIS1 and CIS2 sequences, the yTREX-pig construct was assembled carrying the 21.1 kb prodigiosin biosynthetic gene cluster consisting of pigA-pigN.
Fig. 2.
Heterologous metabolite production in P. putida via chromosomal integration of biosynthetic gene clusters. Recombinant transposon assembly schemes are shown together with metabolite production levels and HPLC-PDA analyses of prodigiosin (A), violaceins (B,C,D), and PCA (E) in extracts of production strains. Recombinant transposons consisting of yTREX elements and biosynthetic genes were assembled via yeast recombinational cloning as indicated, before yTREX-mediated integration in the P. putida chromosome. Metabolite production levels are given as titers or metabolite-specific absorption. HPLC chromatograms, recorded at appropriate wavelengths, are shown together with representative PDA-spectra corresponding to labeled peaks. Identified compounds are indicated.
The yTREX-pig plasmid was transferred into P. putida by conjugation, and strains containing the yTREX-pig transposon could be directly selected using tetracycline-containing medium. Clones with the gene cluster integrated behind a native promoter, where biosynthetic genes were expressed and the respective pathway was thus established, were identified by their red colored phenotype. This phenotype was caused by prodigiosin accumulation reaching titers between 25 and 150 mg/L during cultivation in liquid medium, as determined photometrically and via HPLC-PDA analysis in three exemplary clones pig21, pig22 and pig25 (Fig. 2A), thus verifying yTREX functionality. In addition, the suitability of P. putida as a potential prodigiosin production platform was corroborated.
3.4. Application of yTREX for violacein pathway engineering
Since in a biosynthetic gene cluster, often each gene reflects a part of the biosynthetic pathway, manipulations can direct biosynthetic routes to a certain desired product. To demonstrate yTREX applicability for rapid pathway engineering, the well-described 7.4 kb violacein biosynthesis (vio) gene cluster from Chromobacterium violaceum was used which consists of five genes encoding the branched multi-step bisindole biosynthetic pathway [30], [41] (Fig. S4). Violacein and pathway-related compounds, here summarized as violaceins, have gained interest e.g. for their antibiotic and anticancer bioactivities [42]. The gene cluster was employed in its native form comprising genes vioABCDE and two engineered variants vioABCE (lacking the vioD encoded oxygenase, thus directing the pathway to deoxyviolacein formation) and vioABE (lacking both vioD and vioC encoded oxygenases, thus blocking any enzymatic conversion of the precursor protodeoxyviolaceinic acid and directing the pathway to prodeoxyviolacein). The full length gene cluster was obtained by PCR using genomic DNA as template and oligonucleotides adding a sequence homologous to the CIS of the yTREX vector at each end of the cluster to construct yTREX-vio. To assemble deletion variants, the vio cluster was amplified via PCR in two segments of vioABC and vioE, as well as vioAB and vioE, respectively. Suitable overhangs were introduced at the 5′-end of vioA and the 3′-end of vioE to facilitate recombination into the CIS sequences of the yTREX vector. In addition, PCR fragments were designed to enable recombination between the 3′-end of vioC or vioB, respectively, and the 5′-end of vioE (Fig. 2B).
After yeast recombinational cloning, the resulting vectors yTREX-vio, yTREX-vioABCE and yTREX-vioABE were introduced into P. putida. Clones expressing the gene clusters were identified by their colored phenotype. Notably, isolated strains plated on selection medium exhibited occasional clones with a differing coloration which may point to a certain genetic instability of the violacein gene cluster in the host. Such clones were omitted in the further characterization. In selected P. putida strains vio11, vio12 and vio18 with the full length cluster, photometric and HPLC-PDA analysis verified accumulation of violacein together with smaller amounts of its natural byproduct deoxyviolacein at titers between 36 and 105 mg/L (Fig. 2B). As expected, introduction of the engineered gene cluster variant vioABCE redirected the pathway to the formation of deoxyviolacein as the final product, resulting in titers ranging between 10 and 21 mg/L produced by selected clones dvio4, dvio6 and dvio9 (Fig. 2C). In strains pdvio2, pdvio3 and pdvio4 with the gene cluster variant vioABE, neither violacein nor deoxyviolacein but prodeoxyviolacein was detected as expected. In addition, further pathway metabolites derived from prodeoxyviolacein by non-enzymatic reactions such as deoxychromoviridans appeared in increased amounts as a result of this rerouting (Fig. 2D). Titers may be roughly estimated to be in the mg/L range. These results thus demonstrate the applicability of the yTREX system for easy gene cluster manipulation for e.g. pathway engineering. Although we did not perform a thorough characterization of the newly constructed strains, the biosynthesis of violaceins did not result in any conspicuous phenotype – other than coloration – nor did we observe inhibited growth or negative influence on biomass formation during our experiments, thus suggesting P. putida as a suitable host strain for the production of these compounds.
