Abstract
Seed plants are sessile organisms that have developed a plethora of strategies for sensing, avoiding, and responding to stress. Several proteins, including the glycine-rich protein (GRP) superfamily, are involved in cellular stress responses and signaling. GRPs are characterized by high glycine content and the presence of conserved segments including glycine-containing structural motifs composed of repetitive amino acid residues. The general structure of this superfamily facilitates division of GRPs into five main subclasses. Although the participation of GRPs in plant stress response has been indicated in numerous model and non-model plant species, relatively little is known about the key physiological processes and molecular mechanisms in which those proteins are engaged. Class I, II, and IV members are known to be involved in hormone signaling, stress acclimation, and floral development, and are crucial for regulation of plant cells growth. GRPs of class IV [RNA-binding proteins (RBPs)] are involved in alternative splicing or regulation of transcription and stomatal movement, seed, pollen, and stamen development; their accumulation is regulated by the circadian clock. Owing to the fact that the overexpression of GRPs can confer tolerance to stress (e.g., some are involved in cold acclimation and may improve growth at low temperatures), these proteins could play a promising role in agriculture through plant genetic engineering. Consequently, isolation, cloning, characterization, and functional validation of novel GRPs expressed in response to the diverse stress conditions are expected to be growing areas of research in the coming years. According to our knowledge, this is the first comprehensive review on participation of plant GRPs in the response to diverse stress stimuli.
Keywords: GRPs, conserved segments, temperature stress, wounding, osmotic stress, drought, oxidative stress, biotic stress
Introduction
Plants have evolved a number of adaptive stress responses that operate at morphological, physiological, biochemical, and molecular levels. These strategies depend on a plethora of diverse signaling pathways, involving a broad spectrum of molecules, including enzymes, transcription factors, and other proteins associated with plant defense systems. Such strategies seem to be crucial because plants are sessile organisms constantly exposed to environmental stimuli (Al-Whaibi, 2011; Smékalová et al., 2014; Pan et al., 2016). Under the influence of multiple stressors, distinct adaptive mechanisms are more active than is observed in the case of a single stressor (Miao et al., 2015; Pandey et al., 2015).
Abiotic stress, such as decreased or elevated temperature, can seriously limit plant growth and productivity and, in consequence, global food production (Miao et al., 2015; Guo et al., 2016; Sah et al., 2016; Wang et al., 2016; Rihan et al., 2017). It is increasingly important to reduce losses in food production. To understand the complex mechanisms affecting plant growth and development under stress-inducing conditions, a number of studies on economically and ecologically important species are currently underway. The characterization of novel stress-responsive proteins, including glycine-rich proteins (GRPs), will provide substantial insight into stress tolerance mechanisms and will facilitate plant genetic improvement through the production of genotypes with enhanced stress tolerance or rapid growth and development under sub-optimal environmental conditions (Miao et al., 2015; Pandey et al., 2015; Wang et al., 2016; Rihan et al., 2017).
In this review, various classes of GRPs are described, with special focus on class IV GRPs, which are RNA-binding proteins (RBPs) known to be involved in plant defense mechanisms. To our knowledge, this is the first complex compilation on the participation of plant GRPs in various stress responses. Such proteins are involved in the regulation of diverse steps in RNA post-transcriptional processing, including splicing and polyadenylation, which are believed to play a crucial role in responses to a variety of detrimental conditions (Yang et al., 2014). GRPs also belong to the most important proteins involved in plant defense pathways; despite this, little is known about their function or the relationship between their structure and function. In the current review, we will focus on the structure and crucial functions of GRPs. Despite some gaps in current knowledge, this study presents novel and interesting evidence for specific involvement of GRPs in stress responses and highlights the broad spectrum of activities performed by GRPs and their modes of action.
Characteristics of the GRP superfamily
GRPs are known to be involved in plant defense systems induced by abiotic and biotic stress, and are characterized by a high glycine content (up to 70%) and the presence of glycine-containing motifs composed of repetitive amino acid residues (Ortega-Amaro et al., 2015). The general structure of GRPs makes it possible to divide them into five classes (Figure 1, Table 1). This categorization is based on the arrangement of glycine repeats and presence of conserved motifs within particular GRPs (Mangeon et al., 2010).
Figure 1.
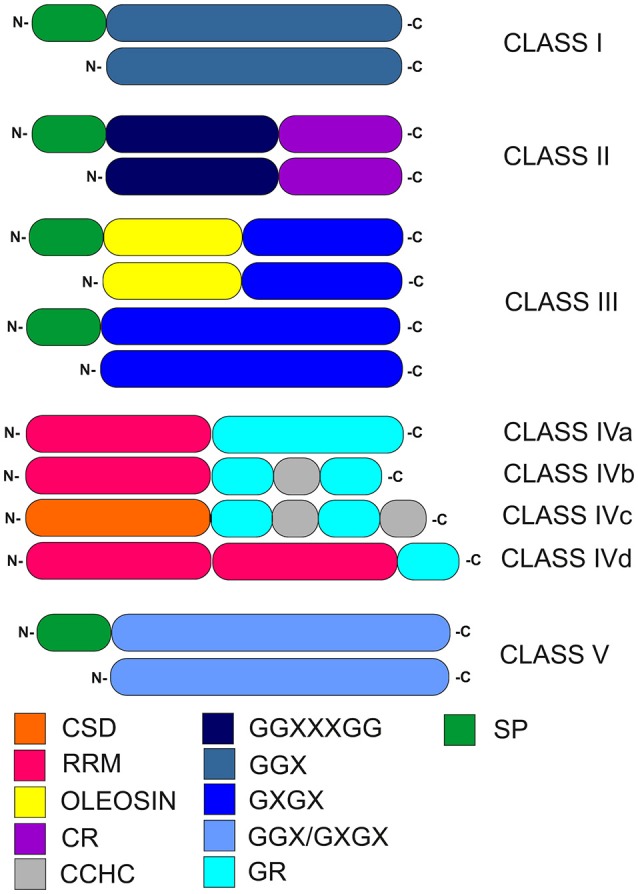
Classification of plant glycine-rich proteins. CSD, cold-shock domain; RRM, RNA-recognition motif; oleosin, oleosin-conserved domain; CR, cysteine-rich domain; CCHC, zinc-finger; GR, glycine-rich domain; GGX, GGXXXGG, GXGX, GGX/GXGX, glycine-rich repeats (G-Gly, X-any amino acid); SP, signal peptide.
Table 1.
Classification of glycine-rich proteins basing on structural features.
| GRP classes | Characteristic features |
|---|---|
| Class I | Signal peptide followed by high glycine-content region with (GGX)n repeats |
| Class II | Signal peptide and presence of a characteristic cysteine-rich C-terminal domain |
| Class III | Signal peptide and contain lower glycine content (in comparison to other GRPs classes), the oleosin domain is the signature motif for their sub-group |
| Class IV | RNA-binding GRPs, glycine-rich domain with RNA-recognition motif (RRM) or a cold shock domain (CSD), CCHC zinc-fingers might be also present in their structure, four sub-groups: (IVa) RRM motif besides the glycine-rich domain, (IVb) single RRM and CCHC zinc-finger motif, (IVc) cold shock domain and two or even more zinc-fingers, (IVd) two RRM motifs |
| Class V | Signal peptide followed by GGX/GXGX motif or only GGX/GXGX motif without signal peptide |
Selected members of GRP classes I–III contain N-terminal signal peptide. Notably, such signal peptide in class I GRPs is followed by a region extremely rich in glycine and containing (GGX)n repeats. In class II, two important regions are present, the first one containing [GG(X)3GG]n glycine-rich repeats and the second one composed of a specific cysteine-rich motif at the C-terminus. The structurally heterogeneous class III is characterized by a substantially lower glycine content, compared to other GRP classes. It contains (GXGX)n repeats, which are accompanied (in selected members) by an adjacent N-terminal oleosin-conserved domain. Class IV, which are also described as RNA-binding GRPs, contains numerous glycine-rich domains. A characteristic feature of class IV of GRPs is presence of a glycine-rich region at the C-termini and up to three N-terminal RNA recognition motifs (RRMs; Xu et al., 2014). This class is further subdivided into four subclasses (denoted IVa, IVb, IVc, and IVd). Members of subclasses IVa, IVb, and IVd contain RRMs, while the cold-shock domain (CSD) is present only in IVc subclass. In subclasses IVa, IVb, and IVd, one of RRMs (like the CSD motif within IVc subclass) is N-terminal. Subclasses IVb and IVc contain CCHC zinc-finger motifs in their secondary structure. Class V is characterized by mixed patterns of (GGX)n/(GXGX)n repeats and a relatively high glycine content (Mangeon et al., 2010). Recently, distinct GRP superfamily members named RZs proteins, were described as a subgroup harboring characteristic internal CCHC-type-zinc finger motifs (Xu et al., 2014).
The role of glycine-rich domains in class IV GRPs has not been precisely established; on the physiological level, it is presumably involved in the cold tolerance and acclimation development. The length of glycine-rich domain is positively associated with stress tolerance. This domain containing zinc finger play a key role in nucleic acid-binding activity as well as RNA chaperoning (Nomata et al., 2004; Kim J. S et al., 2007). Hanano et al. (1996) performed deletion analysis of the C-terminal glycine-rich domain which showed that it is essential for RNA binding. Glycine-rich domain is more important than other functional domains of GRPs, which control protein conformation that are key for GRPs activity. It was predicted that the glycine-rich domain might be involved in interactions with numerous partners (Hanano et al., 1996). Further data on the diverse cellular functions of GRPs will be presented in section Cellular Activity of GRPs.
