Abstract
Genetic etiology of psychopathology symptoms and cognitive performance in schizophrenia is supported by candidate gene and polygenic risk score (PRS) association studies. Such associations are reported to be dependent on several factors - sample characteristics, illness phase, illness severity etc. We aimed to examine if schizophrenia PRS predicted psychopathology symptoms and cognitive performance in patients with chronic schizophrenia. We also examined if schizophrenia associated autosomal loci were associated with specific symptoms or cognitive domains.
Case-only analysis using data from the Clinical Antipsychotics Trials of Intervention Effectiveness-Schizophrenia trials (n = 730). PRS was constructed using Psychiatric Genomics Consortium (PGC) leave one out genome wide association analysis as the discovery data set. For candidate region analysis, we selected 105-schizophrenia associated autosomal loci from the PGC study.
We found a significant effect of PRS on positive symptoms at p-threshold (PT) of 0.5 (R2 = 0.007, p = 0.029, empirical p = 0.029) and negative symptoms at PT of 1e-07 (R2 = 0.005, p = 0.047, empirical p = 0.048). For models that additionally controlled for neurocognition, best fit PRS predicted positive (p-threshold 0.01, R2 = 0.007, p = 0.013, empirical p = 0.167) and negative symptoms (p-threshold 0.1, R2 = 0.012, p = 0.004, empirical p = 0.329). No associations were seen for overall neurocognitive and social cognitive performance tests. Post-hoc analyses revealed that PRS predicted working memory and vigilance performance but did not survive correction. No candidate regions that survived multiple testing corrections were associated with either symptoms or cognitive performance. Our findings point to potentially distinct pathogenic mechanisms for schizophrenia symptoms.
1. Introduction
Schizophrenia is a highly complex and disabling disorder with heritability estimates as high as 80% (Sullivan et al., 2003). The genetic risk of schizophrenia established from twin/family studies, and genome wide association studies denote a complex polygenic architecture with several common (108 candidate regions) and rare variants contributing to schizophrenia susceptibility (Rees et al., 2015). Clinically, schizophrenia is heterogeneous with wide variations in symptom presentation. Despite advances in technology that facilitate biological inquiry, pathogenic mechanisms of schizophrenia still remain largely unknown. Heterogeneity at the clinical level and complexity at the molecular level have been cited as strong reasons for this (Rasetti and Weinberger, 2011). In an attempt to address these challenges, some studies have focused on a sub-phenotype approach to examine molecular mechanisms of symptoms (Fanous et al., 2012). This is a validated approach and there is growing evidence for its utility in advancing the field (Jones et al., 2016; Sengupta et al., 2017).
Though there are multiple approaches to study biological and molecular mechanisms of symptoms, two common genetic approaches seen in the literature are: (1) association studies that examine single nucleotide polymorphism's (SNP) effect on symptoms and (2) polygenic risk score (PRS) associations which utilize an aggregate measure of genetic susceptibility to symptoms by taking into account the additive effects of all significant variants across multiple genes and regulatory areas in the entire genome. When SNP associations point to potential functional pathways, PRS is a taken as evidence for cumulative genetic risk. Since it is an indicator of potentially true genetic risk, examining PRS correlations for symptoms can help inform which symptoms are a result of stronger genetic liability.
Studies that have examined the molecular basis of symptoms have found genetic evidence for specific symptom dimensions in schizophrenia. Evidence from candidate gene association studies identify the possibility of shared (neurotransmitter systems, neuronal development and maintenance) and unique associations to positive and negative symptom dimensions (Xavier and Vorderstrasse, 2017), though functional mechanisms are still unclear. Polygenic scores for schizophrenia have shown associations with symptom dimensions, though with inconsistencies. PRS correlations are reported in the literature for negative/disorganized dimensions not just in schizophrenia patients (Fanous et al., 2012) but also in adolescents in the general population where it predicted negative and anxiety symptoms (Jones et al., 2016). But a recent study had negative findings and the association of PRS with negative symptoms was not replicated in first episode psychosis; instead a polygenic loading was found for general psychopathology dimension and anxiety symptoms (Sengupta et al., 2017). Though in this study a PRS association with negative symptoms was reported in the Caucasian only sub-group (Sengupta et al., 2017).
There is extensive evidence for early onset and persistent cognitive impairments in schizophrenia (Green et al., 2004; Keefe et al., 2005). Such deficits are considered to be a major cause of functional impairment and a quest to understand pathogenic mechanisms of cognitive deficits remains a priority. Though the association between cognitive ability and psychosis is known (Morgan et al., 2014) with evidence supporting a shared genetic basis for cognitive ability and neuropsychiatric illnesses such as schizophrenia (Hagenaars et al., 2016; Hill et al., 2016), the exact nature of this relationship remains obscure (Johnson et al., 2016).
General cognitive ability in the general population has shown to be substantially heritable with large consortium studies identifying multiple candidate associations indicating a polygenic inheritance (Davies et al., 2015). In a recent GWAS meta-analysis of 35,298 healthy individuals, rs76114856 in the CENPO gene and rs6669072 on chromosome 1 were found to be associated with cognitive performance (Trampush et al., 2017). In schizophrenia and psychotic illnesses, several candidate genes (ex: COMT, DTNBP1, NRG1, DISC1, ERBB4) implicating neurotransmitter pathways involving dopaminergic (Green et al., 2014) and glutamatergic signaling (Greenwood et al., 2011, Greenwood et al., 2013, Greenwood et al., 2016) have found to be associated with cognitive performance and/or cognitive deficits in patients, though such findings have not produced knowledge that is translatable to clinical practice (Ehrenreich and Nave, 2014).
The genetic etiology of cognition is also supported by PRS studies which associate a higher polygenic burden with greater cognitive decline between the ages of 11 and 70 and a general lower cognitive ability at the age of 70 (McIntosh et al., 2013). In the Philadelphia neurodevelopmental cohort, investigators also found schizophrenia PRS associations for speed of emotion identification, an aspect of social cognition (Germine et al., 2016). Earlier studies utilizing genetic risk scores did not find associations with cognitive domains (Yeo et al., 2014) or IQ in schizophrenia (Van Scheltinga et al., 2013), though the lack of association could be explained by smaller sample sizes.
