Abstract
Micro RNAs (miRNAs) have emerged as a critical regulator of several biological processes in both animals and plants. They have also been associated with regulation of immune responses in many human diseases during recent years. Visceral leishmaniasis (VL) is the most severe form of leishmaniasis, which is characterized by impairment of both innate and adaptive immune responses. In the present study, we observed that Leishmania establishes hypoxic environment in host macrophages that induces the expression of hypoxia inducible factor-1α (HIF-1α) and miRNA-210. Further, the expression of miRNA-210 was found to be dependent on activation of HIF-1α expression. The HIF-1α silencing by siRNA resulted in significantly (p < 0.001) decreased expression of miR-210 in parasites infected macrophages. We also observed that in siHIF-1α or antagomir-210 treated L. donovani infected macrophages, the parasitic load and percentage infectivity were significantly (p < 0.001) decreased. Furthermore, we found that inhibition of miR-210 leads to activation of NF-κB subunit p50, and it forms heterodimer with p65 and translocates into the nucleus from the cytoplasm. This significantly (p < 0.05) induced the transcription of pro-inflammatory cytokines genes such as TNF-α and IL-12 in miRNA-210 inhibited macrophages compared to uninhibited macrophages whereas the level of IL-10, an anti-inflammatory cytokine, was found to be significantly decreased (p < 0.001). These findings suggested that L. donovani infection induces hypoxic environment inside the macrophages that activates HIF-1α. Further, HIF-1α upregulates miR-210, which eventually establishes a suitable environment for the survival of parasite inside the host macrophages by downregulating NF-κB mediated pro-inflammatory immune responses.
Keywords: HIF-1α, miR-210, NF-κB, cytokines, visceral leishmaniasis, macrophages
Introduction
In 1993, a new era in basic research was started after the discovery of micro RNAs (miRNAs) in Caenorhabditis elegans (Lee et al., 1993; Wightman et al., 1993). miRNAs are single stranded molecules having length 19–25 nucleotides, which mediate post-transcriptional gene silencing by pairing of bases with the untranslated region (UTR) of target genes. miRNA regulates various function of cells, such as immune response, stress response, apoptosis proliferation, and differentiation (Xiao and Rajewsky, 2009; Liston et al., 2010). They also affect transcription factors, histone acetylation or DNA methylation and a single miRNA may target more than hundreds of genes (Bartel and Chen, 2004).
Leishmaniasis is included among 13 neglected tropical parasitic diseases by the World Health Organization Tropical Disease Research (WHO TDR). The disease is prevalent in more than 98 countries and common in Indian subcontinent and East Africa. About 200,000–400,000 active cases detected each year out of which 90% of the cases are reported mainly in India, Ethiopia, Brazil, Somalia, Sudan, and South Sudan (WHO, 2017). The causative agent Leishmania is an obligate intracellular protozoan parasite, which is transmitted by bite of different species of infected sand fly to the mammalian host. Traditionally, the disease has three clinical forms, i.e., visceral leishmaniasis (VL), cutaneous leishmaniasis (CL), and mucosal leishmaniasis (MCL). Visceral disease is the most common in Indian subcontinent and fatal if not treated (Singh et al., 2006). Approximately, 0.1 million cases of VL are estimated to occur annually in India and of these, the state of Bihar accounts for more than 70% of the cases (National Vector Borne Disease Control Programme [NVBDCP], 2014).
Hypoxia inducible factor-1α (HIF-1α) is a transcription factor activated under hypoxic condition that regulates all cellular responses to hypoxia and ensures optimal functional, metabolic, and vascular adaptation to O2 shortages (Semenza, 2011). It is widely expressed in innate and adaptive immune responses regulating cell populations including macrophages and lymphocytes (Cramer et al., 2003; Walmsley et al., 2005; Jantsch et al., 2008; McNamee et al., 2013). Its activation was reported as a general phenomenon in infections with human pathogens (Werth et al., 2010). In CL, the role of HIF-1α has been reported where it helps in the survival of parasites by stabilizing the hypoxic environment (Arrais-Silva et al., 2005; Degrossoli et al., 2011; Schatz et al., 2016). In addition, it has been documented that hypoxia controls the inflammatory response in human dendritic cells after Leishmania infection (Bosseto et al., 2010). Further, drugs like echinomycin and resveratrol have shown to target HIF-1α to reduce the growth of L. amazonensis (Dal’Bó Pelegrini et al., 2016).
NF-κB transcription factor family comprises five members, i.e., p65 (Rel A), Rel B, c-Rel, p50, and p52 that functions as homo and heterodimers. NF-κB is normally remained as inactive in the cytoplasm by IkB. After activation, IkB is phosphorylated by IKK that degrades IkB resulting in the release of the NF-κB. After release, it translocates into the nucleus where it activates the transcription of various pro-inflammatory cytokines by binding to their promoter region (Senftleben et al., 2001; Lawrence, 2009). Studies have demonstrated a cross signaling between the HIF-1α and NF-κB (D’Ignazio and Rocha, 2016) but direct linkage between these two molecules has yet to be investigated. Since HIF-1α is a macrophage associated factor and the parasite Leishmania resides in the host macrophages, we planned this study to elucidate the role of HIF-1α in parasite survival.
The increasing data suggested that several classes of pathogens can manipulate the miRNA networking system of affected cells which are key regulators of host immune response and disease pathogenesis. For example, miR-155 creates suitable immunological environments for the survival of Mycobacterium tuberculosis inside the host macrophages and also works as a marker of the disease (Wu et al., 2012). Let-7 families of miRNAs are shown to act as first line of defense against Salmonella typhimurium in the host (Schulte et al., 2011). In the protozoan infection such as Cryptosporidium parvum, miR-221, and miR-27b have been found to play significant role in disease pathogenesis (Gong et al., 2011). In Leishmania donovani infection, miR-30A-5P has been shown to play a significant role in regulation of autophagy (Singh et al., 2016). In addition, upregulation of miR-294 and 721 modulates the nitric oxide synthase 2 (NOS2) and L-arginine in Leishmania amazonensis infection in the favor of parasite survival (Muxel et al., 2017).
HypoxamiRs (miR-24-1, miR-26, miR-103, miR-181, miR-213, and miR-210) are a class of miRNAs, which are activated under low oxygen tension and induced by HIF-1α. miRNA-210 is one of the first hypoxamiR reported, which is targeted by HIF-1α (Kulshreshtha et al., 2007). Emerging data have also proved the role of miR-210 in various human diseases (Zhang et al., 2012; Grosso et al., 2013) and also found to act as a regulator of immune responses (Qi et al., 2012; Zhang et al., 2012; Zhao et al., 2014). In Leishmania infection, the manifestation of the disease occurs through impairment of host immune responses, and in our previous study, we found that miR-210 is greatly upregulated in macrophages during L. donovani infection (Tiwari et al., 2017). We further hypothesized that the hypoxic conditions inside the host macrophages may regulate inflammatory cytokines production through HIF-1α induced miR-210/NF-κB mediated mechanisms. Herein, we provide a clue that HIF-1α controls the NF-κB mediated regulatory pathways of macrophages by upregulating miR-210 expression and helps in survival of parasites inside the host.
Materials and Methods
Ethics Statement
This study was carried out in accordance with the recommendations of “Institutional Animal Ethical Committee” of the Rajendra Memorial Research Institute of Medical Sciences (Patna, India). The protocol was also approved by the “Institutional Animal Ethical Committee” of the Rajendra Memorial Research Institute of Medical Sciences (Patna, India, Ref No. 364/GO/R/S/2001/CPCSEA).
Animals
Female BALB/c mice of age 6–8 weeks were used in this study. The animals were kept in polypropylene cages and bedding material was chopped wheat straw with the temperature around 20–30°C. All the experimental mice were fed a standard chow and water ad libitum.
Culture of L. donovani
A cloned line of L. donovani strain (AG83) was used in this study. The motile form of promastigotes was cultured in complete Dulbecco’s Modified Eagle Medium (DMEM, pH 7.2; Gibco, United States), which contained 10% heat-inactivated fetal bovine serum (FBS; Gibco, United States), 2 mM L-glutamine, sodium bicarbonate (3.7 gm/L), penicillin (100 U/mL), streptomycin (100 μg/mL), and gentamicin (20 μg/mL) at 26°C in a BOD incubator. In all stages of experiments, metacyclic promastigotes were used and obtained from late log phage to stationary stage using standard protocol (Späth and Beverley, 2001).
Isolation of Macrophages (Mφ) From Mice
A 4% starch solution was injected to peritoneal cavity of mice and after 48 h macrophages were isolated by standard protocol. Cells were thoroughly washed and suspended in RPMI medium supplemented with 10% FBS and antibiotics. Further, macrophages (106 cells per well) were plated in six well culture plates and incubated in CO2 incubator in 5% CO2 atmosphere. After overnight incubation, the non-adherent cells were removed and fresh medium was added to each wells.
Infection of L. donovani in Macrophages
The macrophages were infected with Leishmania parasites for 6 h at a cells/parasite ratio of 1:10. After 6 h, the unbound parasites were removed by washing with RPMI without FBS and incubated up to 24 h as per our experimental conditions. The cells were then fixed with formaldehyde followed by staining with May-Grünwald Giemsa and observation under bright field microscope at 100× with oil immersion objective.