Recently, it has been suggested that GRPs in fact do not represent a distinct protein superfamily, but a group of proteins sharing only some repetitive structural motifs. This suggestion is based on their broad structural diversity, wide variety of modes of action, and their differing subcellular localization (Kar et al., 2012). According to The UniProt Consortium (2017), and SUBA databases predictions (Tanz et al., 2013) members of classes I and II were either predicted to be present in the cytosol and nucleus, or secreted and localized in the extracellular space. Class III was predicted to be secreted or present in the vicinity of the cell membrane. Class IVa is expected to be present in the cytosol, cell wall, Golgi, organelles, while classes IVb and IVc—seems to be restricted mainly to the nucleus. However, some Arabidopsis GRPs, particularly class IVa members (Supplementary Table 1a) could putatively be targeted to the mitochondria.
Expression pattern of GRP genes and functional aspects
Plant GRP genes and their co-expression pattern
In Supplementary Table 1a, general characteristics of the most important Arabidopsis GRPs and their genes is shown, based inter alia on TAIR data (Berardini et al., 2015). Genes encoding members of this superfamily are present in a number of seed plant genomes [e.g., eucalyptus (Eucalyptus sp.), rice (O. sativa), and Arabidopsis]. Data from PhytoMine service (Goodstein et al., 2012) and analyses of plant whole genomes and transcriptomes facilitated the identification of more than 150 distinct genes coding for GRPs in sugarcane (Saccharum officinarum), Eucalyptus, Arabidopsis, and rice, among other plant species (Mangeon et al., 2009, 2010; Supplementary Table 3c).
According to the data from ThaleMining repository (for the ARAPORT project) as well as ATTED-II database (Aoki et al., 2016), a plethora of genes co-expressed with Arabidopsis GRP genes as well as potential interacting partners were revealed. Co-expressed genes comprise genes coding for other GRPs as well as for other stress-related proteins, including LEA proteins and cellular chaperones and chaperonins, cell-wall associated kinases, leucine-rich receptor-like protein kinases, and other proteins (Supplementary Tables 3a,b).
Cellular activity of GRPs
Owing to interaction patterns (chapter Plant GRP Genes and Their Co-Expression Pattern), GRPs are expected to participate in post-transcriptional regulation of gene expression; however, they display higher in vitro affinity to particular ribohomopolymers (Kim et al., 2005). In general, GRPs exhibit chaperonin activity toward nucleic acids, primarily RNA (Kwak et al., 2011).
Emerging evidence suggests that class I and II members are crucial for the positive regulation of plant cell and organ growth; they are also active components of the plant cell wall. Park et al. (2001) reported that class II of GRPs interact with cell wall-associated kinases, thereby initiating the recognition of environmental stimuli and participating in signal transduction. It has been demonstrated that GRPs are responsible for blocking tobamovirus infection (Mangeon et al., 2010). Moreover, GRPs are known for their anti-fungal and anti-bacterial activity, and this activity may allow them to protect plants from the cellular damage resulting from the action of the biotic stress. For instance, pumpkin (Cucurbita pepo) cucurmoschin peptide displayed antifungal activity against Botrytis cinerea, Fusarium oxysporum, and Mycosphaerella oxysporum. Another example of the specific biological activity of GRPs has been provided, namely their activity against Gram-negative bacteria (Cândido Ede et al., 2011). Evidence for the participation of those proteins in plant defense came from the analysis of molecular signals exchanged during interactions between Arabidopsis and Pseudomonas syringae (Fu et al., 2007).
In developmental processes such as pollen hydration and its recognition, class III GRPs with particular emphasis on Arabidopsis GRP17, are important. Class IV GRPs have a strong RNA binding moiety. Henceforth they are involved in molecular processes such as alternative splicing via spliceosome activity, or the regulation of transcription, and in circadian rhythm. Proteins from class IV play a role in the regulation of stomatal movement, seed development, and stamen development [Supplementary Tables 1a,b; GENEVESTIGATOR and eFP Browser data (Winter et al., 2007)].
Basing on the effects on cell division and cell elongation in an Arabidopsis mutant line with a grp3-1 knockout, Mangeon et al. (2016) concluded that AtGRP3 is involved in root size determination. The authors support evidence that, curiously, this mutant line presented enhanced Al tolerance (probably through the specific involvement of AtWAK1-mediated signaling). Other data suggest that different GRP genes can have opposing roles in the cell elongation process, e.g., AtGRP3 expression represses this process, while AtGRP5 enhances it (Mangeon et al., 2009). Overall, these findings indicate the pleiotropic roles of GRPs in plant growth, development and stress responses (Table 2, Supplementary Tables 1, 2).
Table 2.
Participation of the identified plant glycine-rich proteins in stress response.
| GRP gene | Gene source | Stress response | Localization | References |
|---|---|---|---|---|
| AtGRP1 | Arabidopsis thaliana | Overexpression of AtGRP1 protein improve stress response in high-salt conditions | Nucleus/cytoplasm | Wang et al., 2012 |
| AtGRP2 | Arabidopsis thaliana | Under cold stress GRP2 protein positively affects seed germination, cold inducible higher level of expression, predicted role in stabilization and modulation of mRNA in cold acclimation, up-regulation, GRP2 knockout mutants showed enhancement of germination after mannitol treatment, up-regulation enhances tolerance to drought stress | Nucleus/cytoplasm | Fusaro et al., 2007; Kim J. Y et al., 2007; Yang et al., 2014; Ciuzan et al., 2015 |
| AtGRP4 | Arabidopsis thaliana | Up-regulation of abundance during cold adaptation process, overexpressed in high-salt conditions connected with the retarded seed germination | Nucleus/cytoplasm | Kwak et al., 2005, 2011; Kim J. Y et al., 2007 |
| AtGRP7 | Arabidopsis thaliana | Up-regulation during cold adaptation process, involvement in the plant response under osmotic stress, influence on two stress-inducible genes RD29A and RAB18, under drought stress GRP7 knockout mutants showed suppression of germination, up-regulation enhances tolerance to drought stress, upregulated also in response to peroxide-induced oxidative stress, increased level of GRP7 transcripts enhances tolerance to biotic stress such as Pectobacterium carotovorum, tobacco mosaic virus and Pseudomonas syringae | Nucleus/cytoplasm | Cao et al., 2006; Fu et al., 2007; Kim J. S et al., 2007; Schmidt et al., 2010; Kwak et al., 2011; Lee et al., 2012; Yang et al., 2014; Ciuzan et al., 2015 |
| AtGRP7 | Arabidopsis thaliana | RNA chaperone activity during the cold adaptation process, increased in abundance at low temperature, decreased in high temperature | Nucleus/cytoplasm | Kim J. S et al., 2007; Wienkoop et al., 2008; Cramer et al., 2011 |
| AtGRP8 | Arabidopsis thaliana | Up-regulation under oxidative stress induced by the peroxide | Nucleus/cytoplasm | Schmidt et al., 2010 |
| AtRZ-1b | Arabidopsis thaliana | Bacterial cells overexpressing AtRZ-1b grew well at low temperature, RNA chaperone activity | Nucleus/cytoplasm | Kim et al., 2010b |
| CsGRP7-a | Camelina sativa | Plants with overexpression of GRP7 protein are more tolerant to cold and freeze treatment, negative role under high salinity conditions | Nucleus/cytoplasm | Kwak et al., 2016 |
| HvGRP2, HvGRP3 | Hordeum vulgare | Higher expression at low temperature (cold induced), involvement in the response to fungal pathogens Erysiphe graminis and Rhynchosporium secalis | Nucleus/cytoplasm | Molina et al., 1997 |
| LbGRP1 | Limonium bicolor | Elevated proline content in Nicotiana tabacum plants under overexpression and the enhanced tolerance to salt stress | Nucleus/cytoplasm | Wang et al., 2012 |
| LpGRP1 | Lolium perenne | Involvement in cold and freezing tolerance | Nucleus/cytoplasm | Shinozuka et al., 2006 |
| MsGRP | Medicago sativa | Plants with overexpression of GRPs are more sensitive to high salinity | Nucleus/cytoplasm | Long et al., 2013 |
| NgRBP | Nicotiana glutinosa | Induction by biotic stress and possible positive role in plant-pathogen interaction | Nucleus/cytoplasm | Naqvi et al., 1998 |
| NtGRP1 | Nicotiana tabacum | NtGRP1 transcripts were up-regulated after cold treatment in 40 day-old plants (without changes in expression in younger plants), up-regulation of these transcripts was also observed after wounding stress | Nucleus/cytoplasm | Khan et al., 2013 |
| NtGRP-1a, NtGRP3 | Nicotiana tabacum | Up-regulation under cold stress, under high temperature treatment and after wounding, under drought stress plants showed accumulation of NtGRP-1a transcripts and were more tolerant to these conditions | Nucleus/cytoplasm | Chen et al., 2010; Khan et al., 2013 |
| OsGRP1, OsGRP4, OsGRP6 | Oryza sativa | Plants with overexpression of those proteins are visibly more tolerant to the low temperature-related stress | Nucleus/cytoplasm | Kim et al., 2010a |
| PpGRP3 | Physcomitrella patens | Under low temperature stress PpGRP3 protein is probably associated with post-transcriptional processing of mitochondrial RNA | Mitochondria | Nomata et al., 2004 |
Tissue- and developmentally-specific expression of Arabidopsis GRP genes
GRP genes are expressed in a tissue- and organ-specific manner in Arabidopsis development. Class I accumulates mostly in seeds, siliques, roots, leaves, and also in the vegetative rosette and in the rosette after transition to the generative phase. The expression profile of GRPs from class II is similar; however, this is higher in cotyledons, hypocotyls, and cauline leaves. Class III is highly expressed in seeds, siliques, shoots, vegetative rosette, and flowers. This is associated with role of those proteins in lipid storage, sexual reproduction and pollen hydration or its recognition. However, the members of class IV are most highly expressed in shoot apex, vegetative rosette, seeds, and flowers. On the contrary, other GRP genes are lowly expressed in the most tested tissues and up-regulated in inflorescences. The expression of GRP genes is modulated in the key developmental stages, e.g., pollen and embryo development; however, most of them are expressed at low levels in pollen and relatively high levels in the most of tissues [eFP Browser (Winter et al., 2007) and RNA-seq data from Araport11 portal (Krishnakumar et al., 2015); Supplementary Tables 1b, 2a,b].