Data driven approaches have validated the veracity of the polygenic risk score approach (Chen et al., 2017), but PRS results vary across studies and are reported to be dependent on factors such as the sample, stage and/or severity of the disease (Cooke Bailey and Igo, 2016). One study reported PRS to be associated with treatment resistance (Frank et al., 2015), though another study found the evidence to be inconclusive (Martin and Mowry, 2016). PRS has also been reported to be associated with frequent hospitalizations in patients, with a suggested possibility that the association likely comes from the common variants involved in deterioration during the course of the illness (Meier et al., 2016). But PRS predictions of symptoms and cognitive dimensions have not been specifically examined in patients with chronic schizophrenia.
Our study aimed to examine whether schizophrenia PRS is associated with psychopathology symptoms and cognitive dimensions (neurocognition and social cognition) in a sample of patients with chronic schizophrenia. We also examined schizophrenia associated candidate regions (Ripke et al., 2014) for region specific associations with symptoms and cognitive dimensions in this sample.
2. Method
2.1. Subjects
This study is a secondary analysis of data from 741 subjects with genetic data in the Clinical Antipsychotics Trials of Intervention Effectiveness (CATIE)-schizophrenia trial. CATIE (2001–2004) was a United States based multisite trial designed to assess differences in antipsychotic efficacy. We refer you to Sullivan et al. (2007) for details on the study design, patient selection criteria, sample details and genotyping. For the CATIE study, a total of 1493 patients with chronic schizophrenia were recruited from several sites to participate in the trial and of these patients, 741 provided a DNA sample. The study excluded first episode patients, treatment resistant patients, patients with mood disorders or features and patients whose symptoms were directly a result of substance use or abuse. Study diagnoses for patients were determined by CATIE investigators using the Structured Clinical Interview for DSM-IV (First, 1995). In addition to the consent for the main trial, patients also provided a separate consent for genetic studies (Sullivan et al., 2007).
2.2. Genetic data
Genotyping for CATIE genetic samples were done by Perlegen Sciences (Mountain View, CA, USA) using Affymetrix 500K ‘A’ and custom 164K chipsets. We obtained the genetic data set from the National Institutes of Mental Health (NIMH) Repository and Genomics Resource (https://www.nimhgenetics.org) after IRB and relevant data use approvals. Data received from the NIMH repository included information on 741 CATIE subjects and 751 controls. Information was available for a total of 495,172 single nucleotide polymorphisms (SNPs) in coordinates mapped in human genome build 17 (hg17, University of California, Santa Cruz). The data we received were pre-processed with some quality control filters applied by both Perlegen and CATIE investigators. SNPs were removed for low quality, minor allele frequency (maf) <0.01 and hardy-weinberg equilibrium (hwe) <1e-05. Subjects with >5% missing data were also removed. We did additional data processing by removing 21 subjects from the total sample (18 controls and 3 cases) with sex discrepancies in phenotypic and genotypic data and 12 subjects (8 cases and 4 controls) for relatedness. After extracting only cases from the sample and performing LiftOver (https://genome.ucsc.edu/cgi-bin/hgLiftOver) to update to a recent human genome build (hg19) we were left with 730 cases and 486,895 SNPs.
2.3. Measures
2.3.1. Psychopathology symptom variables
Psychopathology symptoms were measured by the Positive and Negative Syndrome Scale (PANSS) which is an extensively validated 30 item scale in which each Likert type item is rated from 1 to 7 with higher scores indicating more severe symptoms (Kay et al., 1987). We used the NIMH consensus model to construct our psychopathology symptom variables as a previous study done on CATIE data has shown that this model fit the data the best (Stefanovics et al., 2014). The NIMH consensus model uses 20 out of the 30 items in PANSS and constructs five symptom dimensions – positive, negative, disorganized, excited and depressed. Please see Table 1 for details on these variables.
Table 1.
Variables analyzed.
| Variable | Description | Measures/tests | |
|---|---|---|---|
| Psycho-pathology symptoms | Positive | Sum of PANSS items P1, P3, P5 and G9 | PANSS |
| Negative | Sum of PANSS items N1, N2, N3, N4, N6, G7 | ||
| Disorganized | Sum of PANSS items P2, N5 and G11 | ||
| Excited | Sum of PANSS items P4, P7, G8, G14 | ||
| Depressed | Sum of PANSS items G2, G3 AND G6 | ||
| Cognitive domains | Neurocognition | Composite score of standardized domain scores of: | |
|
Computerized test of visuo-spatial working memory | ||
|
Hopkins Verbal Learning Test | ||
|
Continuous Performance Test | ||
|
−Controlled oral word association test −Category Instance −Grooved peg board Wechsler adult intelligence scale |
||
|
−Wisconsin card sorting test −Wechsler intelligence scale for children |
||
| Social cognition | Total score is number of correct responses on facial emotion recognition task. Rescaled from 0-30, to 15-30 with all scores below 15 assigned a value of 15 | Facial Emotion Recognition Task | |
PANSS - Positive and Negative Syndrome Scale; PN - Nth item on positive subscale of PANSS; NN - Nth item on negative subscale of PANSS; GN - Nth item on general psychopathology subscale of PANSS.
2.3.2. Cognitive variables
We tested two cognitive variables – neurocognition and social cognition (Table 1). The neurocognition variables were the composite scores computed from predefined domains-working memory, verbal memory, processing speed, reasoning and vigilance. Details on the composite score, domain scores, neurocognitive tests used to compute domain scores, and measurement characteristics are provided in Keefe et al. (2006). Social cognition was measured by the facial emotion recognition task (Kerr and Neale, 1993). Since the task measures facial emotion recognition, is only one component of social cognition from here on we refer to it as emotion recognition. Briefly, in this task subjects are asked to discriminate if two faces presented have the same emotional expression. The total score is the number of correctly discriminated faces and ranges between 0 and 30. The data distribution for this variable was skewed with 16 subjects scoring <15. Subsequently, we rescaled the variable to a lowest score of 15 and all subjects who scored 15 or below were assigned a value of 15.