Measurement of HIF-1α Expression
We evaluated the expression of HIF-1α mRNA in L. donovani infected and uninfected macrophages by semi-quantitative PCR and further validated by real-time PCR (ABI 7500, Applied Biosystem) using HIF-1α specific primers. The cycling conditions for qPCR were as follows: 1 cycle at 95°C for 3 min and 40 cycles of 95°C for 15 s (denaturation), 56°C for 30 s (annealing), and 72°C for 30 s (extension). Results are the target/reference ratios of each sample, normalized by the target/reference ratio of the calibrator. Here, the target/reference values of uninfected macrophages were used as the calibrator and the GAPDH was used as the reference.
Measurement of Cellular Hypoxia
Pimonidazole is a novel marker for confirmation of cellular hypoxia (Varia et al., 1998). The cellular hypoxia was estimated by detection of pimonidazole adducts by western blotting using HypoxyprobeTM-1 kit (Hypoxyprobe-1, Chemicon). Briefly, 250 μM of pimonidazole hydrochloride was added to L. donovani infected and uninfected macrophage culture. After 24 h, proteins were isolated from macrophages and quantified by Lowry’s method (Lowry et al., 1951). The protein (40 μg/lane) was then subjected to SDS–PAGE and transferred on to the nitrocellulose membrane. Further, protein bands on membrane were blocked by TBST buffer with [5% (w/v) non-fat dry milk containing 0.1% (v/v) Tween20] for 2 h. After three times washing the membrane with TBST buffer, membrane was incubated with mouse anti-pimonidazole antibody diluted 1:50 (Chemicon) for 2 h at room temperature (RT). After routine wash with TBST, membrane was incubated with horseradish peroxidase-conjugated secondary antibody, diluted in the ratio of 1:400 for 2 h. Protein bands on the blot were developed by 0.03% (v/v) DAB, 0.01% (v/v) H2O2 solution. β-actin was used as house-keeping protein.
Next-Generation Sequencing, Real-Time Validation, and Selection of miRNA
Small RNA library preparation, TruSeq small RNA library preparation, and miRNAs’ expression are reported in our previous study (Tiwari et al., 2017). In this study, we selected differentially expressed upregulated and downregulated miRNAs for real-time validation. For real-time validation, total RNA was isolated using TRIzol method and quantified by Nanodrop spectrophotometer and cDNA was prepared using miRNA specific primers and following the standard protocol. Briefly, 1 μg of total RNA was taken and reverse transcribed using 20U M-MLV reverse transcriptase (Fermentas, Germany), 1× RT buffer, 20 mM dNTPs (New England Biolabs, United States), 20U RNasin (Fermentas, Germany), 100 ng of random hexamers (Fermentas, Germany), 0.1 M DTT, and DEPC treated water. The expression level was quantified on ABI 7500 Fast system as per manufacturer instructions (Applied Biosystem) using 5 pmol/μL of each miRNA specific primer. Briefly, 20 μL of real-time mixture was prepared which contained 1 μL cDNA, 6 μL MilliQ water, 10 μL of Power SYBER green master mix (Applied Biosystem), and 1.5 μL of forward and reverse primers. Temperatures’ setting for PCR was as follows: initial incubation of 50°C for 2 min, denaturation at 95°C for 10 min and 40 cycles at 95°C for 15 s, 60°C for 1 min, and 72°C for 40 s. The comparative Ct method (ΔΔCt) was used to determine the level of expression of miRNA. The calibrator used for miRNA analysis was non-infected macrophage. Sno miRNA-202 was used as endogenous controls for expression analysis (Choi et al., 2013). We were calculated the fold change (2-ΔΔCt) using the formula normalized gene expression: (2-ΔCt) in the infected macrophage divided the normalized gene expression (2-ΔCt) in the uninfected macrophages as described elsewhere (Livak and Schmittgen, 2001).
Silencing of HIF-1α Gene
For evaluating the role of HIF-1α in regulation of miR-210 expression in L. donovani infected macrophage, HIF-1α was silenced by HIF-1α specific siRNAs or scramble control (Santa Cruz Biotechnology, United States). The silencing of HIF-1α was done using the company specific protocol and reagents. Briefly, macrophages (106 cells/well) were re-suspended in 500 μL transfection medium and transfected with 25 nM siRNA or scramble control. After 6 h of incubation, an additional 500 μL RPMI 1640 complete medium was added and cells were cultured overnight in a six-well culture plate. Next day, cells were washed and further experiments were executed. Silencing efficiency of HIF-1α was evaluated by semi-quantitative PCR. β-actin gene was used as loading control. Further, expression of miR-210 in HIF-1α silenced macrophages was evaluated by qPCR using the miRNA-210 specific primers as described earlier. Sno202 miRNA was taken as an endogenous control.
Bioinformatics Analysis and Luciferase Reporter Assay
We predicted the target of miR-210 by online software miRDB and Target Scan based on target rank and score. Top 20 genes were functionally correlated with activation of NF-κB. Binding site of miR-210 and TNF receptor family was detected by Target Scan 6.0. The 3′-UTR of genes of interest containing the putative miRNA target site(s) WT-TNF-α receptor family (wild-type) and Mut-TNF-α receptor family (mutant) was cloned into the XbaI site of the pGL3 control vector containing firefly luciferase gene linked to the 3′-UTR of gene (Promega, United States). Macrophages were transfected by constructed pGL3 vector with either controls or mimics of miR-210 using Lipofectamine 2000 (Invitrogen, United States). After 24 h, cells were lyzed and luciferase activity was measured by Dual-Glo luciferase Assay System (Promega, United States) according to manufacturer’s instruction on luminometer. Firefly luciferase reading was normalized by renilla luciferase reporter.
Evaluation of the Role of miRNA-210 in NF-κB Mediated Pro-inflammatory Immune Responses
The role of miR-210 in NF-κB mediated pro-inflammatory immune responses was evaluated by silencing with antagomir. The miR-210 antagomir (designed in our laboratory and was synthesized by Integrated DNA Technologies, New Delhi) using sequence, CAGUGUGCGGUGGGCAGUGGCU with the following modification. The 2′-OMe modified bases (2′-hydroxyl of the ribose was replaced with a methoxy group), phosphorothioate (phosphodiester linkages were changed to phosphorothioate) on the first two and last four bases, and an addition of cholesterol motif at 3′ end through a hydroxyproline modified linkage. Antagomir scramble (mismatched miR-210) was also obtained from the same company and used as negative control. The expression of miR-210 was silenced by antagomir according to the standard protocol described previously with required modification (Krützfeldt et al., 2005; Haftmann et al., 2015). Briefly, macrophages (106 cells/well) were suspended in 500 μL of serum-free media for 2 h at 37°C and 5% CO2 supplemented with 1 μM of antagomir-210 or antagomir scramble. After incubation, 150 μL of media containing serum and antibiotics were added and culture for 24 h in CO2 incubator. For evaluating the functional role of miR-210, four experimental groups were plotted as follows: (1) uninfected Mφ, (2) Mφ infected with L. donovani, (3) miR-210 silenced Mφ infected with L. donovani, and (4) antagomir scramble treated Mφ infected with L. donovani. Macrophages were incubated in CO2 incubator with 5% CO2 for 24 h. After 24 h of incubation, supernatants were collected for measurement of cytokines, NOx and reactive oxygen species (ROS).
In another experiment to the study of the role of HIF-1α and miR-210 in parasite infectivity and survival inside the macrophages, parasitic load was measured in siHIF-1α and antagomir treated L. donovani infected macrophages along with negative control at 6, 12, and 24 h of infection. Percent infectivity was calculated by counting the number of infected cells and parasite load was determined by counting the number of amastigotes per 100 macrophages. The results were expressed as mean ± standard deviation (SD) and performed in quadruplicate.
NF-κB Transcription Factor (p50) Activation Assay
Macrophages (106 cells/well) were harvested from culture plates, centrifuged, and pellets were suspended in 1 mL cold PBS. The suspension was further centrifuged at 6000 rpm for 5 min in a cold room and supernatant was removed. Tubes were immediately kept on ice and 5X cytoplasmic proteins extraction buffer (1 M HEPES, 5 M NaCl, 0.5 M EDTA, 100% glycerol, and protease inhibitor) was added and incubated on ice with occasional vortexing from time to time. The suspension was centrifuged at 3000 rpm for 5 min and the supernatant containing cytoplasmic proteins was collected.
Prior to nuclear protein extraction, pellets obtained after cytoplasmic protein were washed with 100 μL of cytoplasmic buffer for four to five times by repeated centrifugation at 3000 rpm for 5 min. Further, 100 μL nuclear protein extraction buffer (1 M HEPES, 2 M KCl, 0.5 M EDTA, 100% NP-40, and protease) was added to the pellet and incubated on ice for 10 min with vortexing occasionally time to time. Samples were further centrifuged at 14,000 rpm for 5 min at 4°C and the supernatant containing nuclear protein was collected. The proteins were quantified and stored at -80°C for execute of further experiments. NF-κB transcription factor (p50) activation assay in cytoplasmic and nuclear proteins extract of all experimental groups was performed using NF-κB activation assay kit (Abcam, ab207217) as per protocol provided by the manufacturer. Optical density was determined at 450 nm using a spectrophotometer.