Hormonal regulation of the expression of GRP genes
The regulation of adaptive responses at the level of gene expression can involve a plethora of molecular mechanisms. Some of the crucial steps in these processes are mediated by RBPs (Cao et al., 2006). GRPs from class I, II, and IV (notably subclass IVc) are involved in mediating responses to hormones, including abscisic acid (ABA), salicylic acid, or ethylene. ABA governs many important aspects of plant development and physiology; GRP expression often increases upon ABA treatment (Cao et al., 2006; Fusaro et al., 2007; Mangeon et al., 2010; Fu et al., 2016). Aneeta et al. (2002) reported that ABA (at 10 μM concentration) induced sbGR-RNP gene expression in Sorghum bicolor. Another gene involved in ABA signaling is AtGRDP1. 35S:AtGRDP1 overexpressing lines were ABA-resistant whereas the disruption of AtGRDP1 gene resulted in ABA hypersensitivity. Under ABA treatment Atgrdp1-null mutant seedlings showed higher level of ABI3 and ABI5 transcripts in contrast to 35S:AtGRDP1 overexpressing lines in which these transcripts were repressed (Rodríguez-Hernández et al., 2014).
The altered expression of GRP genes under ABA treatment is connected with phenotypic modifications aiding stress responses. Cao et al. (2006) reported the repression of AtGRP7 by ABA. In T-DNA mutants they observed hypersensitive responses to ABA evidenced both by the root growth and seed germination pattern. Notably, the authors found that atgrp7-1 mutant plants accumulated RD29A and RAB18 transcripts (both ABA- and stress-inducible) at higher abundance in comparison to wild-type plants. Therefore, AtGRP7 is suspected of being involved in the regulation of ABA signaling in addition to stress responses. AtGRP7 and AtGRP8 respond to oxidative stress, and due to the fact that ABA results in both genes repression, negative feedback may be expected to occur between reactive oxygen species (ROS) and ABA signaling in this case (see section GRPs and Oxidative Stress Response; Carpenter et al., 1994; Cao et al., 2006). In addition, an Arabidopsis zinc finger containing GRPs (atRZ-1a) is involved in both stress responses and ABA signaling. Kim Y. O et al. (2007) used transgenic Arabidopsis plants, either overexpressing atRZ-1a or loss-of-function mutants, and observed that the overexpression of this gene resulted in retarded seedling germination and growth under salinity and drought in comparison to wild-type plants. In contrast, plants with loss-of-function mutations germinated earlier and grew faster than wild-type plants under the same stress conditions. However, the germination pattern of investigated transgenic and mutant plants was influenced by ABA or glucose; the results therefore indicate, that atRZ-1a affects germination in an ABA-dependent manner (Kim Y. O et al., 2007). Contrary to this, GRP2 affects seed germination through a ABA-independent pathway (Kim J. Y et al., 2007). No differences in germination between wild-type and transgenic plants following ABA or glucose addition (without cold exposure in parallel with ABA treatment) were observed. Such relationships between GRP transcript levels, hormone signaling, as well as plant growth and development were also supported by the findings of Ortega-Amaro et al. (2015). The results they obtained suggested that the deregulation of AtGRDP2 expression in Arabidopsis Atgrdp2 mutants and 35S:AtGRDP2 overexpression lines results in plant developmental aberrations. AtGRDP2 is a gene encoding a small GRP containing a DUF1399 domain and a putative RRM motif. AtGRDP2 mRNAs mainly accumulate in flowers. Transgenic lettuce (Lactuca sativa) plants overexpressing AtGRDP2 gene also manifest increased growth rate and early flowering time. The authors compared flowering time under long- and short-day conditions and observed faster development and early flowering in AtGRDP2 overexpression lines in comparison with wild-type plants. Notably, under the same cultivation conditions, lines with GRDP2 knockdown exhibited a late-flowering phenotype. This is clear evidence for the involvement of this gene during key developmental stages (Ortega-Amaro et al., 2015).
AtGRDP2 expression influenced the expression of other genes, namely genes coding for auxin (IAA) response factors (ARF2, ARF6, ARF8) and at-MIR167 involved in the control of floral development and auxin signaling. ARF gene expression is regulated by auxins and various IAA concentrations might be responsible for the more rapid growth of lines overexpressing AtGRDP2. Two of the genes mentioned earlier (ARF6 and ARF8) as well as AUX1, which encodes an auxin transporter, were induced in 35S::AtGRDP2-OE3 line; however, they were repressed in the Atgrdp2-1 mutant. In contrast, ARF2 expression was induced in Atgrdp2-1 line. The level of miR167, which is a negative regulator of ARF6 and ARF8, was increased in Atgrdp2-1 mutant and decreased in AtGRPD2 overexpressing line (Ortega-Amaro et al., 2015). These results reveal the important roles of AtGRDP2 in development and the stress response; interestingly, they suggested a connection between AtGRDP2 gene expression pattern and hormone signaling. However, the involvement of GRP gene expression in the action of hormones and signaling pathways needs to be further investigated.
Expression of GRP genes across stress treatments-general remarks
GRPs from classes I, II, and IV (especially subclass IVc) are involved in various stress responses (Kar et al., 2012); subclass IVc participates notably in cold acclimation. The important role of the distinct RZ class of GRPs in such responses has been also established in Arabidopsis, rice and wheat (Triticum aestivum) (Kim et al., 2005, 2010a,b; Xu et al., 2014). According to the GENEVESTIGATOR and Arabidopsis eFP Browser data (Supplementary Tables 1a, 2c,d; Winter et al., 2007), selected Arabidopsis GRP genes (e.g., MCO15_15) respond to ozone, UV irradiation and Fe deficiency (e.g., MCO15_15 and AtGRP3S) by increasing of the abundance of their respective mRNAs. Under stress, up-regulation of GRP genes exceeds down-regulations. In Supplementary Tables 1, 2 the various expression patterns of GRPs from different classes are compiled according to hormonal regulation, developmental stage as well as to abiotic and biotic stimuli.
Despite the findings discussed above, data on the expression patterns of GRP genes under unfavorable conditions are nor comprehensive. This is discussed further in subsequent sections The Role of GRPs in Temperature Stress, GRPs in Mechanical Stress, Participation of GRPs in Salinity/Osmotic Stress Responses, Participation of GRPs in Drought Stress, GRPs and Oxidative Stress Response, and GRPs in Biotic Stress Responses.
The role of GRPs in temperature stress
According to the literature, three distinct subtypes of temperature-related stress acting on plant cells at the physiological and molecular level can be distinguished. The first occurs at temperatures below freezing, the second acts at low (but above freezing) temperatures (0–20°C), and the third refers to high-temperature treatments (Iba, 2002; Sinha et al., 2015). To date, some GRPs have been described mainly as proteins that enhance plant tolerance to low temperatures. Moreover, it has been suggested that they are involved in cold acclimation and may improve plant growth at low temperatures. In recent years, however, a plethora of novel data on the participation of GRPs in responses to altered temperature has been reported and is presented below.
GRPs and low temperature treatments
Low temperature treatments influence growth, development, productivity, and geographical distribution of crops (Sanghera et al., 2011; Zhou et al., 2011; Yang et al., 2015; Fu et al., 2016). In general, most plant species adapted to tropical and subtropical zones are highly intolerant to chilling and freezing; however, plants grown in temperate climates can adapt to lower temperatures (“cold acclimation”; Xin et al., 2013; Zhan et al., 2016).
GRPs were reported to be involved in acclimation to various temperatures (Table 2). Notably, in Arabidopsis leaves and roots, the expression of GRP genes responded sensitively to cold treatment (Supplementary Table 2; Kim J. S et al., 2007; Khan et al., 2013; Mangeon et al., 2016). According to Kim Y. O et al. (2007), several members of the GRP superfamily confer a significant impact on Arabidopsis seed germination and seedling growth under stress, including not only suboptimal temperature treatments, but also salinity or water deficiency. However, the upregulation of GRP1 gene, which was well visible after cold stress in tobacco plants [Nicotiana tabacum; 4°C treatment at 16:8-h light:dark (L:D) photoperiod, 20, 30, and 40 days after germination] cannot indicate the true relationship between GRP1 and decreased temperature treatment, as it was applied in laboratory conditions to a subtropical plant species and thus without any particular environmental relevance (Khan et al., 2013). In response to cold, the authors showed variations in NtGRP1 mRNA abundance across tobacco plant development (the highest was in 40-day-old plants).