2.3.3. Genetic variables-PRS
To examine if schizophrenia PRS was predictive of symptom and cognitive variables in chronic schizophrenia we first performed imputation on the genotyped data. Autosomes were imputed using 1000 Genomes Project Phase 3 data as the reference set. Imputation was done in following steps: (1) Applied strict quality filters using thresholds for hwe 1e-06 and removed individuals who had >2% missing data after which 721 subjects and 421,802 SNPs remained. (2) Pre-phased the filtered data in SHAPEIT software package by first checking for strand alignment issues, removing strand inconsistencies and then estimating haplotypes (Howie et al., 2012). (3) Conducted imputation in chunks of 5 million bases (Mb) using IMPUTE2 software package (Marchini and Howie, 2010). (4) Applied post imputation quality control filters (the same filters used in the Psychiatric Genomics Consortium analysis: maf 0.1 and info score cutoff of 0.9) to select the list of markers (Ripke et al., 2014).
We constructed PRS using the methods suggested by the International Schizophrenia Consortium (Purcell et al., 2009) using the ‘fastscore’ option in PRSice software package (Euesden et al., 2015). To curate a most informative SNP list for PRS calculations the SNPs are first filtered to address linkage disequilibrium (LD). We used the clump option to address LD by applying thresholds of r2 ≥ 0.1 and distance of 500 kb. Since SNPs in the major histocompatibility complex region of the genome have long range LD, we also removed SNPs in the MHC region (26–33 Mb) prior to constructing PRS. PRS was calculated on an a priori set of significance thresholds (PT = 5e-08, 1e-07, 1e-06, 1e-05, 1e-04, 1e-03, 0.01, 0.05, 0.1,0.2,0.3,0.4 and 0.5) to identify the best fit PRS that was most predictive of an association. Multiple comparisons were addressed by applying 106 permutations.
2.3.4. SNPs for set based analysis
Results of GWAS for complex traits suggest that the disease or trait associations are enriched in genomic regions with several risk variants in a single locus and as such, a set based association test assessing the effect of an aggregated set of SNPs has been suggested to be more powerful than single SNP analyses (Bakshi et al., 2016). Hence, to address our second aim we chose a set based analysis. We first extracted SNP sets by using the genomic locations reported for the validated 108 schizophrenia associated loci (Ripke et al., 2014). Excluding three regions in the X-chromosome we had a total of 105 sets.
2.4. Statistical analyses
2.4.1. Polygenic risk scores
Analyses were run in PRSice using PRS as a continuous measure. We tested associations between PRS, symptoms and cognitive variables using linear regression models. We tested two models for symptom variables: (1) the first model controlled for the effects of base covariates (age, sex and ancestry using four multidimensional scaling components). (2) Due to statistically significant correlations observed between neurocognitive variables and symptom dimensions, we also tested a second model adjusting for neurocognition in addition to base covariates. For cognitive variables, we tested one model controlling for the effects of base covariates. Results are presented as change in variance (R2 of model minus R2 of the reduced covariates only model) and overall significance of the model. We report both unadjusted p-values as well as permutation based p-values addressing multiple comparisons issue to identify best fit PRS.
2.4.2. Set based analysis
This analysis was implemented in Plink2 (https://www.cog-genomics.org/plink2/general_usage) (Chang et al., 2015) using the set option. A set based analysis follows a series of steps: (1) Single SNP analysis is run for all SNPs within the sets. (2) Within each set, a specified number of SNP(s) that are below a specified significance threshold are selected based on a set LD criterion. (3) The set is then permuted using the phenotype status to obtain an empirical p-value adjusted to address multiple comparisons within each set (Purcell et al., 2007). We set the SNP selection significance threshold at 0.05, LD criterion at r2 > 0.8, used a top SNP approach selecting only the top hit SNP within each set and permuted the data set 10,000 times. Since we had 105 sets in the analysis, we applied false discovery rate (FDR, Benjamini-Hochberg procedure with an alpha threshold of 0.05) correction. We tested two models for association analyses with symptom variables, the first model adjusting for base covariates and a second model adjusting for neurocognition in addition to base covariates. We tested cognitive variables adjusting for just the base covariates.
3. Results
3.1. PRS associations
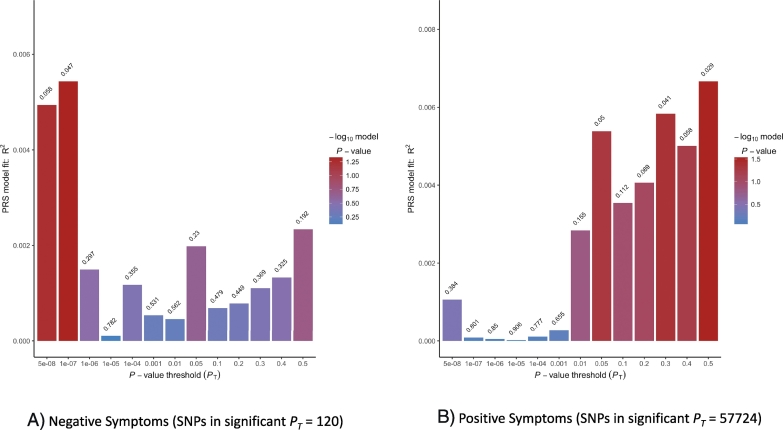
For model 1 which controlled for age, sex and ancestry, we found a significant effect of PRS on positive symptoms at PT of 0.5 (R2 = 0.007, p = 0.029, empirical p = 0.029) and a significant effect on negative symptoms at PT of 1e-07 (R2 = 0.005, p = 0.047, empirical p = 0.048), though the effect on negative symptoms was not in the predicted direction (β = −180.5). Please refer to S1 Table 1 for full results. Fig. 1 shows model 1 bar plots for PRS predictions of positive symptoms and negative symptoms.
Fig. 1.
Model 1 fit for polygenic risk score predictions on positive and negative symptom dimensions. The plots show model 1 results of 13 analyses based on SNP set without linkage disequilibrium. For each PT, SNPs are selected if significant at that threshold and coefficients and effect sizes estimated. Values above each bar are unadjusted p-values of phenotype from regression analyses. For 1A, the best fit PRS for negative symptoms is at the PT of 1e-07 and explains roughly 0.5% of the variance. For 1B, the best fit PRS for positive symptoms is at PT of 0.5 and roughly explains 0.7% of the variance.