3.5. yTREX phenazine production strain identification via reporter expression
To demonstrate feasibility of reporter gene employment for straightforward expression strain identification, the phenazine biosynthesis (phz) gene cluster from Pseudomonas aeruginosa encoding the multi-step conversion of chorismic acid to diverse phenazine compounds [43] (Fig. S5) was employed together with the β-galactosidase gene from E. coli as a reporter. Both the main end product pyocyanin and other pathway-related phenazines including phenazine-1-carboxylic acid, or short PCA, display antibacterial and antifungal properties [44], [45], [46]. The genes phzA-G, spanning 6.4 kb and encoding PCA biosynthesis, as well as the promoter-less β-galactosidase encoding lacZ gene (3.1 kb) were PCR amplified. Employed oligonucleotides enabled yeast homologous recombination, integrating the phz genes, followed by the lacZ gene into the CIS of the yTREX vector, generating plasmid yTREX-phzA-G-lacZ (Fig. 2E).
Transfer of this vector to P. putida enabled generation of clones carrying the yTREX-phz-lacZ transposon, among which those with the gene cluster being transcribed by a native promoter could be identified by their color on appropriate X-Gal containing β-galactosidase indicator medium. Fifteen strains showing a colored phenotype indicating expression and four non-colored strains were selected for further analysis. Conspicuously, strains expressing the reporter were noted to exhibit impaired growth which was not further characterized, but might point toward oxidative stress evoked by electrochemical properties of produced phenazines [47]. HPLC-PDA measurements verified PCA accumulation only in the colored ones, with levels between 24 and 424 mg/L, as exemplarily determined for selected P. putida clones PCA1, PCA2 and PCA8 (Fig. 2E).
Feasibility of reporter gene employment for straightforward expression strain identification was thus demonstrated. In addition, P. putida was verified as a suitable host for PCA biosynthesis, despite certain constraints in strain viability during production which may be overcome by adaption of cultivation conditions [48].
4. Discussion
We demonstrate here a straightforward approach for the rapid generation of bacterial secondary metabolite producers, using the yTREX system to facilitate yeast recombinational cloning and gene cluster transposition into the chromosome of P. putida KT2440 as heterologous host. This strategy yielded production strains for six different bioactive secondary metabolites of the prodiginine, violacein and phenazine family, achieved by transfer and engineering of three gene cluster-encoded biosynthetic pathways from different original hosts.
The yTREX system is uniquely easy in application. It is based on only one vector that is used for gene cluster cloning and transfer of biosynthetic genes. The employment of yeast recombinational cloning not only enabled one-step reconstitution of whole gene clusters from several PCR fragments but also allowed the introduction of targeted manipulations with ease, as here demonstrated by adaption of the vio gene cluster and thus re-routing of the respective violacein pathway. Therefore, the DNA assembly method is a highly useful means for gene cluster cloning and rearrangement. At the same time, the technique is well-described [34] and easily established in molecular biology laboratories. Recent application examples include yeast recombinational cloning of the 36 kb grecocycline biosynthetic gene cluster for functional expression in Streptomyces albus [49], or cloning and refactoring of silent gene clusters encoding biosynthesis of lazarimides A, B and C (22.5 kb), and taromycin A (67 kb) for activation in S. albus and Streptomyces coelicolor, respectively [50], [51]. Assembly of an artificial bacterial chromosome in yeast documents the potential of this method [52], and its increasingly widespread use, including the protocols presented in this study, shall catalyze advances in natural products research. We demonstrate here the implementation of yeast recombinational cloning for the assembly of plasmids carrying native or engineered gene clusters of interest, together with the yTREX cassettes.
