Abstract
This paper describes an origami-inspired self-folding method to form three-dimensional (3D) microstructures of co-cultured cells. After a confluent monolayer of fibroblasts (NIH/3T3 cells) with loaded hepatocytes (HepG2 cells) was cultured onto two-dimensional (2D) microplates, degradation of the alginate sacrificial layer in the system by addition of alginate lyase triggered NIH/3T3 cells to self-fold the microplates around HepG2 cells, and then 3D cell co-culture microstructures were spontaneously formed. Using this method, we can create a large number of 3D cell co-culture microstructures swiftly with ease in the same time. We find that HepG2 cells confined in the 3D cell co-culture microstructures have an ability to enhance the secreted albumin compared to 2D system in a long culture period. The result indicates that the origami-based cell self-folding technique presented here is useful in regenerative medicine and the preclinical stage of drug development.
Introduction
A challenge for regenerative medicine and drug development is to fabricate in vitro 3D structures that mimic tissues in vivo. Several strategies have been developed to fabricate in vitro 3D cell-laden structures using a bottom-up technique1–6, which involves micro-sized 3D cell-laden microstructures such as blocks2, fibers4–6 and spheroids3,7. This approach allows one to control the size and shape of these microstructures, so that they can be easily handled and assembled to mimic in vivo tissue.
3D microstructures with different types of cells have been intensively investigated to mimic in vivo tissues with a heterogeneous structure3,8–11. In this research, we applied an origami based-technique called cell origami12 to produce many 3D cell co-culture microstructures swiftly with ease at the same time. The process of producing 3D cell co-culture microstructures using the cell origami is as simple as that for conventional cell culture in 2D dishes (Fig. 1). The cells are grown on engineered microplates fixed to a flat surface. The microplates are then detached from the surface by degrading an alginate sacrificial layer under the plates using alginate lyase. This allows the cells to pull the plates using their traction force and self-fold around other types of cells and create a 3D culture condition. Unlike other techniques such as microfluidic devices, any extra equipment including tubes and micro pumps, is not necessary in the cell origami technique.
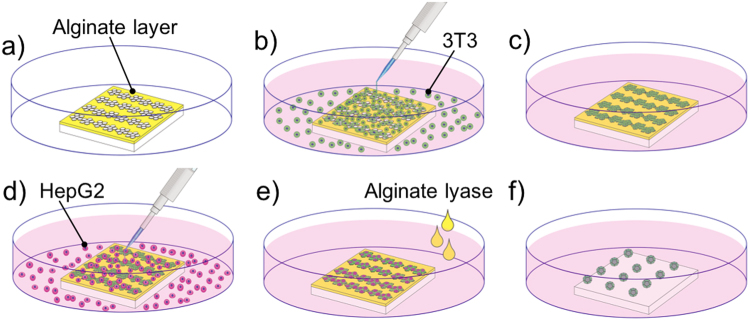
Figure 1.
Processes of seeding and culturing cells on the microplates. (a) The glass substrate with microplates was placed in a petri dish. (b) NIH/3T3 cells were seeded on the microplates, and non-adherent cells were washed away. (c) Adherent NIH/3T3 cells were cultured for 24 h. (d) HepG2 cells were then seeded onto plates and non-adherent cells were washed away. (e) The attached HepG2 cells were cultured 4 h on the NIH/3T3 cells which loaded on the microplates. (e,f) After adding alginate lyase, the microplates were folded, and a number of 3D cell co-culture microstructures were formed.
Other advantages of using the cell origami technique for forming 3D cell co-culture microstructures are that it can provide both flat and 3D culture conditions depending on the cell types and increase the area of interaction between co-culture cells. No other technique with these advantages has been previously developed. It is important to consider different culture conditions to retain the functions of different cell types during co-culture13,14. Previous researches showed that fibroblasts and endothelial cells can proliferate and retain their function on a flat substrate. Conversely, hepatocytes and pancreatic cells prefer 3D culture conditions such as in spheroids. It has also shown that interactions between different types of cells facilitates an increase in their functions4,15–18. A successful co-culture technique, therefore, requires the ability to i) culture one type of cell on a flat substrate, ii) culture another type of cell in 3D conditions, and iii) provide sufficient interactions between these two types of cells. These can be achieved using the cell origami technique.