Expression of NF-κB Transcription Factor (p50)
Expression of NF-κB transcription factor (p50 and p65) was studied by western blot in both cytoplasmic and nuclear proteins of macrophages. Equal amount of protein (40 μg/lane) was loaded and separated on 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE). Total separated protein bands were transferred on nitrocellulose membrane (Millipore, United States). The membranes containing protein bands were blocked in 1× TBST buffer (5% (w/v) non-fat dry milk containing 0.1% (v/v) Tween20) for 1 h at RT. After blocking, membranes were washed with TBST for three times and further incubated with p50 and p65 specific primary antibodies (Santa Cruz Biotechnology, United States) for 4 h (1:200 dilutions). After incubation, membranes were washed with TBST and incubated with horseradish peroxidase-conjugated secondary antibody, diluted in the ratio of 1:500 for 2 h. Protein bands on the blots were developed by 0.03% (v/v) DAB, 0.01% (v/v) H2O2 solution. Lamin B and β-actin (Santa Cruz Biotechnology, United States) proteins were used as reference for nuclear and cytoplasmic protein, respectively. Reference proteins were used to analyze the relative band intensity by Image Analysis Software of gel documentation system (Bio-Rad). The data were represented as mean ±SD of band density ratio of the experiments.
Estimation of Cytokines
The cytokine levels (TNF-α, IL-12, and IL-10) in cell culture supernatant of all the experimental groups were measured by MAXTM standard set enzyme-linked immunosorbent assay (ELISA) kit as per manufacturer’s instructions (BioLegend, United States). The results were represented in pg/mL.
Analysis of Cytokines Expression by RT-PCR
Total RNA was isolated from macrophages of all experimental groups using TRI Reagent® (Sigma Chemicals, United States) following manufacturer’s instruction. Briefly, cells were pelleted by centrifugation at 5,000 rpm and pellet was washed thrice with PBS (0.02 M, pH 7.2). Further, cells were lyzed in 300 μL of TRIzol and 120 μL chloroform. The cells were centrifuged at 10,000 rpm for 10 min. The upper aqueous layer was transferred into another centrifuge tube and twice volume of isopropanol was added. The mixture containing aqueous layer and isopropanol was centrifuged again at 10,000 rpm for 10 min and RNA pellets were collected. In the last step, RNA pellets were washed with 70% (v/v) ethanol treated with RNAse-free DNase to avoid DNA contamination. In all step of centrifugation, 4°C temperature was maintained. For analysis of cytokine mRNA expression, 20 μL cDNA was prepared following the protocol describing elsewhere. Briefly, equal amount of total RNA (1 μg) was taken and reverse transcribed using 1× RT buffer, 20 mM dNTPs (New England Biolabs, United States), 0.1 M DTT with DEPC-treated water, 200 ng of random hexamers (Fermentas, Germany), 20U RNasin (Fermentas, Germany), and 20U M-MLV reverse transcriptase (Fermentas, Germany). The cDNA was subsequently amplified using mRNA specific cytokines primers. The β-actin was used as house-keeping control gene. The 25 μL of reaction mixture was prepared containing 2 μL cDNA templates, 1× polymerase chain reaction buffer, 200 μM dNTPs, 0.5 mM MgCl2, 1U Taq DNA polymerase (New England Biolabs, United States), and 3.2 μM mice mRNA specific forward and reverse primers. Amplification was performed for 22 cycles and temperature and time of PCR were set as follows: denaturation at 94°C for 30 s, annealing at 46–55°C (depended on the Tm of the primers) for 30 s, and extension at 72°C for 30 s. Before the start of PCR cycles, initial denaturation at 94°C for 5 min and after completion of cycles, final extension for 7 min at 72°C was performed. The amplified PCR products were separated on 2% agarose gel containing ethidium bromide (0.5% w/v) and visualized under UV illumination in a gel documentation system (Bio-Rad, United States). The relative mRNA expression levels were analyzed by Image Analysis Software. List of primers used in this study is given in Supplementary Table S1.
Measurement of Total Nitric Oxide (NOx)
In culture supernatant, total nitric oxide (NOx) was estimated by measuring the amount of nitrite using Griess reagent as described elsewhere (Ding et al., 1988). The cultured supernatants (100 μL) were incubated with Griess reagent in the ratio of 1:1 for 10 min at RT. The absorbance was measured at 540 nm. The concentration of total nitrite was determined by comparing with a standard curve plotted using sodium nitrite (50–1.56 μM) as standard and expressed in micromolar.
Measurement of Superoxide Anion (O2-) Level
The super oxide anion content was estimated in cell culture supernatant by the method described elsewhere (Johnstan et al., 1978). The principle of this method is based on the reduction of ferricytochrome c into ferrocytochrome c in the presence of O2- that directly correlates the level of O2- production to the cytochrome c reduced. In brief, 2 mL reaction mixture contained 100 μL of culture supernatant and 0.05 mM ferricytochrome c in PBS. Reaction mixture was incubated for 15 min at 37°C and reactions were terminated by placing the tubes on ice. Absorbance of the supernatant fractions was measured at 550 nm. The concentration of cytochrome c reduced was determined using extinction coefficient 2.1 × 104 M-1 cm-1 and expressed as nmoles of O2- liberated per mg of protein.
Statistical Analysis
The data were analyzed by one-way analysis of variance (ANOVA) using Student’s–Newman–Keuls test by Sigma Stat 3.5 software. The p-value less than 0.05 was considered to be significant. All the experiments were performed in quadruplicate and the data represented as mean ±SD.
Results
L. donovani Increased HIF-1α Expression Levels in Infected Macrophages
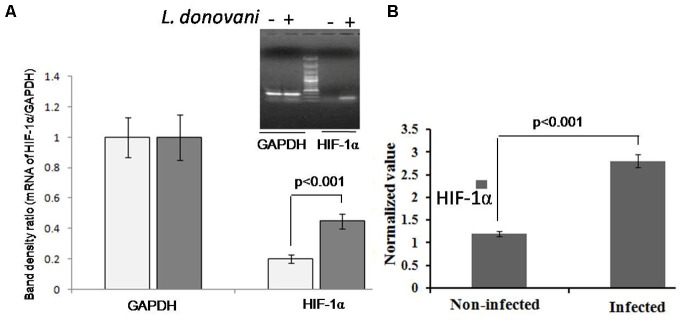
Leishmania donovani infection in macrophages increased the expression of HIF-1α gene, which was significantly higher (p < 0.001) compared to uninfected macrophages. Figure 1A shows the band densitometry of the expression of HIF-1α mRNA and Figure 1B is the real-time validation in infected and uninfected macrophages.
FIGURE 1.
Expression of hypoxia inducible factor-1α (HIF-1α) in macrophages infected with Leishmania donovani. The expression of HIF-1α was evaluated by semi-quantitative (A) or quantitative real-time PCR (B). GAPDH was taken as loading control. Each experiment was performed in quadruplicate and the values were expressed as mean ± SD. The level of HIF-1α in infected macrophages was significantly (p < 0.001) increased compared to uninfected macrophages.
Confirmation of Cellular Hypoxia by Western Blotting
Cellular hypoxia was confirmed by detecting pimonidazole adducts in protein lysate of macrophages by western blotting (Figure 2). In protein lysates of L. donovani infected macrophages, multiple bands were observed since pimonidazole forms adduct with molecules in hypoxic cells.
FIGURE 2.
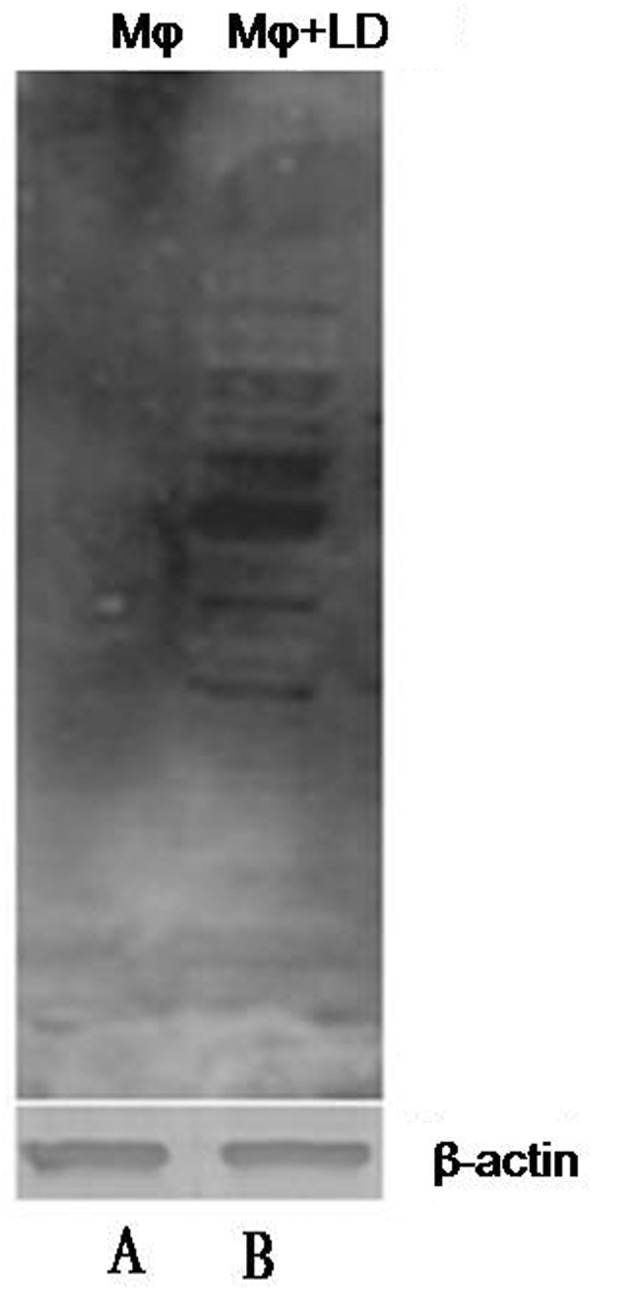
Confirmation of cellular hypoxia in L. donovani infected macrophages by western blotting. We detected multiple bands on blot incubated with mouse anti-pimonidazole antibody diluted 1:50 (Chemicon, United States) for 2 h at room temperature. Lane A shows only blot of macrophage protein, and Lane B shows the blot of L. donovani infected macrophages. The β-actin was taken as control.