The genetic elements of the yTREX cassettes enable conjugational plasmid transfer and random chromosomal integration via Tn5 transposition. Remarkably, random transposition as a key feature of the procedure not only leads to the effective generation of strains stably carrying the gene cluster of interest in their genome, but also to the regular emergence of individual P. putida clones exhibiting immediate expression of biosynthetic genes by chromosomal promoters. The principle of exploiting randomly targeted chromosomal locations in P. putida with promoters suitable for gene cluster expression was previously established with the prodigiosin gene cluster [17] and is in the present study verified as broadly applicable for diverse gene clusters. Note that success of this approach can only be expected if the gene cluster consists of unidirectionally organized genes. Consequently, complex gene clusters with multiple transcription units arranged in different orientations need to be re-constructed into unidirectional organization in the cloning step to follow this strategy. Using the here presented tool, the respective changes in cluster architecture can be introduced easily via one-step yeast recombinational cloning.
Effective metabolite production after introduction of biosynthetic genes at a random position in the bacterial host chromosome requires straightforward identification of expressing clones. Here, in the case of red prodiginines and purple or green violaceins, clones expressing inserted genes were readily identified by their color. Moreover, as demonstrated by addition of promoter-less lacZ at the 3′-end of the phenazine gene cluster, a reporter for gene cluster transcription can be included, enabling facile identification of clones exhibiting gene cluster expression from a chromosomal promoter. The implementation of other reporter genes, e.g. if an additional enzyme activity is undesirable, is likewise feasible due to the versatile adaptability of the yTREX tool.
The here presented strategies allow for rapid generation and identification of recombinant P. putida expression strains in under two weeks (Table S4). Especially for novel biosynthetic pathways, the approach of random chromosomal integration of biosynthesis genes together with a transcription reporter should enable fast pathway investigation, and additionally provide insights if the host and novel pathway are compatible for metabolite production. P. putida represents an especially promising host for heterologous secondary metabolite production [12], [13], [53]. In agreement with previous reports, we corroborate the bacterium's versatile applicability for the biosynthesis of diverse natural products. Maximal titers obtained here for prodigiosin (150 mg/L) and PCA (424 mg/L) are in similar ranges and higher as in previous studies reporting on heterologous metabolite production in the host (94 mg/L prodigiosin in P. putida KT2440 [17], 27 mg/L PCA in P. putida KT2440 [48]). Maximal violacein titers found here (105 mg/L) are tenfold higher than those obtained previously by vio gene expression in P. putida KT2440 (10 mg/L [54]), whereas those of deoxyviolacein (21 mg/L) are two orders of magnitude below previously reported possible outcomes (1.5 g/L deoxyviolacein in P. putida mt-2 [55]). In addition, we describe here for the first time the biosynthesis of prodeoxyviolacein together with deoxychromoviridans in P. putida KT2440. In all cases, very different product levels were found in the individual strains tested, presumably resulting from individual transposon integration sites with differentially active promoters upstream. Notably, here described product accumulation represents the result from chromosomal gene cluster integration and standard cultivation without any further optimization approaches regarding the strains or production processes. With other host systems, such optimization has been undertaken successfully, producing metabolite titers at gram scale using e.g. native prodigiosin producer Serratia marcescens [56], and Corynebacterium glutamicum [57] or E. coli [58] as heterologous hosts for production of violaceins, as well as titers of 660 mg/L PCA using native producer Pseudomonas chlororaphis [59]. Further studies are necessary to elucidate the potential of P. putida-based metabolite production in comparison to these platforms.
