Abstract
Tree peony, one of the most valuable horticultural and medicinal plants in the world, has to go through winter to break dormancy. Growing studies from molecular aspects on dormancy release process have been reported, but inadequate study has been done on miRNA-guided regulation in tree peony. In this study, high-throughput sequencing was employed to identify and characterize miRNAs in three libraries (6 d, 18 d and 24 d chilling treatments). There were 7,122, 10,076 and 9,097 unique miRNA sequences belonging to 52, 87 and 68 miRNA families, respectively. A total of 32 conserved miRNAs and 17 putative novel miRNAs were identified during dormancy release. There were 771 unigenes as potential targets of 62 miRNA families. Total 112 known miRNAs were differentially expressed, of which 55 miRNAs were shared among three libraries and 28 miRNAs were only found in 18 d chilling duration library. The expression patterns of 15 conserved miRNAs were validated and classified into four types by RT-qPCR. Combining with our microarray data under same treatments, five miRNAs (miR156k, miR159a, miR167a, miR169a and miR172a) were inversely correlated to those of their target genes. Our results would provide new molecular basis about dormancy release in tree peony.
Introduction
Tree peony (Paeonia suffruticosa Andrews) is one of the earliest and most well-known horticultural and medicinal plants in the world. Since flower buds of tree peony must go through a period of bud dormancy before bud sprouting in spring, the common adopted measure in agriculture is breaking dormancy by artificial chilling treatment for forcing culture. According to Lang and Martin1, the dormancy in tree peony belongs to endo-dormancy similar to some temperate fruit plants like grape, apple, peach, kiwifruit and so on. A sufficient chilling duration during winter is the main mean to break dormancy and induce growth in the following spring by appropriate warmer temperature2.
Release of dormancy physiological status was controlled through cooperation of large number of genes in woody plants3. Liu et al. obtained 190 significantly differentially expressed genes associated with bud dormancy in pear4. In Chinese cherry, totally 1,644 significantly differentially expressed genes were identified based on RNA-seq transcriptome5. To discover transcriptional pathways associated with dormancy release in Prunus persica, Fu et al. explored the chilling-dependent expressions of 11 genes involved in endoplasmic reticulum stress and the unfolded protein response signal pathways6. Yordanov et al. suggested that EARLY BUD-BREAK 1 (EBB1) have a major and integrative role in reactivation of meristem activity after winter dormancy in poplar trees7. In tea, sequence and gene ontology analysis of 3,500 clones associated with dormancy were analyzed8. In recent years, growing studies from molecular aspects on tree peony endo-dormancy release process have been reported, such as Huang et al. screened 31 unigenes associated with dormancy release in tree peony by SSH (suppression subtractive hybridization)9. Gai et al. obtained over 15,000 high quality unigenes by RNA sequencing during chilling requirement fulfillment through Roche 454 GS FLX platform10, of which 3,174 genes were significantly differentially expressed during endo-dormancy release in tree peony11. More recently, Zhang et al. obtained 20 differentially expressed protein spots (P < 0.05) during dormancy release by combination of two-dimensional gel electrophoresis (2-DE) and matrix-assisted laser desorption/ionisation time of flight/time of flight mass spectrometry (MALDI-TOF/TOF MS)12. In addition, Zhang et al. found a MADS-box member (SUPPRESSOR OF OVEREXPRESSION OF CONSTANS, PsSOC1) that not only accelerates flowering, but also promotes dormancy release in tree peony13. However, endo-dormancy mechanisms in tree peony are still unclear despite many efforts.
Eukaryotic gene expression is mainly regulated at the transcriptional and post-transcriptional levels. MicroRNAs (miRNAs) are a specific class of small non-coding RNA of commonly 19–24 nucleotides involved in post-transcriptional gene expression regulation14. The mature miRNAs negatively regulate gene expression through complementary binding to the opening reading frame (ORF) or UTR regions of specific target genes. In plants, miRNAs generally interact with their targets through near-perfect complementarily, which leads to gene silencing by endonucleolytic cleavage or translational inhibition15–17. Recent studies indicate that miRNAs play important roles in plant developments including organ separation, leaf development and polarity, lateral root formation, floral organ identity and reproduction, etc.18–21. Zhang et al. identified differentially expressed miRNAs responding to cold stress in tea22. Jeyaraj et al. analyzed the expression pattern of tea miRNAs in active and dormant bud using stem-loop pulse RT-qPCR method23. In poplar, ptr-miR169 was found to repress ptrHAP2 at the level of post-transcription during vegetative bud dormancy period24. In tree peony, some of conserved and novel miRNAs were identified under copper stress25. However, no miRNAs have been reported especially during chilling endo-dormancy release in tree peony.
In this study, we aimed to identify and characterize miRNAs by high-throughput sequencing technology in tree peony during the period of bud dormancy release after 6 d, 18 d and 24 d chilling requirement fulfilling, which included three physiological status, endo-dormancy, endo-dormancy release and eco-dormancy10. Our results increase the available information on miRNA-guided regulation mechanism and physiological changes during chilling induced dormancy release in tree peony.
Results
Deep sequencing of Paoenia ostii sRNAs
To investigate small RNA expression profiles in Paoenia ostii during physiological dormancy stages based on the results of morphologic observations11, three small RNA libraries of flower buds were constructed after 6 d, 18 d and 24 d chilling enduring. For each library, small RNAs were collected, pooled together and sequenced. A total of 19,762,599 reads with lengths of 16 bp to 30 bp were obtained from the three libraries, and average 3.8 million (range: 2.69–5.05 million) clean small RNA reads were acquired from each library after removing adapters and low-quality reads. The average number of unique reads per library was 2.06 million ranging from 1.63 to 2.68 million (Table 1). Most obtained sRNA sequences were 21–24 nt in all of the three libraries, of which 24 nt long sRNAs were the most abundant, accounting for approximately 52.3% on average (Fig. 1).
Table 1.
Statistics of small RNA sequence reads.
| Different treatments Title |
6 d | 18 d | 24 d | |||
|---|---|---|---|---|---|---|
| Number | percent | Number | percent | Number | percent | |
| Total Tags number | 5,338,004 | 100% | 8,092,580 | 100% | 6,332,015 | 100% |
| Average quality < 13 Tags | 1,469,084 | 27.52% | 1,577,909 | 19.50% | 1,224,509 | 19.34% |
| Length < 16 | 74,930 | 1.4% | 204,694 | 2.53% | 212,955 | 3.36% |
| Length > 30 | 615,543 | 11.53% | 427,666 | 5.28% | 507,468 | 8.01% |
| Clean number | 2,686,857 | 50.33% | 5,047,088 | 62.37% | 3,770,180 | 59.54% |
| Unique number | 1,629,348 | 30.52% | 2,683,551 | 33.16% | 1,895,066 | 29.93% |
Figure 1.
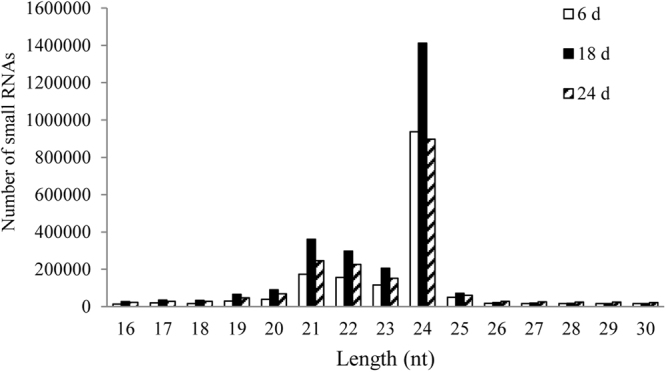
Length distribution of small RNAs in three libraries from tree peony buds after 6 d, 18 d and 24 d chilling treatments.
Clean data were searched against Rfam databases to annotate sRNAs, and known miRNAs were identified by alignment to sequences in miRBase 20.0 with no mismatch. siRNAs, ribosomal RNAs (rRNAs), tRNAs, snRNAs and snoRNAs were annotated by BLASTn to NCBI Genbank database and Rfam database. In order to eliminate the possibility of degraded mRNAs in three libraries, we aligned them through intron/exon alignment with unigenes in tree peony cDNA libraries11. The remaining unannotated sRNAs were used to predict novel miRNAs and potential miRNA seeds (Table 2). It is noticeable that the miRNAs represented 19.62% of the total sRNA in 6 d chilling treatments, but only 14.01% and 14.78% in 18 d and 24 d chilling treatments, which may as a result of many genes associated with endo- and eco-dormancy release are activated during dormancy release. There are about more 9,000 unique miRNAs at the physiological stage of dormancy release and eco-dormancy than at the status of dormancy, which indicates that the miRNA populations in flower buds after dormancy release are more diversified, as well as biological processes are more complex.
Table 2.