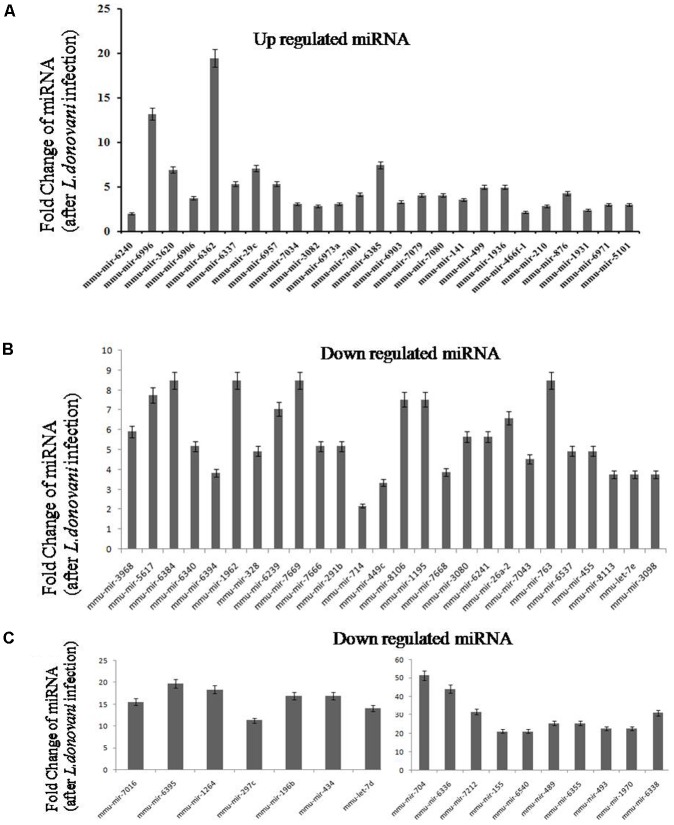
Quantitative PCR for Confirmation of Differentially Expressed miRNA
Out of 150 identified miRNA as shown in heat map (Supplementary Figure S1), we selected 25 highly upregulated and 43 downregulated miRNA for qPCR confirmation. Tables 1, 2 show the fold change of miRNA expression in Leishmania infected macrophages. Graphical representation of qPCR confirmation of differentially expressed miRNAs is shown in Figure 3.
Table 1.
List of upregulated micro RNA (miRNA) in Leishmania donovani infected macrophages.
| Name of miRNA | Fold change | p-Value |
|---|---|---|
| mmu-mir-6240 | 1.986 | 0.035 |
| mmu-mir-6996 | 13.18 | 0.04 |
| mmu-mir-3620 | 6.906 | 0.04 |
| mmu-mir-6906 | 3.744 | 0.048 |
| mmu-mir-6362 | 19.48 | 0.04 |
| mmu-mir-6337 | 5.313 | 0.036 |
| mmu-mir-29c | 7.084 | 0.045 |
| mmu-mir-6957 | 5.313 | 0.04 |
| mmu-mir-7034 | 3.07 | 0.044 |
| mmu-mir-3082 | 2.833 | 0.049 |
| mmu-mir-6973a | 3.086 | 0.049 |
| mmu-mir-7001 | 4.132 | 0.04 |
| mmu-mir-6385 | 7.438 | 0.046 |
| mmu-mir-6903 | 3.276 | 0.048 |
| mmu-mir-7079 | 4.073 | 0.028 |
| mmu-mir-7080 | 4.073 | 0.0368 |
| mmu-mir-141 | 3.542 | 0.041 |
| mmu-mir-499 | 4.958 | 0.046 |
| mmu-mir-1936 | 4.958 | 0.036 |
| mmu-mir-466f-1 | 2.125 | 0.0486 |
| mmu-mir-210 | 2.823 | 0.049 |
| mmu-mir-876 | 4.25 | 0.036 |
| mmu-mir-1931 | 2.391 | 0.045 |
| mmu-mir-6971 | 3.011 | 0.03 |
| mmu-mir-5101 | 3.011 | 0.04 |
Table 2.
List of downregulated miRNA macrophages infected with Leishmania donovani.
| Name of miRNA | Fold change | p-Value |
|---|---|---|
| mmu-mir-704 | 51.387 | 0.006 |
| mmu-mir-6336 | 143.995 | 0.015 |
| mmu-mir-7212 | 31.764 | 0.023 |
| mmu-mir-3968 | 5.916 | 0.018 |
| mmu-mir-7016 | 15.529 | 0.018 |
| mmu-mir-5617 | 7.764 | 0.013 |
| mmu-mir-6384 | 8.47 | 0.012 |
| mmu-mir-6340 | 5.176 | 0.015 |
| mmu-mir-155 | 21.176 | 0.01 |
| mmu-mir-6540 | 21.176 | 0.006 |
| mmu-mir-6395 | 19.764 | 0.01 |
| mmu-mir-6394 | 3.82 | 0.023 |
| mmu-mir-1264 | 18.352 | 0.017 |
| mmu-mir-1962 | 8.47 | 0.013 |
| mmu-mir-6338 | 31.058 | 0.013 |
| mmu-mir-297c | 11.294 | 0.01 |
| mmu-mir-328 | 4.941 | 0.025 |
| mmu-mir-489 | 25.411 | 0.01 |
| mmu-mir-6355 | 25.411 | 0.009 |
| mmu-mir-493 | 22.588 | 0.014 |
| mmu-mir-1970 | 22.588 | 0.014 |
| mmu-mir-6239 | 7.059 | 0.033 |
| mmu-mir-7669 | 8.47 | 0.015 |
| mmu-mir-7666 | 5.176 | 0.027 |
| mmu-mir-291b | 5.176 | 0.046 |
| mmu-mir-714 | 2.186 | 0.039 |
| mmu-mir-449c | 3.337 | 0.047 |
| mmu-mir-8106 | 7.529 | 0.044 |
| mmu-mir-1195 | 7.529 | 0.029 |
| mmu-mir-7668 | 3.882 | 0.0297 |
| mmu-mir-196b | 16.941 | 0.02 |
| mmu-mir-434 | 16.941 | 0.039 |
| mmu-mir-3080 | 5.647 | 0.042 |
| mmu-mir-6241 | 5.647 | 0.04 |
| mmu-mir-26a-2 | 6.588 | 0.014 |
| mmu-mir-7043 | 4.518 | 0.032 |
| mmu-mir-763 | 8.47 | 0.042 |
| mmu-let-7d | 14.117 | 0.034 |
| mmu-mir-6537 | 4.941 | 0.037 |
| mmu-mir-455 | 4.941 | 0.035 |
| mmu-mir-8113 | 3.765 | 0.044 |
| mmu-let-7e | 3.765 | 0.036 |
| mmu-mir-3098 | 3.765 | 0.043 |
FIGURE 3.
Real-time validation of upregulated and downregulated micro RNAs (miRNAs). Macrophages were infected with L. donovani with parasite to macrophage ratio of 10:1. After 6 h, cells were washed to remove non-ingested parasites. Infected macrophages were incubated at 37°C up to 24 h and total RNA was isolated. After next generation sequencing, 25 upregulated (A) and 43 downregulated (B,C) micro-RNA was selected for real-time validation (qPCR). The comparative Ct method (ΔΔCt) was used to determine the levels of expression for the miRNA. The calibrator used for miRNA analysis was non-infected macrophage. Sno miRNA 202 was used as an endogenous control for expression analysis. Relative quantity values or normalized levels expression were obtained by the formula 2-ΔΔCt.
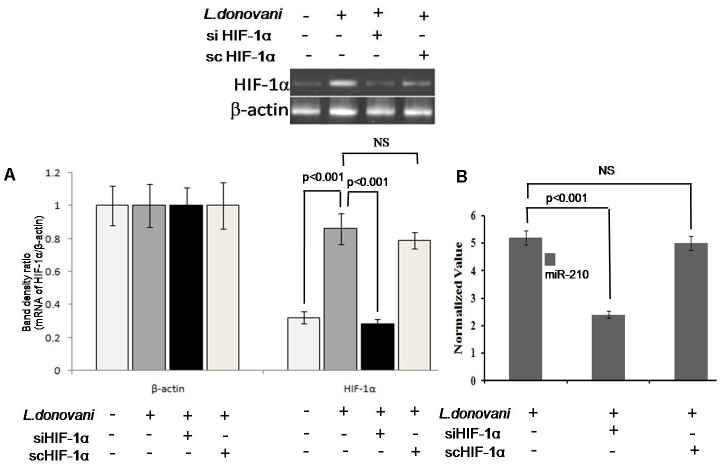
Upregulation of miRNA-210 in Leishmania Infected Macrophage Was HIF-1α Dependent
To find out the role of HIF-1α in regulation of miRNA-210 expression, we silenced HIF-1α gene in macrophages by HIF-1α specific siRNA (Supplementary Figure S2). The HIF-1α expression was found significantly (p < 0.001) increased in Leishmania infected macrophages. However, after silencing of HIF-1α (Figure 4A), the expression of miR-210 was significantly (p < 0.001) downregulated (Figure 4B), which confirmed that the upregulation of miR-210 was HIF-1α dependent. Control scramble siRNA did not affect the HIF-1α either in L. donovani infected or uninfected macrophages.