The level of GRP1 transcripts in crowns, roots and leaves of perennial ryegrass (Lolium perenne) was significantly increased in the course of cold acclimation (6/2°C 8:16-h L:D photoperiod). Over the next 1 week of cold acclimation, the abundance of GRP1 messengers rose further. However, the increased level of LpGRP1 transcripts remained unchanged under prolonged cold stress. When plants were transferred to the control temperature, de-acclimation of freezing tolerance accompanied by the reversion to control level of GRP1 mRNAs was observed (Table 2; Shinozuka et al., 2006). Notably, perennial ryegrass belongs to important pasture grasses having ability to acquire cold tolerance during exposure to low, but non-lethal temperatures.
Kim J. S et al. (2007) reported that GRP2 gene was induced and that GRP4 and GRP7 were also upregulated by Arabidopsis chilling (4°C for 1–4 days under a 16-h photoperiod; Table 2); they also found enhanced freezing tolerance of Arabidopsis plants with GRP2 overexpression compared with the wild type and grp2-knockout mutants. Under cold stress, GRP2 enhances seed germination at low temperature (11°C) and influences Arabidopsis growth in ABA-independent manner (Kim J. Y et al., 2007). Fusaro et al. (2007) observed a strong induction of AtGRP2 gene expression after 6 h of cold treatment (6°C, 16/8-h L/D photoperiod). A higher level of expression was detected till the end of 48-h-long treatment. In this study an early cold-response was noted as the upregulation of AtGRP2 gene was associated with its cold- inducible activity. Fusaro et al. (2007) suggested that plant GRPs are involved in a specific mechanism active in the cold response that acts by stabilizing various transcripts under cold treatment (GRP2 has a strong in vitro affinity to ssRNA and DNA, including homopolymers).
Furthermore, Kim et al. (2010) demonstrated the complementation of a cold-sensitive E. coli mutant by AtRZ-1b during cold treatment (18°C) of bacterial cells. E. coli overexpressing AtRZ-1b grew well at the lowered temperature; colony forming ability was comparable to that of cells overexpressing bacterial RNA chaperone CspA (Table 2), which suggests that AtRZ-1b possesses RNA chaperone activity. In contrast, AtRZ-1c appeared unable to complement cold sensitivity in the same bacterial strain (Kim et al., 2010b).
The ability to suppress cold stress appeared not to be equal for all the investigated GRP genes. For instance, the GRP7 gene appeared to have a greater protective effect during the cold response than GRP2. The GRP7 protein also has the strongest ability to increase E. coli viability or growth rate under cold shock at 4 or 17°C, respectively, and its RNA chaperone activity during the cold adaptation process was shown (Kim J. S et al., 2007). The results obtained by the authors suggest that plant and bacterial cold shock proteins (CSPs) play similar roles in the positive regulation of cell growth at low temperature. Interestingly, the activity of GRPs under low-temperature conditions may depend on the number and size of the C-terminal glycine-rich region (GRP4 has a shorter region in comparison with GRP7 or GRP2 and displays the lowest chaperone activity). Kwak et al. (2011) suggested a crucial role for the N-terminal sequence of AtGRP4 and AtGRP7 in RNA chaperoning; the overall secondary structure of GRPs (including the presence of all RRMs) may be critical for cold adaptation. However, more data is needed to confirm this relationship.
Further interesting results were obtained from the complementation of CSP knockouts in the E. coli BX04 cold-sensitive mutant with GRP genes from Arabidopsis and rice. The BX04 strain has 4 genes coding for CSPs deleted; thus, it is extremely intolerant to low-temperature treatments. Notably, Kim et al. (2010a) concluded that specific types of OsGRPs are capable of increasing of cold-tolerance of this strain (Table 2). The authors clearly demonstrated RNA chaperone activity during cold adaptation for the investigated GRPs.
Kim et al. (2010a) showed that rice GRPs (OsGRP1, OsGRP4, and OsGRP6) have ability to rescue grp7-Arabidopsis knockout plants from cold, and Arabidopsis plants overexpressing these proteins were more tolerant to low temperatures (Arabidopsis seedlings were cold-stressed at 8-11°C). It is worth mentioning that the authors determined the expression profiles of 6 diverse OsGRP genes, which correlated well with the ability to complement cold sensitivity in E. coli mutant. OsGRP2 appeared to exhibit the lowest accumulation and did not provide cold sensitivity complementation. In contrast, OsGRP1 and OsGRP6 showed the highest complementation ability and at the same time displayed higher mRNA accumulation than OsGRP2.
Another example of the involvement of GRPs in stress responses during cold treatment might be NtGRP-1a and NtGRP-3 transcripts (Table 2). These transcripts appeared to increase in abundance 1h after the induction of cold stress (4°C treatment of 4-week-old plants), then slightly decreased after 2 h, then showed continuous upregulation after 4 h (Chen et al., 2010). The same conditions applied to 13-day-old barley (Hordeum vulgare) seedlings grown at 22/18°C L:D thermal regime and 16:8 h L:D photoperiod also led to a significant increase in the abundance of selected GRP mRNAs. In this case, a higher level of expression of cold-inducible Hvgrp2 and Hvgrp3 genes was also noted (Molina et al., 1997).
Kwak et al. (2016) presented further evidence supporting the important role of GRPs in low temperature stress responses; transgenic Camelina sativa plants with CsGRP7a overexpression appeared to be cold-tolerant. C. sativa is a fast growing crop with low requirements for fertilization. After the transgenic 14-day-old C. sativa plants were exposed to cold stress (2°C, 16:8-h L:D photoperiod; Kwak et al., 2016), they had improved survival parameters compared to wild-type plants or grp7 knockout plants (Table 2), supporting the important role of GRP7 during cold responses and cold acclimation.
Notably, GRP7 accumulation is regulated by the circadian clock via the autosplicing of Atgrp7 pre-mRNA (a feedback loop). Staiger et al. (2003) suggested a role of AtGRP7 in splice site selection. In transgenic Arabidopsis plants, the constitutive AtGRP7 overexpression is strictly connected with the accumulation of low amounts of alternatively spliced Atgrp7 mRNAs containing a premature stop codon (due to the usage of 5′cryptic splice site in the intron by AtGRP7). The alternatively spliced transcripts do not accumulate at high levels due to their instability; henceforth, they do not produce functional GRP7 protein. When AtGRP7 is overexpressed, it interacts with 3′UTR and the intron of Atgrp7 messenger. AtGRP7 influences also the choice of the splice site within the Atgrp8 transcript encoding a glycine-rich RNA-binding protein related to AtGRP7. This is attributed to the favorable accumulation of the unstable Atgrp8 transcript, which is alternatively spliced. Such results unambiguously indicate that the mentioned regulatory mechanism is crucial for controlling of GRP levels and that AtGRP7 can efficiently regulate the abundance of target messengers (Staiger et al., 2003).
Further insights into functions and interacting partners of some GRPs were provided by a pioneering study by Meyer et al. (2017) involving the application of an individual nucleotide resolution crosslinking and immunoprecipitation (iCLIP) approach for the identification in vivo targets of AtGRP7. Arabidopsis plants overexpressing GRP7 fused with GFP were used for iCLIP experiments that allowed the identification of 858 messengers with enriched crosslink sites. They were absent in plants containing variants of GRP7 with dysfunctional RRMs or in control lines expressing GFP only. Moreover, an RNA immunoprecipitation (RIP)-sequencing strategy led to the validation of 452 highly-confident binders (constituting for 53% of iCLIP targets). The iCLIP and RIP pools of targets were thus non-identical, but partially overlapped. The abundance and splicing some of some of those transcripts were regulated by AtGRP7. Overall, the results of Meyer et al. (2017) highlight the importance of the RNA binding motif in controlling multiple AtGRP7 targets. It appeared that AtGRP7, which is controlled by the circadian clock, preferentially binds to 3′UTRs. AtGRP7 overexpression notably represses the expression of target genes and affects circadian oscillations (e.g., CCL and DRM2 targets), alternative splicing, as well as the polyadenylation of selected messengers. In addition, AtGRP7 could also bind diverse regions of the same transcripts, which suggests its diverged function. Among the high-confidence binders of AtGRP7, cold-, salinity-, and pathogen defense-responsive transcripts were distinctive. However, the role of GRP7 in tolerance to such stress conditions may depend on various regulatory mechanisms without a direct relationship with mRNA abundance. These results pinpoint the necessity of studying the reconfiguration of post-transcriptional networks (in which GRP7 is involved) under adverse conditions.
Finally, mitochondrial GRPs seem to be also involved in responses to cold stress. Owing to their function, mitochondrial GRPs represents a unique subgroup of plant GRPs. Under low temperature, Physcomitrella patens GRP3 transcripts and mitochondria-targeted GRP3 protein accumulated markedly; it was speculated that this protein may be associated with RNA splicing and editing during stress. The experiment was performed on 7-day-old protonemata of P. patens which had grown at 22°C under continuous light then transferred to 4°C for 12, 24, 36, and 48 h (Nomata et al., 2004). Due to the fact that some GRPs are present in P. patens, it might be obvious that mitochondrial GRPs (many of which remain unidentified) must have separated from another plant GRP superfamily at an early stage of plant evolution (Nomata et al., 2004).