Annotations of sRNAs against Rfam database and Unigenes.
| Type | 6 d | 18 d | 24 d | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unique | Percent (%) | Total | Percent (%) | Unique | Percent (%) | Total | Percent (%) | Unique | Percent (%) | Total | Percent (%) | |
| Exon_antisense | 94,368 | 32.25 | 260,542 | 26.46 | 137,004 | 26.46 | 493,402 | 29.67 | 118,471 | 29.84 | 419,569 | 26.77 |
| Exon_sense | 84,011 | 28.71 | 173,996 | 17.67 | 124,042 | 17.67 | 371,326 | 22.33 | 102,616 | 25.84 | 244,283 | 15.58 |
| miRNA | 7,122 | 2.434 | 193,210 | 19.62 | 10,076 | 19.63 | 232,971 | 14.01 | 9,097 | 2.29 | 231,665 | 14.78 |
| rRNA | 59,554 | 20.35 | 171,166 | 17.38 | 80,480 | 17.39 | 280,605 | 16.87 | 98,087 | 24.70 | 390,132 | 24.89 |
| tRNA | 11,327 | 3.87 | 47,986 | 4.87 | 15,948 | 4.87 | 44,297 | 2.66 | 15,367 | 3.87 | 51,773 | 3.3 |
| snoRNA | 8,995 | 3.07 | 26,961 | 2.73 | 16,174 | 2.74 | 55,757 | 3.35 | 12,911 | 3.25 | 57,613 | 3.68 |
| snRNA | 4,485 | 1.53 | 7,182 | 0.72 | 9,327 | 0.73 | 16,811 | 1.01 | 6,373 | 1.61 | 11,609 | 0.74 |
| unannotated | 22,737 | 7.77 | 103,457 | 10.5 | 45,640 | 10.51 | 167,553 | 10.01 | 34,122 | 8.59 | 160,894 | 10.26 |
| Total | 285,997 | 100 | 984,500 | 100 | 429,269 | 100 | 1,662,722 | 100 | 388,585 | 100.00 | 1,567,538 | 100 |
Nucleotide Preference of 19–24 nucleotide small RNAs
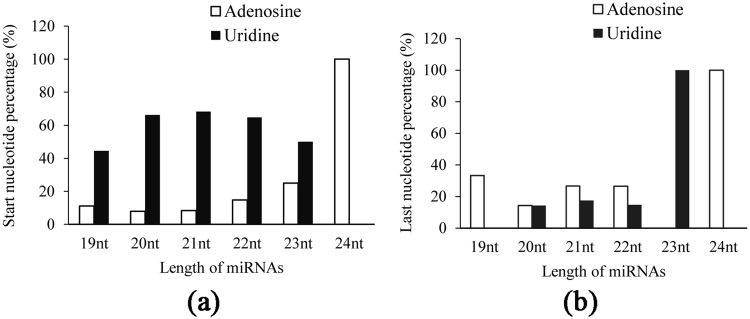
Previous studies have shown that most miRNA sequences start with uridine (U), whereas the majority of 24-nucleotide siRNAs have adenosine (A) as their 5′ first nucleotide in plants26–29. In our result, the same trends were observed among cloned tree peony small RNAs, about 67.1% of small RNAs sequences started with uridine, and all 24-nt siRNAs had start-nucleotide preference with adenosine (Fig. 2b). Besides, we found that about 45.5% of total small RNAs also had a clear preference for adenosine as the last nucleotide, of which all 23-nt had a clear preference for uridine and all 24-nt for adenosine as the last nucleotide (Fig. 2b). In order to investigate whether the last nucleotide preference from 19 nt to 24 nt small RNAs also exist in other model plants like Arabidopsis, we downloaded 427 Arabidopsis small RNA deep sequencing datasets from miRBase database (http://www.mirbase.org/ftp.shtml) and analyzed their nucleotide compositions (Supplementary Figure S1). In all 427 Arabidopsis datasets, strong last nucleotide preference for uridine was observed in 23 and 24 nt small RNAs, indicating that difference of nucleotide preference might exist among species.
Figure 2.
Nucleotide preference of small RNAs. (a) Percentage of adenosine or uridine at the start position of 19 to 24-nucleotide (nt) small RNAs. (b) Percentage of adenosine or uridine at the last position of 19 to 24-nucleotide (nt) small RNAs.
Identification of conserved and novel miRNAs in tree peony
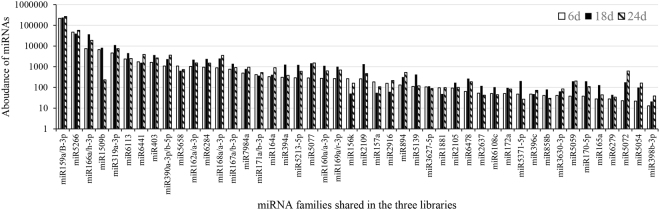
To identify conserved miRNAs in tree peony, but its genome is not available, known plant miRNAs (miRNA precursors and mature miRNAs) registered in miRBase 21.0 were used as reference for miRNA mapping. Clean data that aligned to known miRNAs allowing two mismatches and had no less than 5 reads per million (RPM) in at least one library were regarded as conserved miRNAs. In three libraries, total 112 known miRNAs belonging to 99 miRNA families were identified in the three libraries, of which there were 7,122, 10,076 and 9,097 unique miRNA sequences belonging to 52, 87 and 68 miRNA families, respectively (Additional file 1, Table 3). Of which, there was 32 conserved miRNAs (Table 4). In our data, 15 miRNAs sequences were found anchored in the 5p-arm and 17 miRNAs anchored in the 3p-arm (Addition file 1, Table 4). Unexpectedly, one less-conserved miRNA (miR5072) was obtained, which was previously found only in monocots30. Furthermore, there were 55 miRNAs belonging to 46 miRNA families shared in the three libraries (Fig. 3), and the most abundant miRNA identified by sequence homology was PsmiR159 with more than 500,000 actual sequencing reads, accounting for approximately 69% of the total conserved miRNA reads, following by PsmiR5266 with more than 100,000 actual sequencing reads, PsmiR166, PsmiR319, PsmiR1509 with more than 10,000 actual sequencing reads, and miR398 showed the minimum amount (Table 4). At the same time, the frequencies of miRNAs read varied from 8 (PsmiR2111a-5p) to 709,087 (PsmiR159a), which indicate that miRNAs displayed drastically different expression level in tree peony during dormancy release (Fig. 3). After normalization, more than half of the conserved miRNAs were less than 100 times. In addition, the relative abundance of certain members within same miRNA family varied widely (Table 4). For instance, the normalized number of PsmiR167a was 255, but that for PsmiR167b was only 12. Furthermore, the normalized reads of different members in three treatments were significant different, for example, the abundances of members in miR159 (miR159a and miR159b-3p) ranged from 18,716, 180 (6 d), 20,170, 496 (18 d) to 22,752 and 218 (24 d) reads in three libraries, respectively (Table 4). These results suggest that members showed different expression trends within same miRNA family, probably because their expressions are development-stage specific or either induced or suppressed during dormancy release in tree peony.
Table 3.
Summary of small RNA deep sequencing data.
| Libraries | No. of sequences generateda | No. of non-redundant sequencesa | No. of sequences with perfect matches to the miRBase | Unique miRNA number |
Family number |
|---|---|---|---|---|---|
| 6 d | 2,686,857 | 1,629,348 | 193,210 | 7,122 | 52 |
| 18 d | 5,047,088 | 2,683,551 | 232,971 | 10,076 | 87 |
| 24 d | 3,770,180 | 1,895,066 | 231,665 | 9,097 | 68 |
aLengths between 16–30 nt.
Table 4.
Known miRNAs identified from tree peony flower bud after different chilling treatments.
| Family | miRNA IDs | sequences | Actual sequencing reads /Normalized sequencing reads | zma | ath | osa | vvi | ptc | ||
|---|---|---|---|---|---|---|---|---|---|---|
| 6d | 18d | 24d | zma | ath | osa | vvi | ptc | |||
| well-conserved | ||||||||||
| miR156 | PsmiR156k | UGACGGAGAGAGAGAGCACAC | 263/23 | 51/4 | 161/14 | 0 | 0 | 0 | 0 | 0 |
| PsmiR156f-3p | GCUCACUCUCUAUCUGUCACC | 0/0 | 23/2 | 0/0 | 0 | 0 | 0 | 0 | 0 | |
| miR159 | PsmiR159a | UUUGGAUUGAAGGGAGCUCUA | 215,316/18,716 | 232,033/20,170 | 261,738/22,752 | + | 0 | + | 0 | 0 |