FIGURE 4.
Expression of miR-210 and HIF-1α in infected macrophages. Four different experimental conditions (Mφ, Mφ+LD, siHIF-1α+Mφ+LD, and scHIF-1α+Mφ+LD) were used for expression analysis of HIF-1α by semi-quantitative PCR (A). The expression of miR-210 in L. donovani infected macrophages in the presence of siHIF-1α or scHIF-1α was estimated by qPCR (B). The sno-202 was taken as endogenous control. The application of siHIF-1α decreased (p < 0.001) the level of miR-210 expression in infected macrophages (B).
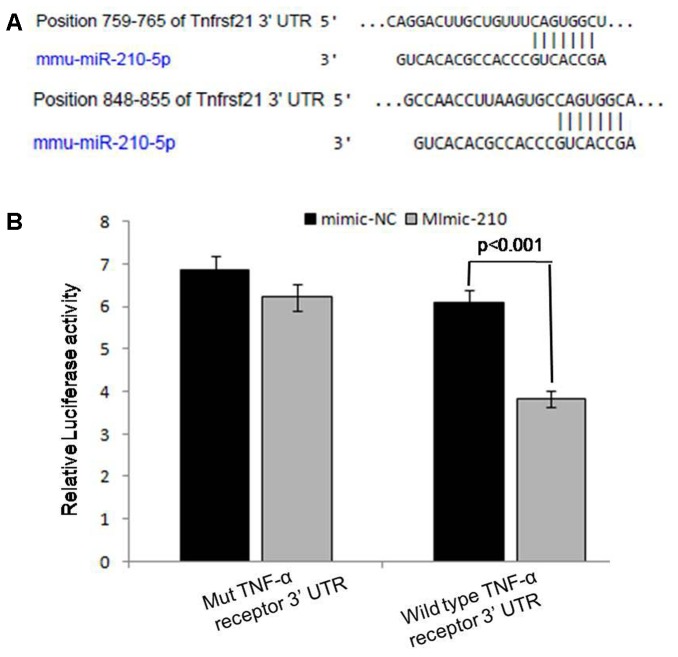
Out of top 20 targeted genes (Supplementary Figure S3) of miRNA-210, we found that sixth rank of its targeted gene, i.e., tumor necrosis factor receptor super family plays an important role in activation of NF-κB (p50) subunit by gene ontology and KEGG (data not shown) since TNF-α is the main inducer of this signaling protein. Further, for validation of miR-210 target genes, we performed the luciferase reporter assay. The transfection with mimic of miR-210 significantly (p < 0.001) reduced the luciferase activities of genes fused to TNF-α receptor family 3′ UTR. Mutated 3′ UTR seed sequences of TNF-α receptor family did not show any significant luciferase activities treated with mimic of miR-210 (Figure 5).
FIGURE 5.
Prediction and validation of TNF-α receptor as target gene of miRNA-210. Schematic representation of miRNA-210 sequence bound to TNF-α receptor (A). The luciferase constructs fused to 3′ untranslated region (UTR) (or Mut) of TNF-α receptor were co-transfected in macrophages cells with mimic-210 or mimic-NC. The luciferase activities were expressed as ratio of firefly luciferase over renilla luciferase. The macrophages transfected with mimic of miR-210 significantly reduced the luciferase activities (p < 0.001) of genes fused to TNF-α receptor family 3′ UTR (B).
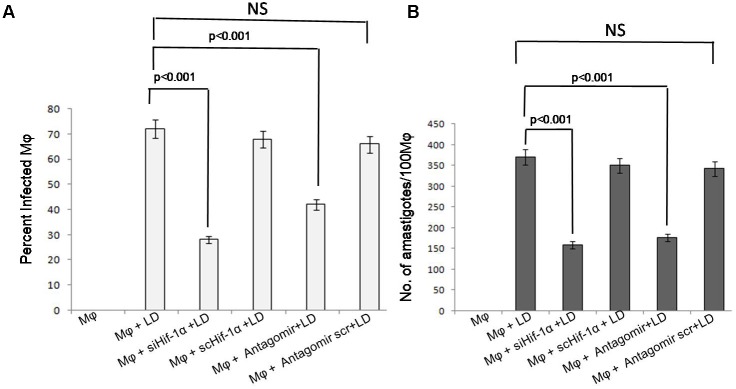
HIF-1α and miR-210 Altered the Parasite Infectivity and Survival in the Macrophages
The role of HIF-1α and miR-210 was validated in the following four experiment groups, i.e., (1) uninfected Mφ, (2) Mφ infected with L. donovani, (3) antagomir treated Mφ infected with L. donovani, and (4) Antagomir scramble treated Mφ infected with L. donovani to validate the role of miR210 on parasitic infectivity and survival.
We determined the percentage of infected macrophages and parasitic load in siHIF-1α and antagomir treated macrophages with negative control. We observed that after silencing HIF-1α, parasite infectivity and parasitic load were significantly (p < 0.001) reduced at 24 h of infection (Figure 6A). A similar pattern was also observed in antagomir-210 treated macrophages where parasite infectivity and parasitic load were found to be significantly (p < 0.001) arrested (Figure 6B). Further, at 12 h of infection, parasite infectivity and parasitic load also found significantly (p < 0.05) decreased (Supplementary Figures S4C,D). However, after 6 h of infection, parasite infectivity and parasitic load did not show significant changes (Supplementary Figures S4A,B).
FIGURE 6.
Effect of siHIF-1α and antagomir-210 on infectivity and survival of Leishmania parasites in the macrophages. The percentage infectivity in siHIF-1α or antagomir-210 treated macrophages was significantly (p < 0.001) decreased compared to untreated macrophages (A). A similar pattern was also observed in parasite survival in macrophages and the amastigotes’ number per 100 macrophages was significantly (p < 0.001) reduced (B). HIF-1α or antagomir scramble did not affect the percentage infectivity or parasite survival.
The miR-210 Regulates the Expression of NF-κB Transcription Factor p50
The role of miR-210 in TNF-α mediated NF-κB p50 activation was investigated. In this study, we performed the NF-κB p50 activation assay in cytoplasmic and nuclear extract in protein lysates in all experimental groups. The optical density in cytoplasmic or nuclear protein lysates was not found significant before antagomir treatment. However, after treatment of macrophages with antagomir, optical density in nuclear extract protein showed optimal level and significantly (p < 0.001) increased (Figure 7).
FIGURE 7.
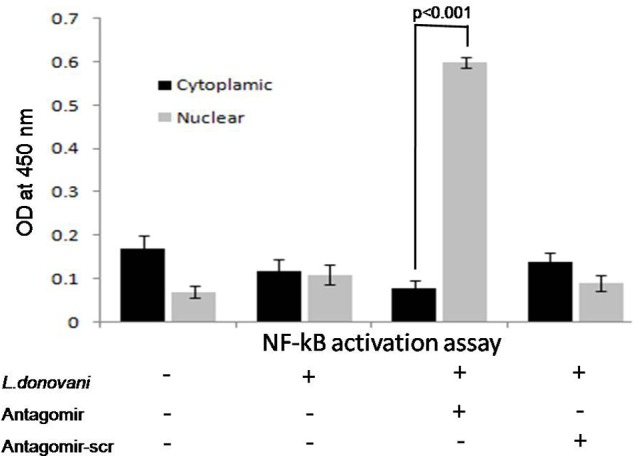
Effect of antagomir-210 in activation of NF-κB p50 subunit in cytoplasmic and nuclear protein extracts of macrophages. The optical density in nuclear protein extracts of antagomir treated macrophage was significantly (p < 0.001) increased. In cytoplasmic protein extract, optical density for NF-κB activation was found normal level. Antagomir scramble did not affect the NF-κB activation in either cytoplasmic or nuclear protein extracts.
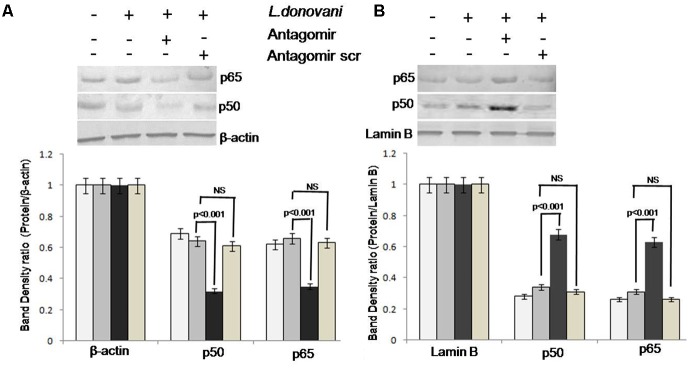
The miR-210 Regulates the NF-κB p50 for Pro-inflammatory Responses in L. donovani Infection
The NF-κB p50 and p65 expression in cytoplasmic and nuclear protein extracts was evaluated by western blotting (Supplementary Figures S5, S6). The expression levels of p50 and p65 in all groups of Mφ cells are depicted in Figures 8A,B. In cytoplasm, before and after L. donovani infection, p50 and p65 showed normal level of expression (lanes 1 and 2 of Figure 8A); however, after miR-210 silencing, the p50 protein forms heterodimer with p65 and translocated to the nucleus and its expression was significantly (p < 0.001) lowered in cytoplasmic protein content (lane 3 of Figure 8A). In a similar manner, before and after L. donovani infection, the expression of p50 and p65 subunit shows normal expression level in nuclear proteins (lane 1 and 2 of Figure 8B); however, after miR-210 silencing, the expression of p50 and p65 proteins was significantly (p < 0.001) higher in nuclear proteins (lane 3 of Figure 8B). Antagomir scramble did not affect the NF-κB p50 and p65 expression in either cytoplasmic or nuclear protein extracts. These findings clearly showed that miR-210 regulates the activation of NF-κB p50 and hypoxia induced miR-210 targets the TNF-α receptor super family gene.