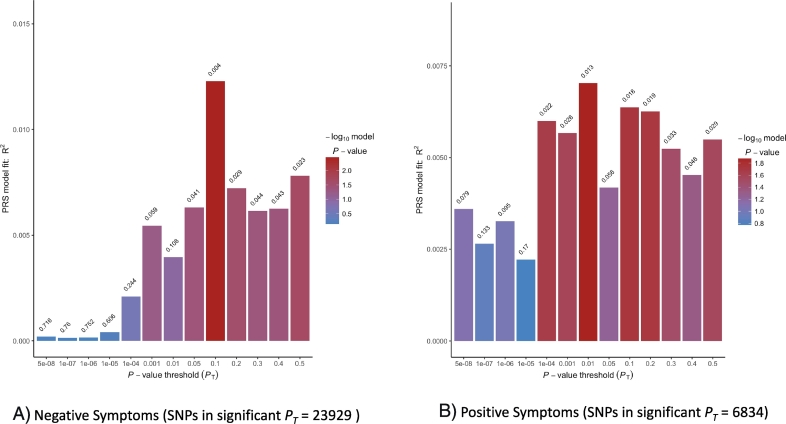
Model 2 which additionally controlled for neurocognition showed findings in a different direction. We found a significant effect of PRS on positive and negative symptoms at higher SNP selection thresholds. Best fit PRS at PT of 0.01 predicted positive symptoms (R2 = 0.007, p = 0.013, empirical p = 0.167) and a significant effect on negative symptoms at PT of 0.1 (R2 = 0.012, p = 0.004, empirical p = 0.329) and did not survive permutation correction. Please refer to S2 Table 2 for full results. Fig. 2 shows model 2 bar plots for PRS predictions of positive symptoms and negative symptoms.
Fig. 2.
Model 2 fit for polygenic risk score predictions on positive and negative symptom dimensions. The plots show model 2 results of 13 analyses based on SNP set without linkage disequilibrium. For each PT, SNPs are selected if significant at that threshold and coefficients and effect sizes estimated. Values above each bar are unadjusted p-values of phenotype from regression analyses. For 2A, the best fit PRS for negative symptoms is at PT of 0.1 and explains roughly 1.2% of the variance. For 2B, the best fit PRS for positive symptoms is at PT of 0.01 and roughly explains 0.7% of the variance.
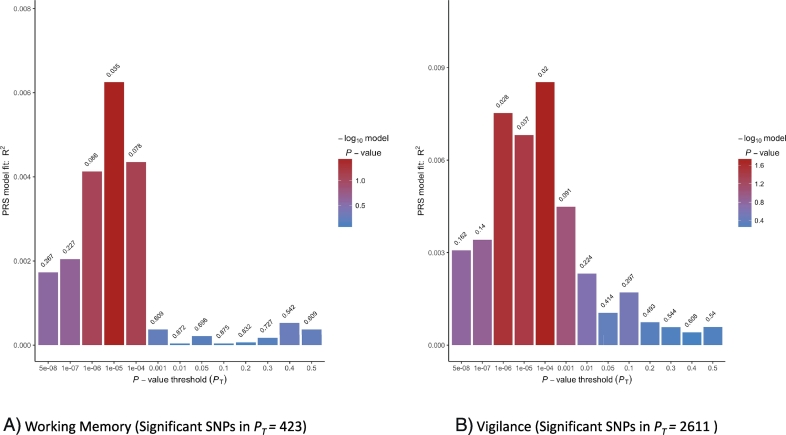
There were no significant effects of PRS on disorganized, depressed or excited symptom dimensions. We did not find evidence for PRS predictions of the neurocognitive composite measure or emotion recognition. Post hoc analyses of neurocognitive domains revealed a significant association of PRS with working memory (PT = 1e-05, R2 = 0.006, p = 0.035, empirical p = 0.15) and vigilance (PT = 1e-04, R2 = 0.008, p = 0.020, empirical p = 0.09) (Fig. 3), but they did not survive permutation correction.
Fig. 3.
Model fit for polygenic risk score predictions of neurocognitive domains (working memory and vigilance) from post hoc analyses. The plots show the results of 13 analyses per variable based on SNP set without linkage disequilibrium. For each PT, SNPs are selected if significant at that threshold and coefficients and effect sizes estimated. Values above each bar are unadjusted p-values of phenotype from regression analyses. For 3A, the best fit PRS for working memory is at PT of 1e-05 and explains roughly 0.6% of the variance. For 3B, the best fit PRS for vigilance is at PT of 1e-04 and roughly explains 0.8% of the variance.
3.2. Set based associations
Results from our set based analysis are presented in Tables 2 and 3. Though we had a few sets with variants that were significantly associated with symptom dimensions, no loci survived correction for multiple set comparisons. Similarly, no variants associated with neurocognition or emotion cognition survived correction. Results from our post hoc analyses of set based associations with neurocognitive sub-domains are presented in supplementary Table 3. We found that after multiple set comparison correction was applied, variant rs12706998, an intergenic variant, was significantly associated with working memory (FDR p = 0.031) and variant rs17269617, also an intergenic variant, (FDR p = 0.020) was associated with verbal memory.
Table 2.
Associations (p < 0.05 with 10,000 permutations within each set) of quantitative psychopathology symptom phenotypes.