GRPs and heat stress
Elevated temperatures exert a negative impact on plant growth, development and productivity (Zhu et al., 2013; Hu et al., 2015; Zhai et al., 2016). Heat stress during the generative phase of food crop growth might be particularly destructive. Recent results have supported the important role of these proteins in the acquisition of heat tolerance. Zhu et al. (2013) identified four down-regulated protein spots containing GRPs in heat-stressed Pinellia ternata (38°C, 1 day), which indicated possible metabolic impairment under stress. The level of GRP transcripts in P. ternata leaves after various durations of stress was not reflected by the protein accumulation profile. At the beginning of heat stress, GRP transcripts were slightly down-regulated, but after 12 h they rose significantly, which may be associated with post-transcriptional and post-translational events. In tobacco Chen et al. (2010) observed a remarkable increase in the abundance of NtGRP-1a and NtGRP-3 transcripts, with a peak at after 8 h of heat stress. Under the same conditions, curiously, the expression of NtGRP-1b gene was unaffected.
Wienkoop et al. (2008) also demonstrated the involvement of AtGRP7 in temperature stress responses (Table 2). The level of AtGRP7 protein was high at low temperatures, compared with elevated temperature (32°C, 3 days) (Supplementary Table 2). AtGRP7 abundance correlated with proline and glutamine content. Interestingly, the temperature response of AtGRP7 as well as osmolyte accumulation appeared independently of the responses of other markers characteristic of temperature stress.
GRPs in mechanical stress
GRPs perform essential roles in plant survival and respond to mechanical stress, including wounding after pathogen infection. Such responses may be associated with increased membrane permeabilization and, consequently, to pathogen cell death (Cândido Ede et al., 2011; Boyd et al., 2013; Savatin et al., 2014). Khan et al. (2013) measured NtGRP1 mRNA expression levels after leaf wounding by scraping with pins and observed a 2.5-fold increase 12 h after treatment. However, NtGRP1 expression levels decreased to control values 24 h after the cessation of stress. Similar results in tobacco were reported by Chen et al. (2010). NtGRP-1a and NtGRP-3 genes were rapidly upregulated at 1 h after wounding (cutting leaves with forceps), then their expression increased until reaching a peak at 4 h-long stress and gradually reducing (Table 2).
In summary, the exact role of GRPs during heat and mechanical stress has to be elucidated and the mode of its action remains unclear.
Participation of GRPs in salinity/osmotic stress responses
Plants respond to salinity stress at various levels. This type of abiotic stress can be particularly detrimental, because of protein denaturation, ionic imbalance, and toxicity, ROS production and subsequent osmotic stress, loss of turgor, as well as membrane modifications and other cellular effects (Conde et al., 2011; Long et al., 2013; Joshi et al., 2014; AbdElgawad et al., 2016; Kushwaha et al., 2016). Salinity stress can cause abnormalities during enzymatic reactions, due to Na+ toxicity and elevated K+ uptake (Suzuki et al., 2016). Elevated levels of salt in the soil may initiate drought stress; moreover, the coexistence of salinity and drought is very frequent. GRPs have a suggested influence on Arabidopsis seed germination under salinity (75, 100, 125, 150, and 175 mM NaCl), but strikingly without any apparent influence on seedling growth. GRP2 seems to play a significant role in seed germination and seedling growth of Arabidopsis plants under osmotic stress (Kim J. Y et al., 2007).
Under high-salt conditions (250 mM NaCl), the overexpression of GRP4, another GRP family member identified in Arabidopsis, retarded seed germination in this species (Table 2). The same was observed by Kwak et al. (2005) under dehydration (4-week-old plants were placed on a filter paper for 1 h at room temperature to remove residual water completely, and then were transferred to a growth chamber at 23°C).
Another GRP involved in the osmotic stress response is GRP7 (Tables 2, 3). The expression of the AtGRP7 gene was repressed by 300 mM NaCl; the application of very high concentrations of NaCl in Kwak et al. (2005) and Cao et al. (2006) studies may result in sub-lethal effects associated with the massive repression of gene expression, however salinity stress in the Cao et al. (2006) report lasted for 6 h only. On the contrary, increased levels of expression of AtGRP7 observed during seed germination suggested hypersensitive responses to osmotic stress conditions. Moreover, the atgrp7-1 mutant plants lacking AtGRP7 gene, presented significantly higher RD29A and RAB18 transcripts level in comparison to wild-type plants. This evidence supports the involvement of AtGRP7 in plant osmotic stress response (Cao et al., 2006). Kwak et al. (2016) also proposed a negative role of CsGRP7 under high salinity. During salt stress, the growth of transgenic C. sativa plants was retarded in comparison with wild-type plants. Using specific Medicago sativa expressed-sequence tag data, Long et al. (2013) cloned and characterized a novel MsGRP gene (accession no. JQ340083) and suggest its negative role in salinity responses as Arabidopsis plants overexpressing M. sativa protein appeared to be more sensitive to high salinity than wild-type plants.
Ortega-Amaro et al. (2015) compared salt stress tolerance in Arabidopsis wild-type plants, Atgrdp2 mutants and 35S:AtGRDP2 overexpression lines. Arabidopsis lines with GRDP2 overexpression displayed enhanced tolerance to high salinity conditions. After 7 days of 150 mM NaCl treatment they observed that more than 50% plants with AtGRDP2 overexpression recovered in comparison with only 20% in the case of Atgrdp2-1 mutant.
Additional clues about the involvement of GRPs in salinity and osmotic stress tolerance were obtained by Rodríguez-Hernández et al. (2014). AtGRDP1, like AtGRDP2, contains DUF1399 domain, a putative ribonucleoprotein (RNP) motif and a short glycine-rich domain. The authors showed that the expression of AtGRDP2 is modulated in response to mannitol, sorbitol, glucose, NaCl, LiCl, and ABA treatment. The AtGRDP1 gene expression level was greatly increased depending on the quality and duration of treatment in comparison to control plants. On the contrary, Atgrdp1-null mutant line displays increased sensitivity to salt and osmotic stress during cotyledon development and germination.
During stress, plants have the ability to accumulate diverse osmolytes controlling osmotic potential inside cells. There is evidence for a positive correlation between proline accumulation and the participation of GRPs in salinity responses. Wang et al. (2012) demonstrated that the overexpression of LbGRP1 (from Limonium bicolor) in transgenic tobacco plants can markedly increase proline content under salt stress (Table 2). They showed that the transgenic lines displayed higher salt tolerance than wild-type plants indicating a significant improvement in stress responses under high salinity that was attributable to AtGRP1 overexpression. Aneeta et al. (2002) measured the level of SbGR-RNP (GRP from Sorghum bicolor containing the conserved ribonucleoprotein motif) after 1 day treatments with 1 M and 500 mM NaCl. They observed a notable increase in SbGR-RNP transcripts abundance in S. bicolor seedlings subjected to all tested stress conditions.
Taken together, these findings markedly enhanced current knowledge of the functional roles of GRPs in response to salinity and osmotic stress.
Participation of GRPs in drought stress
Drought often results in stomatal closure reduction, decrease in stomatal density and/or increased water uptake (Yu et al., 2008, 2013; Zhu et al., 2016). Dehydration and subsequent rehydration might induce “transcriptional memory,” helping plants to mitigate harmful effects (Chinnusamy and Zhu, 2009; Ding et al., 2012; Hu et al., 2015).
Ciuzan et al. (2015) tested the influence of drought stress on Arabidopsis seed germination and showed that GRP2 knockouts (contrary to GRP7) showed enhanced germination after mannitol treatment (Table 2). Yang et al. (2014) also examined the influence AtGRP2 and AtGRP7 overexpression on drought tolerance in transgenic rice. Contrary to the findings of Ciuzan et al. (2015), Yang et al. (2014) indicated that rice plants overexpressing the mentioned GRPs were more tolerant to water deficit than wild-type plants, which may illustrate species-specific effects of GRP overexpression. Chen et al. (2010) measured the accumulation of NtGRP-1a transcripts under drought stress (seedlings without watering in a growth chamber with 50% humidity and 16:8-h L:D photoperiod) and demonstrated that they were continuously accumulated for 3–6 days (Table 2). Notably, plants died if drought time lengthened beyond 6 days.
GRPs and oxidative stress response
Biotic and abiotic stress factors can lead to the rapid production of ROS in plant tissues, which can cause extensive damage to cell membranes and other cellular components. The disproportionate production of ROS relative to scavenging rate results in oxidative stress and represents one of the most common causes of stress-induced injuries (Tuteja et al., 2014). Plants have evolved a number of effective mechanisms for ROS detoxification (Lehmann et al., 2012).
The impact of oxidative stress on the expression patterns of GPR genes has not been extensively investigated. Schmidt et al. (2010) noticed alterations in the expression profiles of AtGRP7 and AtGRP8 genes during oxidative stress (Table 2). Both genes were rapidly upregulated in response to peroxide-induced oxidative stress in Arabidopsis plants, which is in line with results obtained for these genes in response to other stresses (see above). The negative relationship between oxidative stress and ABA signaling may be particularly expected in this case as Cao et al. (2006) and Carpenter et al. (1994) demonstrated that AtGRP7 and AtGRP8 expression is downregulated (even 2- to 3-fold) by ABA treatment.
GRPs in biotic stress responses
Biotic stress, including bacterial, fungal, nematode, and virus infections might be also fatal for plants. Climate change influences pathogen spread and infection intensity. Biotic and abiotic stress conditions (drought, in particular) acting together result in particularly destructive effects on plant growth and development; they also accelerate further pathogen invasion (Suzuki et al., 2014; Sinha et al., 2017).