Our findings show that the yTREX tool contributes to overcoming the lack of advanced expression strategies for biosynthetic gene clusters which is currently accepted as a major bottleneck limiting the versatile applicability of P. putida [5]. This tool, applied with the here presented strategy of exploiting chromosomal promoters for gene cluster expression may thus in the future serve to identify further compounds for which this bacterium represents an ideal production host. In principle, utilization of conjugational transfer and Tn5 transposition should allow the application of here presented yTREX strategies likewise with other host organisms.
6. Conclusions
We show here that the yTREX system combines vector assembly via homologous recombination in yeast, enabling fast and easy one-step gene cluster cloning, advantageous e.g. in pathway engineering or reporter integration, with the features of the TREX system facilitating transfer, integration and expression. As demonstrated in this study, effective gene cluster expression can be achieved in P. putida simply by random genomic integration of unidirectional gene clusters, a strategy allowing a highly straightforward workflow and fast assessment of the host's suitability for the production of a certain compound. In summary, the presented synthetic biology tool effectively enables the rapid generation of secondary metabolite producing bacteria by activation of heterologous gene clusters, applicable for natural compound discovery and combinatorial biosynthesis.
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Acknowledgments
The scientific activities of the Bioeconomy Science Center were financially supported by the Ministry of Innovation, Science and Research of the German federal state of North Rhine-Westphalia MIWF within the framework of the NRW Strategieprojekt BioSC (No. 313/323-400-00213). Moreover, we gratefully acknowledge the MIWF and the Heinrich-Heine-University Düsseldorf for a scholarship within the CLIB Graduate Cluster Industrial Biotechnology for AD. The authors gratefully acknowledge Dennis Binder and Fabienne Hilgers for very fruitful discussions on violacein biosynthesis.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.synbio.2017.11.001.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Katz L., Baltz R.H. Natural product discovery: past, present, and future. J Ind Microbiol Biotechnol. 2016;43:155–176. doi: 10.1007/s10295-015-1723-5. [DOI] [PubMed] [Google Scholar]
- 2.Ziemert N., Alanjary M., Weber T. The evolution of genome mining in microbes – a review. Nat Prod Rep. 2016;0:1–18. doi: 10.1039/c6np00025h. [DOI] [PubMed] [Google Scholar]
- 3.Wohlleben W., Mast Y., Stegmann E., Ziemert N. Antibiotic drug discovery. Microb Biotechnol. 2016;9:541–548. doi: 10.1111/1751-7915.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breitling R., Takano E. Synthetic biology advances for pharmaceutical production. Curr Opin Biotechnol. 2015;35:46–51. doi: 10.1016/j.copbio.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang M.M., Wang Y., Ang E.L., Zhao H. Engineering microbial hosts for production of bacterial natural products. Nat Prod Rep. 2016;33:963–987. doi: 10.1039/c6np00017g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim E., Moore B.S., Yoon Y.J. Reinvigorating natural product combinatorial biosynthesis with synthetic biology. Nat Chem Biol. 2015;11:649–659. doi: 10.1038/nchembio.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu J., Wenzel S.C., Perlova O., Wang J., Gross F., Tang Z. Efficient transfer of two large secondary metabolite pathway gene clusters into heterologous hosts by transposition. Nucleic Acids Res. 2008;36:1–14. doi: 10.1093/nar/gkn499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu J., Bian X., Hu S., Wang H., Huang F., Seibert P.M. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat Biotechnol. 2012;30:440–446. doi: 10.1038/nbt.2183. [DOI] [PubMed] [Google Scholar]