| Symptom pheno-types | Loci | Top variant in set | p-Val in set | Beta | Gene | Relevant biological functiona |
|---|---|---|---|---|---|---|
| Positive | chr7:137039644–137085244 | rs16874961 | 0.0028 | −0.94 | DGKI (Diacylglycerol kinase iota) | Intracellular signal transduction |
| chr1:30412551–30437271 | rs1009080b | 0.0158 | 0.76 | c | ||
| chr1:177247821–177300821 | rs3176443 | 0.0241 | 0.71 | FAM5B (BMP/retinoic acid inducible neural specific protein 2) | Positive regulation of neuronal differentiation, protein forms the cell component of neuronal cell body | |
| Negative | chr11:133808069–133852969 | rs3758927 | 0.003 | 1.75 | IGSF9B (protein turtle homolog B) | G-protein coupled receptor, cell component of postsynaptic membrane, dendrite, neuronal cell body. |
| chr11:113317794–113423994 | rs7115090 | 0.0111 | −1.05 | c | ||
| chr18:53453389–53804154 | rs1960054 | 0.0228 | 1.54 | c | ||
| chr7:110034393–110106693 | rs211814 | 0.0243 | 4.26 | c | ||
| chr2:185601420–185785420 | rs16826183 | 0.0274 | −2.44 | ZNF804A (zinc finger protein 804A) | Implicated in several neurocognitive processes | |
| chr1:149998890–150242490 | rs11205334 | 0.0281 | −3.42 | PLEKHO1 (Pleckstrin homolog domain containing family O member 1) | NA | |
| chr1:97792625–98559084 | rs10783063 | 0.0284 | 1.44 | DPYD (Dihydropyrimidine dehydrogenase) | NA | |
| chr15:84661161–85153461 | rs17158441 | 0.0312 | −1.41 | ADAMTSL3 (ADAMTS like protein 3) | NA | |
| chr2:146416922–146441832 | rs13006403b | 0.0413 | 0.87 | c | ||
| Dis-organized | chr7:104598064–105063064 | rs10249746 | 0.01 | 0.67 | c | |
| chr7:86403226–86459326 | rs2214011b | 0.024 | −1.05 | GRM3(Metabotropic glutamate receptor 3) | G-protein coupled receptor signaling pathway, ionotropic glutamate pathway, neuro system process, neuron-neuron synaptic transmission | |
| chr6:73132701–73171901 | rs2815712b | 0.034 | −0.51 | c | ||
| chr3:52541105–52903405 | rs1573815b | 0.0495 | −0.56 | TMEM110(Transmembrane protein 110 -MUSTN1 related) | Calcium channel regulator activity | |
| Excited | chr6:84279922–84407274 | rs16871626 | 0.0092 | 1.54 | SNAP91 (Clathrin coat assembly protein AP180) | Intracellular protein transport, receptor mediated endocytosis, chemical synaptic transmission |
| chr17:2095899–2220799 | rs749240b | 0.0473 | −0.42 | SMG6 (Telomerase binding protein EST1A) | Telomere maintenance, RNA stability | |
| chr11:123394636–123395986 | rs4936823 | 0.0477 | 0.038 | GRAMD1B (Gram domain containing protein 1) | NA | |
| Depressed | chr19:50067499–50135399 | rs2288920 | 0.0052 | 0.61 | PRR12 (proline rich protein 12) | NA |
| chr2:193848340–194028340 | rs17662626 | 0.0212 | −1.08 | c | ||
| chr16:58669293–58682833 | rs9922575b | 0.0459 | −0.41 | c |
NA-information not available.
Source: PANTHER database (http://pantherdb.org).
Variant non-significant when controlled for neurocognition.
Intergenic variants.
Table 3.
Associations (p < 0.05 with 10,000 permutations within each set) of cognitive phenotypes.
| Cognitive pheno-types | Loci | Top variant in set | p-Val in set | Beta | Gene | Relevant biological functiona |
|---|---|---|---|---|---|---|
| Neuro-cognition | chr8:60475469–60954469 | rs17279957 | 0.0059 | −0.47 | b | |
| chr15:40566759–40602237 | rs3784397 | 0.021 | 0.15 | PLCB2 (1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta) | G-protein coupled receptor binding signaling pathway, intracellular signal transduction; Thyrotropin, oxytocin, WNT signaling, alpha adrenergic, 5HT2 receptor signaling etc. | |
| chr2:146416922–146441832 | rs10496985 | 0.0258 | −0.26 | b | ||
| chr1:149998890–150242490 | rs11205334 | 0.0309 | −0.55 | PLEKHO1 | NA | |
| Social cognition | chr8:27412627–27453627 | rs17466684 | 0.0065 | −0.82 | CLU (Clusterin) | CCKR signaling map |
| chr12:2321860–2523731 | rs11062160 | 0.0081 | −1.22 | CACNA1C (Voltage dependent L type ca channel subunit alpha 1C) | 5HT2, Alzheimer’s disease amyloid secretase, Nicotinic acetyl choline receptor, oxytocin receptor signaling pathways | |
| chr19:50067499–50135399 | rs2288920 | 0.0319 | 0.48 | PRRG2 (Transmembrane gamma-carboxyglutamic acid protein 2) | Immune system process, proteolysis, response to stimulus | |
| chr1:149998890–150242490 | rs12073359 | 0.0498 | 0.66 | b |
NA-information not available.
Source: PANTHER database (http://pantherdb.org).
Intergenic variants.
4. Discussion
The current study aimed to examine if a schizophrenia PRS was associated with psychopathology symptom and cognitive dimensions in addition to potential candidate associations in a sample of patients with chronic schizophrenia. The biological basis of symptom dimensions and performance in cognitive domains (Glahn et al., 2014) are deemed to be less complex than the schizophrenia phenotype itself and as such we evaluated for overlap with schizophrenia genetic liability. In our chronic sample, we found a significant PRS association with negative symptoms for both models (with and without neurocognition). Model that controlled for the effects of neurocognition explained a significant amount of variance (1.2%) in negative symptoms, though this did not survive permutation testing and could likely be explained by our smaller sample size. PRS associations with negative symptoms have been somewhat inconsistent in the literature with both positive (Jones et al., 2016; Fanous et al., 2012) and negative findings (Sengupta et al., 2017). Such inconsistencies could be a reflection of the heterogeneity of the sample or the sample size itself. PRS prediction accuracies are found to be strongly linked to sample size (Dudbridge, 2013).
As with negative symptoms, we also found evidence for PRS predicting positive symptoms in the two models tested. Though the amount of variance in positive symptoms (0.7%) explained by both models were the same (potentially suggesting a negligible effect of neurocognition on positive symptoms), model 2 controlling for neurocognition did not survive permutation correction and again could likely be explained by our smaller sample size. PRS association of positive symptoms is contradictory to previous studies that did not show a polygenic loading on positive symptoms (Fanous et al., 2012). This could likely be explained by the differences in sample characteristics. We had a less heterogeneous group (only chronic patients) in comparison to previous studies. It's been suggested that positive symptoms that emerge in adolescence may be strongly influenced by environmental factors such as childhood trauma and/or cannabis use, hence a lack of association in adolescence or first episode patients; PRS associations with positive symptoms may therefore emerge in patients with a later onset of such symptoms (Jones et al., 2016). Our findings could be suggestive of this difference in the pathogenicity and require further study.