In Nicotiana glutinosa, the expression of a gene encoding glycine-rich RNA-binding protein (NgRBP) containing two conserved N-terminal RNP motifs as well as C-terminal glycine-rich domain, was negatively regulated under tobacco mosaic virus (TMV) infection; at 24 h after TMV infection, an increased level of ngRBP mRNAs was observed (Table 2). This observation suggests that NgRBP protein is induced by a specific biotic stressor and may play an important role in plant-pathogen interactions (Naqvi et al., 1998). The expression of Hvgrp2 and Hvgrp3 genes in barley was also altered during infection by fungal pathogens Erysiphe graminis and Rhynchosporium secalis (Molina et al., 1997).
Lee et al. (2012) provided further evidence for the participation of AtGRP7 in pathogen response and defense (Table 2). Due to its role in the regulation of RNA metabolism, AtGRP7 is generally important for plant innate immunity; in addition, this protein seems to be a specific “defense regulator.” AtGRP7 transcript levels were significantly elevated in Arabidopsis plants 48 h after Pectobacterium carotovorum infection. Interestingly, AtGRP7 transcript levels upon B. cinerea infection were not substantially altered. This might suggest a species-specific mode of action and different effects of this protein, depending on the pathogen type (Lee et al., 2012).
In summary, it has been shown that AtGRP7 plays a positive role in defense against P. carotovorum in Arabidopsis and in transgenic tobacco plants against TMV. Wild-type Arabidopsis plants exhibited greater resistance to the growth of P. carotovorum than in the case of grp7 mutant plants. For transgenic tobacco lines with overexpressed AtGRP7 protein, necrotic lesion formation was delayed and the total number and size of these lesions was significantly less than in wild-type plants (Lee et al., 2012). However, contrasting results were obtained by this research group regarding the role of AtGRP7 in defense against B. cinerea, because this protein enhances the susceptibility of Arabidopsis plants to this fungus. This result is correlated with stable AtGRP7 transcript levels and confirms that GRP7 protein is a negative “defense regulator” (Lee et al., 2012).
Other interesting results confirming a positive role for AtGRP7 in plant pathogen defense were obtained by Fu et al. (2007). They showed that Arabidopsis grp7 mutants (with AtGRP7 suppressed expression) were much more susceptible to P. syringae than wild-type plants (Fu et al., 2007).
Future directions and concluding remarks
Plants are organisms without any active ability to change their environment under unfavorable conditions. Consequently, plants have evolved various adaptive mechanisms that enable them to cope with stress. In the natural environment, plants rarely encounter single stressors; in fact, plant stress responses depend on stressor combinations. Glycine-rich RBPs are among the most crucial proteins responding to such conditions. To date, data available in the literature supported their significant role in response to various stress conditions.
There is a growing body of evidence indicating that protein engineering and biotechnology is an important tool for the production of plant cultivars with improved stress resistance. In the future, the isolation, cloning, characterization and functional validation of diverse GRPs that are expressed in response to various stresses might be helpful in achieving this goal. As discussed in the present review, the experimental results describing involvement of GRPs in plant stress responses suggest that the overexpression of these proteins may play a positive role in plant adaptation response to a range of stressors. Plant species displaying increased levels of GRP transcripts were more stress-resistant than wild-type plants. According to functional analyses, a role for GRPs in the stress response in many plant species, including model and non-model ones, is indicated. However, relatively little is known about the mechanisms underlying these processes, and this gap in knowledge should be promptly bridged. For instance, the question posed by Mangeon et al. (2010) on the participation of GRPs in multifunctional protein complexes as potent interactors is still open. To what extent could GRPs be replaced by other proteins assisting in signaling pathways within the plant cell? How do the functions of GRPs differ between monocotyledons and dicotyledons? In addition, the relevance of additional structural domains found among GRPs is far from understood. Can they replace glycine-rich domains functionally? It would be also valuable to better understand how glycine-rich domains affect the function of GRPs. Current research regarding GRPs (especially in the functional context) is still growing, and this superfamily indeed seems to have a prospective role in agriculture and consequently in worldwide food production.
Author contributions
MC: Designed the review, deposited and analyzed all literature data, prepared Figure 1, Tables 1, 2 and wrote the manuscript; MR: Substantially assisted in the designation of the review concept, prepared supplementary materials, reviewed the data quality and soundness as well as updated the entire manuscript. Both authors have accepted the final version of the manuscript and agreed to be accountable for all aspects of the work.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer IW and handling Editor declared their shared affiliation.
Acknowledgments
This work was supported by the KNOW RNA Research Centre in Poznań (no. 01/KNOW2/2014).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00302/full#supplementary-material
References
- AbdElgawad H., Zinta G., Hegab M. M., Pandey R., Asard H., Abuelsoud W. (2016). High salinity induces different oxidative stress and antioxidant responses in maize seedlings organs. Front. Plant Sci. 7:276. 10.3389/fpls.2016.00276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Whaibi M. H. (2011). Plant heat-shock proteins: a mini review. J. King Saud Univ. Sci. 23, 139–150. 10.1016/j.jksus.2010.06.022 [DOI] [Google Scholar]
- Aneeta, Sanan-Mishra N., Tuteja N., Kumar Sopory S. (2002). Salinity- and ABA-induced up-regulation and light-mediated modulation of mRNA encoding glycine-rich RNA-binding protein from Sorghum bicolor. Biochem. Biophys. Res. Commun. 296, 1063–1068. 10.1016/S0006-291X(02)02050-8 [DOI] [PubMed] [Google Scholar]
- Aoki Y., Okamura Y., Tadaka S., Kinoshita K., Obayashi T. (2016). ATTED-II in 2016: a plant coexpression database towards lineage-specific coexpression. Plant Cell Physiol. 57:e5. 10.1093/pcp/pcv165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini T. Z., Reiser L., Li D., Mezheritsky Y., Muller R., Strait E., et al. (2015). The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis 53, 474–485. 10.1002/dvg.22877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd L. A., Ridout C., O'Sullivan D. M., Leach J. E., Leung H. (2013). Plant-pathogen interactions: disease resistance in modern agriculture. Trends Genet. 29, 233–240. 10.1016/j.tig.2012.10.011 [DOI] [PubMed] [Google Scholar]
- Cândido Ede S., Pinto M. F., Pelegrini P. B., Lima T. B., Silva O. N., Pogue R., et al. (2011). Plant storage proteins with antimicrobial activity: novel insights into plant defense mechanisms. FASEB J. 25, 3290–3305. 10.1096/fj.11-184291 [DOI] [PubMed] [Google Scholar]
- Cao S., Jiang L., Song S., Jing R., Xu G. (2006). AtGRP7 is involved in the regulation of abscisic acid and stress responses in Arabidopsis. Cell. Mol. Biol. Lett. 11, 526–535. 10.2478/s11658-006-0042-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter C. D., Kreps J. A., Simon A. E. (1994). Genes encoding glycine-rich Arabidopsis thaliana proteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadian rhythm. Plant Physiol. 104, 1015–1025. 10.1104/pp.104.3.1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zeng Q., Lu X., Yu D., Li W. (2010). Characterization and expression analysis of four glycine-rich RNA-binding proteins involved in osmotic response in tobacco (Nicotiana tabacum cv. Xanthi). Agric. Sci. China 9, 1577–1587. 10.1016/S1671-2927(09)60254-6 [DOI] [Google Scholar]
- Chinnusamy V., Zhu J. K. (2009). Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 12, 133–139. 10.1016/j.pbi.2008.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciuzan O., Lazar S. L., Lung M. L., Pop O. L., Pamfil D. (2015). Involvement of the glycine-rich RNA-binding proteins (GRP) in plant abiotic stress response: a comparison between GRP 2 and GRP 7. Bull. Univ. Agric. Sci. Vet. Med. Cluj Napoca Hortic. 72, 61–67. 10.15835/buasvmcn-hort:10912 [DOI] [Google Scholar]
- Conde A., Silva P., Agasse A., Conde C., Gerós H. (2011). Mannitol transport and mannitol dehydrogenase activities are coordinated in Olea europaea under salt and osmotic stresses. Plant Cell Physiol. 52, 1766–1775. 10.1093/pcp/pcr121 [DOI] [PubMed] [Google Scholar]
- Cramer G. R., Urano K., Delrot S., Pezzotti M., Shinozaki K. (2011). Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol. 11:163. 10.1186/1471-2229-11-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Fromm M., Avramova Z. (2012). Multiple exposures to drought “train” transcriptional responses in Arabidopsis. Nat. Commun. 3:740. 10.1038/ncomms1732 [DOI] [PubMed] [Google Scholar]