FIGURE 8.
Western blot analysis of NF-κB p50 and p65 in cytoplasmic and nuclear protein extract of macrophages. Proteins were quantified by Lowry methods and equal amount of protein (40 μg/lane) was subjected to SDS–PAGE followed by western blotting using NF-κB p50 and p65 specific monoclonal antibodies. The expression of NF-κB p50 and p65 in cytoplasmic and nuclear proteins was observed by gel documentation system and the bands’ intensity was quantified by densitometry by Quantity One software. The NF-κB p50 and p65 expression in cytoplasmic protein of antagomir treated macrophages was significantly (p < 0.001) decreased compared to untreated macrophages (A). Further, in nuclear protein, the NF-κB p50 and p65 expression in antagomir treated macrophages was significantly (p < 0.001) increased (B). Antagomir scramble did not affect the either cytoplasmic or nuclear protein expression.
Estimation of Cytokines
Release of cytokines in cultured supernatant was measured by cytokine ELISA and is depicted in Figure 9. The level of pro-inflammatory cytokines, i.e., TNF-α and IL-12, was significantly higher (p < 0.05) in antagomir treated macrophages compared to untreated macrophages. However, the level of anti-inflammatory cytokines, i.e., IL-10 in antagomir treated macrophages was found lower (p < 0.001) compared to untreated macrophages. The level of pro-inflammatory cytokines mRNAs showed a similar pattern of result and their expression in antagomir treated macrophages was higher compared to untreated macrophages (Supplementary Figures S7–S9). The expression level of IL-10 was lower in antagomir treated macrophages compared to untreated cells. The macrophages treated with antagomir scramble did not show significant change in pro- or anti-inflammatory cytokines level compared to untreated macrophages. These results indicated that miR-210 has an important role in IL-10 cytokine production during Leishmania infection and its control can upregulate the production of inflammatory cytokines.
FIGURE 9.
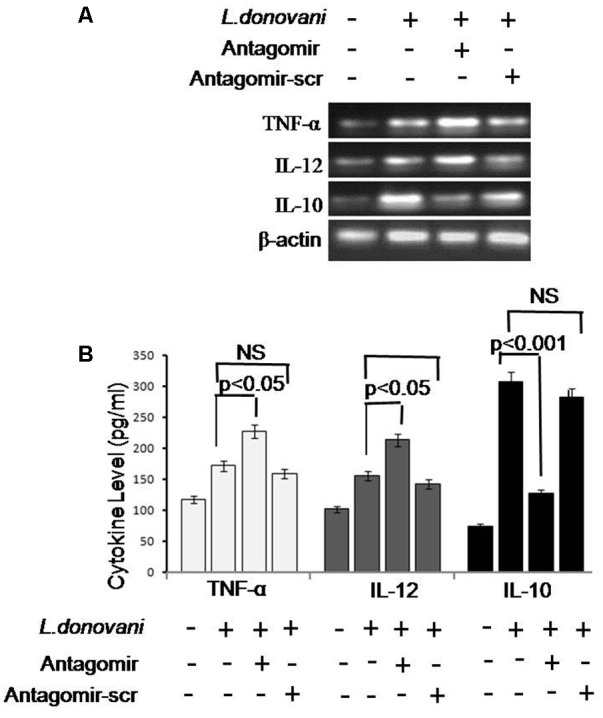
Expression of pro-inflammatory (TNF-α and IL-12) and anti-inflammatory cytokines (IL-10) cytokines. Cytokines mRNA expression levels were quantified by semi-quantitative PCR. β-actin was used as house keeping control gene (A). We observed that mRNA expression of TNF-α and IL-12 was significantly upregulated after antagomir treatment; however, expression of IL-10 was significantly downregulated. Antagomir scramble did not alter the mRNA expression of cytokines. A similar pattern was also observed by enzyme-linked immunosorbent assay (ELISA) (B). The infected cells expressed low levels of pro-inflammatory cytokines and increased level of IL-10. In antagomir treated macrophages, the level of pro-inflammatory cytokines was increased (p < 0.05) and IL-10 level was decreased (p < 0.001). Antagomir scramble did not affect the level of pro or anti-inflammatory cytokines.
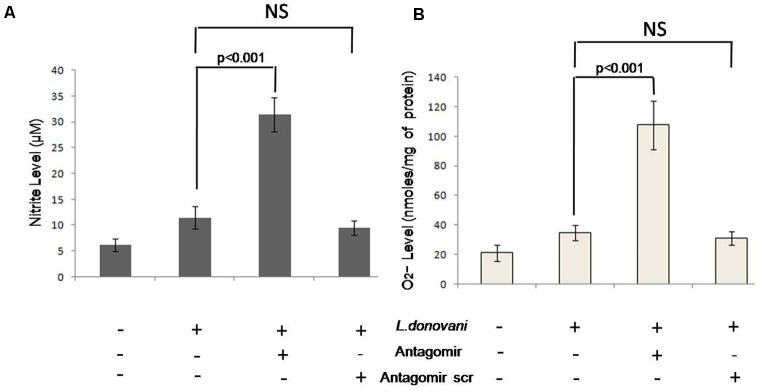
Estimation of O2- and NOx Level
Respiratory burst activity was measured in terms of O2- level after 2 h and NOx levels after 24 h of infection in culture supernatants and results are depicted in Figures 10A,B, respectively. The levels of O2- and NOx in antagomir treated macrophages were 107.64 ± 15.26 nmoles/per mg of protein and 31.45 ± 3.12 μM, respectively. However, in untreated macrophages, their levels were 34.67 ± 5.41 nmoles/per mg and 11.53 ± 2.21 μM, respectively. The O2- and NOx levels were found increased (p < 0.001) in antagomir treated macrophages compared to untreated macrophages. The antagomir scramble treated macrophages did not show significant change in either O2- or NOx levels. These results confirmed the regulatory role of miR-210 in macrophage effector functions in terms of respiratory burst activity.
FIGURE 10.
Production of nitrite and superoxide by macrophages. The level of nitrite was measured using Griess reagent and the absorbance was recorded in ELISA plate reader at 540 nm and the results were expressed in μM (A). The superoxide anion absorbance was recorded at 550 nm and the results were expressed in nmoles/mg of protein (B). The nitrite and O2- levels in antagomir treated macrophages culture supernatant were significantly (p < 0.001) higher than non-treated groups. Antagomir scramble did not affect their level of production.
Discussion
Low oxygen condition or hypoxia is a physiological stress that is created in affected cells in many human diseases (Semenza, 2007). After stress of low oxygen, a transcription factor HIF-1α is induced inside the affected cells and has been to shown to regulate host immune response in either the favor or the elimination of pathogens. Previously, it has been reported that after infection, several classes of pathogens such as Salmonella typhimurium, Pseudomonas aeruginosa, Streptococcus pyogenes, and HIF-1α promote killing of pathogens by modulating immune response of the host (Peyssonnaux et al., 2005; Nizet and Johnson, 2009). However, in Toxoplasma gondii infection, the activation of HIF-1α in affected host cells promotes survival and growth of this protozoan parasite albeit exact mechanisms are not known (Wiley et al., 2010). The role of HIF-1α activation and its consequences on host immune mechanisms in protozoan parasites are poorly understood. The study on Leishmania amazonensis infection confirmed that HIF-1α promotes survival of parasites but the mechanisms are largely unknown (Arrais-Silva et al., 2005). This study confirmed that L. donovani infection activates HIF-1α expression, which eventually alters the production of inflammatory cytokines in the favor of parasite survival through upregulation of miR-210 expression.
HypoxamiR is a class of miRNAs that is induced under influence of hypoxia in different types of primary and transformed cells (Devlin et al., 2011). MicroRNA-210 is the most important hypoxamiR, found in many cell types including macrophages. The HIF-1α binds to highly conserved sequences, i.e., hypoxia responsive element (HRE) on the proximal miR-210 promoter, which is located 400 bp upstream on the chromosome 11p15.5 (Huang et al., 2010). The miRNA profiles of macrophages have been done in response to different species of Leishmania infection (Lemaire et al., 2013; Geraci et al., 2015; Singh et al., 2016). In our present study, we found that during course of L. donovani infection, miR-210 expression was upregulated that helps in survival of parasites by suppression of host pro-inflammatory immune response. Further, the regulation of miR-210 expression in Leishmania infection was found to be HIF-1α dependent as silencing of HIF-1α, downregulated the miR-210 expression. This study suggests that L. donovani exploits activation of HIF-1α and miR-210 in mammalian host for its survival and growth. The findings of this study may open many lock and be useful in exploring immunological regulation during VL.