| PsmiR159b-3p | UAUUGGAGUGAAGGGAGCUCC | 2,067/180 | 5,704/496 | 2,505/218 | + | + | 0 | + | + | |
| miR160 | PsmiR160a | UGCCUGGCUCCCUGUAUGCCA | 216/19 | 1,071/93 | 577/50 | + | + | 0 | 0 | 0 |
| PsmiR160a-3p | GCGUAUGAGGAGCCAAGCAUA | 60/5 | 45/4 | 49/4 | 0 | 0 | 0 | 0 | 0 | |
| miR162 | PsmiR162-3p | UCGAUAAACCUCUGCAUCCA | 551/48 | 1,236/107 | 913/79 | NA | 0 | 0 | 0 | 0 |
| PsmiR162a | UCGAUAAACCUCUGCAUCCAG | 489/43 | 954/83 | 602/52 | 0 | 0 | 0 | 0 | 0 | |
| PsmiR162b | UGCCUGGCUCCCUGUAUGCCA | 112/10 | 4/0 | 740/64 | 0 | 0 | 0 | 0 | 0 | |
| miR164 | PsmiR164a | UGGAGAAGGGGAGCACGUGCA | 332/29 | 409/36 | 900/78 | + | ++ | + | 0 | 0 |
| PsmiR164b | UGGAGAAGCAGGGCACAUGCU | 2/0 | 10/1 | 740/64 | + | ++ | + | 0 | 0 | |
| PsmiR164c | UGGAGAAGCAGGGCACGUGCU | 524/46 | 8/1 | 2/0 | 0 | + | 0 | + | + | |
| miR166 | PsmiR166a | UCGGACCAGGCUUCAUUCCCC | 3,665/319 | 19,051/1,656 | 7,933/690 | NA | NA | NA | NA | NA |
| PsmiR166h-3p | UCUCGGACCAGGCUUCAUUCC | 3761/330 | 17,903/1,556 | 11,027/959 | 0 | 0 | 0 | 0 | 0 | |
| miR167 | PsmiR167a | UGAAGCUGCCAGCAUGAUCUGA | 714/63 | 1,339/116 | 884/77 | 0 | 0 | 0 | 0 | 0 |
| PsmiR167b-3p | GGUCAUGCUCUGACAGCCUCACU | 41/4 | 56/5 | 33/3 | 0 | 0 | 0 | 0 | 0 | |
| miR168 | PsmiR168a | UCGCUUGGUGCAGGUCGGGAA | 738/64 | 2,375/206 | 3,210/279 | ++ | 0 | ++ | 0 | 0 |
| PsmiR168a-3p | CCCGCCUUGCAUCAACUGAAU | 151/13 | 140/12 | 295/26 | ++ | 0 | ++ | 0 | 0 | |
| miR169 | PsmiR169a | CAGCCAAGGAUGACUUGCCGA | 50/4 | 170/15 | 77/7 | 0 | 0 | 0 | 0 | 0 |
| PsmiR169r-3p | GCAAGUUGUCUUUGGCUACA | 218/19 | 837/73 | 615/53 | 0 | 0 | 0 | 0 | 0 | |
| miR171 | PsmiR171a | UGAUUGAGCCGCGCCAGUAUC | 277/24 | 234/20 | 346/30 | 0 | 0 | 0 | 0 | + |
| PsmiR171b-3p | UUGAGCCGCGUCAAUAUCUCU | 141/12 | 136/12 | 179/16 | + | + | + | 0 | ++ | |
| miR172 | PsmiR172a | AGAAUCUUGAUGAUGCUGCAU | 51/4 | 96/8 | 84/7 | 0 | 0 | 0 | + | 0 |
| PsmiR172a-3p | GCAGCGUCCUCAAGAUUCACA | 2/0 | 6/1 | 10/1 | NA | NA | NA | NA | NA | |
| miR319 | PsmiR319a | UUGGACUGAAGGGAGCUCCCU | 0/0 | 4,519/393 | 10,486/911 | + | 0 | + | 0 | 0 |
| PsmiR319a-3p | UUGGACUGAAGGGAGCUCCC | 4,587/403 | 11,200/974 | 7,432/646 | NA | 0 | NA | 0 | NA | |
| PsmiR319i | UUGGACUGAAGGGGGCUCCC | 1,408/122 | 0/0 | 0/0 | 0 | 0 | NA | 0 | NA | |
| miR390 | PsmiR390a-3p | CGCUAUCCAUCCUGAGUCUCA | 26/2 | 139/12 | 136/12 | ++ | + | ++ | + | 0 |
| PsmiR390b-5p | AAGCUCAGGAGGGAUAGCACC | 1,062/92 | 2,193/191 | 3,466/301 | 0 | 0 | 0 | 0 | 0 | |
| miR394 | PsmiR394a | UUGGCAUUCUGUCCACCUCC | 307/27 | 1,266/110 | 385/33 | 0 | 0 | 0 | 0 | 0 |
| miR396 | PsmiR396b-3p | GCUCAAGAAAGCUGUGGGAAA | 0/0 | 17/1 | 18/2 | ++ | NA | NA | NA | NA |
| PsmiR396c | UUCAAGAAAGUCGUGGGAGA | 48/4 | 48/4 | 73/6 | ++ | ++ | ++ | ++ | + | |
| Less-conserved | ||||||||||
| miR5266 | PsmiR5266 | CGGGGGACUGCUCGGGCC | 46,659/4,056 | 39,563/3,439 | 57,262/4,978 | NA | NA | NA | NA | NA |
| miR4414 | PsmiR4414a-5p | AGCUGCUGACUCGUUCAUUCA | 0/0 | 23/2 | 0/0 | NA | NA | NA | NA | NA |
| PsmiR4414b | UGUGAAUGAUGCGGGAGACAA | 70/6 | 0/0 | 102/9 | NA | NA | NA | NA | NA | |
| miR403 | PsmiR403 | UUAGAUUCACGCACAAACCCA | 1,629/142 | 3,659/318 | 2,662/231 | NA | ++ | NA | ++ | ++ |
| miR5054 | PsmiR5054 | GCCCCACGGUGGGCGCCA | 22/2 | 99/9 | 162/14 | NA | NA | NA | NA | NA |
| miR5059 | PsmiR5059 | UCCUGGGCAGCAACACCA | 38/3 | 200/17 | 206/18 | NA | NA | NA | NA | NA |
| miR5077 | PsmiR5077 | UUCACGUCGGGUUCACCA | 288/25 | 1,483/129 | 1,529/133 | NA | NA | NA | NA | NA |
| miR5139 | PsmiR5139 | AACCUGGCUCUGAUACCA | 113/10 | 425/37 | 119/10 | NA | NA | NA | NA | NA |
| miR5213 | PsmiR5213-5p | UGCGUGUGUCUUCACCUCUGA | 293/25 | 1,228/107 | 606/53 | NA | NA | NA | NA | NA |
| miR5371 | PsmiR5371-5p | UUGGAAUCUAGUCGACUCAGAC | 48/4 | 205/18 | 27/2 | NA | NA | NA | NA | NA |
| miR5658 | PsmiR5658 | AUGAUGAUGAUGCUGAGAC | 1,085/94 | 640/56 | 723/63 | NA | ++++ | NA | NA | NA |
| miR6108c | PsmiR6108c | AAUCGUAAGAAGAAUGCUGAAGCC | 51/4 | 103/9 | 45/4 | NA | NA | NA | NA | NA |
| miR6113 | PsmiR6113 | UGAAACUCAAGAAAACGUCG | 2,367/206 | 4,552/396 | 2,504/218 | NA | NA | NA | NA | NA |
| miR6279 | PsmiR6279 | UAGAAAGUAAUUCCAUGACACC | 28/2 | 44/4 | 34/3 | NA | NA | NA | NA | NA |
| miR6284 | PsmiR6284 | UACUUGGACCCUGAAUGAAGAUU | 954//83 | 2,398/208 | 1,505/131 | NA | NA | NA | NA | NA |
| miR6441 | PsmiR6441 | AAUUGACGGAAGGGCACA | 1,738/151 | 1,553/135 | 3,920/341 | NA | NA | NA | NA | ++++ |
| miR6478 | PsmiR6478 | CCGACCUUAGCUCAGUUGGUAGA | 64/6 | 270/23 | 187/16 | NA | NA | NA | NA | + |
| miR7984a | PsmiR7984a | UCCGACUUUGUGAAAUGACUU | 492/43 | 778//68 | 953/83 | NA | NA | NA | NA | NA |
| miR858b | PsmiR858b | UUCGUUGUCUGUUCGACCUUG | 41/4 | 80/7 | 30/3 | NA | 0 | NA | NA | NA |
| miR894 | PsmiR894 | UGUUCGUUUCACGUCGGGUUCACCA | 131/11 | 320/28 | 530/46 | NA | NA | NA | NA | NA |
| miR6300 | PsmiR6300 | GUCGUUGUAGUAUAGUGG | 10,410/905 | 0/0 | 0/0 | NA | NA | NA | NA | NA |
| miR398 | PsmiR398b-3p | UUGUGUUCUCAGGUCACCCCU | 13/1 | 21/2 | 39/3 | NA | NA | 0 | NA | 0 |
| miR5072 | PsmiR5072 | AACGACUCCCCAGCAGAGUCGCC | 23/2 | 178/15 | 622/54 | 0 | NA | 0 | NA | + |
0 represents no mismatch, + represents one mismatch, ++ represents tow mismatches, and so on. zma, Zea mays; ath, Arabidopsis thaliana; osa, Oryza sativa; vvi, Vitis vinifera; ptc, Populus trichocarpa.
Figure 3.
Abundance of most conserved miRNA families in three libraries from tree peony buds after 6 d, 18 d and 24 d chilling treatments.
Based on the miRNA annotation criteria31, novel miRNAs could be predicted by mapping the remaining non-annotated sRNAs to Populus genome. In our data, seventeen novel miRNAs were obtained and named as PsmiR1 to PsmiR17 (Table 5). All precursors of novel miRNAs had regular stem-loop structures and the predicted hairpin structures. To investigate the conservation of these 17 novel miRNAs in other plant species including Malus domestica, Physcomitrella patens and Populus trichocarpa, they were used to perform BLAST searches against miRBase databases. PsmiR5, PsmiR7 and PsmiR16 matched genomes of other plant species (Table 5). Reverse transcript PCR (RT-PCR) was performed to validate the expression of some new predicted miRNAs in flower buds after chilling treatments. The primer sequences were listed in Supplementary Table S1. We found five of the 17 predicted miRNAs including PsmiR9, PsmiR3, PsmiR1, PsmiR4 and PsmiR13 expressed in tree peony flower buds (Fig. 4). PsU6 was amplified as a positive control. We found that these novel miRNAs could be detected in flower buds after 18 d chilling treatments.
Table 5.