- Fu J., Miao Y., Shao L., Hu T., Yang P. (2016). De novo transcriptome sequencing and gene expression profiling of Elymus nutans under cold stress. BMC Genomics 17:870. 10.1186/s12864-016-3222-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z. Q., Guo M., Jeong B., Tian F., Elthon T. E., Cerny R. L., et al. (2007). A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature 447, 284–288. 10.1038/nature05737 [DOI] [PubMed] [Google Scholar]
- Fusaro A. F., Bocca S. N., Ramos R. L., Barrôco R. M., Magioli C., Jorge V. C., et al. (2007). AtGRP2, a cold-induced nucleo-cytoplasmic RNA-binding protein, has a role in flower and seed development. Planta 225, 1339–1351. 10.1007/s00425-006-0444-4 [DOI] [PubMed] [Google Scholar]
- Goodstein D. M., Shu S., Howson R., Neupane R., Hayes R. D., Fazo J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucl. Acids Res. 40, D1178–D1186. 10.1093/nar/gkr944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Liu J.-H., Ma X., Luo D.-X., Gong Z.-H., Lu M.-H. (2016). The plant heat stress transcription factors (HSFs): structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 7:114. 10.3389/fpls.2016.00114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanano S., Sugita M., Sugiura M. (1996). Isolation of a novel RNA-binding protein and its association with a large ribonucleoprotein particle present in the nucleoplasm of tobacco cells. Plant Mol. Biol. 31, 57–68. 10.1007/BF00020606 [DOI] [PubMed] [Google Scholar]
- Hu T., Liu S.-Q., Amombo E., Fu J.-M. (2015). Stress memory induced rearrangements of HSP transcription, photosystem II photochemistry and metabolism of tall fescue (Festuca arundinacea Schreb.) in response to high-temperature stress. Front. Plant Sci. 6:403. 10.3389/fpls.2015.00403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iba K. (2002). Acclimative response to temperature stress in higher plants: approaches of gene engineering for temperature tolerance. Annu. Rev. Plant Biol. 53, 225–245. 10.1146/annurev.arplant.53.100201.160729 [DOI] [PubMed] [Google Scholar]
- Joshi R., Ramanarao M. V., Lee S., Kato N., Baisakh N. (2014). Ectopic expression of ADP ribosylation factor 1 (SaARF1) from smooth cordgrass (Spartina alterniflora Loisel) confers drought and salt tolerance in transgenic rice and Arabidopsis. Plant Cell Tissue Organ Cult. 117, 17–30. 10.1007/s11240-013-0416-x [DOI] [Google Scholar]
- Kar B., Nayak S., Joshi R. K. (2012). Classification and comparative analysis of Curcuma longa L. expressed sequences tags (ESTs) encoding glycine-rich proteins (GRPs). Bioinformation 8, 142–146. 10.6026/97320630008142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan F., Sultana T., Deeba F., Naqvi S. M. (2013). Dynamics of mRNA of glycine-rich RNA-binding protein during wounding, cold and salt stresses in Nicotiana tabacum. Pak. J. Bot. 45, 297–300. [Google Scholar]
- Kim J. S., Park S. J., Kwak K. J., Kim Y. O., Kim J. Y., Song J., et al. (2007). Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in Escherichia coli. Nucleic Acids Res. 35, 506–516. 10.1093/nar/gkl1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. Y., Kim W. Y., Kwak K. J., Oh S. H., Han Y. S., Kang H. (2010a). Glycine-rich RNA-binding proteins are functionally conserved in Arabidopsis thaliana and Oryza sativa during cold adaptation process. J. Exp. Bot. 61, 2317–2325. 10.1093/jxb/erq058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. Y., Kim W. Y., Kwak K. J., Oh S. H., Han Y. S., Kang H. (2010b). Zinc finger-containing glycine-rich RNA-binding protein in Oryza sativa has an RNA chaperone activity under cold stress conditions. Plant Cell Environ. 33, 759–768. 10.1111/j.1365-3040.2009.02101.x [DOI] [PubMed] [Google Scholar]
- Kim J. Y., Park S. J., Jang B., Jung C.-H., Ahn S. J., Goh C. H., et al. (2007). Functional characterization of a glycine-rich RNA-binding protein 2 in Arabidopsis thaliana under abiotic stress conditions. Plant J. 50, 439–451. 10.1111/j.1365-313X.2007.03057.x [DOI] [PubMed] [Google Scholar]
- Kim W. Y., Kim J. Y., Jung H. J., Oh S. H., Han Y. S., Kang H. (2010). Comparative analysis of Arabidopsis zinc finger-containing glycine-rich RNA-binding proteins during cold adaptation. Plant Physiol. Biochem. 48, 866–872. 10.1016/j.plaphy.2010.08.013 [DOI] [PubMed] [Google Scholar]
- Kim Y. O., Kim J. S., Kang H. (2005). Cold-inducible zinc finger-containing glycine-rich RNA-binding protein contributes to the enhancement of freezing tolerance in Arabidopsis thaliana. Plant J. 42, 890–900. 10.1111/j.1365-313X.2005.02420.x [DOI] [PubMed] [Google Scholar]
- Kim Y. O., Pan S., Jung C. H., Kang H. (2007). A zinc finger-containing glycine-rich RNA-binding protein, atRZ-1a, has a negative impact on seed germination and seedling growth of Arabidopsis thaliana under salt or drought stress conditions. Plant Cell Physiol. 48, 1170–1181. 10.1093/pcp/pcm087 [DOI] [PubMed] [Google Scholar]
- Krishnakumar V., Hanlon M. R., Contrino S., Ferlanti E. S., Karamycheva S., Kim M., et al. (2015). Araport: the Arabidopsis information portal. Nucl. Acids Res. 43, D1003–D1009. 10.1093/nar/gku1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushwaha H. R., Joshi R., Pareek A., Singla-Pareek S. L. (2016). MATH-domain family shows response toward abiotic stress in Arabidopsis and rice. Front. Plant Sci. 7:923. 10.3389/fpls.2016.00923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak K. J., Kim H.-S., Jang H. Y., Kang H., Ahn S.-J. (2016). Diverse roles of glycine-rich RNA-binding protein 7 in the response of camelina (Camelina sativa) to abiotic stress. Acta Physiol. Plant. 38:129 10.1007/s11738-016-2144-4 [DOI] [Google Scholar]
- Kwak K. J., Kim Y. O., Kang H. (2005). Characterization of transgenic Arabidopsis plants overexpressing GR-RBP4 under high salinity, dehydration, or cold stress. J. Exp. Bot. 56, 3007–3016. 10.1093/jxb/eri298 [DOI] [PubMed] [Google Scholar]
- Kwak K. J., Park S. J., Han J. H., Kim M. K., Oh S. H., Han Y. S., et al. (2011). Structural determinants crucial to the RNA chaperone activity of glycine-rich RNA-binding proteins 4 and 7 in Arabidopsis thaliana during the cold adaptation process. J. Exp. Bot. 62, 4003–4011. 10.1093/jxb/err101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J., Kim J. S., Yoo S. J., Kang E. Y., Han S. H., Yang K.-Y., et al. (2012). Different roles of glycine-rich RNA-binding protein7 in plant defense against Pectobacterium carotovorum, Botrytis cinerea, and tobacco mosaic viruses. Plant Physiol. Biochem. 60, 46–52. 10.1016/j.plaphy.2012.07.020 [DOI] [PubMed] [Google Scholar]
- Lehmann M., Laxa M., Sweetlove L. J., Fernie A. R., Obata T. (2012). Metabolic recovery of Arabidopsis thaliana roots following cessation of oxidative stress. Metabolomics 8, 143–153. 10.1007/s11306-011-0296-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long R., Yang Q., Kang J., Zhang T., Wang H., Li M., et al. (2013). Overexpression of a novel salt stress-induced glycine-rich protein gene from alfalfa causes salt and ABA sensitivity in Arabidopsis. Plant Cell Rep. 32, 1289–1298. 10.1007/s00299-013-1443-0 [DOI] [PubMed] [Google Scholar]
- Mangeon A., Junqueira R. M., Sachetto-Martins G. (2010). Functional diversity of the plant glycine-rich proteins superfamily. Plant Signal. Behav. 5, 99–104 10.4161/psb.5.2.10336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeon A., Magioli C., Menezes-Salgueiro A. D., Cardeal V., de Oliveira C., Galvão V. C., et al. (2009). AtGRP5, a vacuole-located glycine-rich protein involved in cell elongation. Planta 230, 253–265. 10.1007/s00425-009-0940-4 [DOI] [PubMed] [Google Scholar]
- Mangeon A., Pardal R., Menezes-Salgueiro A. D., Duarte G. L., de Seixas R., Cruz F. P., et al. (2016). AtGRP3 is implicated in root size and aluminum response pathways in Arabidopsis. PLoS ONE 11:e0150583. 10.1371/journal.pone.0150583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K., Köster T., Nolte C., Weinholdt C., Lewinski M., Grosse I., et al. (2017). Adaptation of iCLIP to plants determines the binding landscape of the clock-regulated RNA-binding protein AtGRP7. Genome Biol. 18:204. 10.1186/s13059-017-1332-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z., Xu W., Li D., Hu X., Liu J., Zhang R., et al. (2015). De novo transcriptome analysis of Medicago falcata reveals novel insights about the mechanisms underlying abiotic stress-responsive pathway. BMC Genomics 16:818. 10.1186/s12864-015-2019-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina A., Mena M., Carbonero P., García-Olmedo F. (1997). Differential expression of pathogen-responsive genes encoding two types of glycine-rich proteins in barley. Plant Mol. Biol. 33, 803–810 10.1023/A:1005712803130 [DOI] [PubMed] [Google Scholar]
- Naqvi S. M., Park K. S., Yi S. Y., Lee H. W., Bok S. H., Choi D. (1998). A glycine-rich RNA-binding protein gene is differentially expressed during acute hypersensitive response following Tobacco Mosaic Virus infection in tobacco. Plant Mol. Biol. 37, 571–576. [DOI] [PubMed] [Google Scholar]
- Nomata T., Kabeya Y., Sato N. (2004). Cloning and characterization of glycine-rich RNA-binding protein cDNAs in the moss Physcomitrella patens. Plant Cell Physiol. 45, 48–56. 10.1093/pcp/pch005 [DOI] [PubMed] [Google Scholar]