- 9.Niu G., Li L., Wei J., Tan H. Cloning, heterologous expression, and characterization of the gene cluster required for gougerotin biosynthesis. Chem Biol. 2013;20:34–44. doi: 10.1016/j.chembiol.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 10.Gibson D.G. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 2011;498:349–361. doi: 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nah H.-J., Pyeon H.-R., Kang S.-H., Choi S.-S., Kim E.-S. Cloning and heterologous expression of a large-sized natural product biosynthetic gene cluster in Streptomyces species. Front Microbiol. 2017;8:1–10. doi: 10.3389/fmicb.2017.00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nikel P.I., Chavarría M., Danchin A., de Lorenzo V. From dirt to industrial applications: Pseudomonas putida as a synthetic biology chassis for hosting harsh biochemical reactions. Curr Opin Chem Biol. 2016;34:20–29. doi: 10.1016/j.cbpa.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 13.Loeschcke A., Thies S. Pseudomonas putida—a versatile host for the production of natural products. Appl Microbiol Biotechnol. 2015;99:6197–6214. doi: 10.1007/s00253-015-6745-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zobel S., Benedetti I., Eisenbach L., de Lorenzo V., Wierckx N., Blank L.M. A Tn7-based device for calibrated heterologous gene expression in Pseudomonas putida. ACS Synth Biol. 2015;12:1341–1352. doi: 10.1021/acssynbio.5b00058. [DOI] [PubMed] [Google Scholar]
- 15.Martínez-García E., Aparicio T., Goñi-Moreno A., Fraile S., De Lorenzo V. SEVA 2.0: an update of the Standard European Vector Architecture for de-/re-construction of bacterial functionalities. Nucleic Acids Res. 2015;43:D1183–D1189. doi: 10.1093/nar/gku1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loeschcke A., Markert A., Wilhelm S., Wirtz A., Rosenau F., Jaeger K.E. TREX: a universal tool for the transfer and expression of biosynthetic pathways in bacteria. ACS Synth Biol. 2013;2:22–33. doi: 10.1021/sb3000657. [DOI] [PubMed] [Google Scholar]
- 17.Domröse A., Klein A.S., Hage-Hülsmann J., Thies S., Svensson V., Classen T. Efficient recombinant production of prodigiosin in Pseudomonas putida. Front Microbiol. 2015;6:1–10. doi: 10.3389/fmicb.2015.00972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 19.Simon R., Priefer U., Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat Biotechnol. 1983;1:784–791. [Google Scholar]
- 20.Nelson K.E., Weinel C., Paulsen I.T., Dodson R.J., Hilbert H., Martins dos Santos V.A.P. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol. 2002;4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- 21.Kouprina N., Annab L., Graves J., Afshari C., Barrett J.C., Resnick M.A. Functional copies of a human gene can be directly isolated by transformation-associated recombination cloning with a small 3′ end target sequence. Proc Natl Acad Sci U. S. A. 1998;95:4469–4474. doi: 10.1073/pnas.95.8.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noskov V., Kouprina N., Leem S.-H., Koriabine M., Barrett J.C., Larionov V. A genetic system for direct selection of gene-positive clones during recombinational cloning in yeast. Nucleic Acids Res. 2002;30:1–7. doi: 10.1093/nar/30.2.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- 24.Kaiser C., Michaelis S., Mitchell A. Cold Spring Harbor Laboratory Press; 1994. Methods in yeast genetics. [Google Scholar]
- 25.Green M.R., Sambrook J. fourth ed. vol. 1. Cold Spring Harbor Laboratory Press; 2012. (Molecular cloning: a laboratory manual). [Google Scholar]
- 26.Gietz R.D., Schiestl R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- 27.Wang H., Stillman D.J. Transcriptional repression in Saccharomyces cerevisiae by a SIN3-LexA fusion protein. Mol Cell Biol. 1993;13:1805–1814. doi: 10.1128/mcb.13.3.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Binder D., Bier C., Grünberger A., Drobietz D., Hage-Hülsmann J., Wandrey G. Photocaged arabinose: a novel optogenetic switch for rapid and gradual control of microbial gene expression. ChemBioChem. 2016;17:296–299. doi: 10.1002/cbic.201500609. [DOI] [PubMed] [Google Scholar]