Candidate novel miRNAs in tree peony.
| novel miRNA | mature sequence (5′-3′) |
other species’ ID in miRbase |
MFE (kcAl/mol) |
MFEI | Predicted precursors | p-value | Predicted target ID | Normalized miRNA abundance | ||
|---|---|---|---|---|---|---|---|---|---|---|
| 6 d | 18 d | 24 d | ||||||||
| PsmiR1-5p | AGGGACTCCTTTCACTCCACT | — | −81.9 | 1 | JI448260:47.0.214:+ | 0.99 | — | 7,624.35 | 9,555.42 | 23,848.02 |
| PsmiR2-5p | CATACTTCTGGATAACG | — | −11.2 | 0.4 | JI455606:1198..1247:+ | 0 | JI455773 | 0 | 4.78 | 2.65 |
| PsmiR3-5p | GGTGGACTGCTCGAGCC | — | −27.8 | 0.9 | JI443786:93..136:- | 0.99 | JI450527 | 23,0084.13 | 159,333.51 | 223,257.97 |
| PsmiR 4-3p | TATGAGACTTGGACGAGGCAC | — | −37.9 | 1 | JI451506:405..481:- | 0.99 | JI445930 | 3,384.18 | 2,348.2 | 2,987.29 |
| PsmiR 5-5p | AGAGAATTGAAGATGAGCACCT | ppt-miR1023b-3p | −41.2 | 0.7 | ContiG02457:19..227:- | 0 | JI446191 | 0 | 0 | 2.65 |
| PsmiR 6-3p | ATCTCTTTGAGCTGCAAGAAGGCC | — | −66.4 | 0.5 | JI458593:131..405:- | 0 | JI458593 | 0 | 0 | 2.65 |
| PsmiR 7-3p | AAGCCATGGATGAAGCTAT | ptc-miR169y | −82 | 0.7 | JI448255:1023..1305:- | 0 | JI450838 | 0 | 4.78 | 0 |
| PsmiR 8-5p | TGTACTACAGGGTAGGAAAGA | — | −24.3 | 0.9 | JI456175:234..311:+ | 0.99, | JI452932 | 79.63 | 45.43 | 108.48 |
| PsmiR 9-5p | CGGTGGACTGCTCGAGCCG | — | −28.9 | 0.9 | JI443786:93..137:- | 0 | JI443786 | 378,494.16 | 410,756.3 | 374,225.73 |
| PsmiR 10-5p | AGCCTTCTTTGGGTTGCGACC | — | −73.2 | 1.3 | JI448950:74..188:- | 0 | JI448950 | 23.89 | 253.47 | 58.21 |
| PsmiR 11-5p | AGCTTTTGTATGTTCTCCGTTA | — | −57.7 | 0.6 | JI452338:1477..1738:+ | 0 | — | 0 | 2.39 | 0 |
| PsmiR 12-5p | GGTGGATGTATGAACCCAGCCT | −47.6 | 0.6 | JI454613:413..564:+ | 0 | JI453131 | 0 | 2.39 | 0 | |
| PsmiR 13-5p | TTGTTTGAATTCTTGCAACAGA | — | −63.9 | 1.5 | JI444316:158..242:+ | 0 | JI444316 | 1,282.01 | 4,421.41 | 828.18 |
| PsmiR 14-3p | CGACTGGGAAGGATTGGGGA | — | −80 | 0.6 | JI454686:1279..1546:- | 0 | JI454686 | 0 | 0 | 2.65 |
| PsmiR 15-5p | AGGGCATGTCCATGGGCTCT | — | −49.4 | 0.5 | JI458732:712..921: - | 0 | JI458732 | 0 | 2.39 | 0 |
| PsmiR 16-3p | AGAAGAGAAGAGAGAGGA | mdm-miR169e | −29.9 | 0.6 | JI452660:25..140: - | 0 | JI457551 | 0 | 0 | 2.65 |
| PsmiR 17-3p | CCAAGTTAAGCTCGGCGAG | −10 | 0.4 | JI450973:261..299:+ | 0 | JI453625 | 0 | 2.39 | 0 | |
| PsmiR 13-3p | TGTTGCAGAATTCAAACAAA | — | −63.9 | 1.5 | JI444316:158..242:+ | 0 | JI444316 | 557.39 | 2,529.94 | 494.79 |
ppt: Physcomitrella patens; ptc: Populus trichocarpa; mdm: Malus domestica.
Figure 4.
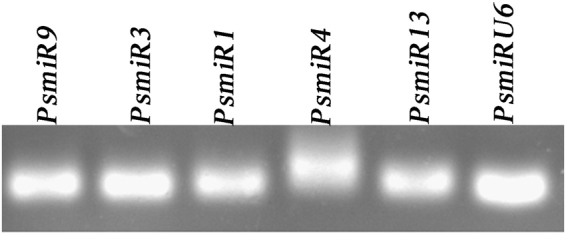
Reverse transcript PCR (RT-PCR) electrophoresis results for expression identification of novel miRNAs in flower buds after 18 d chilling treatments. In total, 5 of 17 novel miRNAs were confirmed by Reverse transcript PCR (RT-PCR) with 40 cycle-amplification. The sizes of PCR products were about 100 bp. PsU6 was used as positive control.
We found 469 mature miRNAs could be aligned to other species’ precursors (precursors data from miRbase r21), mostly in Glycine max (36.7%), Oryza sativa (30.7%), and Arabidopsis thaliana (30.06%). The 17 precursors of novel miRNAs derived from tree peony transcriptome was listed in Table 5, only 1 precursors coded both 5p and 3p side mature miRNAs, others only possessed 1-side mature sequence.
Prediction of miRNA targets in tree peony
Previous study found that plant miRNA target sites mainly situate at opening reading frames (ORFs)32. To understand possible biological functions of the identified miRNAs in tree peony, we predicted the miRNA targets using the mRNA transcriptome of Paoenia ostii flower buds as a reference sequence since the genome of Paoenia ostii is not publicly available10. A total of 771 unigenes were predicted as potential targets of 62 known miRNA families (Additional file 2), and the majority of target proteins and corresponding annotations were shown in Table 6. Most miRNAs had more than one predicted target proteins, and some of the miRNAs have more than 10. Based on GO annotation, more than half of the predicted target genes were involved in biological process (metabolic process, regulation of transcription, signal transduction, transport and regulation of act polymerization) and molecular function (binding and methyltransferase activity) (Fig. 5). However, there were many conserved miRNAs target genes that had no functional annotation. Novel miRNAs targets were also predicted, but only two of them have been found target relationship with two unigenes (Table 5).
Table 6.
Majority of the predicted target genes and corresponding annotation of known miRNAs in tree peony.
| miRNA family | Target ID | Targets annotation | miRNA family | Target ID | Targets annotation |
|---|---|---|---|---|---|
| miR1509 | JI451099 | Peroxiredoxin | miR319 | JI449827 | AP2 domain-containing transcription factor |
| miR156 | JI447102 | DNMT2 (DNA METHYLTRANSFERASE-2) | miR319 | JI447690 | Polygalacturonase precursor |
| miR156 | JI446831 | SQUAMOSA promoter-binding protein-like | miR395 | JI453154 | beta galactosidase |
| miR159 | JI446967 | RAB6A; GTP binding/protein binding | miR396 | JI445772 | Chitin-inducible gibberellin-responsive protein |
| miR159 | JI446401 | asparagine synthetase | miR396 | JI444318 | glyceraldehyde 3-phosphate dehydrogenase |
| miR162 | JI449996 | ubiquitin | miR397 | JI455403 | lipoxygenase |
| miR164 | JI458131 | PID2 (PINOID2); ATP binding/protein kinase | miR414 | JI453820 | zinc finger protein |
| miR164 | JI449546 | dtdp-glucose 4-6-dehydratase | miR414 | JI445308 | phosphoesterase |
| miR166 | JI446960 | pentatricopeptide repeat-containing protein | miR414 | JI444902 | Phospho-2-dehydro-3-deoxyheptonate aldolase 1 |
| miR167 | JI458177 | transmembrane protein | miR5059 | JI445796 | CAM7 (CALMODULIN 7); calcium ion binding |
| miR167 | JI452318 | trytophan synthase alpha subunit | miR5083 | JI446932 | COP1-interacting protein-related |
| miR167 | JI451976 | Serine/threonine-protein kinase PBS1 | miR5658 | JI455720 | Serine/threonine-protein kinase SAPK10 |
| miR168 | JI451707 | GRAS family transcription factor | miR5658 | JI454156 | dolichyl glycosyltransferase |
| miR169 | JI454583 | similar to Protein kinase | miR5658 | JI451464 | Stromal cell-derived factor 2 precursor |
| miR169 | JI450321 | Acetyl glucosaminyl transferase | miR6113 | JI450180 | Lactoyl glutathione lyase |
| miR169 | JI447773 | F-box family protein | miR6300 | JI450877 | 3-dehydroquinate synthase |
| miR169 | JI445119 | calcium-dependent protein kinase | miR6300 | JI450707 | DNA damage checkpoint protein |
| miR171 | JI449485 | endoglucanase | miR6300 | JI448956 | ankyrin repeat domain protein |
| miR172 | JI445772 | Transcription factor GRAS | miR8175 | JI444623 | aminobutyrate aminotransferase |
| miR172 | JI446524 | AP2 domain-containing transcription factor | miR845 | JI448900 | cell division protein |
Figure 5.
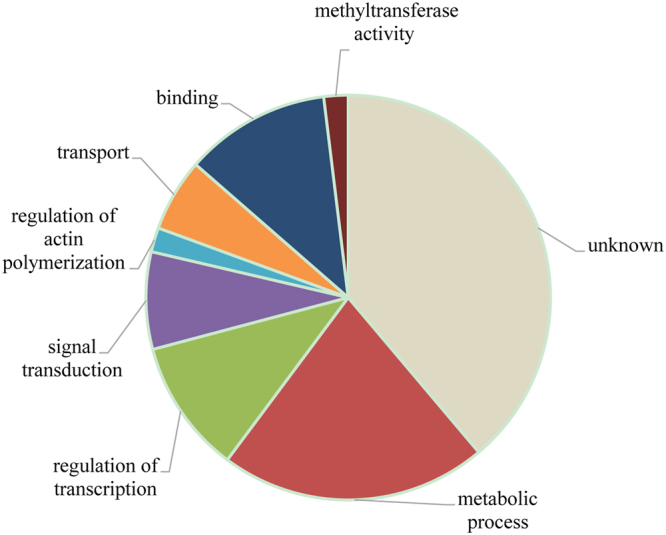
Go analysis of targets of known miRNAs in this study.