Here, we produced the 3D cell co-culture microstructures with fibroblasts (NIH/3T3) and hepatoma cells (HepG2) simply and rapidly using the cell origami technique. This 3D cell co-culture microstructure provides both flat and 3D culture conditions for NIH/3T3 and HepG2 cells, respectively. We then performed a viability assay and examined the hepatic function of the co-culture cells in the 3D microstructures by analysis of secreted albumin.
Results and Discussion
Determination of initial NIH/3T3 cell concentration
To wrap HepG2 cells completely, two conditions are required for NIH/3T3 cells. First, the NIH/3T3 cells have to bridge the neighbouring microplates (depicted by the arrows in Fig. 2a) in order to behave as hinges and fold the microplates by their traction force12. Second, NIH/3T3 cells have to be cultured in a confluent monolayer. Thus, we first determined the initial NIH/3T3 cell concentration, CN, for satisfying these conditions.
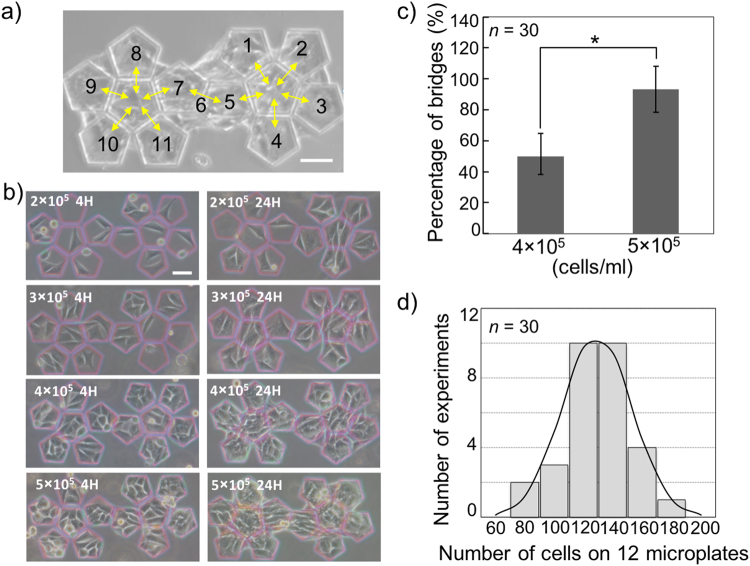
Figure 2.
Determination of CN. (a) In this research, one unit included 12 pieces of microplates to form a 3D dodecahedron microstructure. The total area of each unit is 0.0516 mm2. The bridges of cells between the neighbouring microplates are shown with the direction of traction force by the arrows. (b) Examination of the occupied condition of the unit after seeding various CN at 4 h and 24 h cultivation. (c) Quantification of the number of cell bridges after 24 h with CN of 4 × 105 and 5 × 105 cells/ml. *p < 0.005. (d) After seeding 5 × 105 cells/ml, the numbers of NIH/3T3 cells were counted on each unit after 24 h, scale bar = 50 µm.
We observed that the CN values of 2 × 105 cells/ml and 3 × 105 cells/ml were insufficient for the cells to bridge the neighbouring microplates after 24 h (Fig. 2b). However, with CN of 4 × 105 cells/ml, half of the bridge regions were occupied by the cells, and with CN of 5 × 105 cells/ml, almost all of the bridge regions were occupied by the cells (Fig. 2b,c). When CN was 5 × 105 cells/ml, we found that the cells occupied all microplates as a confluent monolayer within 24 h (Fig. 2b) and that the average number of cells was approximately 130 after 24 h (Fig. 2d). Therefore, we used CN of 5 × 105 cells/ml as an optimized condition to ensure a quick formation of bridges and confluent monolayers on the microplates. Using this method, the cell numbers on the microplates could be easily controlled.