- 29.Rodrigues A.L., Göcke Y., Bolten C., Brock N.L., Dickschat J.S., Wittmann C. Microbial production of the drugs violacein and deoxyviolacein: analytical development and strain comparison. Biotechnol Lett. 2012;34:717–720. doi: 10.1007/s10529-011-0827-x. [DOI] [PubMed] [Google Scholar]
- 30.Sánchez C., Braña A.F., Méndez C., Salas J.A. Reevaluation of the violacein biosynthetic pathway and its relationship to indolocarbazole biosynthesis. ChemBioChem. 2006;7:1231–1240. doi: 10.1002/cbic.200600029. [DOI] [PubMed] [Google Scholar]
- 31.Lee M.E., Aswani A., Han A.S., Tomlin C.J., Dueber J.E. Expression-level optimization of a multi-enzyme pathway in the absence of a high-throughput assay. Nucleic Acids Res. 2013;41:10668–10678. doi: 10.1093/nar/gkt809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katzke N., Knapp A., Loeschcke A., Drepper T., Jaeger K.-E. Novel tools for the functional expression of metagenomic DNA. Metagenomics Methods Protoc. Methods Mol Biol. 2017;1539:159–196. doi: 10.1007/978-1-4939-6691-2_10. [DOI] [PubMed] [Google Scholar]
- 33.Oldenburg K.R., Vo K.T., Michaelis S., Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997;25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shao Z., Luo Y., Zhao H. Rapid characterization and engineering of natural product biosynthetic pathways via DNA assembler. Mol Biosyst. 2011;7:1056–1059. doi: 10.1039/c0mb00338g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brigulla M., Wackernagel W. Molecular aspects of gene transfer and foreign DNA acquisition in prokaryotes with regard to safety issues. Appl Microbiol Biotechnol. 2010;86:1027–1041. doi: 10.1007/s00253-010-2489-3. [DOI] [PubMed] [Google Scholar]
- 36.Reznikoff W.S. Transposon Tn5. Annu Rev Genet. 2008;42:269–286. doi: 10.1146/annurev.genet.42.110807.091656. [DOI] [PubMed] [Google Scholar]
- 37.Williamson N.R., Fineran P.C., Leeper F.J., Salmond G.P.C. The biosynthesis and regulation of bacterial prodiginines. Nat Rev Microbiol. 2006;4:887–899. doi: 10.1038/nrmicro1531. [DOI] [PubMed] [Google Scholar]
- 38.Harris A.K.P., Williamson N.R., Slater H., Cox A., Abbasi S., Foulds I. The Serratia gene cluster encoding biosynthesis of the red antibiotic, prodigiosin, shows species- and strain-dependent genome context variation. Microbiology. 2004;150:3547–3560. doi: 10.1099/mic.0.27222-0. [DOI] [PubMed] [Google Scholar]
- 39.Pérez-Tomás R., Viñas M. New insights on the antitumoral properties of prodiginines. Curr Med Chem. 2010;17:2222–2231. doi: 10.2174/092986710791331103. [DOI] [PubMed] [Google Scholar]
- 40.Danevčič T., Borić Vezjak M., Tabor M., Zorec M., Stopar D. Prodigiosin induces autolysins in actively grown Bacillus subtilis cells. Front Microbiol. 2016;7:1–10. doi: 10.3389/fmicb.2016.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoshino T. Violacein and related tryptophan metabolites produced by Chromobacterium violaceum: biosynthetic mechanism and pathway for construction of violacein core. Appl Microbiol Biotechnol. 2011;91:1463–1475. doi: 10.1007/s00253-011-3468-z. [DOI] [PubMed] [Google Scholar]
- 42.Choi S.Y., Yoon K.H., Lee J Il, Mitchell R.J. Violacein: properties and production of a versatile bacterial pigment. Biomed Res Int. 2015:1–8. doi: 10.1155/2015/465056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mavrodi D.V., Bonsall R.F., Delaney S.M., Soule M.J., Phillips G., Thomashow L.S. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol. 2001;183:6454–6465. doi: 10.1128/JB.183.21.6454-6465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sudhakar T., Karpagam S. Int. Conf. Green technol. Environ. Conserv. GTEC-2011. 2011. Antifungal efficacy of pyocyanin produced from bioindicators of nosocomial hazards; pp. 224–229. [Google Scholar]