5′-RNA ligase-mediated (RLM)- RACEs were carried out to confirm the miRNA-guided cleavage and cleavage sites of predicted target transcripts. Squamosa-promoter-binding protein-like (SPL) family genes and APETALA2 (AP2) had been reported that they were the predicted targets of miR156 and miR172, respectively15,33. Our results showed that PsAP2 (JI446524) could be cleaved at the site between bases 12 (T) and 13 (C) within the complementary region to PsmiR172a (Fig. 6). PsSPL (JI446831) could be cleaved by PsmiR156 at the site between 10 (C) and 11 (T), which was also identified as a miRNA cleavage site in rose33.
Figure 6.
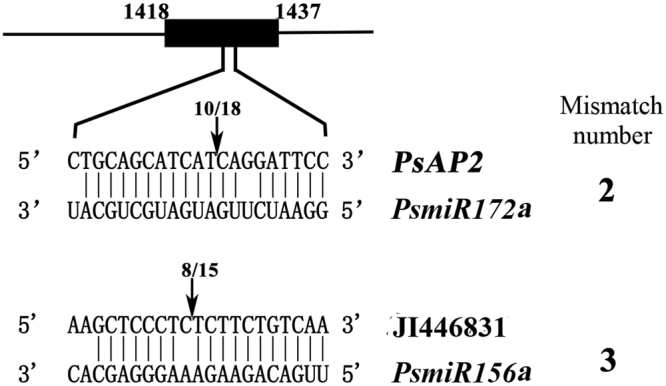
Validation of miRNA predicted targets by 5′ RLM-RACE in tree peony bud. Positions of the cleavage sites are indicated by arrows with the proportion of sequenced clones.
Expression profile of Paoenia ostii miRNAs during dormancy release
To identify miRNAs that were responsive during dormancy release, we compared miRNA expression level among three libraries. All the conserved candidate miRNAs with no less than 10 reads in each library were analyzed. Differentially expressed miRNAs that exhibited more than a 2-fold change were selected between each two treatments. There were 112 known miRNAs differentially expressed among three libraries (Additional file 3), and of which 55 miRNAs were shared among three libraries (See Additional file 3-shared miRNAs in three libraries). There existed 28 miRNAs (including miR8126-3p, miR6479, miR2949b, miR5057, miR6144, miR7743-3p, miR6483, miR5083, etc.) only in 18 d chilling duration library, which might play important roles in regulation dormancy release (Additional file 4). Based on the method of Audic and Man, the shared miRNAs among three libraries were normalized (Table 7). There were 11 down-regulated and 43 up-regulated miRNAs from 6 d to 18 d chilling treatment. Among them, PsmiR5072 showed the highest degree of induction (7.7-fold), PsmiR390a-3p, PsmiR5059, PsmiR170-5p, PsmiR166a, PsmiR2109 and PsmiR5077 were also clearly up-regulated (>5-fold). In addition, PsmiR5519 was specifically and significantly induced during dormancy release. To further elucidate the potential regulatory roles of miRNAs in the transition from dormancy to eco-dormancy, we made a comparative analysis of miRNA expression between 18 d chilling and 24 d chilling. There were 31 down-regulated and 24 up-regulated miRNAs, among them PsmiR1509b was the most significantly down-regulated (>35-fold). Similar to the report in poplar34, eight known development-related miRNA families were also detected differentially expressed during dormancy release in our data, including PsmiR164, PsmiR396, PsmiR168, PsmiR319, PsmiR171, PsmiR166, PsmiR156 and PsmiR172. These miRNAs mainly acted on cell proliferation (miR164, miR396 and miR319)28,35, vascular development (miR166)36 and miRNA biogenesis (miR168)37. PsmiR164 and PsmiR168a were continuously induced from dormancy to eco-dormancy stage, which were different from that in poplar34. PsmiR166 was up-regulated during dormancy release and repressed in eco-dormancy stage, similar results were detected in poplar during chilling induced dormancy-release34. Members of the PsmiR171 and PsmiR166 families showed the same expression patterns, but distinct differences of expression levels were also observed within other families during the same process. For example, PsmiR159a was up-regulated from 18 d to 24 d chilling treatments, while PsmiR159b-3p was down-regulated (>2-fold). PsmiR168a-3p was repressed during dormancy release (from 6 d chilling to 18 d chilling treatments), but PsmiR168a was continuously induced, indicating that members from same miRNA family might play different roles during this process. In addition, there were 28 miRNAs detected only in 18 d chilling treatment library and 7 miRNAs (PsmiR5227, PsmiR5665, PsmiR1886.1, PsmiR774b-5p, PsmiR4357, PsmiR1217-5p and PsmiR5224b) only in 24 d chilling treatment library (Additional file 4), which might mainly function in the transition from dormancy to dormancy release stage and eco-dormancy stage, respectively.
Table 7.
Differentiated expressions of shared miRNAs from tree peony flower bud after different chilling treatments.
| miRNA | Dormancy release | Endo-dormancy | miRNA | Dormancy release | Endo-dormancy | miRNA | Dormancy release | Endo-dormancy |
|---|---|---|---|---|---|---|---|---|
| 18 d vs 6 d | 24 d vs 18 d | 18 d vs 6 d | 24 d vs 18 d | 18 d vs 6 d | 24 d vs 18 d | |||
| PsmiR 5072 | 7.73913 | 3.494382 | PsmiR 159b -3p | 2.759555 | −2.27705 | PsmiR 167b -3p | 1.365854 | −1.69697 |
| PsmiR 390a -3p | 5.346154 | −1.02206 | PsmiR 6284 | 2.513627 | −1.59336 | PsmiR 1509b | 1.239167 | −35.2232 |
| PsmiR 5059 | 5.263158 | 1.03 | PsmiR 894 | 2.442748 | 1.65625 | PsmiR 164a | 1.231928 | 2.200489 |
| PsmiR 170 -5p | 5.263158 | −1.83486 | PsmiR 319a -3p | 2.441683 | −1.507 | PsmiR 3627 -5p | 1.084906 | −1.29213 |
| PsmiR 166a | 5.19809 | −2.40149 | PsmiR 403 | 2.246163 | −1.37453 | PsmiR 159a | 1.077639 | 1.128021 |
| PsmiR 2109 | 5.153846 | −2.88793 | PsmiR 2637 | 2.245283 | −2.83333 | PsmiR 396c | 1 | 1.520833 |
| PsmiR 5077 | 5.149306 | 1.031018 | PsmiR 162 -3p | 2.243194 | −1.35378 | PsmiR 171b -3p | −1.03676 | 1.316176 |
| PsmiR 160a | 4.958333 | −1.85615 | PsmiR 390b -5p | 2.064972 | 1.580483 | PsmiR 168a -3p | −1.07857 | 2.107143 |
| PsmiR 166h -3p | 4.76017 | −1.62356 | PsmiR 6108c | 2.019608 | −2.28889 | PsmiR 6441 | −1.11912 | 2.524147 |
| PsmiR 165a | 4.607143 | −2.93182 | PsmiR 858b | 1.95122 | −2.66667 | PsmiR 5266 | −1.17936 | 1.447362 |
| PsmiR 5054 | 4.5 | 1.636364 | PsmiR 162a | 1.95092 | −1.58472 | PsmiR 171a | −1.18376 | 1.478632 |
| PsmiR 5371 -5p | 4.270833 | −7.59259 | PsmiR 6113 | 1.923109 | −1.81789 | PsmiR 160a -3p | −1.33333 | 1.088889 |
| PsmiR 6478 | 4.21875 | −1.44385 | PsmiR 172a | 1.882353 | −1.14286 | PsmiR 5658 | −1.69531 | 1.129688 |
| PsmiR 5213 -5p | 4.191126 | −2.0264 | PsmiR 167a | 1.87535 | −1.51471 | PsmiR 1881 | −1.95918 | 2 |
| PsmiR 394a | 4.123779 | −3.28831 | PsmiR 2105 | 1.797872 | −1.67327 | PsmiR 2916 | −2.56452 | 3.516129 |
| PsmiR 169r -3p | 3.83945 | −1.36098 | PsmiR 398b -3p | 1.615385 | 1.857143 | PsmiR 157a | −3.38182 | 1.963636 |
| PsmiR 5139 | 3.761062 | −3.57143 | PsmiR 3630 -3p | 1.585366 | 1.307692 | PsmiR 156k | −5.15686 | 3.156863 |
| PsmiR 169a | 3.4 | −2.20779 | PsmiR 7984a | 1.581301 | 1.224936 | PsmiR 5519 | 2.25 | 0 |
| PsmiR 168a | 3.218157 | 1.351579 | PsmiR 6279 | 1.571429 | −0.29412 |
Note: + and − indicate the induction and repression of miRNA, respectively.
The sequencing results showed that the abundance of novel miRNAs was relatively less than that of conserved miRNAs (Table 5). Among these 17 predicted novel miRNAs, PsmiR1 was dramatically up-regulated during dormancy release. PsmiR3 and PsmiR4 were sharply reduced from 6 d to 18 d chilling treatments, PsmiR9, PsmiR10 and PsmiR13 showed opposite pattern with that of PsmiR3 and PsmiR4. PsmiR7, PsmiR11, PsmiR12, PsmiR15 and PsmiR17 were detected only in flower buds after 18 d chilling treatments.
Expression Validation of tree peony miRNAs
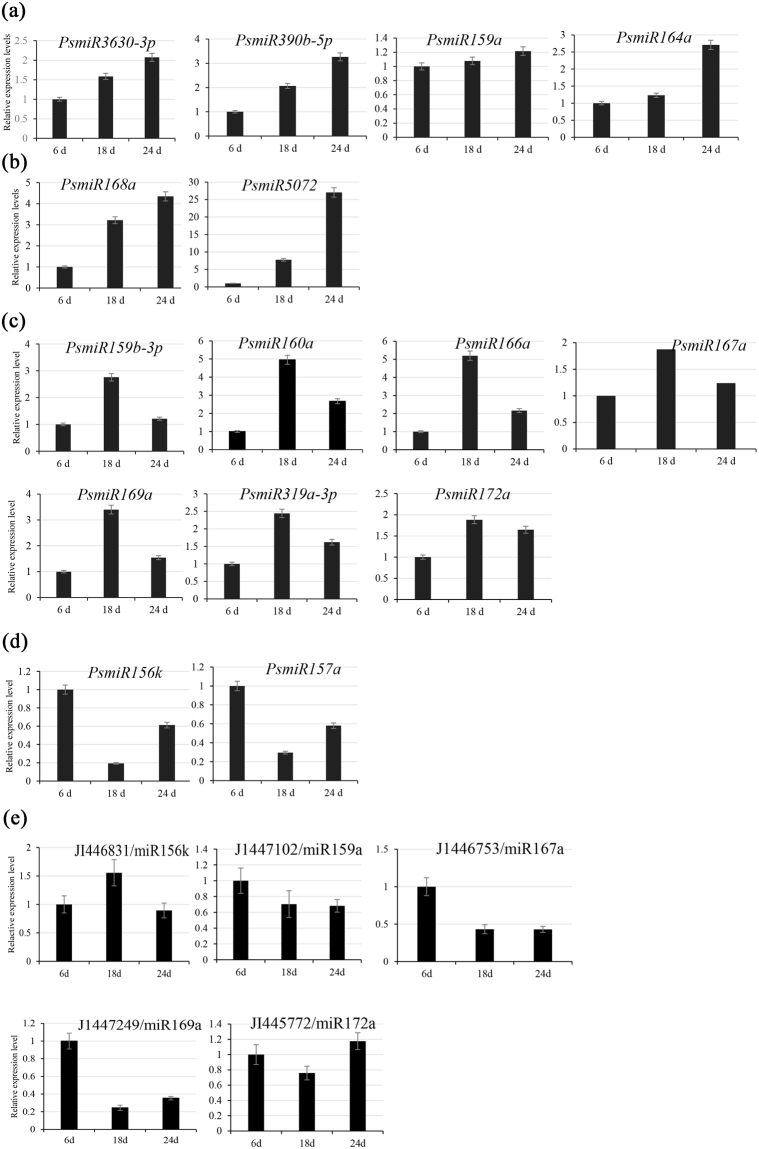
To confirm the expression patterns of miRNAs, as well as detect their responses to chilling treatments at three physiological stages, the expression of 15 conserved miRNAs, whose sequencing counts altered significantly after treatment, were analyzed by RT-qPCR. It is showed that the expression levels of miRNAs were a constant change process with the time of treatment. We classified them into four types (Fig. 7). Type a: slowly increased (PsmiR3630, PsmiR390b-5p, PsmiR159a and PsmiR164a); type b: suddenly increased (PsmiR168a and PsmiR5072); type c: first increased and then decreased (PsmiR159b-3p, PsmiR160a, PsmiR166a, PsmiR167a, PsmiR169a, PsmiR319-3p and PsmiR172a); type d: first decreased and then increased (PsmiR156k and PsmiR157a). These results suggest that miRNAs belonging to type c were early stage response miRNAs, those belonging to type a and type b might accelerate endo-dormancy release. Notably, the expression trends of two PsmiR159 family members, PsmiR159a and PsmiR159b-3p were different, indicating that the expression of miRNAs is a multiform process with the altered time of chilling treatments. Combining with our microarray data under same treatments11, five target genes of five miRNAs (miR156k, miR159a, miR167a, miR169a and miR172a) showed inverse expression patterns (Fig. 7e).
Figure 7.
Validation of miRNAs expression patterns by Reverse transcriptase quantitative PCR (RT-qPCR) and expression patterns of partially corresponding target genes in our microarray results at three physiological stages. Type (a) slowly increased; type (b) suddenly increased; type (c) first increased and then decreased; type (d) first decreased and then increased. (e) Expression patterns of partially corresponding target genes in our microarray results.
Discussion
Tree peony is an important horticultural crop worldwide of great ornamental, medical and economic value. Native to China, tree peony is regarded as “King of flower” and have deep botanical history in Chinese culture. It is crucial to understand the molecular mechanism of dormancy, which is a main obstacle for tree peony forcing culture. Based on morphological changes of Paoenia ostii ‘Feng Dan’ and global mRNA expression profiling, the physiological status of flower buds receiving less than 18 d chilling treatment are regarded as endo-dormancy, and that receiving more than 18 d are defined as eco-dormancy11. miRNAs are paid more and more attention as key regulators of gene activity in animals and plants26,38,39. In this study, we adapt high-throughput sequencing technology to identify sRNAs from Paeonia ostii and analyze their response to dormancy release. This work will provide useful information to deepen our understanding of the miRNA regulatory mechanisms during dormancy release.
There are 243, 511 and 207 miRNAs annotated in Arabidopsis, rice and soybean according to the miRBase database, respectively40–42. In this study, we first completed construction of sRNA libraries (6 d, 18 d and 24 d chilling treatments) in tree peony and obtained over 19 million 16–30 nt reads. The size distribution of sRNAs revealed that 21, 22, 23 and 24 nt sRNAs were relatively abundant, of which 24 nt sRNAs were significantly higher than others. Similar results were observed from most plants, such as Arabidopsis, rice, tomato43, cucumber44, apple45, peach46 and rose33. However, 21 nt-long sRNAs were the second enriched in this study, which was different with previous reports in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa)30,47,48. But in poplar, the 21 nt sRNAs are the most abundant49. Most of 21 nt sRNAs in our data started with uridine, which was consistent with the observation that ARGONAUTE1 (AGO1) usually harbors miRNAs with a 5′ terminal uridine. 24 nt sRNAs had start-nucleotide preference of adenosine, which was also reported in previous work26,27,29.
Tree peony endo-dormancy transcriptome database was employed to predict putative targets of tree peony miRNAs. The well-known conserved miRNAs including miR156, miR159, and miR164 have been identified. However, nearly half of known miRNAs and three novel miRNAs were not predicted homologous to any proteins in the Genbank nr database, which might because of the incomplete tree peony genome and limited number of transcript data in public database. Hundreds of miRNAs have been surveyed since high-throughput sequencing technology is widely used, but little has been done on identifying and analyzing miRNAs in tree peony and their response during dormancy release. Total 112 known miRNAs belonging to 99 families were identified in the three libraries. Two miRNAs families, PsmiR159 and PsmiR166, were relatively abundant. PsmiR166a increased continuously until dormancy release (6 d –18 d) and had a very low expression level at eco-dormant stage (24 d), which was consistent with recent studies in poplar34,50. For the expression level of 17 novel miRNAs, PsmiR1 was continuously up-regulated from dormancy to eco-dormancy stage, PsmiR9, PsmiR10 and PsmiR13 (up-regulation) and PsmiR3 and PsmiR4 (down-regulation) had converse expression pattern at the early stage of dormancy release.
Cold-responsive and auxin-related miRNAs
A continuous chilling accumulation is an effective natural or artificial way to release dormancy in tree peony. The endo-dormant bud can respond to chilling treatment, which stimulates growth-promoting respond signals including auxin or appropriate outside conditions (such as exogenous GA). In our study, there were 112 conserved miRNAs differentially expressed between 6 d and 18 d chilling library and between 18 d and 24 d chilling library. Among them, PsmiR160a was highly expressed in endo-dormancy release (18 d chilling) and quickly decreased in eco-dormancy (24 d chilling). MiR160 targeted auxin response factor 10/16/17 (ARF10/ARF16/ARF17) and negatively regulated auxin signaling51,52. Ding et al. found that miR160 was highly expressed in eco-dormancy (five weeks cold treatment)34. In our case, the high expression of PsmiR160 in endo-dormancy release might because of the difference of dormancy mechanism exist in tree peony and poplar. On the other hand, the expression of miR160 in endo-dormancy release may strengthen the auxin signal by inhibition of its target genes, and this hypothesis is also consistent with earlier report that exogenous GA could effectively promote endo-dormancy release10. Ding et al. reported other two auxin-related miRNAs, miR390 and miR167, increased during active growth34. In our study, the expression of PsmiR390b was steadily increased during the transition from endo-dormancy to eco-dormancy, and PsmiR167a was significantly induced during dormancy release, which suggest that auxin signal pathway participated in the process of dormancy release. We also found that PsmiR168a was continuously up-regulated from endo-dormancy stage to eco-dormancy stage, the same trend was found in poplar34. Similarly, miR168, member of the Csn-miR168 family, was found to be a cold-responsive miRNA, which was induced in two tea cultivars after 12 h of cold treatment22. MiR168 regulates its target ARGONAUTE1 (AGO1) to participate in miRNA biogenesis37. The high expression level of PsmiR168a would lead to the repression of AGO1, which would cause a reduction in the miRNA expression levels. The up-regulation of PsmiR168a suggested that cold-responsive miRNA participated in the release of endo-dormancy, their inductions were also consistent with its function of miRNA biogenesis37.
MiRNA targets
Since the genome of Paoenia ostii is not publicly available, the mRNA transcriptome of Paoenia ostii flower buds11 were employed as a reference to predict the putative miRNA targets. Based on GO annotation, more than half of the predicted targets in tree peony were involved in binding, catalytic activity, metabolic process and cellular process. For example, miR5141 targets gene encoding ATP synthase, which have been reported to be involved in ATP synthesis and ATP utilization during seed dormancy breaking53. In pear, specific enrichment of unigenes was observed for 15 pathways involved in metabolic processes including oxidative phosphorylation4. Several other target transcripts, which encode alpha N-terminal protein methyltransferase, Endoglucanase, GTPase-activating protein and F-box domain associated with various biological processes and cellular activities were also detected. For instance, PsmiR395 targets genes encoding enzyme beta galactosidase, which recently have been reported to be involved in cell wall modification during the transition from dormancy to eco-dormancy in onion bulbs54. PsmiR171 targets genes coding endoglucanase, which had been shown to be antifreeze proteins during seed germination in sunflower55. F-box proteins, the target of PsmiR169, have been identified previously as a key regulator of karrikin signaling and seed dormancy in Arabidopsis56. Novel miRNAs target genes were also predicted, but only two of them have not been found target relationship.