Formation of 3D cell co-culture microstructures
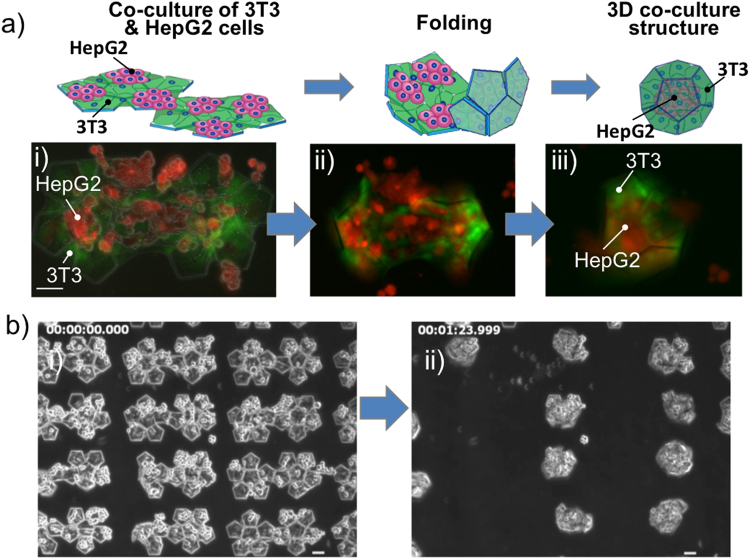
Figure 3a–i shows an image of the HepG2 cells (red) and the NIH/3T3 cells (green) on the microplates after 4 h and 28 h culturing, respectively. NIH/3T3 cells adhered and proliferated on all areas of the microplates, and HepG2 cells were on the top of NIH/3T3 cells. As the alginate layer was degreased by the addition of alginate lyase, the microplates started to self-fold by the traction force of NIH/3T3 cells (Fig. 3a–ii, a–iii and b–i). Many dodecahedrons in which NIH/3T3 cells covered HepG2 cells were formed simultaneously (Fig. 3b–ii, Supplementary Movie 1). The self-folding process was completed within 2 min, which is much faster than 3 days in the previous study using gelatine as a sacrificial layer12. Our present results demonstrate that the use of the alginate layer with the cell origami folding technique can simply and rapidly create a large number of 3D cell co-culture microstructures.
Figure 3.
Formation of 3D cell co-culture microstructures. (a) Top, schematic of folding process. Bottom, NIH/3T3 and HepG2 cells dyed by CellTracker™ Green CMFDA Dye and CellTracker™ Red CMTPX Dye, respectively, were seeded on and folded the plates. (a-i) NIH/3T3 and HepG2 cells cultured on microplates. (a-ii) After addition of alginate lyase, the microplates began to fold by cell traction force. (a-iii) The 3D microstructure is eventually formed with the HepG2 cells covered by NIH/3T3 cells inside. (b) The formation of many 3D microstructures within 2 min are detected by microscopy after the addition of alginate lyase, scale bar = 50 µm.
Confocal microscopic images show that HepG2 cells (red) were covered by NIH/3T3 cells (green) inside the dodecahedron after 3 days incubation in the folded state (Fig. 4, Supplementary Movie 2). This result suggested that the co-culture technique using the cell origami method is suitable for culturing one type of cell (NIH/3T3) on a flat substrate, culturing another type of cell (HepG2) in the 3D conditions, and providing sufficient area of interactions between these two types of cells.
Figure 4.
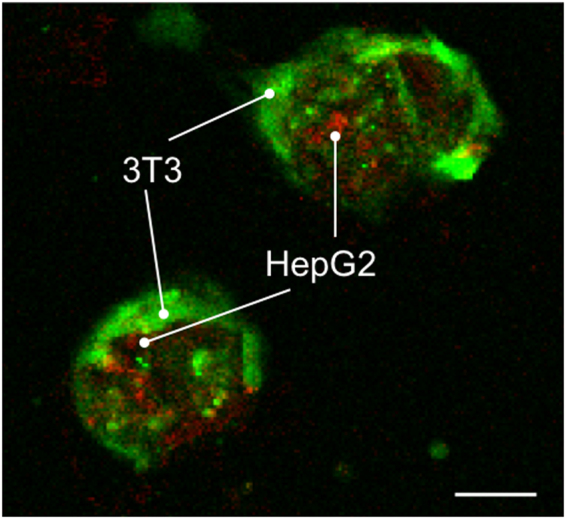
Co-cultured cells inside 3D microstructures after 3 days. HepG2 cells covered by NIH/3T3 cells were detected inside of dodecahedrons at 3 days after initial generation of microstructures, scale bar = 50 µm.