HIF-1α dependent activation of NF-κB has also been demonstrated in cancer cell lines (Bandarra et al., 2015). It is well established that HIF-1α directly or indirectly regulates NF-κB activation but the intermediate molecules between these two are not yet identified. Further, the activation of the transcription factor NF-κB in L. donovani infection is poorly understood (Guizani-Tabbane et al., 2004). In the present study, we found that miR-210 targets many genes of inflammatory immune response that was observed by online software miRDB. TNF receptor family is sixth rank target gene of miR-210 that was further validated by luciferase activity in our study and helps in NF-κB activation. TNF-α is one of the main inducers of NF-κB family and our results pointed its miR-210 mediated immune-regulation during VL that favors survival of parasites.
The NF-κB activated by the action of several stimuli such as inflammatory cytokines, bacterial products, viruses, physical and physiological stresses, and the pathway regulating TNF-α mediated NF-κB activation has been well established (Zhou et al., 2003). It has been previously reported that active NF-κB p50 forms heterodimer with p65 or c-Rel and translocates into nucleus where it binds to consensus sequences and activates the transcription of various genes (Chen and Ghosh, 1999). This study showed that inhibition of miR-210 by antagomir activates NF-κBp50 and augments the pro-inflammatory immune response. The NF-κBp50 has been shown to play an important role in pro-inflammatory immune responses (Tak and Firestein, 2001). In addition, it has also been shown to be involved in activation of inflammatory cytokine TNF-α in human monocytes by blocking the anti-inflammatory cytokine, i.e., IL-10 production (Schottelius et al., 1999). This study adds a new insight that TNF-α production is NF-κB dependent, which is broadly controlled by miR-210 during course of L. donovani infection.
After infection, Leishmania struggles for survival inside the macrophages that is directly linked to its combat with pro-oxidants like O2 and H2O2. After phagocytosis of parasites, bacteria, and other pathogens by a phagocytic cell, the ROS are rapidly produced by NADPH oxidase and help in the generation of anti-pathogenic environment (Horta et al., 2012). These toxic radicals, especially produced by macrophages and neutrophils, react with pathogens proteins, lipids, and nucleic acids, which eventually lead to their killing (Iles and Forman, 2002; Winyard et al., 2005). These radicals also augment the inflammatory response by playing the regulatory role of tyrosine and mitogen-activated protein kinases and activation of transcription factors like NF-κB, AP-1 leading to the production of pro- or anti-inflammatory cytokines (Closa and Folch-Puy, 2004). Leishmania species are known to downregulate the production of toxic free radicals that are considered as major macrophage effector molecules to control infection (Gantt et al., 2001). This study showed that HIF-1α induces miR-210 expression, which also controls respiratory burst activities of macrophages during Leishmania infection. After silencing of miR-210, the increased levels of NOx and ROS were observed that might be due to pro-inflammatory cytokines activation, which influenced the production of these free radicals and helped in the killing of parasites.
Conclusion
To conclude, the HIF-1α helps L. donovani survival through mir-210 upregulation. Further, the upregulated miR-210 inhibits TNF-α receptor family leading to decreased synthesis of various pro-inflammatory cytokines, which helps the parasite survival inside the macrophages. However, after silencing the miR-210 with antagomir, TNF-α stimulates NF-κB p50, which forms heterodimer with subunit of NF-κB and translocates into the nucleus where it binds to consensus sequences and promotes transcription of pro-inflammatory cytokines genes. This cascade further augments the ROS and NO production leading to killing of parasites.
Author Contributions
VK and AjK designed and performed the experiments. AsK, KA, SD, SV, and AM helped in the design of the study and statistical analysis. VK, RS, and PD designed and co-wrote the manuscript. PD conceived, designed, directed, and supervised the complete study.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
VK is very thankful to Science and Engineering Research Board, New Delhi, for providing research grant under Young Scientist Scheme (YSS/2015/000687). RS is very thankful to Science and Engineering Research Board, Department of Science and Technology, New Delhi, for providing research grant (SB/SO/HS/0091/2013).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00385/full#supplementary-material
References
- Arrais-Silva W. W., Paffaro V. A., Jr., Yamada A. T., Giorgio S. (2005). Expression of hypoxia-inducible factor-1alpha in the cutaneous lesions of BALB/c mice infected with Leishmania amazonensis. Exp. Mol. Pathol. 78 49–54. 10.1016/j.yexmp.2004.09.002 [DOI] [PubMed] [Google Scholar]
- Bandarra D., Biddlestone J., Mudie S., Müller H. A., Rocha S. (2015). HIF-1α restricts NF-κB-dependent gene expression to control innate immunity signals. Dis. Model. Mech. 8 169–181. 10.1242/dmm.017285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P., Chen C. Z. (2004). Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 5 396–400. 10.1038/nrg1328 [DOI] [PubMed] [Google Scholar]
- Bosseto M. C., Palma P. V., Covas D. T., Giorgio S. (2010). Hypoxia modulates phenotype, inflammatory response, and leishmanial infection of human dendritic cells. J. Pathol. Microb. Immunol. 118 108–114. 10.1111/j.1600-0463.2009.02568.x [DOI] [PubMed] [Google Scholar]
- Chen F. E., Ghosh G. (1999). Regulation of DNA binding by Rel/NF-kappaB transcription factors: structural views. Oncogene 18 6845–6852. 10.1038/sj.onc.1203224 [DOI] [PubMed] [Google Scholar]
- Choi J. W., Kang S. M., Lee Y., Hong S. H., Sanek N. A., Young W. S., et al. (2013). MicroRNA profiling in the mouse hypothalamus reveals oxytocin-regulating microRNA. J. Neurochem. 126 331–337. 10.1111/jnc.12308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Closa D., Folch-Puy E. (2004). Oxygen free radicals and the systemic inflammatory response. IUBMB Life 56 185–191. 10.1080/15216540410001701642 [DOI] [PubMed] [Google Scholar]
- Cramer T., Yamanishi Y., Clausen B. E., Förster I., Pawlinski R., Mackman N., et al. (2003). HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell 7 645–657. 10.1016/S0092-8674(03)00154-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal’Bó Pelegrini M., Pereira J. B., Dos Santos Costa S., Salazar Terreros M. J., Degrossoli A. (2016). Evaluation of hypoxia inducible factor targeting pharmacological drugs as antileishmanial agents. Asian Pac. J. Trop. Med. 9 652–657. 10.1016/j.apjtm.2016.05.018 [DOI] [PubMed] [Google Scholar]
- Degrossoli A., Arrais-Silva W. W., Colhone M. C., Gadelha F. R., Joazeiro P. P., Giorgio S. (2011). The influence of low oxygen on macrophage response to Leishmania infection. Scand. J. Immunol. 74 165–175. 10.1111/j.1365-3083.2011.02566.x [DOI] [PubMed] [Google Scholar]
- Devlin C., Greco S., Martelli F., Ivan M. (2011). miR-210: more than a silent player in hypoxia. IUBMB Life 63 94–100. 10.1002/iub.427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ignazio L., Rocha S. (2016). Hypoxia induced NF-κB. Cells 5:10. 10.3390/cells5010010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding A. H., Nathan C. F., Stuehr D. J. (1988). Release of reactive nitrogen intermediates and reactive oxygen intermediates from mouse peritoneal macrophages: comparison of activating cytokines and evidence for independent production. J. Immunol. 141 2407–2412. [PubMed] [Google Scholar]
- Gantt K. R., Goldman T. L., McCormick M. L., Miller M. A., Jeronimo S. M., Nascimento E. T., et al. (2001). Oxidative responses of human and murine macrophages during phagocytosis of Leishmania chagasi. J. Immunol. 167 893–901. 10.4049/jimmunol.167.2.893 [DOI] [PubMed] [Google Scholar]
- Geraci N. S., Tan J. C., McDowell M. A. (2015). Characterization of microRNA expression profiles in Leishmania infected human phagocytes. Parasite Immunol. 37 43–51. 10.1111/pim.12156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong A. Y., Hu G., Zhou R., Liu J., Feng Y., Soukup G. A., et al. (2011). MicroRNA-221 controls expression of intercellular adhesion molecule-1 in epithelial cells in response to Cryptosporidium parvum infection. Int. J. Parasitol. 41 397–403. 10.1016/j.ijpara.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosso S., Doyen J., Parks S. K., Bertero T., Paye A., Cardinaud B. (2013). MiR-210 promotes a hypoxic phenotype and increases radioresistance in human lung cancer cell lines. Cell Death Dis. 14:e544. 10.1038/cddis.2013.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizani-Tabbane L., Ben-Aissa K., Belghith M., Sassi A., Dellagi K. (2004). Leishmania major amastigotes induce p50/c-Rel NF-kappa B transcription factor in human macrophages: involvement in cytokine synthesis. Infect. Immun. 72 2582–2589. 10.1128/IAI.72.5.2582-2589.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haftmann C., Riedel R., Porstner M., Wittmann J., Chang H. D., Radbruch A. (2015). Direct uptake of Antagomirs and efficient knockdown of miRNA in primary B and T lymphocytes. J. Immunol. Methods 426 128–133. 10.1016/j.jim.2015.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horta M. F., Mendes B. P., Roma E. H., Noronha F. S., Macêdo J. P., Oliveira L. S., et al. (2012). Reactive oxygen species and nitric oxide in cutaneous leishmaniasis. J. Parasitol. Res. 2012:203818. 10.1155/2012/203818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Le Q. T., Giaccia A. J. (2010). MiR-210–micromanager of the hypoxia pathway. Trends Mol. Med. 16 230–237. 10.1016/j.molmed.2010.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iles K. E., Forman J. A. (2002). Macrophage signaling and respiratory burst. Immunol. Res. 26 95–105. 10.1385/IR:26:1-3:095 [DOI] [PubMed] [Google Scholar]