MiR169 and miR166 regulated cellular process and biological process by acting on their target genes. MiR166 function mainly in vascular development36, and the down-regulation of PsmiR166a at eco-dormant stage might help to increase the expression level of its target gene. In addition, Potkar et al. found that ptrmiR169a and its target gene PtrHAP2-5 showed inverse expression patterns during the dormancy period, which suggests that miR169 mediate attenuation of the target HAP2-5 transcript at this process24. Jeyaraj et al. found that CsmiR169 targeted COBRA-like protein encoding gene and regulated cellulose synthase, which suggests that miR169 have possible role in cell cycle and other biological function during the bud development23. In our study, PsmiR169a was highly expressed at the early stage of dormancy release and steadily down regulated at eco-dormant stage, and similar results were obtained during vegetative bud dormancy period of aspen24.
MiR156 and miR172 regulate and control the juvenile-to adult vegetative transition by targeting transcription factors SQUAMOSA promoter-binding protein-like (SPL) and APETALA2 (AP2) genes in both annual herbs17,57 and woody perennial plants58, showing converse expression patterns and regulatory relationships57. It is noteworthy that in our study we found the expression levels of PsmiR156k and PsmiR172a during the transition from endo-dormancy to eco-dormancy also had the converse expression patterns. Similarly, Ding et al. also found miR156 and miR172 showed completely converse expression patterns during the dormancy-active growth transition34. Our results showed that target genes (JI446524 and JI446831, putative AP2 and SPL genes) had cleavage sits of PsmiR172a and PsmiR156a, respectively, which suggested that miR156 and miR172 might play an important role during dormancy transition, which need to be further confirmed by experiments.
Materials and Methods
Plant materials
Four-year-old tree peony plants (Paoenia ostii ‘Feng Dan’) were potted and moved to refrigeration house with temperature 0–4 °C from 5 November to 30 December, 2014 in Qingdao, Shandong, China. The morphologic observation indicated flower buds receiving less than 18 d chilling treatment are in physiological status of endo-dormancy, while those receiving more than 18 d chilling treatment were in eco-dormancy physiological status11. Therefore, in order to decrease individuality, more than 5 plants were mixed buds-three buds for each individual were collected after 6 d, 18 d and 24 d chilling requirement fulfilling. Three replicates samples were harvested and immediately frozen in liquid nitrogen and stored at −80 °C until further use.
Small RNA library construction and sequencing
Total RNA from tree peony flower buds after chilling treatments (6 d, 18 d and 24 d) was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instruction and separated on 15% denaturing polyacrylamide gels. The 16–30 nt sRNAs were excised and recovered. The adapters (5′ and 3′) were ligated to the isolated sRNAs, which were sequentially reverse transcribed and amplified by PCR. The purified PCR products were sequenced using an Illumina Genome Analyzer (Illumina, USA) at Beijing Biomarker Technologies, Beijing, China.
Analysis of sequencing data
Raw sequence reads were produced by Illumina Genome Analyzer at Biomarker-Beijing, China and processed into clean full length reads by the Biomarker small RNA pipeline. During this procedure, all low-quality reads including 3′ adapter reads and 5′ adapter contaminants were removed. The remaining high-quality sequences were trimmed of their adapter sequences. Sequences larger than 30 nt and smaller than 16 nt were discarded. All high-quality sequences were considered as significant and further analyzed.
All matched sRNA sequences were categorized into classes including miRNAs, siRNAs, ribosomal RNAs (rRNAs), tRNAs, snRNAs, snoRNAs, repeat-associated sRNAs and degrade tags of extrons of introns, etc. Then, the clean sequences were annotated by performing BLASTn searches against the Rfam (http://www.Sanger.ac.uk/Software/Rfam) and NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi) databases, the detailed processes were following: the clean data were aligned with known miRNAs (miRNA precursors and mature miRNAs) registered in miRBase 21.0 (http://microran.sanger.ac.uk/sequence/index.html/) because of the difference among species, this process allowed two mismatches and free gaps to get temporary miRNA sequences and count of miRNA families; The highest-expression miRNA for each temporary mature miRNA family were selected to form a temporary miRNA database of Paoenia ostii. Finally, alignment of clean data to temporary miRNA database to identify conserved miRNAs in Paoenia ostii, only those perfect matching (≤two mismatches) were considered as conserved miRNAs. Potential novel miRNAs were identified using criteria that were previously developed for plant miRNA prediction59. The unique fold back structures of miRNA precursors can be utilized to predict novel miRNAs using MIREAP program (http://sourceforge.net/projects/mireap/). Potential targets for both known and novel miRNAs were identified on TAPIR and Target Finder based on Paeonia ostii transcriptome sequencing data10 according to the search algorithm that only three or fewer mismatches and no gap are allowed to be present in the complementarily between miRNAs and their corresponding targets32. The biological function category of the predicted targets was annotated against the Universal Protein Resource (http://www.uniprot.org).
Differential expression analysis of miRNA and Reverse Transcriptase quantitative PCR (RT-qPCR) and 5′ RLM-RACE
Differential expression analysis of miRNAs was performed based on sequence reads generated from three libraries after different chilling treatments according to the method described by Ren49. In detail, the expression of miRNAs was normalized to obtain the number of miRNAs per million reads [normalized expression = (number of miRNA reads/total number of clean reads) × 1,000,000]. Normalized miRNA reads with values less than one in three libraries were excluded. The remaining miRNAs were used to calculate differences in expression by fold change (normalized miRNA reads in 18 d or 24 d chilling treatment/normalized miRNA reads in 6 d chilling treatment) and significant P-values60,61.
To validate miRNA expression, sRNAs were isolated from flower buds after different chilling treatments using an RNAiso for small RNA (TaKaRa, Dalian, China) following the manufacturer’s instructions. Then, the sRNA was polyadenylated by poly (A) polymerase, and first-strand cDNA was obtained using SYBR® Primescript miRNA RT-PCR Kit (TaKaRa, Dalian, China). Briefly, the polyA was added to the 3′ of total RNA, then the RNA was reverse-transcribed with an oligo-dT adaptor. Quantitative PCR was performed in a total volume of 25 μL, containing 2 μL cDNA, 0.4 μM PCR forward primer (1 μL), 0.4 μM Uni-miR RT-qPCR primer (1 μL), 12.5 μL of 2 × SYBR premix Ex Taq ΙI, and 8.5 μL dd H2O. The reactions were completed using Roche Light Cycler 480 (Roche, Mannheim, Germany) with the following program: 95 °C for 10s and 40 cycles of 95 °C for 5s, 55 °C for 30s and 72 °C for 15s. The reactions were run in triplicate and the 2−ΔΔCt relative quantification method was used to calculate the relative changes in gene expression62. Small nuclear RNA U6 was used as endogenous reference, primers used in this study were listed in Supplementary Table S1.
To conform whether the predicted targets were cut by miRNAs and cleavage sites, the 5′ RLM-RACE were carried out using the FirstChoice RLM-RACE Kit (Ambion). Specifically, one microgram total RNA was firstly ligated to 5′ RACE oligo adaptor without calf intestine alkaline phosphatase and tobacco acid pyrophosphatase treatments. Then, the ligated RNA was used to synthesize the cDNA. The primers of miR172a target gene (JI446524) (5′-TCGGAGAAATGCTTTGTCCATGGCCAT-3′) and miR156a target gene (JI446831) (5′-TTGCGAGGTTCTGGGTTTGGAG-3′) for 5′ RLM-RACE were designed by Primer premier 5.0 software (Supplementary Table S1). PCR was carried out according to the manufacturer instructions, and the PCR products were purified by 1.0% agarose gel electrophoresis and cloned into the pMD18-T vector (Takara, Dalian, China) for sequencing.
Availability of Data and Materials
Our data have been presented in the main paper or additional supporting files.
Electronic supplementary material
Acknowledgements
This work was supported by grants from National Natural Science Foundation of China (31372104 and 31471908).
Author Contributions
Conceived and designed the experiments: Shupeng Gai. Designed, analyzed data, drafted and revised the manuscript: Yuxi Zhang and Shupeng Gai. Collected materials: Yanyan Wang. Analyzed the data: Yanyan Wang and Xuekai Gao. Gave valuable advice: Chunying Liu.
Competing Interests
The authors declare no competing interests.
Footnotes
Yuxi Zhang and Yanyan Wang contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-22415-5.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lang GA, Early JD, Martin GC, Darnell RL. Endo-, para-, and ecodormancy: physiological terminology and classification for dormancy research. Hortscience. 1987;22:371–377. [Google Scholar]
- 2.Rallo L, Martin GC. The role of chilling in releasing olive floral buds from dormancy. Hortscience. 1991;26:1058–1062. [Google Scholar]
- 3.Rohde, A. et al. Molecular Aspects of Bud Dormancy in Trees, 89–134, (Springer Netherlands, 2000).