Viability of the co-culture cells inside the 3D microstructures
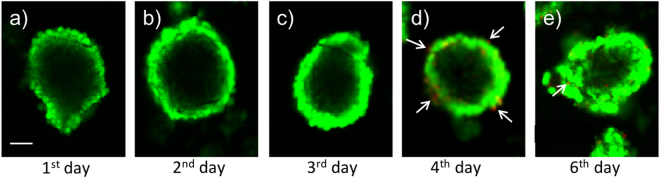
We investigated the viability of co-culture cells embedded in the microstructures. Figure 5 shows the representative images of co-culture cells inside the dodecahedrons from the 1st day to the 6th day. During the first three days (Fig. 5a–c), all the cells inside survived (green), indicating that NIH/3T3 and HepG2 cells were not damaged during the folding process. From the 4th day to the 6th day, most of the co-culture cells remained alive inside the dodecahedrons (Fig. 5d,e), even though a few dead cells (red) were observed (Fig. 5d,e, arrows). This result suggests that cells inside the 3D microstructures can survive at least 6 days. This may be because the gaps of 5 μm between the microplates allowed the medium and necessary nutrition to pass into the structures and reach the cells inside, supporting cell survival.
Figure 5.
Viability of cells inside of 3D microstructures. Cells were stained by the LIVE/DEAD Viability/Cytotoxicity kit, and living and dead cells were distinguished by green and red colours, respectively. (a–c) The co-cultured cells inside 3D microstructures were alive during the first 3 days. (d) On the 4th day, several dead cells were detected (arrows). (e) At the 6th day, most of cells were still alive, with a few dead cells detectable (arrow), scale bar = 50 µm.
Albumin secretion of co-culture cells inside the 3D microstructures
To assess the function of HepG2 cells under these culture conditions, we measured the amounts of secreted albumin from the co-culture cells in two groups, named the folding group (Fig. 6a–i) and the unfolding group (Fig. 6a–ii). The folding group is that in which the microplates with alginate layer were detached from the glass substrate and folded to form a number of dodecahedrons with co-cultured cells. The unfolding group is that in which the microplates without the alginate layer could not fold.
Figure 6.
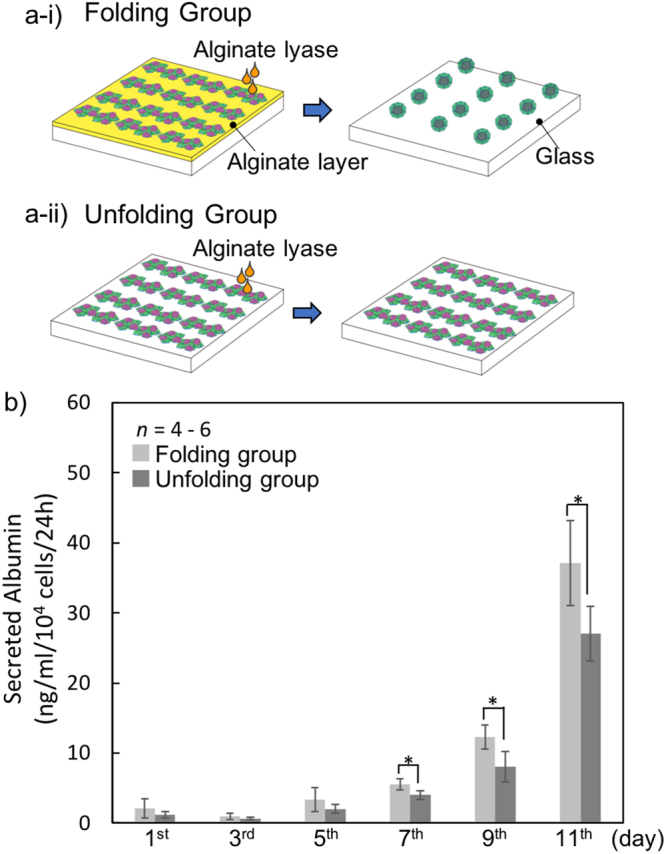
Secretion of albumin in folding and unfolding groups. (a-i) Folding group: microplates with alginate sacrificial layer. After adding alginate lyase, the microplates were detached from the glass substrate and folded to form a number of dodecahedrons with co-cultured cells. (a-ii) Unfolding group: microplates without the alginate sacrificial layer. After adding alginate lyase, the microplates could not undergo folding. (b) Secreted albumin levels were measured in the folding and unfolding groups from the 1st day to the 11th day after co-culture. CN and CH were 5 × 105 cells/ml. Levels in each group were measured as described in Methods. *p < 0.05.