- Jantsch J., Chakravortty D., Turza N., Prechtel A. T., Buchholz B., Gerlach R. G., et al. (2008). Hypoxia and hypoxia-inducible factor-1 alpha modulate lipopolysaccharide-induced dendritic cell activation and function. J. Immunol. 180 4697–4705. 10.4049/jimmunol.180.7.4697 [DOI] [PubMed] [Google Scholar]
- Johnstan R. B., Godzik C. A., Cohn Z. A. (1978). Increased superoxide anion production by immunologically activated and chemically elicited macrophages. J. Exp. Med. 48 115–129. 10.1084/jem.148.1.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krützfeldt J., Rajewsky N., Braich R., Rajeev K. G., Tuschl T., Manoharan M. (2005). Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438 685–689. 10.1038/nature04303 [DOI] [PubMed] [Google Scholar]
- Kulshreshtha R., Ferracin M., Wojcik S. E., Garzon R., Alder H., Agosto-Perez F. J. (2007). A microRNA signature of hypoxia. Mol. Cell. Biol. 27 1859–1867. 10.1128/MCB.01395-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence T. (2009). The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 1:a001651. 10.1101/cshperspect.a001651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L., Ambros V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75 843–854. 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- Lemaire J., Mkannez G., Guerfali F. Z., Gustin C., Attia H., Sghaier R. M., et al. (2013). MicroRNA expression profile in human macrophages in response to Leishmania major infection. PLoS Negl. Trop. Dis. 3:e2478. 10.1371/journal.pntd.0002478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A., Linterman M., Lu L. F. (2010). MicroRNA in the adaptive immune system, in sickness and in health. J. Clin. Immunol. 30 339–346. 10.1007/s10875-010-9378-5 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lowry O. H., Rosebrough N. J., Farr A. L., Randal R. J. (1951). Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193 265–275. [PubMed] [Google Scholar]
- McNamee E. N., Korns Johnson D., Homann D., Clambey E. T. (2013). Hypoxia and hypoxia-inducible factors as regulators of T cell development, differentiation, and function. Immunol. Res. 55 58–70. 10.1007/s12026-012-8349-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muxel S. M., Laranjeira-Silva M. F., Zampieri R. A., Floeter-Winter L. M. (2017). Leishmania (Leishmania) amazonensis induces macrophage miR-294 and miR-721 expression and modulates infection by targeting NOS2 and L-arginine metabolism. Sci. Rep. 7:44141. 10.1038/srep44141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Vector Borne Disease Control Programme [NVBDCP] (2014). Available at: http://nvbdcp.gov.in/Doc/Road-map-KA_2014.pdf [Google Scholar]
- Nizet V., Johnson R. S. (2009). Interdependence of hypoxic and innate immune responses. Nat. Rev. Immunol. 9 609–617. 10.1038/nri2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyssonnaux C., Datta V., Cramer T., Doedens A., Theodorakis E. A., Gallo R. L. (2005). HIF-1alpha expression regulates the bactericidal capacity of phagocytes. J. Clin. Invest. 115 1806–1815. 10.1172/JCI23865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J., Qiao Y., Wang P., Li S., Zhao W., Gao C. (2012). microRNA-210 negatively regulates LPS induced production of proinflammatory cytokines by targeting NF-κB1 in murine macrophages. FEBS Lett. 586 1201–1207. 10.1016/j.febslet.2012.03.011 [DOI] [PubMed] [Google Scholar]
- Schatz V., Strüssmann Y., Mahnke A., Schley G., Waldner M., Ritter U., et al. (2016). Myeloid cell-derived HIF-1α promotes control of Leishmania major. J. Immunol. 15 4034–4041. 10.4049/jimmunol.1601080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schottelius A. J., Mayo M. W., Sartor R. B., Baldwin A. S., Jr. (1999). Interleukin-10 signaling blocks inhibitor of kappaB kinase activity and nuclear factor kappaB DNA binding. J. Biol. Chem. 274 31868–31874. 10.1074/jbc.274.45.31868 [DOI] [PubMed] [Google Scholar]
- Schulte L. N., Eulalio A., Mollenkopf H. J., Reinhardt R., Vogel J. (2011). Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J. 30 1977–1989. 10.1038/emboj.2011.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza G. L. (2007). Hypoxia and human diseases. J. Mol. Med. 85 1293–1294. 10.1007/s00109-007-0285-z [DOI] [PubMed] [Google Scholar]
- Semenza G. L. (2011). Hypoxia-inducible factor 1: regulator of mitochondrial metabolism and mediator of ischemic preconditioning. Biochim. Biophys. Acta 1813 1263–1268. 10.1016/j.bbamcr.2010.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senftleben U., Cao Y., Xiao G., Greten F. R., Krähn G., Bonizzi G., et al. (2001). Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science 293 1495–1499. 10.1126/science.1062677 [DOI] [PubMed] [Google Scholar]
- Singh A. K., Pandey R. K., Shaha C., Madhubala R. (2016). microRNA expression profiling of Leishmania donovani infected host cells uncovers the regulatory role of MIR30A-3p in host autophagy. Autophagy 2 1817–1831. 10.1080/15548627.2016.1203500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. K., Pandey H. P., Sundar S. (2006). Visceral leishmaniasis (kala-azar): challenges ahead. Ind. J. Med. Res. 123 331–344. [PubMed] [Google Scholar]
- Späth G. F., Beverley S. M. (2001). A lipophosphoglycan-independent method for isolation of infective Leishmania metacyclic promastigotes by density gradient centrifugation. Exp. Parasitol. 99 97–103. 10.1006/expr.2001.4656 [DOI] [PubMed] [Google Scholar]
- Tak P. P., Firestein G. S. (2001). NF-κB: a key role in inflammatory diseases. J. Clin. Invest. 107 7–11. 10.1172/JCI11830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari N., Kumar V., Gedda M. R., Singh A. K., Singh V. K., Singh S. P., et al. (2017). Identification and characterization of miRNAs in response to Leishmania donovani infection: delineation of their roles in macrophage dysfunction. Front. Microbiol. 8:314 10.3389/fmicb.2017.00314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varia M. A., Calkins-Adams D. P., Rinker L. H., Kennedy A. S., Novotny D. B., Fowler W. C., et al. (1998). Pimonidazole: a novel hypoxia marker for complementary study of tumor hypoxia and cell proliferation in cervical carcinoma. Gynecol. Oncol. 71 270–277. 10.1006/gyno.1998.5163 [DOI] [PubMed] [Google Scholar]
- Walmsley S. R., Print C., Farahi N., Peyssonnaux C., Johnson R. S., Cramer T., et al. (2005). Hypoxia-induced neutrophil survival is mediated by HIF-1alpha-dependent NF-kappaB activity. J. Exp. Med. 201 105–115. 10.1084/jem.20040624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werth N., Beerlage C., Rosenberger C., Yazdi A. S., Edelmann M., Amr A., et al. (2010). Activation of hypoxia inducible factor 1 is a general phenomenon in infections with human pathogens. PLoS One 5:e11576. 10.1371/journal.pone.0011576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2017). Community-driven Programme is Key to Defeating Visceral Leishmaniasis in Bangladesh. Available at: http://www.who.int/leishmaniasis/en/ [Google Scholar]
- Wightman B., Ha I., Ruvkun G. (1993). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75 855–862. 10.1016/0092-8674(93)90530-4 [DOI] [PubMed] [Google Scholar]
- Wiley M., Sweeney K. R., Chan D. A., Brown K. M., McMurtrey C., Howard E. W., et al. (2010). Toxoplasma gondii activates hypoxia-inducible factor (HIF) by stabilizing the HIF- 1alpha subunit via type I activin-like receptor kinase receptor signaling. J. Biol. Chem. 285 26852–26860. 10.1074/jbc.M110.147041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winyard P. G., Moody C. J., Jacob C. (2005). Oxidative activation of antioxidant defence. Trends Biochem. Sci. 30 453–461. 10.1016/j.tibs.2005.06.001 [DOI] [PubMed] [Google Scholar]
- Wu J., Lu C., Diao N., Zhang S., Wang S., Wang F., et al. (2012). Analysis of microRNA expression profiling identifies miR-155 and miR-155 as potential diagnostic markers for active tuberculosis: a preliminary study. Hum. Immunol. 73 31–37. 10.1016/j.humimm.2011.10.003 [DOI] [PubMed] [Google Scholar]
- Xiao C., Rajewsky K. (2009). MicroRNA control in the immune system: basic principles. Cell 136 26–36. 10.1016/j.cell.2008.12.027 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Fei M., Xue G., Zhou Q., Jia Y., Li L. (2012). Elevated levels of hypoxia-inducible microRNA-210 in pre-eclampsia: new insights into molecular mechanisms for the disease. J. Cell. Mol. Med. 16 249–259. 10.1111/j.1582-4934.2011.01291.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Wang L. T., Liang G. P., Zhang P., Deng X. J., Tang Q. (2014). Up-regulation of microRNA210 induces immune dysfunction via targeting FOXP3 in CD4+ T cells of psoriasis vulgaris. Clin. Immunol. 150 22–30. 10.1016/j.clim.2013.10.009 [DOI] [PubMed] [Google Scholar]
- Zhou A., Scoggin S., Gaynor R. B., Williams N. S. (2003). Identification of NF-κB-regulated genes induced by TNFalpha utilizing expression profiling and RNA interference. Oncogene 22 2054–2064. 10.1038/sj.onc.1206262 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.