- 4.Liu G, et al. Transcriptomic analysis of ‘Suli’ pear (Pyrus pyrifoliawhite pear group) buds during the dormancy by RNA-Seq. BMC Genomics. 2012;13:700. doi: 10.1186/1471-2164-13-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu Y, et al. RNA-Seq-based transcriptome analysis of dormant flower buds of Chinese cherry (Prunus pseudocerasus) Gene. 2015;555:362–376. doi: 10.1016/j.gene.2014.11.032. [DOI] [PubMed] [Google Scholar]
- 6.Fu XL, et al. Roles of endoplasmic reticulum stress and unfolded protein response associated genes in seed stratification and bud endodormancy during chilling accumulation in Prunus persica. Plos One. 2014;9:e101808. doi: 10.1371/journal.pone.0101808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yordanov YS, Ma C, Strauss SH, Busov VB. EARLY BUD-BREAK 1 (EBB1) is a regulator of release from seasonal dormancy in poplar trees. Proc. Natl. Acad. Sci. USA. 2014;111:10001–10006. doi: 10.1073/pnas.1405621111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thirugnanasambantham K, Prabu G, Palanisamy S, Chandrabose SR, Mandal AK. Analysis of dormant bud (Banjhi) specific transcriptome of tea (Camellia sinensis (L.) O. Kuntze) from cDNA library revealed dormancy-related genes. Appl. Biochem. Biotech. 2013;169:1405–1417. doi: 10.1007/s12010-012-0070-5. [DOI] [PubMed] [Google Scholar]
- 9.Huang X, et al. Genes associated with the release of dormant buds in tree peonies (Paeonia suffruticosa) Acta Physiol Plant. 2008;30:797–806. doi: 10.1007/s11738-008-0184-0. [DOI] [Google Scholar]
- 10.Gai S, et al. Transcriptome analysis of tree peony during chilling requirement fulfillment: Assembling, annotation and markers discovering. Gene. 2012;497:256–262. doi: 10.1016/j.gene.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 11.Gai S, Zhang Y, Liu C, Zhang Y, Zheng G. Transcript profiling of Paoenia ostii during artificial chilling induced dormancy release identifies activation of GA pathway and carbohydrate metabolism. Plos One. 2013;8:e55297. doi: 10.1371/journal.pone.0055297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang YX, et al. Differential expression proteins associated with bud dormancy release during chilling treatment of tree peony (Paeonia suffruticosa) Plant Biol. 2015;17:114–122. doi: 10.1111/plb.12213. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, et al. Isolation and characterization of a SOC1 -Like gene from tree peony (Paeonia suffruticosa) Plant Mol. Bio.l Rep. 2015;33:855–866. doi: 10.1007/s11105-014-0800-7. [DOI] [PubMed] [Google Scholar]
- 14.Bartel, D. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. (2004). [DOI] [PubMed]
- 15.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Llave C, Carrington JC. Cleavage of scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 17.Lauter N, Kampani A, Carlson S, Goebel M, Moose S. P. microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc. Natl. Acad. Sci. USA. 2005;102:9412–9417. doi: 10.1073/pnas.0503927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang JW, Wang LJ, Xue HW, Chen XY. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell. 2005;17:2204–2216. doi: 10.1105/tpc.105.033076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo HS, Xie Q, Fei JF, Chua NH. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell. 2005;17:1376–1386. doi: 10.1105/tpc.105.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J, et al. microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J. 2005;42:84–94. doi: 10.1111/j.1365-313X.2005.02354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, et al. Identification and characterization of cold-responsive microRNAs in tea plant (Camellia sinensis) and their targets using high-throughput sequencing and degradome analysis. BMC Plant Biol. 2014;14:271. doi: 10.1186/s12870-014-0271-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeyaraj A, Chandran V, Gajjeraman P. Differential expression of microRNAs in dormant bud of tea [Camellia sinensis (L.) O. Kuntze] Plant Cell Rep. 2014;33:1053–1069. doi: 10.1007/s00299-014-1589-4. [DOI] [PubMed] [Google Scholar]
- 24.Potkar R, Recla J, Busov V. ptr-MIR169 is a posttranscriptional repressor of PtrHAP2 during vegetative bud dormancy period of aspen (Populus tremuloides) trees. Biochem. Biophys. Res. Commun. 2013;431:512–518. doi: 10.1016/j.bbrc.2013.01.027. [DOI] [PubMed] [Google Scholar]
- 25.Jin Q, et al. Identification and characterization of microRNAs from tree peony (Paeonia ostii) and their response to copper stress. Plos One. 2015;10:e117584. doi: 10.1371/journal.pone.0117584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 27.Mi S, et al. Sorting of small RNAs into arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubio-Somoza I, Cuperus JT, Weigel D, Carrington JC. Regulation and functional specialization of small RNA-target nodes during plant development. Curr. Opin. Plant Biol. 2009;12:622–627. doi: 10.1016/j.pbi.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat. Rev. Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sunkar R, Zhou X, Yun Z, Zhang W, Zhu JK. Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol. 2008;8:25. doi: 10.1186/1471-2229-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lelandaisbrière C, et al. Genome-wide Medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant Cell. 2009;21:2780–2796. doi: 10.1105/tpc.109.068130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srivastava PK, Moturu TR, Pandey P, Baldwin IT, Pandey SP. A comparison of performance of plant miRNA target prediction tools and the characterization of features for genome-wide target prediction. BMC Genomics. 2014;15:348. doi: 10.1186/1471-2164-15-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pei H, et al. Integrative Analysis of miRNA and mRNA profiles in response to ethylene in rose petals during flower opening. Plos One. 2013;8:e64290. doi: 10.1371/journal.pone.0064290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding Q, Zeng J, He XQ. Deep sequencing on a genome-wide scale reveals diverse stage-specific microRNAs in cambium during dormancy-release induced by chilling in poplar. BMC Plant Biol. 2014;14:267. doi: 10.1186/s12870-014-0267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raman S, et al. Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J. 2010;55:65–76. doi: 10.1111/j.1365-313X.2008.03483.x. [DOI] [PubMed] [Google Scholar]
- 36.Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans M. C. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature. 2004;428:84–88. doi: 10.1038/nature02363. [DOI] [PubMed] [Google Scholar]
- 37.Vaucheret H, Mallory A, Bartel D. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell. 2006;22:129–136. doi: 10.1016/j.molcel.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA. Conservation and divergence of plant microRNA genes. Plant J. 2006;46:243–259. doi: 10.1111/j.1365-313X.2006.02697.x. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, et al. Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol. 2010;152:2222–2231. doi: 10.1104/pp.109.151803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griffithsjones S, Saini HK, Dongen SV, Enright A. J. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright A. J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song QX, et al. Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol. 2011;11:5. doi: 10.1186/1471-2229-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuo J, et al. Sculpting the maturation, softening and ethylene pathway: The influences of microRNAs on tomato fruits. BMC Genomics. 2012;13:7. doi: 10.1186/1471-2164-13-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martínez G, Forment J, Llave C, Pallás V, Gómez G. High-throughput sequencing, characterization and detection of new and conserved Cucumber miRNAs. Plos One. 2011;6:e19523. doi: 10.1371/journal.pone.0019523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rui X, Hong Z, An YQ, Beers EP, Liu Z. Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol. 2012;13:R47. doi: 10.1186/gb-2012-13-6-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao Z, et al. Identification and validation of potential conserved microRNAs and their targets in peach (Prunus persica) Mol. Cells. 2012;34:239–249. doi: 10.1007/s10059-012-0004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu QH, et al. A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res. 2008;18:1456–1465. doi: 10.1101/gr.075572.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao YT, et al. Small RNA profiling in two Brassica napus cultivars identifies microRNAs with oil production- and development-correlated expression and new small RNA classes. Plant Physiol. 2012;158:813–823. doi: 10.1104/pp.111.187666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren Y, et al. Differential profiling analysis of miRNAs reveals a regulatory role in low N stress response of Populus. Funct. Integr. Genomic. 2015;15:1–13. doi: 10.1007/s10142-014-0408-x. [DOI] [PubMed] [Google Scholar]
- 50.Ko JH, Prassinos C, Han KH. Developmental and seasonal expression of PtaHB1, a Populus gene encoding a class III HD-Zip protein, is closely associated with secondary growth and inversely correlated with the level of microRNA (miR166) New Phytol. 2006;169:469–478. doi: 10.1111/j.1469-8137.2005.01623.x. [DOI] [PubMed] [Google Scholar]
- 51.Liu PU, et al. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007;52:133–146. doi: 10.1111/j.1365-313X.2007.03218.x. [DOI] [PubMed] [Google Scholar]
- 52.Mallory AC, Bartel DP, Bartel B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005;17:1360–1375. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perl M. ATP synthesis and utilization in the early stage of seed germination in relation to seed dormancy and quality. Physiol. Plantarum. 2010;66:177–182. doi: 10.1111/j.1399-3054.1986.tb01253.x. [DOI] [Google Scholar]
- 54.Chope GA, Cools K, Terry LA, Hammond JP, Thompson AJ. Association of gene expression data with dormancy and sprout suppression in onion bulbs using a newly developed onion microarray. Acta Horticulturae. 2012;969:169–174. doi: 10.17660/ActaHortic.2012.969.22. [DOI] [Google Scholar]
- 55.Kumar A, Bhatla SC. Polypeptide markers for low temperature stress during seed germination in sunflower. Biol. Plantarum. 2006;50:81–86. doi: 10.1007/s10535-005-0078-6. [DOI] [Google Scholar]
- 56.Nelson DC, et al. F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2011;108:8897–8902. doi: 10.1073/pnas.1100987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133:3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang JW, et al. miRNA control of vegetative phase change in trees. Plos Genet. 2011;7:e1002012. doi: 10.1371/journal.pgen.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meyers BC, et al. Criteria for annotation of plant MicroRNAs. Plant Cell. 2008;20:3186–3190. doi: 10.1105/tpc.108.064311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 61.Man MZ, Wang X, Wang Y. POWER_SAGE: comparing statistical tests for SAGE experiments. Bioinformatics. 2000;16:953–959. doi: 10.1093/bioinformatics/16.11.953. [DOI] [PubMed] [Google Scholar]
- 62.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Our data have been presented in the main paper or additional supporting files.