We next investigated how the initial HepG2 cell concentration, CH, affected the secreted albumin in the co-culture system. We observed that the secreted albumin in both folding and unfolding groups at CH = 5 × 105 cells/ml was much higher than that at CH = 1 × 105 cells/ml on the 5th and 7th days (Supplementary Figure 3). For CH = 5 × 105 cells/ml, the secreted albumin in both groups increased with increasing culture days (Fig. 6b). Thus, we adopted CH = 5 × 105 cells/ml for the co-culture with NIH/3T3 cells using the cell origami technique. Using this CH condition, we found that the folding caused significantly higher albumin secretion compared with the unfolding from the 7th day to the 11th day (p < 0.05) (Fig. 6b). In the folding group, HepG2 cell clusters were surrounded by NIH/3T3 cells inside the dodecahedrons, and the cells proliferated and were tightly packed in the confined conditions. This indicates that the 3D microstructure may provide a large contact area between these two kinds of cells. In contrast, in the unfolding group, HepG2 cells formed a spheroidal structure on the NIH/3T3 cell monolayer, which reduced the contact area between HepG2 and NIH/3T3 cells and thereby restricted the cell-cell interaction between these cells. Previous studies have established that increasing cell-cell interactions between hepatocytes and feeder cells such as NIH/3T3 cells leads to increased function of hepatocytes18,19. Therefore, the large contact areas in the folding group may be one of the important factors that increase the function of HepG2 cells.
Conclusion
Our findings demonstrate that many 3D cell co-culture microstructures with NIH/3T3 cells covering HepG2 cells were rapidly with ease produced by the cell origami technique using an alginate sacrificial layer. This 3D cell co-culture microstructure provides both flat and 3D culture conditions for NIH/3T3 and HepG2 cells, respectively, with sufficient interactions between these two types of cells. We confirmed that the cells survived inside of the 3D microstructures and showed that the albumin secretion function of HepG2 cells co-cultured with NIH/3T3 cells inside of the 3D microstructures was increased in comparison with cells co-cultured on the microplates. Our findings suggest that these 3D cell co-culture microstructures that have maintained the cellular functions can be applied not only in regenerative medicine and cell therapy but also in the preclinical stage of drug development.
The cell origami technique can also be applied to the production of 3D microstructures with various shapes and cell types. The shapes of the microplates can easily be varied by changing the glass masks with different designs, allowing for easy changes to the shapes of the microstructures. Human tissues that comprise different types of cells show various 3D shapes in vivo, and thus the cell origami technique may be useful for producing artificial human tissues that are more similar to human tissue structures in vivo.
Materials and Methods
Fabrication of cell origami substrate
Two types of microplates were prepared: microplates with and without a sacrificial layer. We used alginate as a sacrificial layer to accelerate the cell self-folding speed. Alginate degrades immediately upon addition of the alginate lyase with no damage to cells; this strategy has been used extensively for many cell-based applications20,21.