- 45.Upadhyay A., Srivastava S. Phenazine-1-carboxylic acid is a more important contributor to biocontrol Fusarium oxysporum than pyrrolnitrin in Pseudomonas fluorescens strain Psd. Microbiol Res. 2011;166:323–335. doi: 10.1016/j.micres.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 46.Zhang L., Tian X., Kuang S., Liu G., Zhang C., Sun C. Antagonistic activity and mode of action of phenazine-1-carboxylic acid, produced by marine bacterium Pseudomonas aeruginosa PA31x, against Vibrio anguillarum in vitro and in a zebrafish in vivo model. Front Microbiol. 2017;8:1–11. doi: 10.3389/fmicb.2017.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Price-Whelan A., Dietrich L.E.P., Newman D.K. Rethinking “secondary” metabolism: physiological roles for phenazine antibiotics. Nat Chem Biol. 2006;2:71–78. doi: 10.1038/nchembio764. [DOI] [PubMed] [Google Scholar]
- 48.Schmitz S., Nies S., Wierckx N., Blank L.M., Rosenbaum M.A. Engineering mediator-based electroactivity in the obligate aerobic bacterium Pseudomonas putida KT2440. Front Microbiol. 2015;6:1–13. doi: 10.3389/fmicb.2015.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bilyk O., Sekurova O.N., Zotchev S.B., Luzhetskyy A. Cloning and heterologous expression of the grecocycline biosynthetic gene cluster. PLoS One. 2016;11:1–17. doi: 10.1371/journal.pone.0158682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamanaka K., Reynolds K.A., Kersten R.D., Ryan K.S., Gonzalez D.J., Nizet V. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc Natl Acad Sci. 2014;111:1957–1962. doi: 10.1073/pnas.1319584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Montiel D., Kang H.-S., Chang F.-Y., Charlop-Powers Z., Brady S.F. Yeast homologous recombination-based promoter engineering for the activation of silent natural product biosynthetic gene clusters. Proc Natl Acad Sci U. S. A. 2015;112:8953–8958. doi: 10.1073/pnas.1507606112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gibson D.G., Benders G.A., Axelrod K.C., Zaveri J., Algire M.A., Moodie M. One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc Natl Acad Sci U. S. A. 2008;105:20404–20409. doi: 10.1073/pnas.0811011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nikel P.I., Martínez-García E., de Lorenzo V. Biotechnological domestication of pseudomonads using synthetic biology. Nat Rev Microbiol. 2014;12:368–379. doi: 10.1038/nrmicro3253. [DOI] [PubMed] [Google Scholar]
- 54.Zhang J.J., Tang X., Zhang M., Nguyen D., Moore B.S. Broad-host-range expression reveals native and host regulatory elements that influence heterologous antibiotic production in Gram-negative bacteria. MBio. 2017;8 doi: 10.1128/mBio.01291-17. e01291–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xing X, Jiang P. Recombinat bacteria for producing deoxyviolacein and uses thereof. United States patent US 20110183384A1. 2011 Jul 28.
- 56.Chen W.-C., Yu W.-J., Chang C.-C., Chang J.-S., Huang S.-H., Chang C.-H. Enhancing production of prodigiosin from Serratia marcescens C3 by statistical experimental design and porous carrier addition strategy. Biochem Eng J. 2013;78:93–100. [Google Scholar]
- 57.Sun H., Zhao D., Xiong B., Zhang C., Bi C. Engineering Corynebacterium glutamicum for violacein hyper production. Microb Cell Fact. 2016;15:1–9. doi: 10.1186/s12934-016-0545-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodrigues A.L., Becker J., de Souza Lima A.O., Porto L.M., Wittmann C. Systems metabolic engineering of Escherichia coli for gram scale production of the antitumor drug deoxyviolacein from glycerol. Biotechnol Bioeng. 2014;111:2280–2289. doi: 10.1002/bit.25297. [DOI] [PubMed] [Google Scholar]
- 59.Shen X., Wang Z., Huang X., Hu H., Wang W., Zhang X. Developing genome-reduced Pseudomonas chlororaphis strains for the production of secondary metabolites. BMC Genomics. 2017;18:715. doi: 10.1186/s12864-017-4127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.