- Ortega-Amaro M. A., Rodríguez-Hernández A. A., Rodríguez-Kessler M., Hernández-Lucero E., Rosales-Mendoza S., Ibáñez-Salazar A., et al. (2015). Overexpression of AtGRDP2, a novel glycine-rich domain protein, accelerates plant growth and improves stress tolerance. Front. Plant Sci. 5:782. 10.3389/fpls.2014.00782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Li J., Jiao L., Li C., Zhu D., Yu J. (2016). A non-specific Setaria italica lipid transfer protein gene plays a critical role under abiotic stress. Front. Plant Sci. 7:1752. 10.3389/fpls.2016.01752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey P., Ramegowda V., Senthil-Kumar M. (2015). Shared and unique responses of plants to multiple individual stresses and stress combinations: physiological and molecular mechanisms. Front. Plant Sci. 6:723. 10.3389/fpls.2015.00723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park A. R., Cho S. K., Yun U. J., Jin M. Y., Lee S. H., Sachetto-Martins G., et al. (2001). Interaction of the Arabidopsis receptor protein kinase Wak1 with a glycine-rich protein, AtGRP-3. J. Biol. Chem. 276, 26688–26693. 10.1074/jbc.M101283200 [DOI] [PubMed] [Google Scholar]
- Rihan H. Z., Al-Issawi M., Fuller M. P. (2017). Advances in physiological and molecular aspects of plant cold tolerance. J. Plant Interact. 12, 143–157. 10.1080/17429145.2017.1308568 [DOI] [Google Scholar]
- Rodríguez-Hernández A. A., Ortega-Amaro M. A., Delgado-Sánchez P., Salinas J., Jiménez-Bremont J. F. (2014). AtGRDP1 Gene encoding a glycine-rich domain protein is involved in germination and responds to ABA signaling. Plant Mol. Biol. Rep. 32, 1187–1202. 10.1007/s11105-014-0714-4 [DOI] [Google Scholar]
- Sah S. K., Reddy K. R., Li J. (2016). Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 7:571. 10.3389/fpls.2016.00571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghera G. S., Wani S. H., Hussain W., Singh N. B. (2011). Engineering cold stress tolerance in crop plants. Curr. Genomics 12, 30–43. 10.2174/138920211794520178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savatin D. V., Gramegna G., Modesti V., Cervone F. (2014). Wounding in the plant tissue: the defense of a dangerous passage. Front. Plant Sci. 5:470. 10.3389/fpls.2014.00470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F., Marnef A., Cheung M.-K., Wilson I., Hancock J., Staiger D., et al. (2010). A proteomic analysis of oligo(dT)-bound mRNP containing oxidative stress-induced Arabidopsis thaliana RNA-binding proteins ATGRP7 and ATGRP8. Mol. Biol. Rep. 37, 839–845. 10.1007/s11033-009-9636-x [DOI] [PubMed] [Google Scholar]
- Shinozuka H., Hisano H., Yoneyama S., Shimamoto Y., Jones E. S., Forster J. W., et al. (2006). Gene expression and genetic mapping analyses of a perennial ryegrass glycine-rich RNA-binding protein gene suggest a role in cold adaptation. Mol. Genet. Genomics 275, 399–408. 10.1007/s00438-005-0095-3 [DOI] [PubMed] [Google Scholar]
- Sinha R., Gupta A., Senthil-Kumar M. (2017). Concurrent drought stress and vascular pathogen infection induce common and distinct transcriptomic responses in chickpea. Front. Plant Sci. 8:333. 10.3389/fpls.2017.00333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S., Raxwal V. K., Joshi B., Jagannath A., Katiyar-Agarwal S., Goel S., et al. (2015). De novo transcriptome profiling of cold-stressed siliques during pod filling stages in Indian mustard (Brassica juncea L.). Front. Plant Sci. 6:932. 10.3389/fpls.2015.00932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smékalová V., Doskočilová A., Komis G., Samaj J. (2014). Crosstalk between secondary messengers, hormones and MAPK modules during abiotic stress signalling in plants. Biotechnol. Adv. 32, 2–11. 10.1016/j.biotechadv.2013.07.009 [DOI] [PubMed] [Google Scholar]
- Staiger D., Zecca L., Wieczorek Kirk D. A., Apel K., Eckstein L. (2003). The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J. Cell Mol. Biol. 33, 361–371. 10.1046/j.1365-313X.2003.01629.x [DOI] [PubMed] [Google Scholar]
- Suzuki N., Bassil E., Hamilton J. S., Inupakutika M. A., Zandalinas S. I., Tripathy D., et al. (2016). ABA is required for plant acclimation to a combination of salt and heat stress. PLoS ONE 11:e0147625. 10.1371/journal.pone.0147625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Rivero R. M., Shulaev V., Blumwald E., Mittler R. (2014). Abiotic and biotic stress combinations. New Phytol. 203, 32–43. 10.1111/nph.12797 [DOI] [PubMed] [Google Scholar]
- Tanz S. K., Castleden I., Hooper C. M., Vacher M., Small I., Millar H. A. (2013). SUBA3: a database for integrating experimentation and prediction to define the SUBcellular location of proteins in Arabidopsis. Nucl. Acids Res. 41, D1185–D1191. 10.1093/nar/gks1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium (2017). UniProt: the universal protein knowledgebase. Nucl. Acids Res. 45, D158–D169. 10.1093/nar/gkh131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja N., Banu M. S., Huda K. M., Gill S. S., Jain P., Pham X. H., et al. (2014). Pea p68, a DEAD-box helicase, provides salinity stress tolerance in transgenic tobacco by reducing oxidative stress and improving photosynthesis machinery. PLoS ONE 9:e98287. 10.1371/journal.pone.0098287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Hu J., Huang X., Li X., Zhou G., Yan Z. (2016). Comparative transcriptome analysis reveals heat-responsive genes in Chinese cabbage (Brassica rapa ssp. chinensis). Front. Plant Sci. 7:939. 10.3389/fpls.2016.00939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Zhang D.-W., Wang Y.-C., Zheng L., Yang C.-P. (2012). A glycine-rich RNA-binding protein can mediate physiological responses in transgenic plants under salt stress. Mol. Biol. Rep. 39, 1047–1053. 10.1007/s11033-011-0830-2 [DOI] [PubMed] [Google Scholar]
- Wienkoop S., Morgenthal K., Wolschin F., Scholz M., Selbig J., Weckwerth W. (2008). Integration of metabolomic and proteomic phenotypes: analysis of data covariance dissects starch and RFO metabolism from low and high temperature compensation response in Arabidopsis thaliana. Mol. Cell. Proteomics 7, 1725–1736. 10.1074/mcp.M700273-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G. V., Provart N. J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2:e718. 10.1371/journal.pone.0000718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H., Zhu W., Wang L., Xiang Y., Fang L., Li J., et al. (2013). Genome wide transcriptional profile analysis of Vitis amurensis and Vitis vinifera in response to cold stress. PLoS ONE 8:e58740. 10.1371/journal.pone.0058740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Gu L., Choi M. J., Kim R. J., Suh M. C., Kang H. (2014). Comparative functional analysis of wheat (Triticum aestivum) zinc finger-containing glycine-rich RNA-binding proteins in response to abiotic stresses. PLoS ONE 9:e96877. 10.1371/journal.pone.0096877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D. H., Kwak K. J., Kim M. K., Park S. J., Yang K. Y., Kang H. (2014). Expression of Arabidopsis glycine-rich RNA-binding protein AtGRP2 or AtGRP7 improves grain yield of rice (Oryza sativa) under drought stress conditions. Plant Sci. Int. J. Exp. Plant Biol. 214, 106–112. 10.1016/j.plantsci.2013.10.006 [DOI] [PubMed] [Google Scholar]
- Yang Q. S., Gao J., He W. D., Dou T. X., Ding L. J., Wu J. H., et al. (2015). Comparative transcriptomics analysis reveals difference of key gene expression between banana and plantain in response to cold stress. BMC Genomics 16:446. 10.1186/s12864-015-1551-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Chen X., Hong Y.-Y., Wang Y., Xu P., Ke S.-D., et al. (2008). Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20, 1134–1151. 10.1105/tpc.108.058263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Chen X., Wang Z., Wang S., Wang Y., Zhu Q., et al. (2013). Arabidopsis enhanced drought tolerance1/HOMEODOMAIN GLABROUS11 confers drought tolerance in transgenic rice without yield penalty. Plant Physiol. 162, 1378–1391. 10.1104/pp.113.217596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y., Guo M., Wang H., Lu J., Liu J., Zhang C., et al. (2016). Autophagy, a conserved mechanism for protein degradation, responds to heat, and other abiotic stresses in Capsicum annuum L. Front. Plant Sci. 7:131. 10.3389/fpls.2016.00131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X., Yang L., Wang D., Zhu J. K., Lang Z. (2016). De novo assembly and analysis of the transcriptome of Ocimum americanum var. pilosum under cold stress. BMC Genomics 17:209. 10.1186/s12864-016-2507-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M. Q., Shen C., Wu L. H., Tang K. X., Lin J. (2011). CBF-dependent signaling pathway: a key responder to low temperature stress in plants. Crit. Rev. Biotechnol. 31, 186–192. 10.3109/07388551.2010.505910 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Zhu G., Guo Q., Zhu Z., Wang C., Liu Z. (2013). A comparative proteomic analysis of Pinellia ternata leaves exposed to heat stress. Int. J. Mol. Sci. 14, 20614–20634. 10.3390/ijms141020614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Sun B., Xu X., Chen H., Zou L., Chen G., et al. (2016). Overexpression of AtEDT1/HDG11 in Chinese kale (Brassica oleracea var. alboglabra) enhances drought and osmotic stress tolerance. Front. Plant Sci. 7:1285. 10.3389/fpls.2016.01285 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