To form the alginate layer, 10–15 mg/ml sodium alginate (Wako, Japan) was spin-coated at 3000 rpm on the hydrophilic surface of glass substrates (20 × 20 mm, MATSUNAMI GLASS IND., LTD., Japan). The substrates were then dipped into 22 mg/ml calcium chloride (CaCl2, Sigma-Aldrich, USA) solution to create the alginate layer on the glass substrates by the reaction between sodium alginate and Ca2+.
Parylene C (Speedline Technologies, USA) was used to produce the microplates because of its biocompatibility and ease of microfabrication22. The transparency property of the parylene is also useful for allowing observation of the co-cultured cells inside the 3D microstructures. Parylene microplates were made according to the previously described method12. Although microelectromechanical systems (MEMS) techniques are needed to fabricate the microplates for the cell origami technique, only the standard MEMS techniques are required. Briefly, the parylene layer (3 μm thickness) was deposited on the glass substrates with or without the alginate layer by chemical vapour deposition (LABCOTERPDS2010, Specialty Coating Systems, USA). Aluminium (Al) was evaporated (EBX-8C, ULVAC, Japan) on the top of the parylene (Supplementary Figure 1a). Al was patterned to define the region of the microplates using a photolithographic technique (Supplementary Figure 1b,c). The parts of exposed parylene were then removed by O2 plasma (10 ml/min, 25 W, RIE-10NR, SAMCO, Japan) to make the microplates (Supplementary Figure 1d).
The 2-methacryloyloxyethyl phosphorylcholine (MPC) polymer, which exhibits inhibitory properties against protein and cell adsorption, was applied to the glass substrate to pattern cells only on microplates. MPC polymer was spin-coated on the glass substrate with microplates at 2000 rpm and dried in an ethanol atmosphere chamber for 20 min to uniformly form the polymer layer. The substrate was baked at 70 °C for 4 h to form a covalent linkage between the MPC polymer and the glass surface by a dehydration reaction (Supplementary Figure 1e). Before culturing cells, the MPC polymer on the microplates was lifted-off by removing the patterned Al using an alkaline solution of NMD-3 developer (Tokyo Ohka, Japan), revealing the final parylene microplates (Supplementary Figure 1f).
Our previous study showed that we can easily create 3D microstructures with various shapes depending on the shape of the microplates12. In this research, we produced a dodecahedron that consists of 12 microplates. The distance between each microplate was 5 μm, and the length of the microplate was 50 μm.
Cell culture
NIH/3T3 cells (TKG299, RIKEN Cell Bank, Japan) and HepG2 cells (TKG0205, RIKEN Cell Bank, Japan) were cultured in High Glucose Dulbecco’s modified Eagle’s medium (HG-DMEM, Sigma-Aldrich, USA) with 10% fatal bovine serum (FBS, Sigma-Aldrich, USA) and 1% penicillin-streptomycin solution (P/S, Sigma-Aldrich, USA) in 60 mm dishes (Nunclon Delta Surface, Thermo Fisher, USA). Cells were cultured at 37 °C with 5% CO2.
Before culturing the cells, 20 µg/ml fibronectin (Funakoshi, Japan) was coated on the microplates to enhance cell adhesion (Supplementary Figure 2a). We prepared NIH/3T3 suspensions at different concentrations (2 × 105 cells/ml, 3 × 105 cells/ml, 4 × 105 cells/ml and 5 × 105 cells/ml) to determine CN required to form 3D microstructures. NIH/3T3 cell suspension (1 mL) was seeded on the glass substrate with the microplates in a 40 mm non-adherent culture dish. We decided the culturing time of the cells was limited in 24 hours to speed up of a whole process of forming 3D microstructures. Non-adherent cells were washed and removed by changing medium after 4 h of cultivation. HepG2 cell suspension (1 mL) with CH of 1 × 105 cells/ml, 5 × 105 cells/ml or 2.5 × 106 cells/ml was then seeded on NIH/3T3 cells after 24 h (Supplementary Figure 2b). Non-adhered HepG2 cells were also washed and removed after 4 h of cultivation (Supplementary Figure 2c).