We did not find an association with disorganized symptom dimension. The study that previously associated PRS with disorganized symptoms had a different factor structure which clumped negative and disorganized symptoms into a single factor (Fanous et al., 2012). We had separate factor structures for these symptom dimensions which is likely the reason for the absence of association.
Despite the polygenic signal evident in our study we did not find any SNP associations with symptom dimensions that withstood correction which could be due to our sample size. Large sample sizes are required to identify effects of common variants (Ripke et al., 2011). It is very likely that we were underpowered to detect an effect.
We did not find evidence for a strong polygenic association for neurocognition as a single composite factor. This could be due to different genetic susceptibility for different sub-domains. We confirm this in our post hoc analyses where we found that two subdomains (working memory and vigilance) of neurocognition were predicted by PRS, though they did not survive correction for multiple tests. Earlier studies have found PRS associations with working memory in the healthy general population (Hatzimanolis et al., 2015). In addition, PRS has been associated not just with behavioral measures of cognition but also brain based endophenotypes such as prefrontal inefficiency (Walton et al., 2014). Working memory has been strongly associated with prefrontal function (Perlstein et al., 2001) and this offers potential causal mechanisms for further investigation. In our study we also found PRS associations with vigilance measured by the continuous performance test. Vigilance measured by the same test has been associated with schizophrenia PRS in a healthy population (Hatzimanolis et al., 2015).
Literature also supports a polygenic basis for social cognition. Social cognition measured by emotion recognition was found to be associated with PRS in subjects between the ages of 8–21 with findings replicated in a sample of healthy adults (Germine et al., 2016). Of importance here, is that PRS was specifically associated with the speed of emotion recognition and not the ability to recognize emotion per se. Our study did not support a polygenic association and is likely due to measurement differences. Our social cognitive variable assessed for the ability to discriminate emotion and did not measure the speed, which could explain the lack of association.
Taken together, our results are suggestive of a polygenic association for specific symptom and cognitive dimensions in chronic schizophrenia. This study has several limitations; the obvious one is the sample size which is small in comparison to the recommended n of 2000 (Wray et al., 2014). General limitations of the PRS analysis apply to our study-PRS identifies a polygenic signal but cannot provide specific associations that can help elucidate functional mechanisms. PRS is computed from common variants with small effects, so the contributions of rare and structural variants are not known. Additionally, in our study we only used autosomes for PRS calculations, so effects of common variants in sex chromosomes contributing to polygenic burden are not known. For our set based analysis, we utilized a top SNP approach for the loci analyzed which is a limitation as this could potentially exclude several SNPs that could be causal. Some of these loci are known to have several genes (Ripke et al., 2014).
In conclusion, in a sample of patients with chronic schizophrenia we examined if genetic susceptibility to schizophrenia could predict symptom dimensions and cognitive performance. We did not find specific variants associated with symptom or cognitive dimensions. However, we found that PRS predicted a small amount of variance in positive and negative symptom dimensions and performance on working memory and vigilance tests. Some of our findings are consistent with the literature and extend these associations in the population with chronic schizophrenia. Given that these symptoms and deficits in cognitive domains contribute heavily to morbidity in schizophrenia, these findings hold promise to inform further research and potentially guide an objective approach to diagnosis and development of precise therapeutic interventions.
Conflicts of interest
RX, JD and AV have no potential or relevant conflicts of interest to report.
RK is the founder and owner of NeuroCog Trials Inc. No other potential or relevant conflicts of interest to disclose.
Acknowledgements
Data were obtained from the NIMH Repository and Genomics Resource (RGR). Dataset Identifier: NIMH Study 17, Clinical Antipsychotic Trials of Intervention Effectiveness (CATIE)-schizophrenia trial. The principal investigators of the CATIE trial were Jeffrey A. Lieberman, M.D., T. Scott Stroup, M.D., M.P.H., and Joseph P. McEvoy, M.D. The CATIE trial was funded by a grant from the National Institute of Mental Health (N01 MH900001) along with MH074027 (PI PF Sullivan). Genotyping for the CATIE trials were funded by Eli Lilly and Company. This manuscript reflects the views of the authors and may not reflect the opinions or views of the NIMH or of the submitters submitting original data to NIMH RGR.
This work was supported in part by a doctoral training grant to RX by the Robert Wood Johnson Foundation Future of Nursing Scholars program (project number 72099).
We also gratefully acknowledge the support and data provided by the Schizophrenia Working Group of the Psychiatric Genomics Consortium for this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scog.2018.01.001.
Contributor Information
Rose Mary Xavier, Email: rxavier@pennmedicine.upenn.edu.
Jennifer R. Dungan, Email: Jennifer.dungan@duke.edu.
Richard S.E. Keefe, Email: richard.keefe@duke.edu.
Allison Vorderstrasse, Email: allisonvorderstrasse@nyu.edu.
