Abstract
Atrial natriuretic factor and brain natriuretic peptide are two important biomarkers in clinical cardiology. These two natriuretic peptide hormones are encoded by the paralogous genes Nppa and Nppb, which are evolutionary conserved. Both genes are predominantly expressed by the heart muscle during the embryonic and fetal stages, and in particular Nppa expression is strongly reduced in the ventricles after birth. Upon cardiac stress, Nppa and Nppb are strongly upregulated in the ventricular myocardium. Much is known about the molecular and physiological ques inducing Nppa and Nppb expression; however, the transcriptional regulatory mechanisms of the Nppa–Nppb cluster in vivo has proven to be quite complex and is not well understood. In this review, we will provide recent insights into the dynamic and complex regulation of Nppa and Nppb during heart development and hypertrophy, and the association of this gene cluster with the cardiomyocyte-intrinsic program of heart regeneration.
Keywords: Atrial and brain natriuretic peptide, Epigenetics, Gene cluster, Heart development, Heart regeneration, Hypertrophy, Super enhancer
Introduction
Pathological stress in the heart results in physiological changes accompanied by alterations at both the transcriptional and epigenetic level. These stresses include cardiac hypertrophy and ischemic injury (myocardial infarction). During hypertrophy, the myocardium undergoes adverse structural remodeling that can lead to heart failure, the heart being unable to meet the circulatory demands of the body [1]. Myocardial infarction leads to loss of muscle mass, scar formation and compensatory hypertrophy [2]. A commonly observed response during cardiac hypertrophy is reactivation of the “fetal gene program”. Normally, these fetal genes are abundantly expressed in the prenatal heart but become downregulated after birth. Once the heart undergoes pathological stress, the expression of these genes is induced and this response is thought to play a role in the process of cardiac remodeling and compensation [3–6]. The induction, however, may be orchestrated by a stress-induced regulatory mechanism different to that of the developmental regulatory program [7, 8].
Nppa and Nppb, cardiac genes encoding atrial natriuretic factor (ANF) and brain natriuretic peptide (BNP), respectively, belong to this fetal gene program. Both genes are abundantly expressed in the atrial and ventricular myocardium during embryonic and fetal stages. After birth, both genes remain expressed in the heart, however, postnatal expression of Nppa is strongly downregulated in the ventricles [9–11]. Upon stress, the pro-peptides are released by the heart and the ventricular expression of both Nppa and Nppb is strongly increased in the cardiomyocytes [12, 13]. Because of this feature, the gene products, especially NT-pro-BNP that has a longer half-life compared to BNP and ANF, serve as reliable molecular markers to assess cardiac disease and heart failure progression [14, 15]. Additionally, Nppa has also become an important marker for myocardial chamber differentiation and congenital heart defects [16, 17]. The importance of Nppa and Nppb in heart development and disease has initiated in-depth studies on the transcriptional regulatory mechanisms of these genes. Insights into these mechanisms have already substantially increased our understanding of the molecular events underlying heart development and pathological stress of the heart [18].
The paralogous genes Nppa and Nppb are positioned in close proximity to each other and organized in an evolutionary conserved gene cluster [19–21]. The structural organization and regulation of Nppa and Nppb expression have proven to be more complex than was initially thought [7, 8, 22–24]. Therefore, the identification and functional analysis of regulatory sequences of the Nppa–Nppb cluster has been challenging. Nevertheless, current genomic technologies applied to study epigenetic landscapes, chromatin structure and gene regulation (e.g. chromatin immunoprecipitation sequencing and chromatin conformation capturing combined with transgenic reporter mice) has shed light on the regulatory mechanisms of the Nppa–Nppb cluster in vivo [8, 13, 23, 24]. In this review, we will discuss recent progress in deciphering the regulatory landscape of the Nppa–Nppb cluster during heart development and disease.
Gene clusters: conceptual framework of sharing and co-regulation
The spatial and temporal pattern of gene expression is regulated through cis-regulatory DNA elements (e.g. promoters, enhancers, insulators, repressors) that function in strictly context-dependent manners. The transcriptional machinery that drives cell-specific gene expression involves the binding of transcription factors and co-factors at specific locations on the DNA via sequence-dependent affinity. This process is coordinated by epigenetic motifs and signatures, and the three-dimensional arrangement of chromatin, which is responsible for bringing necessary components in spatial proximity [18, 25–27]. During evolution, the natriuretic peptide genes Nppa and Nppb have arisen from the ancestral CNP-3 gene through the process of gene duplication followed by divergence [28]. Nppa and Nppb are positioned in close proximity to each other in the mammalian genome, separated by only several kilo base pairs (kbp) of DNA sequence. Comparative studies have demonstrated that these paralogous genes show very similar expression patterns in the developing atrial and ventricular chamber myocardium of mouse, rat and human. In contrast, birds have lost the Nppa gene, and their Nppb gene is expressed at high levels in both atria and ventricles [9, 29]. Both Nppa and Nppb are upregulated in response to hypertrophy [30] and in the injury border zone after myocardial infarction [13]. These common features of Nppa and Nppb suggest that this gene cluster may contain cis-regulatory sequences shared by both genes, and a topology that facilitates co-regulation during development and stress. Sharing of regulatory sequences and co-regulation of clustered paralogous genes has been proposed previously; however, to date only few examples have been comprehensively described, including the Iroquois (Irx) and Hox gene clusters.
The Irx gene cluster is present in both invertebrates and vertebrates. In mammals, the Irx genes are divided into two paralogous clusters, IrxA (Irx1, Irx2 and Irx4) and IrxB (Irx3, Irx5 and Irx6), located on different chromosomes [31]. In both clusters, the orientation of the three genes is strictly conserved and organized. The developmental expression patterns of clustered genes Irx1 and Irx2, and of Irx3 and Irx5, respectively, are highly similar [32]. All six genes are expressed in specific patterns in the heart, and Irx3, 4 and 5 are involved in cardiac development and conduction [33, 34]. Extensive screening of the genomic regions of the IrxA and IrxB cluster revealed highly conserved non-coding regions with cis-regulatory elements. These cis-regulatory elements physically interact with the promoters of the first two genes of the Irx gene clusters. Furthermore, Irx1/Irx2 and Irx3/Irx5 are engaged in promoter–promoter interaction and this explains why their expression patterns overlap during development. The third genes Irx4 and Irx6, respectively, do not seem to interact with the other two genes of their cluster or their shared regulatory elements and consistently show distinct expression patterns [35].
Hox genes play a crucial role in vertebrate anterior–posterior patterning and limb development [36–38]. In mammals, 39 Hox genes are found organized in four genomic clusters (HoxA, B, C and D) that are localized on different chromosomes. The regulation of Hox genes is controlled by shared, distant regulatory regions. Moreover, the epigenetic state and chromatin organization of the Hox clusters determine the function of regulatory elements in the regulation of the Hox genes [39, 40]. The regulation of HoxD cluster during limb development has been shown to be tightly controlled by a collection of regulatory elements distributed over two gene deserts (a regulatory archipelago) on either side of the HoxD cluster. Through conformational changes in the HoxD locus, these regulatory elements are brought together to regulate HoxD gene transcription and coordinate the transition from early to late limb development [41–43].
The examples of the Irx and Hox gene clusters provide a conceptual framework for co-regulation by shared cis-regulatory elements in the locus or even at long distance. It has been proposed that the structural stability of these clusters throughout evolution is maintained by the sharing of conserved regulatory elements by the genes within the cluster [44]. More recently, other loci harboring clustered paralogous genes that are functionally important for heart development and function have come to our attention, including Tbx3–Tbx5, Scn5a–Scn10a and Nppa–Nppb, and have provided insights into the role of structure and composition of the chromatin in genomic function and gene transcription [8, 45, 46].
Spatial and functional organization of Nppa–Nppb cluster
With the development of new technologies, different approaches are being used to study loci with respect to their regulatory landscapes of gene loci. These include functional testing of regulatory elements [enhancer and bacterial artificial chromosome (BAC) transgenesis], chromosome conformation capturing, analysis of epigenetic states (ChIP-seq, etc.), and have improved our understanding of the regulatory domains controlling the Nppa–Nppb cluster [8, 13].
In gene clusters such as Irx and Hox, the promoters and their shared distal regulatory regions must be brought together physically in order to regulate transcriptional activity. In general, regulatory elements find their target genes within topologically associating domains (TADs). TADs are chromosomal regions, typically about 1 Mbp in size, within which sequences preferentially contact each other. They are separated by boundary regions for CCCTC-binding factor (CTCF) binding sites [47–49]. It has been established that chromatin loops direct enhancers to target genes, thereby creating a three-dimensional regulatory landscape [25, 50, 51]. High-resolution chromatin conformation capturing (4C) revealed that the intergenomic interactions of the Nppa–Nppb cluster are confined to a domain between the two closest CTCF sites, which is a stretch of approximately 60 kbp [8]. Notably, the chromatin conformation of Nppa and Nppb differs only little between heart tissue and other tissues, indicating it is permissive, existing in a pre-formed 3D conformation, and not instructive and cell-type dependent [8, 25]. This phenomenon of pre-formed chromatin loops has been demonstrated for other loci as well, including the Tbx3–Tbx5 cluster [52].
Although the exact role of the CTCF sites in Nppa–Nppb regulation has yet to be investigated, it is thought that CTCF sites maintain the stability of the regulatory domain. Previously it has been described that deletion of CTCF sites in the Hox gene clusters (HoxA and HoxD) disrupted the chromatin conformation and altered the regulatory and transcriptional activities in the TADs [53, 54]. Similarly, changing the orientation of a CTCF site influences DNA-looping interactions, consequently leading to transcriptional misregulation [54, 55]. Recent studies of the functional role of CTCF in chromatin folding and transcriptional regulation describe that CTCF is indeed required for the formation and maintenance of loops between CTCF target sites and architecture of TADs at the genomic level [49, 56, 57]. Conditional depletion of CTCF in mouse embryonic stem cells caused insulation defects at most TAD boundaries and abrogation of chromatin loops between CTCF sites. This resulted in altered enhancer–promoter interactions across the DNA region leading to upregulation of a subset of genes that were previously insulated from neighboring regulatory elements. In addition, it has been suggested that CTCF might also have a direct impact on transcriptional regulation independent of loops and chromatin folding. CTCF sites were often found near transcription start site and were mostly in direct orientation with transcription of the downregulated genes prior to CTCF depletion. In contrast, CTCF depletion did not affect genomic compartments. Restoring CTCF levels reversed the chromatin interactome to its normal state [57].
The accessibility of chromatin relies on structural features, which is tightly controlled by epigenetic processes including DNA methylation, histone modifications and ATP-dependent chromatin remodeling. Particular epigenetic mechanisms are associated with active promoters and cis-regulatory elements. In adult cardiomyopathy, including cardiac hypertrophy and heart failure, epigenetic changes such as histone acetylation and methylation are observed in response to cardiac stress. This can contribute to transcriptional reprogramming in the heart and changes in cardiac gene expression [58]. Genome-wide analysis of the epigenetic signature of hypertrophied hearts of mice showed that multiple genes implicated in hypertrophic cardiomyopathy and associated enhancers are modified through histone-3 lysine-27 acetylation (H3K27ac), a modification associated with activation [59]. In patients with heart failure, reactivation of NPPA and NPPB is correlated with demethylation of H3K9 at their promoter regions, although a modest increase in H3K27ac could also be observed [60]. The cofactor p300, important in acetylation of histones, promotes cardiac remodeling (e.g. left ventricular dilation) in infarcted mouse hearts through interaction with transcription factor Gata4 [61]. Furthermore, p300 is found to be recruited to the Nppa and Nppb promoter, which is associated with increased histone acetylation such as H3K27ac [62]. Within the Nppa-Nppb regulatory domain, physical interactions are found between cis-regulatory regions and the promoters of Nppa and Nppb. These regulatory sequences function to control either developmental or stress-responsive expression of Nppa and Nppb. Analysis of the distribution of H3K27ac and RNA polymerase II (Pol II) across the Nppa–Nppb locus revealed that epigenetic signatures within the regulatory domain change during cardiac stress. During pressure overload-induced cardiac hypertrophy in mice, H3K27ac is enriched near and at the promoters of Nppa and Nppb, whereas Pol II occupation, associated with active promoters and enhancers, changed much less. Even though no significant change in Pol II occupation has been observed, both promoters may still be involved in stress-induced expression of Nppa and Nppb. In the conserved upstream regulatory region that is associated with fetal expression of Nppa, the levels of H3K27ac and Pol II are decreased upon stress [7, 8, 63]. It should be noted that in the normal adult heart, this regulatory region is already highly occupied by H3K27ac and presumably maintains Nppb expression after birth (Fig. 1a) [8, 59].
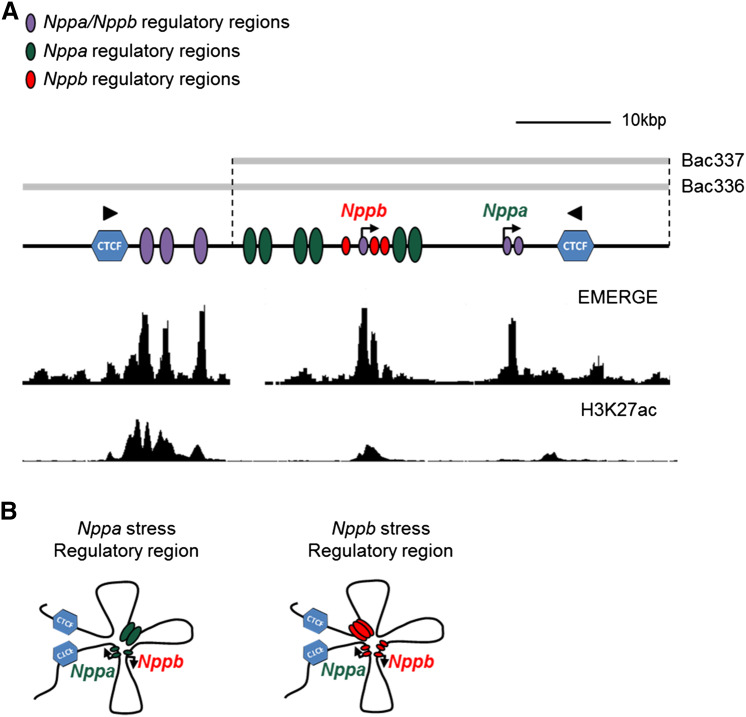
Fig. 1.
The regulatory landscape of the Nppa-Nppb locus. a Developmental and stress response regulatory regions of Nppa and Nppb are located within a 60 kbp domain between two CTCF sites arranged in a convergent orientation [49, 91]. Purple, shared regulatory regions of Nppa/Nppb; green, regulatory regions of Nppa; red, regulatory regions of Nppb. Gray bars, BAC clones. Displaying EMERGE track for heart and H3K27ac track for mouse cardiomyoyctes [92, 93]. b Nppa and Nppb are regulated by different regulatory elements during stress. The Nppa promoter interacts with Nppb promoter and several distal and proximal regulatory elements. Stress-induced expression of Nppb is regulated by a upstream regulatory region and the Nppa/Nppb promoters
Regulation of the Nppa–Nppb gene cluster during development and hypertrophy
Genome-wide association studies have found a correlation between genetic variants identified in the NPPA–NPPB locus and the levels of natriuretic peptides in blood of patients with cardiac dysfunction. A variant (rs5065) in the coding region of NPPA [64] and an intronic variant (rs1023252) in CLCN6 [65] are associated with NT-pro-BNP levels in severe heart failure patients. Furthermore, genetic variants identified upstream and downstream of NPPB has proven to significantly affect levels of BNP [66]. Together, this suggest that variants in the NPPA–NPPB regulatory domain (and in linkage disequilibrium with the reported variants) influence regulatory DNA function. Genetic variants associated with blood pressure and hypertension at the AGTRAP–PLOD1 locus are suggested to influence the expression of multiple genes, including NPPA and NPPB, within this region [67]. These genetic variants are in linkage disequilibrium with the NPPA–NPPB regulatory domain and, therefore, may only report the presence of variants influencing regulatory DNA function of NPPA and NPPB during disease. Indeed, it has been reported that genetic variants positioned within the regulatory domain of NPPA–NPPB locus potentially affect gene expression by a yet undefined mechanism [68]. The biological and clinical relevance of Nppa and Nppb is probably the major reason the transcriptional regulation of these genes has been the subject of several studies [7, 8, 22–24]. Nppa and Nppb are reactivated in the stressed myocardium as part of an induction of a “fetal gene program”. The question remained whether the transcriptional mechanisms involved in hypertrophic stress induction are the same as those governing the fetal gene program. Previously, it has been suggested that the proximal Nppa promoter mediates the developmental expression pattern of Nppa, although later it was found that its capacity to drive ventricular expression was largely absent [7, 22, 69, 70]. Furthermore, this promoter is inducible in cell culture systems, but not sufficient for stress-induced Nppa expression in vivo, suggesting the involvement of other distal regulatory elements [7, 22]. Furthermore, ventricular expression of Nppa was found to be driven by distal sequences, whereas stress induction required more proximal sequences, demonstrating that the transcriptional mechanisms driving fetal expression and stress-induced expression are different [7].
The Nppa promoter has been used as a model for understanding transcriptional gene regulation during cardiac development [9, 69, 71, 72]. It was thought that the Nppa promoter drives embryonic and fetal Nppa expression in the atria and ventricles but different fragment sizes of the promoter could not recapitulate the correct ventricular expression [7, 70]. Several regulatory elements that lie upstream of the proximal Nppa promoter region appeared to be involved in the ventricular expression of Nppa during development. Two reporter BAC clones with 85 kbp of overlapping sequences (BAC336-EGFP and BAC337-EGFP) (Fig. 1a) covering the Nppa–Nppb locus were used in an attempt to define the distal regulatory regions that control the pre- and postnatal expression of Nppa in the ventricles (Fig. 1a). BAC337-EGFP was shown to lack the regulatory sequences necessary for Nppa ventricular activity, and unique sequences located in BAC336-EGFP (− 141 to – 27 kbp relative to Nppa) drove Nppa-like expression patterns during development [7]. The potential of this regulatory region (− 141 to – 27 kbp relative to Nppa) in mediating the developmental expression of Nppa was further supported by analysis of Nkx2–5 occupancy and function in vivo. The transcription factor Nkx2–5 has a major role in the regulation of gene expression in the developing heart. In vivo screening of the regulatory elements within the Nppa–Nppb locus in inducible Nkx2–5 knockout mice showed a diminished expression in the heart, indicating an essential role of Nkx2–5 in the regulation of Nppa. Indeed, 3C analysis showed that these regulatory elements enriched for Nkx2–5 interact with the Nppa promoter. However, stress-induced expression of Nppa did not depend on Nkx2–5 transcriptional regulation [23]. Further studies on BAC336-EGFP and BAC337-EGFP revealed that both were able to induce reporter gene expression upon hypertrophic stress. This demonstrates that both BAC clones contain regulatory sequences that mediate stress-induced Nppa expression. These sequences are thought to be located in the overlapping 85 kbp region and downstream of Nppa (Fig. 1a) [7]. Analysis of both BAC clones revealed that the distal regulatory region is responsible for Nppa expression in the embryonic/fetal and adult heart, whereas the proximal regulatory region is required for stress-induced Nppa expression.
The development and stress-induced regulatory elements of Nppb were less well described compared to Nppa. There is evidence that the promoter constitutively drives weak Nppb expression in the normal and stressed heart [73, 74]. As described below, later studies showed other regulatory elements within the Nppa–Nppb locus are required [8].
Recently, a more extensive characterization of the spatial and functional organization of the Nppa–Nppb cluster in vivo has been provided. Based on H3K27ac and Pol2 ChIP-seq data, heart-specific regulatory regions were defined in the Nppa–Nppb locus (Fig. 1a), which were functionally tested in a transgenic mouse model carrying a BACs with two modifications. The function of the Nppa–Nppb cluster can be monitored simultaneously for both Nppa and Nppb due to the insertion of the Luciferase and Katushka genes at the translation start sites of these genes, respectively, within the BAC. Both reporter genes recapitulate the tissue-specific and developmental pattern of expression and stress response of endogenous Nppa and Nppb [8, 13]. Analyses of the BAC transgenic mice showed that developmental expression of Nppa and Nppb is mediated by shared cis-regulatory elements located approximately 27 kbp upstream of Nppa (Fig. 1a). This regulatory region, roughly 10 kbp, is enriched for epigenetic features including heart-specific DNaseI hypersensitivity sites and histone modifications, and binding sites for various cardiac transcription factors (e.g. Nkx2–5 and Gata4). Furthermore, this regulatory region is being described as a “super enhancer” [75, 76]. According to the conformation of the Nppa–Nppb locus, this region contacts the promoters of both genes, suggesting that regulatory elements within this region drive the fetal ventricular expression of Nppa and Nppb. Furthermore, this region might also contain regulatory elements involved in Nppb expression during hypertrophic stress in the adult heart (Fig. 1a, b) A 650-bp fragment located in the same region was implicated in stress-induced Nppa expression [24]; however, the BAC transgenesis study indicates it may be involved in Nppb regulation. This finding is further supported by analysis of transgenic lines with BAC337 that lacks this region, in which strong EGFP expression (reporting for Nppa) was observed in stressed ventricles [7, 8, 24]. However, it is uncertain whether this 650 bp fragment is involved in induction during hypertrophy as no response has been observed in vitro after stimulation with phenylephrine (PE) or hypertrophic stress in transgenic mice with this fragment (Sergeeva, unpublished data).
Which particular regulatory elements drive stress-induced Nppa expression remains unresolved. There are indications that the Nppb promoter might be involved; its deletion within the double reporter BAC rendered Luciferase/Nppa non-responsive to hypertrophy. However, the Nppb promoter alone was not sufficient to drive Nppa expression upon hypertrophic stress. The Nppb promoter is necessary for the embryonic/fetal and adult expression of Nppb itself, but not required for hypertrophic induction. Nevertheless, the Nppb promoter drives Luciferase expression in rat ventricular cardiomyocytes after PE stimulation. Together, these data suggest that the Nppb promoter is part of a complex of proximal and distal regulatory elements, all required in vivo, whereas several of these elements may drive stress-responsive expression when tested outside their endogenous context (Fig. 1a) [8].
Nppa–Nppb cluster locus containing conserved regulatory elements activated during zebrafish heart regeneration
Myocardial infarction causes loss of heart muscle. In contrast to lower vertebrates like fish and amphibians, the mammalian heart has a highly insufficient capacity to regenerate and restore this muscle tissue. Studies on zebrafish heart regeneration have demonstrated through genetic lineage tracing that proliferating cardiomyocytes are the source of the newly formed cardiomyocytes. These (adult) cardiomyocytes have first undergone dedifferentiation, which is characterized by disassembly of sarcomeric structures and re-expression of genes such as Gata4 involved in heart development and Nppa/Nppb [77–80]. Recently is has been shown that a similar regenerative response is found in neonatal mice, in which the cardiomyocytes retain for a short period of time the ability to proliferate [81]. Cardiomyocyte renewal in adult mice (and humans) is very limited under normal conditions with a less turnover rate of less than 1 percent per year [82, 83]. In adult mouse myocardial infarction models, cardiomyocyte proliferation has been observed, but too low to regenerate the injured heart [82, 84, 85].
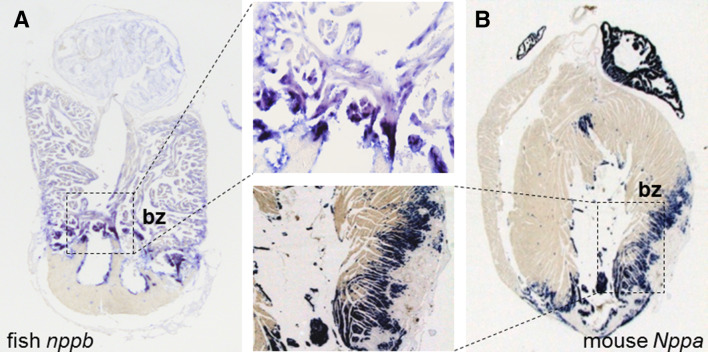
During fetal and neonatal development, cardiomyocytes rapidly proliferate and, therefore, the myocardium can regenerate upon injury. From studies aimed at understanding the regulation of cardiomyocyte proliferation and regeneration it has been suggested that cardiomyocyte-intrinsic programs can promote these regenerative processes upon cardiac injury [86]. Exploiting the transcriptional dynamics during zebrafish heart regeneration suggest that these transcriptional regulatory mechanisms recapitulate the fetal gene program [79, 87]. Furthermore, the spatial gene expression profile of a cryo-injured zebrafish heart revealed the transcriptional activation of nppa and nppb in a district region (the border zone) within the heart where also regeneration occurs (Fig. 2a) [80]. Interestingly, reactivation of Nppa and Nppb is also restricted to the border zone of an injured mouse heart (Fig. 2b) [10]. This raises the question whether conserved stress responsive regulatory elements for Nppa/nppa and Nppb/nppb exists in the mouse and zebrafish heart that are associated with an intrinsic mechanism for cardiomyocyte renewal. Only recently, evidence suggests that conserved regulatory elements may indeed be present that can induce the transcriptional programs for heart regeneration upon tissue damage. In the zebrafish leptin b locus a distal regulatory element has been identified that directs gene expression after injury, including fin amputation and cryo-injury [88]. This regulatory element and response of leptin are not conserved in the mouse, and the regulatory element is active in the endocardium. Nevertheless, the leptin-linked regulatory element was activated in an injured neonatal mouse heart. Furthermore, the leptin-linked regulatory element could activate Nrg1/ErbB2/ErbB4 pathway to promote cardiomyocyte proliferation after re-sectioning of the zebrafish heart [88]. Recent histone H3.3 replacement profiling of regenerative zebrafish hearts uncovered thousands of putative regenerative-responsive enhancers in the fish genome [89]. These findings raise the possibility that the Nppa–Nppb cluster might also harbor conserved regulatory elements which are activated after cardiac injury that can initiate transcriptional programs for dedifferentiation and proliferation of adult cardiomyocytes. Studying the transcriptional regulation of Nppa and Nppb during disease may uncover these regulatory elements.
Fig. 2.
Expression of zebrafish nppa and nppb and of mouse Nppa mRNA in sections of an injured zebrafish and mouse heart. a, b Both fetal genes are reactivated in the border zone (bz) after cryo-injury and myocardial infarction, respectively [10, 80]
Conclusion and future perspectives
The natriuretic peptides ANF and BNP are widely used as biomarkers in various cardiovascular diseases in clinical settings. Studies of the structure and function of the Nppa–Nppb cluster has provided novel insights into the transcriptional regulatory mechanisms of Nppa and Nppb expression during heart development and disease. The transcriptional regulation of Nppa and Nppb has proven to be complex. Nppa, which is highly expressed during ventricular stress, is controlled by several different proximal and distal regulatory elements, including the Nppb promoter, to regulate its dynamic expression in the embryonic/fetal and adult heart. Nppb expression relies on the interaction of its promoter and a conserved large distal regulatory region, classified as a “super enhancer”. Moreover, the Nppa–Nppb cluster shares (developmental) enhancers found in the super enhancer region. The Nppa–Nppb gene cluster provides a conceptual framework for understanding gene cluster function and enhancer sharing that likely applies to other loci that harbor clustered genes. Other interesting gene clusters such as Tbx3–Tbx5 [45], Scn5a–Scn10a [46], Kcne1–Kcne2, Kcnj2–Kcnj16, HoxA and HoxB [90] are being studied or have yet to be studied with respect to transcriptional (co-)regulation and genomic function in the heart. Although the paradigm of heart regeneration in the mammalian adult heart is being debated, evidence suggests that conserved regulatory elements are activated after cardiac injury, which controls the transcriptional programs for heart regeneration in fish. Therefore, an intriguing question is whether the regulatory elements found in the Nppa–Nppb cluster respond to a regenerative mechanism in the stressed myocardium. Future research may focus on the manipulation of the regulatory sequences of the Nppa–Nppb locus in vivo by CRISPR/Cas9 genome editing to determine their physiological relevance in the context of hypertrophic stress or ischemic injury. Furthermore, stress response regulatory elements of the mammalian Nppa–Nppb cluster can be integrated into the zebrafish genome by site-directed transgene integration to assess whether these sequences are transcriptionally activated during zebrafish heart regeneration. The identification of these conserved regulatory elements can provide tools to drive therapeutic genes that promote adult mammalian heart regeneration.
Acknowledgements
This work was supported by CVON HUSTCARE and Foundation Leducq.
Abbreviations
- ANF
Atrial natriuretic factor
- BAC
Bacterial artificial chromosome
- BNF
Brain natriuretic peptide
- BZ
Border zone
- CTCF
CCCTC-binding factor
- Irx
Iroquois
- PE
Phenylephrine
- Pol II
RNA polymerase II
- TADs
Topologically associated domains
References
- 1.Drazner MH. The progression of hypertensive heart disease. Circulation. 2011;123(3):327–334. doi: 10.1161/CIRCULATIONAHA.108.845792. [DOI] [PubMed] [Google Scholar]
- 2.Hellermann JP, Jacobsen SJ, Gersh BJ, Rodeheffer RJ, Reeder GS, Roger VL. Heart failure after myocardial infarction: a review. Am J Med. 2002;113(4):324–330. doi: 10.1016/S0002-9343(02)01185-3. [DOI] [PubMed] [Google Scholar]
- 3.Izumo S, Nadal-Ginard B, Mahdavi V. Protooncogene induction and reprogramming of cardiac gene expression produced by pressure overload. Proc Natl Acad Sci USA. 1988;85:339–343. doi: 10.1073/pnas.85.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chien KR, Knowlton KU, Zhu H, Chien S. Regulation of cardiac gene expression during myocardial growth and hypertrophy: molecular studies of an adaptive physiologic response. FASEB J. 1991;5:3037–3046. doi: 10.1096/fasebj.5.15.1835945. [DOI] [PubMed] [Google Scholar]
- 5.Komuro I, Yazaki Y. Control of cardiac gene expression by mechanical stress. Annu Rev Physiol. 1993;55:55–75. doi: 10.1146/annurev.ph.55.030193.000415. [DOI] [PubMed] [Google Scholar]
- 6.Kuwahara K, Nishikimi T, Nakao K. Transcriptional regulation of the fetal cardiac gene program. J Pharmacol Sci. 2012;119(3):198–203. doi: 10.1254/jphs.12R04CP. [DOI] [PubMed] [Google Scholar]
- 7.Horsthuis T, Houweling AC, Habets PEMH, de Lange FJ, el Azzouzi H, Clout DEW, Moorman AFM, Christoffels VM. Distinct regulation of developmental and heart disease induced atrial natriuretic factor expression by two separate distal sequence. Circ Res. 2008;102:849–859. doi: 10.1161/CIRCRESAHA.107.170571. [DOI] [PubMed] [Google Scholar]
- 8.Sergeeva IA, Hooijkaas IB, Ruijter JM, van der Made I, de Groot NE, van de Werken HJ, Creemers EE, Christoffels VM. Identification of a regulatory domain controlling the Nppa–Nppb gene cluster during heart development and stress. Development. 2016 doi: 10.1242/dev.132019. [DOI] [PubMed] [Google Scholar]
- 9.Houweling AC, van Borren MM, Moorman AFM, Christoffels VM. Expression and regulation of the atrial natriuretic factor encoding gene Nppa during development and disease. Cardiovasc Res. 2005;67:583–593. doi: 10.1016/j.cardiores.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 10.Sergeeva I, Christoffels VM. Regulation of expression of atrial and brain natriuretic peptide, biomarkers for heart development and disease. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbadis.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Cameron VA, Aitken GD, Ellmers LJ, Kennedy MA, Espiner EA. The sites of gene expression of atrial, brain, and C-type natriuretic peptides in mouse fetal development: temporal changes in embryos and placenta. Endocrinology. 1996;137(3):817–824. doi: 10.1210/endo.137.3.8603590. [DOI] [PubMed] [Google Scholar]
- 12.Guo J, Gan XT, Haist JV, Rajapurohitam V, Zeidan A, Faruq NS, Karmazyn M. Ginseng inhibits cardiomyocyte hypertrophy and heart failure via NHE-1 inhibition and attenuation of calcineurin activation. Circ Heart Fail. 2011;4(1):79–88. doi: 10.1161/CIRCHEARTFAILURE.110.957969. [DOI] [PubMed] [Google Scholar]
- 13.Sergeeva IA, Hooijkaas IB, van der Made I, Jong WM, Creemers EE, Christoffels VM. A transgenic mouse model for the simultaneous monitoring of ANF and BNP gene activity during heart development and disease. Cardiovasc Res. 2013 doi: 10.1093/cvr/cvt228. [DOI] [PubMed] [Google Scholar]
- 14.Troughton R, Michael Felker G, Januzzi JL., Jr Natriuretic peptide-guided heart failure management. Eur Heart J. 2014;35(1):16–24. doi: 10.1093/eurheartj/eht463. [DOI] [PubMed] [Google Scholar]
- 15.de Antonio M, Lupon J, Galan A, Vila J, Urrutia A, Bayes-Genis A. Combined use of high-sensitivity cardiac troponin T and N-terminal pro-B type natriuretic peptide improves measurements of performance over established mortality risk factors in chronic heart failure. Am Heart J. 2012;163(5):821–828. doi: 10.1016/j.ahj.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Christoffels VM, Habets PEMH, Franco D, Campione M, de Jong F, Lamers WH, Bao ZZ, Palmer S, Biben C, Harvey RP, Moorman AFM. Chamber formation and morphogenesis in the developing mammalian heart. Dev Biol. 2000;223:266–278. doi: 10.1006/dbio.2000.9753. [DOI] [PubMed] [Google Scholar]
- 17.Bruneau BG. Atrial natriuretic factor in the developing heart: a signpost for cardiac morphogenesis. Can J Physiol Pharmacol. 2011;89(8):533–537. doi: 10.1139/y11-051. [DOI] [PubMed] [Google Scholar]
- 18.Kathiriya IS, Nora EP, Bruneau BG. Investigating the transcriptional control of cardiovascular development. Circ Res. 2015;116(4):700–714. doi: 10.1161/CIRCRESAHA.116.302832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seilhamer JJ, Arfsten A, Miller JA, Lundquist P, Scarborough RM, Lewicki JA, Porter JG. Human and canine gene homologs of porcine brain natriuretic peptide. Biochem Biophys Res Commun. 1989;165(2):650–658. doi: 10.1016/S0006-291X(89)80015-4. [DOI] [PubMed] [Google Scholar]
- 20.Wu JP, Kovacic-Milivojevic B, Lapointe MC, Nakamura K, Gardner DG. cis-Active determinants of cardiac-specific expression in the human atrial natriuretic peptide gene. Mol Endocrinol. 1991;5(9):1311–1322. doi: 10.1210/mend-5-9-1311. [DOI] [PubMed] [Google Scholar]
- 21.Inoue K, Sakamoto T, Yuge S, Iwatani H, Yamagami S, Tsutsumi M, Hori H, Cerra MC, Tota B, Suzuki N, Okamoto N, Takei Y. Structural and functional evolution of three cardiac natriuretic peptides. Mol Biol Evol. 2005;22(12):2428–2434. doi: 10.1093/molbev/msi243. [DOI] [PubMed] [Google Scholar]
- 22.Knowlton KU, Rockman HA, Itani M, Vovan A, Seidman CE, Chien KR. Divergent pathways mediate the induction of ANF transgenes in neonatal and hypertrophic ventricular myocardium. J Clin Investig. 1995;96:1311–1318. doi: 10.1172/JCI118166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warren SA, Terada R, Briggs LE, Cole-Jeffrey CT, Chien WM, Seki T, Weinberg EO, Yang TP, Chin MT, Bungert J, Kasahara H. Differential role of Nkx2–5 in activation of the ANF gene in developing vs. failing heart. Mol Cell. 2011 doi: 10.1128/MCB.05940-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuoka K, Asano Y, Higo S, Tsukamoto O, Yan Y, Yamazaki S, Matsuzaki T, Kioka H, Kato H, Uno Y, Asakura M, Asanuma H, Minamino T, Aburatani H, Kitakaze M, Komuro I, Takashima S. Noninvasive and quantitative live imaging reveals a potential stress-responsive enhancer in the failing heart. FASEB J. 2014;28(4):1870–1879. doi: 10.1096/fj.13-245522. [DOI] [PubMed] [Google Scholar]
- 25.de Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502(7472):499–506. doi: 10.1038/nature12753. [DOI] [PubMed] [Google Scholar]
- 26.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489(7414):109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13(9):613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 28.Inoue K, Naruse K, Yamagami S, Mitani H, Suzuki N, Takei Y. Four functionally distinct C-type natriuretic peptides found in fish reveal evolutionary history of the natriuretic peptide system. Proc Natl Acad Sci USA. 2003;100(17):10079–10084. doi: 10.1073/pnas.1632368100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takei Y, Inoue K, Trajanovska S, Donald JA. B-type natriuretic peptide (BNP), not ANP, is the principal cardiac natriuretic peptide in vertebrates as revealed by comparative studies. Gen Comp Endocrinol. 2011;171(3):258–266. doi: 10.1016/j.ygcen.2011.02.021. [DOI] [PubMed] [Google Scholar]
- 30.Rockman HA, Ross RS, Harris AN, Knowlton KU, Steinhelper ME, Field LJ, Ross J, Jr, Chien KR. Segregation of atrial-specific and inducible expression of an atrial natriuretic factor transgene in an in vivo murine model of cardiac hypertrophy. Proc Natl Acad Sci USA. 1991;88(18):8277–8281. doi: 10.1073/pnas.88.18.8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peters T, Dildrop R, Ausmeier K, Rüther U. Organization of mouse Iroquois homeobox genes in two clusters suggests a conserved regulation and function in vertebrate development. Genome Res. 2000;10(10):1453–1462. doi: 10.1101/gr.144100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houweling AC, Dildrop R, Peters T, Mummenhoff J, Moorman AFM, Rüther U, Christoffels VM. Gene and cluster-specific expression of the Iroquois family members during mouse development. Mech Dev. 2001;107:169–174. doi: 10.1016/S0925-4773(01)00451-8. [DOI] [PubMed] [Google Scholar]
- 33.Kim KH, Rosen A, Bruneau BG, Hui CC, Backx PH. Iroquois homeodomain transcription factors in heart development and function. Circ Res. 2012;110(11):1513–1524. doi: 10.1161/CIRCRESAHA.112.265041. [DOI] [PubMed] [Google Scholar]
- 34.Christoffels VM, Keijser AGM, Houweling AC, Clout DEW, Moorman AFM. Patterning the embryonic heart: Identification of five mouse Iroquois homeobox genes in the developing heart. Dev Biol. 2000;224:263–274. doi: 10.1006/dbio.2000.9801. [DOI] [PubMed] [Google Scholar]
- 35.Tena JJ, Alonso ME, Calle-Mustienes E, Splinter E, de Laat W, Manzanares M, Gomez-Skarmeta JL. An evolutionarily conserved three-dimensional structure in the vertebrate Irx clusters facilitates enhancer sharing and coregulation. Nat Commun. 2011;2:310. doi: 10.1038/ncomms1301. [DOI] [PubMed] [Google Scholar]
- 36.Mallo M, Wellik DM, Deschamps J. Hox genes and regional patterning of the vertebrate body plan. Dev Biol. 2010;344(1):7–15. doi: 10.1016/j.ydbio.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wellik DM. Hox patterning of the vertebrate axial skeleton. Dev Dyn. 2007;236(9):2454–2463. doi: 10.1002/dvdy.21286. [DOI] [PubMed] [Google Scholar]
- 38.Zakany J, Duboule D. The role of Hox genes during vertebrate limb development. Curr Opin Genet Dev. 2007;17(4):359–366. doi: 10.1016/j.gde.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 39.Garcia-Fernandez J. The genesis and evolution of homeobox gene clusters. Nat Rev Genet. 2005;6(12):881–892. doi: 10.1038/nrg1723. [DOI] [PubMed] [Google Scholar]
- 40.Tschopp P, Duboule D. A regulatory ‘landscape effect’ over the HoxD cluster. Dev Biol. 2011;351(2):288–296. doi: 10.1016/j.ydbio.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 41.Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147(5):1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 42.Tarchini B, Duboule D. Control of Hoxd genes’ collinearity during early limb development. Dev Cell. 2006;10(1):93–103. doi: 10.1016/j.devcel.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 43.Andrey G, Montavon T, Mascrez B, Gonzalez F, Noordermeer D, Leleu M, Trono D, Spitz F, Duboule D. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science. 2013;340(6137):1234167. doi: 10.1126/science.1234167. [DOI] [PubMed] [Google Scholar]
- 44.Spitz F, Duboule D. Global control regions and regulatory landscapes in vertebrate development and evolution. Adv Genet. 2008;61:175–205. doi: 10.1016/S0065-2660(07)00006-5. [DOI] [PubMed] [Google Scholar]
- 45.van Weerd JH, Badi I, van den Boogaard M, Stefanovic S, van de Werken HJ, Gomez-Velazquez M, Badia-Careaga C, Manzanares M, de Laat W, Barnett P, Christoffels VM. A large permissive regulatory domain exclusively controls Tbx3 expression in the cardiac conduction system. Circ Res. 2014;115:432–441. doi: 10.1161/CIRCRESAHA.115.303591. [DOI] [PubMed] [Google Scholar]
- 46.van den Boogaard M, Smemo S, Burnicka-Turek O, Arnolds DE, van de Werken HJ, Klous P, McKean D, Muehlschlegel JD, Moosmann J, Toka O, Yang XH, Koopmann TT, Adriaens ME, Bezzina CR, de Laat W, Seidman C, Seidman JG, Christoffels VM, Nobrega MA, Barnett P, Moskowitz IP. A common genetic variant within SCN10A modulates cardiac SCN5A expression. J Clin Investig. 2014;124(4):1844–1852. doi: 10.1172/JCI73140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nora EP, Dekker J, Heard E. Segmental folding of chromosomes: a basis for structural and regulatory chromosomal neighborhoods? Bioessays. 2013;35(9):818–828. doi: 10.1002/bies.201300040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ong CT, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15(4):234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Wit E, Vos ES, Holwerda SJ, Valdes-Quezada C, Verstegen MJ, Teunissen H, Splinter E, Wijchers PJ, Krijger PH, de Laat W. CTCF binding polarity determines chromatin looping. Mol Cell. 2015;60(4):676–684. doi: 10.1016/j.molcel.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 50.Krijger PH, de Laat W. Regulation of disease-associated gene expression in the 3D genome. Nat Rev Mol Cell Biol. 2016;17(12):771–782. doi: 10.1038/nrm.2016.138. [DOI] [PubMed] [Google Scholar]
- 51.Schwarzer W, Spitz F. The architecture of gene expression: integrating dispersed cis-regulatory modules into coherent regulatory domains. Curr Opin Genet Dev. 2014;27:74–82. doi: 10.1016/j.gde.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 52.van Weerd JH, Koshiba-Takeuchi K, Kwon C, Takeuchi JK. Epigenetic factors and cardiac development. Cardiovasc Res. 2011;91(2):203–211. doi: 10.1093/cvr/cvr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Narendra V, Rocha PP, An D, Raviram R, Skok JA, Mazzoni EO, Reinberg D. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015;347(6225):1017–1021. doi: 10.1126/science.1262088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lupianez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, Santos-Simarro F, Gilbert-Dussardier B, Wittler L, Borschiwer M, Haas SA, Osterwalder M, Franke M, Timmermann B, Hecht J, Spielmann M, Visel A, Mundlos S. Disruptions of topological chromatin domains cause pathogenic rewiring of gene–enhancer interactions. Cell. 2015;161(5):1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, Lu Y, Wu Y, Jia Z, Li W, Zhang MQ, Ren B, Krainer AR, Maniatis T, Wu Q. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell. 2015;162(4):900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137(7):1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell. 2017;169(5):930 e922–944 e922. doi: 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mahmoud SA, Poizat C. Epigenetics and chromatin remodeling in adult cardiomyopathy. J Pathol. 2013;231(2):147–157. doi: 10.1002/path.4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Papait R, Cattaneo P, Kunderfranco P, Greco C, Carullo P, Guffanti A, Vigano V, Stirparo GG, Latronico MV, Hasenfuss G, Chen J, Condorelli G. Genome-wide analysis of histone marks identifying an epigenetic signature of promoters and enhancers underlying cardiac hypertrophy. Proc Natl Acad Sci USA. 2013;110(50):20164–20169. doi: 10.1073/pnas.1315155110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hohl M, Wagner M, Reil JC, Muller SA, Tauchnitz M, Zimmer AM, Lehmann LH, Thiel G, Bohm M, Backs J, Maack C. HDAC4 controls histone methylation in response to elevated cardiac load. J Clin Investig. 2013;123(3):1359–1370. doi: 10.1172/JCI61084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miyamoto S, Kawamura T, Morimoto T, Ono K, Wada H, Kawase Y, Matsumori A, Nishio R, Kita T, Hasegawa K. Histone acetyltransferase activity of p300 is required for the promotion of left ventricular remodeling after myocardial infarction in adult mice in vivo. Circulation. 2006;113(5):679–690. doi: 10.1161/CIRCULATIONAHA.105.585182. [DOI] [PubMed] [Google Scholar]
- 62.Mathiyalagan P, Chang L, Du XJ, El-Osta A. Cardiac ventricular chambers are epigenetically distinguishable. Cell Cycle. 2010;9(3):612–617. doi: 10.4161/cc.9.3.10612. [DOI] [PubMed] [Google Scholar]
- 63.Sayed D, He M, Yang Z, Lin L, Abdellatif M. Transcriptional regulation patterns revealed by high resolution chromatin immunoprecipitation during cardiac hypertrophy. J Biol Chem. 2013;288(4):2546–2558. doi: 10.1074/jbc.M112.429449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vassalle C, Andreassi MG, Prontera C, Fontana M, Zyw L, Passino C, Emdin M. Influence of ScaI and natriuretic peptide (NP) clearance receptor polymorphisms of the NP system on NP concentration in chronic heart failure. Clin Chem. 2007;53(11):1886–1890. doi: 10.1373/clinchem.2007.088302. [DOI] [PubMed] [Google Scholar]
- 65.Del Greco MF, Pattaro C, Luchner A, Pichler I, Winkler T, Hicks AA, Fuchsberger C, Franke A, Melville SA, Peters A, Wichmann HE, Schreiber S, Heid IM, Krawczak M, Minelli C, Wiedermann CJ, Pramstaller PP. Genome-wide association analysis and fine mapping of NT-proBNP level provide novel insight into the role of the MTHFR–CLCN6–NPPA–NPPB gene cluster. Hum Mol Genet. 2011;20(8):1660–1671. doi: 10.1093/hmg/ddr035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lanfear DE, Stolker JM, Marsh S, Rich MW, McLeod HL. Genetic variation in the B-type natriuretic peptide pathway affects BNP levels. Cardiovasc Drugs Ther. 2007;21(1):55–62. doi: 10.1007/s10557-007-6007-5. [DOI] [PubMed] [Google Scholar]
- 67.Flister MJ, Tsaih SW, O’Meara CC, Endres B, Hoffman MJ, Geurts AM, Dwinell MR, Lazar J, Jacob HJ, Moreno C. Identifying multiple causative genes at a single GWAS locus. Genome Res. 2013;23(12):1996–2002. doi: 10.1101/gr.160283.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Newton-Cheh C, Larson MG, Vasan RS, Levy D, Bloch KD, Surti A, Guiducci C, Kathiresan S, Benjamin EJ, Struck J, Morgenthaler NG, Bergmann A, Blankenberg S, Kee F, Nilsson P, Yin X, Peltonen L, Vartiainen E, Salomaa V, Hirschhorn JN, Melander O, Wang TJ. Association of common variants in NPPA and NPPB with circulating natriuretic peptides and blood pressure. Nat Genet. 2009;41(3):348–353. doi: 10.1038/ng.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Habets PEMH, Moorman AFM, Clout DEW, van Roon MA, Lingbeek M, Lohuizen M, Campione M, Christoffels VM. Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: implications for cardiac chamber formation. Genes Dev. 2002;16:1234–1246. doi: 10.1101/gad.222902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de Lange FJ, Moorman AFM, Christoffels VM. Atrial cardiomyocyte-specific expression of Cre recombinase driven by an Nppa gene fragment. Genesis. 2003;37:1–4. doi: 10.1002/gene.10220. [DOI] [PubMed] [Google Scholar]
- 71.Nemer G, Nemer M. Regulation of heart development and function through combinatorial interactions of transcription factors. Ann Med. 2001;33(9):604–610. doi: 10.3109/07853890109002106. [DOI] [PubMed] [Google Scholar]
- 72.Temsah R, Nemer M. Gata factors and transcriptional regulation of cardiac natriuretic peptide genes. Regul Pept. 2005;128:177–185. doi: 10.1016/j.regpep.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 73.Majalahti T, Suo-Palosaari M, Sarman B, Hautala N, Pikkarainen S, Tokola H, Vuolteenaho O, Wang J, Paradis P, Nemer M, Ruskoaho H. Cardiac BNP gene activation by angiotensin II in vivo. Mol Cell Endocrinol. 2007;273(1–2):59–67. doi: 10.1016/j.mce.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 74.He Q, Wang D, Yang XP, Carretero OA, LaPointe MC. Inducible regulation of human brain natriuretic peptide promoter in transgenic mice. Am J Physiol Heart Circ Physiol. 2001;280(1):H368–H376. doi: 10.1152/ajpheart.2001.280.1.H368. [DOI] [PubMed] [Google Scholar]
- 75.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wei Y, Zhang S, Shang S, Zhang B, Li S, Wang X, Wang F, Su J, Wu Q, Liu H, Zhang Y. SEA: a super-enhancer archive. Nucleic Acids Res. 2016;44(D1):D172–D179. doi: 10.1093/nar/gkv1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298(5601):2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 78.Jopling C, Sleep E, Raya M, Marti M, Raya A, Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464(7288):606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, MacRae CA, Stainier DY, Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464(7288):601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu CC, Kruse F, Vasudevarao MD, Junker JP, Zebrowski DC, Fischer K, Noel ES, Grun D, Berezikov E, Engel FB, van Oudenaarden A, Weidinger G, Bakkers J. Spatially resolved genome-wide transcriptional profiling identifies BMP signaling as essential regulator of zebrafish cardiomyocyte regeneration. Dev Cell. 2016;36(1):36–49. doi: 10.1016/j.devcel.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 81.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331(6020):1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, Wu TD, Guerquin-Kern JL, Lechene CP, Lee RT. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493(7432):433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S, Frisen J. Evidence for cardiomyocyte renewal in humans. Science. 2009;324(5923):98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bersell K, Arab S, Haring B, Kuhn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138(2):257–270. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- 85.Steinhauser ML, Lee RT. Regeneration of the heart. EMBO Mol Med. 2011;3(12):701–712. doi: 10.1002/emmm.201100175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Foglia MJ, Poss KD. Building and re-building the heart by cardiomyocyte proliferation. Development. 2016;143(5):729–740. doi: 10.1242/dev.132910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gupta V, Gemberling M, Karra R, Rosenfeld GE, Evans T, Poss KD. An injury-responsive gata4 program shapes the zebrafish cardiac ventricle. Curr Biol CB. 2013;23(13):1221–1227. doi: 10.1016/j.cub.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kang J, Hu J, Karra R, Dickson AL, Tornini VA, Nachtrab G, Gemberling M, Goldman JA, Black BL, Poss KD. Modulation of tissue repair by regeneration enhancer elements. Nature. 2016;532(7598):201–206. doi: 10.1038/nature17644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Goldman JA, Kuzu G, Lee N, Karasik J, Gemberling M, Foglia MJ, Karra R, Dickson AL, Sun F, Tolstorukov MY, Poss KD. Resolving heart regeneration by replacement histone profiling. Dev Cell. 2017;40(4):392 e395–404 e395. doi: 10.1016/j.devcel.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nolte C, Jinks T, Wang X, Martinez Pastor MT, Krumlauf R. Shadow enhancers flanking the HoxB cluster direct dynamic Hox expression in early heart and endoderm development. Dev Biol. 2013;383(1):158–173. doi: 10.1016/j.ydbio.2013.09.016. [DOI] [PubMed] [Google Scholar]
- 91.Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159(7):1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gilsbach R, Preissl S, Gruning BA, Schnick T, Burger L, Benes V, Wurch A, Bonisch U, Gunther S, Backofen R, Fleischmann BK, Schubeler D, Hein L. Dynamic DNA methylation orchestrates cardiomyocyte development, maturation and disease. Nat Commun. 2014;5:5288. doi: 10.1038/ncomms6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.van Duijvenboden K, de Boer BA, Capon N, Ruijter JM, Christoffels VM. EMERGE: a flexible modelling framework to predict genomic regulatory elements from genomic signatures. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1144. [DOI] [PMC free article] [PubMed] [Google Scholar]