Detachment and folding of the NIH/3T3-HepG2 cells
After 4 h cultivation of HepG2 cells, alginate lyase (Sigma-Aldrich, USA) at 40 μg/ml concentration was used for degrading the sacrificial layer of the alginate under the microplates, which triggered the self-folding process (Supplementary Figure 2d,e).
Observation of cell morphology
To observe the co-cultured cells inside of the 3D microstructures, NIH/3T3 and HepG2 cells were stained with CellTracke Green CMFDA Dye and CellTracke Red CMTPX Dye (Invitrogen, USA), respectively, before seeding on the microplates. After formation of the 3D microstructures, the cells were observed by a fluorescence microscope (DP 72, Olympus, Japan) and a confocal microscope (Digital Eclipse C1, Nikon, Japan).
We used the LIVE/DEAD Viability/Cytotoxicity fluorescent imaging kit (Invitrogen, USA) to observe the viability of co-culture cells inside of the 3D microstructures from 1 to 6 days after formation of the microstructures. Living and dead cells were distinguished by green and red colours, respectively. The cells were dyed just before image acquisition by the confocal microscope.
Measurement of albumin secretion
We evaluated the function of HepG2 cells by measuring the levels of secreted albumin in the folding group and the unfolding group. Two dishes of each group were prepared for each time point and the experiment was performed three times. The medium was changed every 24 h. From the 1st to the 11th day, the medium was collected, and albumin levels were measured using the albumin ELISA assay kits (EA3201-1, ASSAYPRO). The cell numbers of each dish were also counted and the amount of albumin per 104 cells in each dish was determined.
Electronic supplementary material
Acknowledgements
We gratefully acknowledge Professor Shoji Takeuchi at The University of Tokyo for supporting to produce a glass mask, and Professor Kazuhiko Ishihara at The University of Tokyo for providing the MPC polymer. We also sincerely acknowledge Masaru Sakuma and Kazumi Nakahata for their technical support in mask design and microplate production. This work was supported by the Grant-in-Aid for Scientific Research (B) [JSPS KAKENHI Grant Number JP26286030] from Japan Society for the Promotion of Science (JSPS) and Shiseido female research foundation. A part of this work was conducted at Hokkaido University, supported by “Nanotechnology Platform” Program of the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
Author Contributions
Q.H. and K.K.S. designed the experiments. Q.H. and K.K.S. performed the experiments and analysed the data. Q.H., T.O., H.O. and K.K.S. wrote the paper. A.S. and K.S. provided technique supports. All authors contributed to the discussion of the paper and approved the manuscript
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-22598-x.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Langer R, Vacanti JP. Tissue engineering. Science. 1993;260:920–926. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 2.Gurkan UA, Tasoglu S, Kavaz D, Demirel MC, Demirci U. Emerging technologies for assembly of microscale hydrogels. Adv. Healthc. Mater. 2012;1:149–158. doi: 10.1002/adhm.201200011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsunaga YT, Morimoto Y, Takeuchi S. Molding cell beads for rapid construction of macroscopic 3D tissue architecture. Adv. Mater. 2011;23:90–94. doi: 10.1002/adma.201004375. [DOI] [PubMed] [Google Scholar]
- 4.Onoe H, et al. Metre-long cell-laden microfibres exhibit tissue morphologies and functions. Nat. Mater. 2013;12:584–90. doi: 10.1038/nmat3606. [DOI] [PubMed] [Google Scholar]
- 5.Yamada M, Sugaya S, Naganuma Y, Seki M. Microfluidic synthesis of chemically and physically anisotropic hydrogel microfibers for guided cell growth and networking. Soft Matter. 2012;8:3122. doi: 10.1039/c2sm07263g. [DOI] [Google Scholar]
- 6.Tamayol A, et al. Fiber-based tissue engineering: Progress, challenges, and opportunities. Biotechnol. Adv. 2013;31:669–687. doi: 10.1016/j.biotechadv.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy SV. Organ printing: tissue spheroids as building blocks. Nat. Biotechnol. 2014;32:773–785. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi A, et al. Preparation of stripe-patterned heterogeneous hydrogel sheets using microfluidic devices for high-density coculture of hepatocytes and fibroblasts. J. Biosci. Bioeng. 2013;116:761–767. doi: 10.1016/j.jbiosc.2013.05.034. [DOI] [PubMed] [Google Scholar]
- 9.Torisawa Y, et al. Microfluidic hydrodynamic cellular patterning for systematic formation of co-culture spheroids. Integr. Biol. (Camb). 2009;1:649–54. doi: 10.1039/b915965g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan HF, Zhang Y, Leong KW. Efficient One-Step Production of Microencapsulated Hepatocyte Spheroids with Enhanced Functions. Small. 2016 doi: 10.1002/smll.201502932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee HW, Kook Y-M, Lee HJ, Park H, Koh W-G. A three-dimensional co-culture of HepG2 spheroids and fibroblasts using double-layered fibrous scaffolds incorporated with hydrogel micropatterns. RSC Adv. 2014;4:61005–61011. doi: 10.1039/C4RA12269K. [DOI] [Google Scholar]
- 12.Kuribayashi-Shigetomi K, Onoe H, Takeuchi S. Cell Origami: Self-Folding of Three-Dimensional Cell-Laden Microstructures Driven by Cell Traction Force. PLoS One. 2012;7:e51085. doi: 10.1371/journal.pone.0051085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okuyama T, et al. Preparation of arrays of cell spheroids and spheroid-monolayer cocultures within a microfluidic device. JBIOSC. 2010;110:572–576. doi: 10.1016/j.jbiosc.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 14.Fukuda J, et al. Micromolding of photocrosslinkable chitosan hydrogel for spheroid microarray and co-cultures. Biomaterials. 2006;27:5259–5267. doi: 10.1016/j.biomaterials.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 15.Yamada M, et al. Controlled formation of heterotypic hepatic micro-organoids in anisotropic hydrogel microfibers for long-term preservation of liver-specific functions. Biomaterials. 2012;33:8304–8315. doi: 10.1016/j.biomaterials.2012.07.068. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda Y, et al. Long-term survival and functional maintenance of hepatocytes by using a microfabricated cell array. Colloids Surfaces B Biointerfaces. 2012;97:97–100. doi: 10.1016/j.colsurfb.2012.04.022. [DOI] [PubMed] [Google Scholar]
- 17.Kojima R, et al. Spheroid array of fetal mouse liver cells constructed on a PEG-gel micropatterned surface: upregulation of hepatic functions by co-culture with nonparenchymal liver cells. Lab Chip. 2009;9:1991. doi: 10.1039/b903388b. [DOI] [PubMed] [Google Scholar]
- 18.Lu HF, et al. Three-dimensional co-culture of rat hepatocyte spheroids and NIH/3T3 fibroblasts enhances hepatocyte functional maintenance. Acta Biomater. 2005;1:399–410. doi: 10.1016/j.actbio.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Tsuda Y, Kikuchi A, Yamato M, Chen G, Okano T. Heterotypic cell interactions on a dually patterned surface. Biochem. Biophys. Res. Commun. 2006;348:937–944. doi: 10.1016/j.bbrc.2006.07.138. [DOI] [PubMed] [Google Scholar]
- 20.Augst AD, Kong HJ, Mooney DJ. Alginate hydrogels as biomaterials. Macromol. Biosci. 2006;6:623–633. doi: 10.1002/mabi.200600069. [DOI] [PubMed] [Google Scholar]
- 21.Dvir-Ginzberg M, Gamlieli-Bonshtein I, Agbaria R, Cohen S. Liver tissue engineering within alginate scaffolds: effects of cell-seeding density on hepatocyte viability, morphology, and function. Tissue Eng. 2003;9:757–766. doi: 10.1089/107632703768247430. [DOI] [PubMed] [Google Scholar]
- 22.Chang TY, et al. Cell and protein compatibility of parylene-C surfaces. Langmuir. 2007;23:11718–11725. doi: 10.1021/la7017049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.