Appendix A. Supplementary data
Supplementary tables
References
- Bakshi A., Zhu Z., Vinkhuyzen A.A., Hill W.D., McRae A.F., Visscher P.M., Yang J. Fast set-based association analysis using summary data from GWAS identifies novel gene loci for human complex traits. Sci. Rep. 2016;6:32894. doi: 10.1038/srep32894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Calhoun V.D., Pearlson G.D., Perrone-Bizzozero N.I., Turner J.A., Ehrlich S., Ho B.C., Liu J. Independent component analysis of SNPs reflects polygenic risk scores for schizophrenia. Schizophr. Res. 2017;181:83–85. doi: 10.1016/j.schres.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke Bailey J.N., Igo R.P. Genetic risk scores. Curr. Protoc. Hum. Genet. 2016;1.29(1-1.29):9. doi: 10.1002/cphg.20. [DOI] [PubMed] [Google Scholar]
- Davies G., Armstrong N., Bis J.C., Bressler J., Chouraki V., Giddaluru S., Hofer E., Ibrahim-Verbaas C.A., Kirin M., Lahti J., Van Der Lee S.J. Genetic contributions to variation in general cognitive function: a meta-analysis of genome-wide association studies in the CHARGE consortium (N = 53,949) Mol. Psychiatry. 2015;20(2):183–192. doi: 10.1038/mp.2014.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich H., Nave K.A. Phenotype-based genetic association studies (PGAS) - towards understanding the contribution of common genetic variants to schizophrenia subphenotypes. Genes. 2014;5:97–105. doi: 10.3390/genes5010097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euesden J., Lewis C.M., O'reilly P.F. 2015. PRSice: Polygenic Risk Score Software v1; p. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanous A.H., Zhou B., Aggen S.H., Bergen S.E., Amdur R.L., Duan J., Sanders A.R., Shi J., Mowry B.J., Olincy A. Genome-wide association study of clinical dimensions of schizophrenia: polygenic effect on disorganized symptoms. Am. J. Psychiatr. 2012;169:1309–1317. doi: 10.1176/appi.ajp.2012.12020218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- First M.B. Wiley Online Library; 1995. Structured Clinical Interview for the DSM (SCID) [Google Scholar]
- Frank J., Lang M., Witt S., Strohmaier J., Rujescu D., Cichon S., Degenhardt F., Nöthen M., Collier D., Ripke S. Identification of increased genetic risk scores for schizophrenia in treatment-resistant patients. Mol. Psychiatry. 2015;20:150. doi: 10.1038/mp.2014.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germine L., Robinson E., Smoller J., Calkins M., Moore T., Hakonarson H., Daly M., Lee P., Holmes A., Buckner R. Association between polygenic risk for schizophrenia, neurocognition and social cognition across development. Transl. Psychiatry. 2016;6 doi: 10.1038/tp.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn D.C., Knowles E.E., Mckay D.R., Sprooten E., Raventós H., Blangero J., Gottesman I.I., Almasy L. Arguments for the sake of endophenotypes: examining common misconceptions about the use of endophenotypes in psychiatric genetics. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2014;165:122–130. doi: 10.1002/ajmg.b.32221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M.F., Kern R.S., Heaton R.K. Longitudinal studies of cognition and functional outcome in schizophrenia: implications for MATRICS. Schizophr. Res. 2004;72:41–51. doi: 10.1016/j.schres.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Green M.J., Chia T.Y., Cairns M.J., Wu J., Tooney P.A., Scott R.J., Carr V.J. Catechol-O-methyltransferase (COMT) genotype moderates the effects of childhood trauma on cognition and symptoms in schizophrenia. J. Psychiatr. Res. 2014;49:43–50. doi: 10.1016/j.jpsychires.2013.10.018. [DOI] [PubMed] [Google Scholar]
- Greenwood T.A., Lazzeroni L.C., Calkins M.E., Freedman R., Green M.F., Gur R.E., Gur R.C., Light G.A., Nuechterlein K.H., Olincy A., Radant A.D. Genetic assessment of additional endophenotypes from the Consortium on the Genetics of Schizophrenia Family Study. Schizophr. Res. 2016;170(1):30–40. doi: 10.1016/j.schres.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood T.A., Lazzeroni L.C., Murray S.S., Cadenhead K.S., Calkins M.E., Dobie D.J., Green M.F., Gur R.E., Gur R.C., Hardiman G., Kelsoe J.R. Analysis of 94 candidate genes and 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. Am. J. Psychiatr. 2011;168(9):930–946. doi: 10.1176/appi.ajp.2011.10050723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood T.A., Swerdlow N.R., Gur R.E., Cadenhead K.S., Calkins M.E., Dobie D.J., Freedman R., Green M.F., Gur R.C., Lazzeroni L.C., Nuechterlein K.H. Genome-wide linkage analyses of 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. Am. J. Psychiatr. 2013;170(5):521–532. doi: 10.1176/appi.ajp.2012.12020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenaars S.P., Harris S.E., Davies G., Hill W.D., Liewald D.C., Ritchie S.J., Marioni R.E., Fawns-Ritchie C., Cullen B., Malik R., Worrall B.B. Shared genetic aetiology between cognitive functions and physical and mental health in UK Biobank (N = 112,151) and 24 GWAS consortia. Mol. Psychiatry. 2016;21(11):1624. doi: 10.1038/mp.2015.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzimanolis A., Bhatnagar P., Moes A., Wang R., Roussos P., Bitsios P., Stefanis C.N., Pulver A.E., Arking D.E., Smyrnis N., Stefanis N.C., Avramopoulos D. Common genetic variation and schizophrenia polygenic risk influence neurocognitive performance in young adulthood. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2015;168:392–401. doi: 10.1002/ajmg.b.32323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W.D., Davies G., Harris S.E., Hagenaars S.P., Davies G., Deary I.J., Debette S., Verbaas C.I., Bressler J., Schuur M., Smith A.V. Molecular genetic aetiology of general cognitive function is enriched in evolutionarily conserved regions. Transl. Psychiatry. 2016;6(12) doi: 10.1038/tp.2016.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B., Fuchsberger C., Stephens M., Marchini J., Abecasis G.R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.R., Shkura K., Langley S.R., Delahaye-Duriez A., Srivastava P., Hill W.D., Rackham O.J., Davies G., Harris S.E., Moreno-Moral A., Rotival M. Systems genetics identifies a convergent gene network for cognition and neurodevelopmental disease. Nat. Neurosci. 2016;19(2):223–232. doi: 10.1038/nn.4205. [DOI] [PubMed] [Google Scholar]
- Jones H.J., Stergiakouli E., Tansey K.E., Hubbard L., Heron J., Cannon M., Holmans P., Lewis G., Linden D.E.J., Jones P.B., Smith G.D., O'donovan M.C., Owen M.J., Walters J.T., Zammit S., Davey Smith G. Phenotypic manifestation of genetic risk for schizophrenia during adolescence in the general population. JAMA Psychiat. 2016;73(221–228):8p. doi: 10.1001/jamapsychiatry.2015.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay S.R., Fiszbein A., Opler L.A. The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophr. Bull. 1987;13:261–276. doi: 10.1093/schbul/13.2.261. [DOI] [PubMed] [Google Scholar]
- Keefe R.S., Bilder R.M., Harvey P.D., Davis S.M., Palmer B.W., Gold J.M., Meltzer H.Y., Green M.F., Miller D.D., Canive J.M., Adler L.W. Baseline neurocognitive deficits in the CATIE schizophrenia trial. Neuropsychopharmacology. 2006;31(9):2033–2046. doi: 10.1038/sj.npp.1301072. [DOI] [PubMed] [Google Scholar]
- Keefe R.S., Eesley C.E., Poe M.P. Defining a cognitive function decrement in schizophrenia. Biol. Psychiatry. 2005;57:688–691. doi: 10.1016/j.biopsych.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Kerr S.L., Neale J.M. Emotion perception in schizophrenia: specific deficit or further evidence of generalized poor performance? J. Abnorm. Psychol. 1993;102:312. doi: 10.1037//0021-843x.102.2.312. [DOI] [PubMed] [Google Scholar]
- Marchini J., Howie B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- Martin A.K., Mowry B. Increased rare duplication burden genomewide in patients with treatment-resistant schizophrenia. Psychol. Med. 2016;46(3):469–476. doi: 10.1017/S0033291715001701. [DOI] [PubMed] [Google Scholar]
- McIntosh A.M., Gow A., Luciano M., Davies G., Liewald D.C., Harris S.E., Corley J., Hall J., Starr J.M., Porteous D.J. Polygenic risk for schizophrenia is associated with cognitive change between childhood and old age. Biol. Psychiatry. 2013;73:938–943. doi: 10.1016/j.biopsych.2013.01.011. [DOI] [PubMed] [Google Scholar]
- Meier S.M., Agerbo E., Maier R., Pedersen C.B., Lang M., Grove J., Hollegaard M.V., Demontis D., Trabjerg B.B., Hjorthøj C., Ripke S. High loading of polygenic risk in cases with chronic schizophrenia. Mol. Psychiatry. 2016;21(7):969–974. doi: 10.1038/mp.2015.130. [DOI] [PubMed] [Google Scholar]
- Morgan V.A., McGrath J.J., Jablensky A., Badcock J.C., Waterreus A., Bush R., Carr V., Castle D., Cohen M., Galletly C., Harvey C. Psychosis prevalence and physical, metabolic and cognitive co-morbidity: data from the second Australian national survey of psychosis. Psychol. Med. 2014;44(10):2163–2176. doi: 10.1017/S0033291713002973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlstein W.M., Carter C.S., Noll D.C., Cohen J.D. Relation of prefrontal cortex dysfunction to working memory and symptoms in schizophrenia. Am. J. Psychiatr. 2001;158:1105–1113. doi: 10.1176/appi.ajp.158.7.1105. [DOI] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., De Bakker P.I., Daly M.J. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S.M., Wray N.R., Stone J.L., Visscher P.M., O'donovan M.C., Sullivan P.F., Sklar P., Ruderfer D.M., Mcquillin A., Morris D.W. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasetti R., Weinberger D.R. Intermediate phenotypes in psychiatric disorders. Curr. Opin. Genet. Dev. 2011;21:340–348. doi: 10.1016/j.gde.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees E., O'donovan M.C., Owen M.J. Genetics of schizophrenia. Current Opinion in Behavioral Sciences. 2015;2:8–14. [Google Scholar]
- Ripke S., Neale B.M., Corvin A., Walters J.T., Farh K.H., Holmans P.A., Lee P., Bulik-Sullivan B., Collier D.A., Huang H., Pers T.H. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S., Sanders A.R., Kendler K.S., Levinson D.F., Sklar P., Holmans P.A., Lin D.Y., Duan J., Ophoff R.A., Andreassen O.A., Scolnick E. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 2011;43(10):969. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S.M., MacDonald K., Fathalli F., Yim A., Lepage M., Iyer S., Malla A., Joober R. Polygenic Risk Score associated with specific symptom dimensions in first-episode psychosis. Schizophr. Res. 2017;184:116–121. doi: 10.1016/j.schres.2016.11.039. [DOI] [PubMed] [Google Scholar]
- Stefanovics E.A., Elkis H., Zhening L., Zhang X.Y., Rosenheck R.A. A cross-national factor analytic comparison of three models of PANSS symptoms in schizophrenia. Psychiatry Res. 2014;219:283–289. doi: 10.1016/j.psychres.2014.04.041. [DOI] [PubMed] [Google Scholar]
- Sullivan P.F., Keefe R.S., Lange L.A., Lange E.M., Stroup T.S., Lieberman J., Maness P.F. NCAM1 and neurocognition in schizophrenia. Biol. Psychiatry. 2007;61:902–910. doi: 10.1016/j.biopsych.2006.07.036. [DOI] [PubMed] [Google Scholar]
- Sullivan P.F., Kendler K.S., Neale M.C. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- Trampush J.W., Yang M.L.Z., Yu J., Knowles E., Davies G., Liewald D.C., Starr J.M., Djurovic S., Melle I., Sundet K., Christoforou A. GWAS meta-analysis reveals novel loci and genetic correlates for general cognitive function: a report from the COGENT consortium. Mol. Psychiatry. 2017;22(3):336–345. doi: 10.1038/mp.2016.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Scheltinga A.T., Bakker S.C., Van Haren N.E.M., Derks E.M., Buizer-Voskamp J.E., Cahn W., Ripke S., Ophoff R.A., Kahn R.S. Schizophrenia genetic variants are not associated with intelligence. Psychol. Med. 2013;43(12):2563–2570. doi: 10.1017/S0033291713000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton E., Geisler D., Lee P.H., Hass J., Turner J.A., Liu J., Sponheim S.R., White T., Wassink T.H., Roessner V., Gollub R.L., Calhoun V.D., Ehrlich S. Prefrontal inefficiency is associated with polygenic risk for schizophrenia. Schizophr. Bull. 2014;40:1263–1271. doi: 10.1093/schbul/sbt174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray N.R., Lee S.H., Mehta D., Vinkhuyzen A.A., Dudbridge F., Middeldorp C.M. Research review: polygenic methods and their application to psychiatric traits. J. Child Psychol. Psychiatry. 2014;55:1068–1087. doi: 10.1111/jcpp.12295. [DOI] [PubMed] [Google Scholar]
- Xavier R.M., Vorderstrasse A. Genetic basis of positive and negative symptom domains in schizophrenia. Biological Research For Nursing. 2017;0 doi: 10.1177/1099800417715907. [DOI] [PubMed] [Google Scholar]
- Yeo R.A., Gangestad S.W., Walton E., Ehrlich S., Pommy J., Turner J.A., Liu J., Mayer A.R., Schulz S.C., Ho B.C., Bustillo J.R. Genetic influences on cognitive endophenotypes in schizophrenia. Schizophr. Res. 2014;156(1):71–75. doi: 10.1016/j.schres.2014.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables