Abstract
Background & Aims
Aberrations in the esophageal proliferation-differentiation gradient are histologic hallmarks in eosinophilic esophagitis (EoE) and gastroesophageal reflux disease. A reliable protocol to grow 3-dimensional (3D) esophageal organoids is needed to study esophageal epithelial homeostasis under physiological and pathologic conditions.
Methods
We modified keratinocyte-serum free medium to grow 3D organoids from endoscopic esophageal biopsies, immortalized human esophageal epithelial cells, and murine esophagi. Morphologic and functional characterization of 3D organoids was performed following genetic and pharmacologic modifications or exposure to EoE-relevant cytokines. The Notch pathway was evaluated by transfection assays and by gene expression analyses in vitro and in biopsies.
Results
Both murine and human esophageal 3D organoids displayed an explicit proliferation-differentiation gradient. Notch inhibition accumulated undifferentiated basal keratinocytes with deregulated squamous cell differentiation in organoids. EoE patient-derived 3D organoids displayed normal epithelial structure ex vivo in the absence of the EoE inflammatory milieu. Stimulation of esophageal 3D organoids with EoE-relevant cytokines resulted in a phenocopy of Notch inhibition in organoid 3D structures with recapitulation of reactive epithelial changes in EoE biopsies, where Notch3 expression was significantly decreased in EoE compared with control subjects.
Conclusions
Esophageal 3D organoids serve as a novel platform to investigate regulatory mechanisms in squamous epithelial homeostasis in the context of EoE and other diseases. Notch-mediated squamous cell differentiation is suppressed by cytokines known to be involved in EoE, suggesting that this may contribute to epithelial phenotypes associated with disease. Genetic and pharmacologic manipulations establish proof of concept for the utility of organoids for future studies and personalized medicine in EoE and other esophageal diseases.
Keywords: Three-Dimensional, Keratinocytes, Eosinophilic Esophagitis, Squamous Cell Differentiation
Abbreviations used in this paper: aDMEM/F12, advanced Dulbecco's Modified Eagle Medium: Nutrient Mixture F-12; BCH, basal cell hyperplasia; DAPI, 4′,6-Diamidino-2-Phenylindole, Dihydrochloride; DNMAML1, dominant negative MAML1; DOX, doxycycline; EGF, epidermal growth factor; EMT, epithelial-mesenchymal transition; EoE, eosinophilic esophagitis; GERD, gastroesophageal reflux disease; GFP, green fluorescent protein; GSI, γ-secretase inhibitor; H&E, hematoxylin and eosin; IF, immunofluorescence; IHC, immunohistochemistry; IL, interleukin; IVL, Involucrin; KSFM, keratinocyte SFM; KSFMC, KSFM containing 0.6 mM Ca2+; MAML1, Mastermind-like protein1; OFR, organoid formation rate; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; 3D, 3-dimensional; TNF-α, tumor necrosis factor-α; Tslp, thymic stromal lymphopoietin
Summary.
We optimized culture conditions for 3-dimensional mouse and human esophageal organoids and used this experimental platform with potential applications toward personalized medicine to identify disruption of notch3-mediated squamous cell differentiation as a mechanism contributing to reactive epithelial changes under inflammatory conditions.
Stratified squamous epithelia comprises basal and suprabasal cell layers, displaying an explicit differentiation gradient. The basal cell layer contains undifferentiated and proliferative basal cells (keratinocytes) that exit cell division cycles and undergo terminal differentiation within the suprabasal cell layers to migrate and desquamate into the lumen, permitting epithelial renewal. Basal cells express defining molecular markers, such as β1-integrin (CD29).1 The basal cell layer harbors stem cells with long-term self-renewal capacity.2, 3 Cell-cell junctions in suprabasal cell layers provide epithelial barrier function. The differentiation gradient is disrupted under esophageal disease conditions, both benign and malignant. Esophageal epithelial homeostasis is influenced by a variety of agents and factors including chemical carcinogens, radiation, acids, growth factors, and inflammatory cytokines. These factors activate multiple signaling pathways, including epidermal growth factor (EGF) receptor, bone morphogenetic protein, Wnt, and Notch, which regulate esophageal epithelial renewal, proliferation, differentiation, transdifferentiation, senescence, apoptosis, or survival.4, 5, 6, 7, 8, 9, 10
The Notch pathway is critical in squamous epithelial homeostasis.11 Notch signaling is activated via cell-cell contact, permitting cell surface ligand-receptor interaction. Activation of Notch receptor leads to a series of proteolytic cleavages and nuclear translocation of its intracellular domain, which physically associates with the common downstream effector transcription factor CSL/RBPJ along with the coactivator Mastermind-like protein 1 (MAML1). Notch-activated CSL-mediated transcriptional targets include Involucrin (IVL), a squamous-cell differentiation marker. Of 4 mammalian paralogs of Notch receptor, Notch1 is the master regulator of squamous cell differentiation. Loss of Notch signaling in the epidermis results in noncell autonomous and cell autonomous effects, inducing basal cell hyperplasia (BCH; expansion of basal keratinocytes without postmitotic terminal differentiation),12 deregulated squamous cell differentiation and hyperkeratosis, and dermal inflammation and eosinophilic infiltrates.13 In murine esophageal keratinocytes, genetic or pharmacologic pan-Notch signaling inhibition via dominant negative MAML1 (DNMAML1) or γ-secretase inhibitors (GSI), impairs squamous cell differentiation with a concurrent downregulation of IVL and other differentiation-related genes in vitro and in vivo.8
Many esophageal diseases, such as gastroesophageal reflux disease (GERD) and eosinophilic esophagitis (EoE), involve esophageal epithelial pathologies. For example, common histopathologic manifestations of EoE and GERD include esophageal BCH and spongiosis (dilated intracellular spaces). Featuring eosinophilic inflammation and lamia propria fibrosis,14 EoE is an emerging food allergen–induced cytokine-mediated inflammatory disorder, affecting children and adults. Long-term disease status leads to irreversible fibrotic esophageal stenosis. Genome-wide association studies and gene expression profiling of endoscopic esophageal biopsies from patients with EoE have implicated several epithelial cell–associated molecules, such as thymic stromal lymphopoietin (Tslp) and Desmoglein-1, the latter a regulator of esophageal epithelial barrier function.15, 16 Transcriptome analysis has revealed differentiation as the most affected biologic process in EoE.17 Additionally, EoE-relevant inflammatory cytokines, such as tumor necrosis factor (TNF)-α and transforming growth factor-β, alter epithelial cell characteristics by inducing epithelial-mesenchymal transition (EMT).18, 19 Esophageal inflammation associated with GERD also involves TNF-α and other cytokines.20 There are currently no reliable epithelial functional assays or models to understand human epithelial biology related to EoE and other esophageal diseases in the context of personalized medicine.
The 3-dimensional (3D) organoid system is a cell-culture based, novel, and physiologically relevant biologic platform.21 With a small number of cells isolated from tissues or cultured cells as starting materials, 3D organoids are grown and passaged in basement membrane matrix. Organoids provide insights into the roles of molecular pathways and niche factors essential in intestinal stem cell renewal, proliferation, and differentiation. Although 3D organoids have been generated from broad tissue types, the application of this technique to human stratified squamous epithelia remains elusive.
Herein, we have established a highly efficient protocol to generate human and murine esophageal organoids from endoscopic biopsies, immortalized normal human esophageal epithelial cell lines, and murine esophagi. In concert with genetic and pharmacologic modifications, our 3D organoids reveal novel functional crosstalk between cytokines and Notch signaling in reactive epithelial changes in the context of inflammatory milieu. Specifically, disruption of Notch3-mediated squamous cell differentiation may serve as a novel mechanism contributing to reactive epithelial changes in EoE, GERD, and potentially other esophageal pathologies.
Methods
Patients and Esophageal Biopsies
In accordance with institutional review board standards and guidelines at the Children’s Hospital of Philadelphia and the Hospital of the University of Pennsylvania, biopsies were procured from the distal esophagus of patients with EoE, GERD, and normal mucosa undergoing diagnostic esophagogastroduodenoscopy (ie, upper endoscopy) (Table 1). Patients who met clinical criteria of EoE22 with histologic presence of ≥15 esophageal mucosal eosinophils (eos) per high-powered field (hpf) were classified as “active EoE.” Patients with EoE who demonstrated resolution of histologic inflammation and eosinophilia (<15 eos/hpf) on follow-up upper endoscopy were designated as “inactive EoE.” Subjects with a history of gastroesophageal reflux who demonstrated mild esophageal inflammation (1–5 eos/hpf) but did not meet histologic criteria for EoE were designated as GERD. “Normal” subjects include patients who reported symptoms warranting upper endoscopy and did not carry a previous diagnosis of EoE but demonstrated no histopathologic abnormalities.
Table 1.
Patients Used to Generate Biopsy-Derived Esophageal 3D Organoids
| # | Age | Sex | Diagnosis | Passage |
|---|---|---|---|---|
| 1 | 15 | M | Normal | n.d. |
| 2 | 5 | M | Inactive | n.d. |
| 3 | 8 | M | Inactive | n.d. |
| 4 | 14 | M | Inactive | n.d. |
| 5 | 14 | F | Normal | n.d. |
| 6 | 11 | F | GERD | n.d. |
| 7 | 11 | M | GERD | n.d. |
| 8 | 13 | M | GERD | n.d. |
| 9 | 8 | M | Active | n.d. |
| 10 | 14 | F | Active | n.d. |
| 11 | 16 | M | PPI-REE | 2a |
| 12 | 6 | M | Inactive | 3b |
| 13 | 8 | F | Inactive | n.d. |
| 14 | 11 | M | Inactive | 3b |
| 15 | 5 | M | Active | 4b |
| 16 | 18 | F | Active | 5b |
| 17 | 16 | F | Normal | 3b |
| 18 | 9 | M | Active | 6a |
| 19 | 10 | F | Normal | n.d. |
| 20 | 8 | F | Normal | n.d. |
| 21 | 6 | M | Inactive | 4b |
| 22 | 5 | M | PPI-REE | 5b |
| 23 | 7 | F | Active | 4b |
| 24 | 14 | M | Inactive | 7a |
| 25 | 8 | M | Normal | 4b |
| 26 | 11 | F | Active | 3a |
| 27 | 10 | M | Inactive | 3b |
| 28 | 6 | M | Inactive | n.d. |
| 29 | 8 | M | Inactive | n.d. |
| 30 | 10 | M | Active | 4a |
| 31 | 18 | M | Active | n.d. |
| 32 | 18 | M | Inactive | n.d. |
| 33 | 18 | F | Normal | 4a |
| 34 | 12 | F | Normal | n.d. |
NOTE. Normal, no pathologic diagnosis; inactive, <15 eos/hpf but with previous diagnosis of EoE; GERD, 1–5 eos/hpf; active, >15 eos/hpf.
n.d., not determined; PPI-REE, proton pump inhibitor–responsive esophageal eosinophilia.
3D organoid culture was not passaged after this passage number.
3D organoids failed to grow after this passage number.
Mice
Murine esophageal epithelial sheets were prepared as described23 from wild-type C57BL/6J mice and Notch1loxP/loxP mice24 (Jackson Laboratory, Bar Harbor, ME). All experiments were done under University of Pennsylvania IACUC-approved protocols.
Monolayer and 3-Dimensional Organoid Cultures With Esophageal Epithelial Cell Lines and Biopsies
All cell culture reagents and supplies were purchased from Thermo Fisher Scientific (Philadelphia, PA) unless otherwise noted. Telomerase-immortalized normal human esophageal epithelial cell line EPC2-hTERT and derivatives carrying GFP, DNMAML1, or DNMAML1Tet-Off 6, 8, 25 were grown in keratinocyte-SFM (KSFM) medium containing 0.09 mM Ca2+ as described previously.23, 26 Cell authentication was externally done by short-tandem repeat DNA profiling at the American Type Culture Collection (Manassas, VA).
Murine esophageal 3D organoids were generated as described27 using advanced Dulbecco's Modified Eagle Medium: Nutrient Mixture F-12 (aDMEM/F12) containing EGF,10 Noggin/R-Spondin1-conditioned medium, Wnt3A (R&D Systems, Minneapolis, MN), A83-01 (Sigma-Aldrich, St. Louis, MO), and SB202190 (Selleck Chemical, Huston, TX), unless the use of KSFM or modified KSFM containing 0.6 mM Ca2+ (KSFMC) was noted (see Table 2 for media compositions). For ex vivo Notch1 deletion in 3D esophageal organoids generated from Notch1loxP/loxP mice, organoids were incubated with Adenovirus expressing Cre recombinase or green fluorescent protein (GFP, control) (University of Iowa Gene Transfer Vector Core). Adenovirus was added at 1:500 at the time of organoid plating.
Table 2.
Media Constituents
| Media |
aDMEM/F12 |
KSFMC |
KSFM |
|
|---|---|---|---|---|
| Base medium | Advanced DMEM/F12 | Keratinocyte-SFM | Keratinocyte-SFM | |
| Supplements | L-glutamine | (+) | (-) | (-) |
| HEPES | (+) | (-) | (-) | |
| Bovine pituitary extract | (-) | 50 μg/mL | 50 μg/mL | |
| Penicillin-streptomycin | (+) | (+) | (+) | |
| Y27632 | 10 μM | 10 μM | 10 μM | |
| N2 supplementa | 1x | (-) | (-) | |
| B27 supplementa | 1x | (-) | (-) | |
| N-acetylcysteinea | 1 mM | (-) | (-) | |
| Noggin/R-Spondin (conditioned media)a |
3% (vol/vol) | (-) | (-) | |
| Wnt3Aa | 100 ng/mL | (-) | (-) | |
| A83-01a | 500 nM | (-) | (-) | |
| SB202190a | 10 μM | (-) | (-) | |
| Gastrina | 10 nM | (-) | (-) | |
| Nicotinamidea | 10 mM | (-) | (-) | |
| EGF | 50 ng/mL | 1 ng/mL | 1 ng/mL | |
| Calcium chloride | 1.0 mM | 0.6 mM | 0.09 mM | |
Growth supplements in Figure 2.
Organoids from human esophageal biopsies were established as described in Figure 1. Briefly, endoscopic biopsy pieces were preserved in KSFM medium and kept on ice until further processing. KSFM was then replaced with 1 mL 10 U/mL Dispase (Corning, Corning, NY) and incubated for 10 minutes at room temperature. After rinsing with 1 mL phosphate-buffered saline to remove Dispase, the biopsy pieces were incubated in 0.5 mL 0.05% trypsin-EDTA at 37°C for 10 minutes and mixed concurrently at 700∼800 rpm using ThermoMixer C (Eppendorf, Hamburg, DE). Using the rubber plunger head taken from a tuberculin syringe, dissociated cells were gently forced through a 70-μm cell strainer and collected in a 50-mL conical tube containing 2 mL 250 μg/mL soybean trypsin inhibitor (Sigma-Aldrich). Cells were filtrated again into a 5-mL round bottom polystyrene tube with a 35-μm cell strainer snap cap (BD Bioscience, Franklin Lakes, NJ). Following centrifugation at 1500 rpm at 4°C for 5 minutes, cells were resuspended in 50–100 μL KSFM and counted by the Countess II FL Automated Cell Counter (Invitrogen, Carlsbad, CA) where cell viability was determined by Trypan Blue dye exclusion test. When EPC2-hTERT and derivatives were used to generate 3D organoids, monolayer cultures were trypsinized to prepare cell suspensions as described.23 Using 24-well plates, 2000 cells were seeded per well in 50 μL of ice cold 1:1 mixture of Matrigel basement membrane matrix (Corning) and KSFMC, and incubated at 37°C for 30 minutes. After solidification, 500 μL of KSFMC containing 10 uM Y27632 (Sigma-Aldrich) was added to each well on the first day, and then wells were replenished with KSFMC without Y27632 every other day. When indicated, the previously described fully supplemented aDMEM/F12 was used instead of KSFMC. Alternatively, KSFM was supplemented with the growth supplements used in aDMEM/F12.27
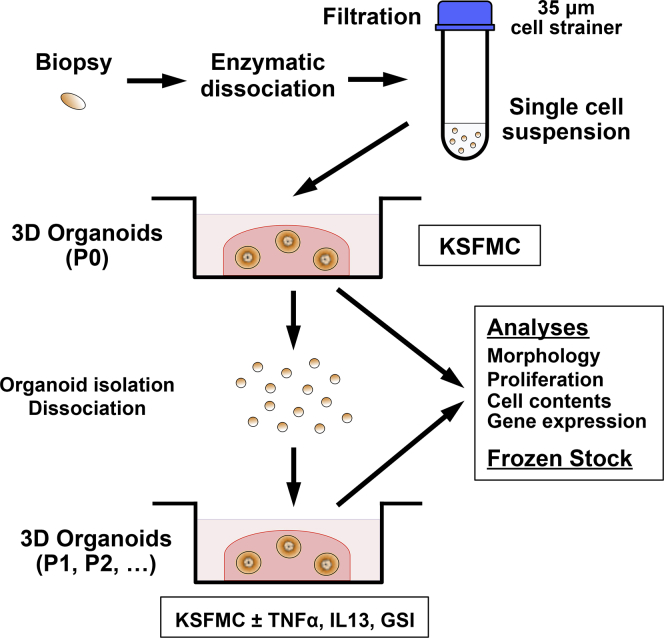
Figure 1.
Generation and analyses of biopsy-derived esophageal 3D organoids. Biopsies from normal mucosa were taken at the time of diagnostic upper endoscopy and enzymatically dissociated and filtrated, yielding in 105–106 cells per biopsy. A single cell suspension is mixed with 50% Matrigel to initiate 3D organoid culture in KSFMC medium. The resulting 3D products are passaged, frozen-stocked, and characterized via morphologic and functional assays at P0 or subsequent passages (P1 or later) where organoids are treated with cytokines and pharmacologic agents.
When indicated, both monolayer and 3D organoid cultures were grown in the presence or absence of 1 μM Compound E, a GSI or 1–40 ng/mL of TNF-α, 1–10 ng/mL interleukin (IL)-4, IL5, or IL13 (R&D) as described.8, 28, 29, 30 We first established nonlethal stimulatory concentration of cytokines by performing dose response (data not shown). 3D organoids were grown for 11 days, unless otherwise noted, and phase-contrast images were acquired to determine their number and size using a Nikon Eclipse E600 microscope (Nikon, Tokyo, Japan). Organoid formation rate (OFR) was defined as the average number of ≥50 μm spherical structures at Day 11 divided by the total number of cells seeded in each well at Day 0. 3D organoids were recovered by digesting Matrigel with Dispase (10 U/mL), dissociated into single cell suspensions by incubation with 0.05% Trypsin-EDTA at 37°C for 10 minutes, and reseeded for secondary and subsequent passages. When indicated, 3D organoid structures recovered from Matrigel were fixed overnight in 4.0% paraformaldehyde and embedded in 2.0% Bacto-Agar: 2.5% gelatin for paraffin embedding and subjected to histologic analyses.
WST1 Assays
To assess metabolically active viable cells, organoid cultures were incubated with WST-1 reagent (Roche, Basel, Switzerland) for 90 minutes and then only reacted media was transferred to a 96-well plate, before colorimetric cell proliferation assays according to the manufacturer's instructions. All experiments were performed in triplicate.
Histology, Immunofluorescence, and Immunohistochemistry
Paraffin-embedded biopsies and 3D organoid products were serially sectioned and subjected to hematoxylin and eosin (H&E) staining, immunofluorescence (IF), and immunohistochemistry (IHC) as described previously.8, 30, 31 H&E-stained slides for 3D organoids and corresponding biopsies were evaluated by a pathologist (A.K.S.) blinded to molecular and clinical data. In brief, sections were incubated overnight at 4°C with anti-IVL (mouse monoclonal anti-IVL I9018, 1:100; Sigma-Aldrich) for IF and anti-Ki-67 (rabbit monoclonal anti-Ki-67 ab16667; 1:200; Abcam, Cambridge, UK) or anti-Notch1 (rabbit monoclonal anti-ICN1Val1744 D3B8, 1:200; Cell Signaling, Danvers, MA) for IHC. Stained objects were captured and imaged with a Nikon Microphot microscope with a digital camera. Ki-67 labeling index was determined by counting at least 6 organoids per group. Basaloid cell content analysis was determined as a percentage of the total height of basaloid cell layers per organoid diameter spanning both basaloid cell layers and the differentiated cell layers including keratinized inner cell mass in H&E-stained organoids with 3 areas measured per organoid in least 6 organoids per group.
Flow Cytometry
Flow cytometry was performed as described using LSR II cytometers (BD Biosciences) and FlowJo software (Tree Star, Ashland, OR) for cells suspended in Hank’s balanced salt solution (Invitrogen) containing 1% bovine serum albumin (Sigma-Aldrich) and stained with FITC-anti-CD29 at 1:25 (MCA1188, Bio-Rad Laboratory, Hercules, CA), APC-anti-CD45 at 1:20 (HI30, BioLegend, San Diego, CA), PE-anti-CD325 at 1:20 (8C11, BioLegend) on ice for 30 minutes. 4′,6-diamidino-2-phenylindole (DAPI) was used to determine cell viability. Flow cytometry was repeated for each genotype and condition at least 3 times.
Immunoblotting
3D organoids isolated from Matrigel and cells in monolayer culture were lysed and subjected to immunoblotting as described.6, 8, 29 Rabbit monoclonal anti-ICN1Val1744 D3B8 (1:1000; Cell Signaling), rabbit monoclonal anti-NOTCH3 D11B8 (1:1000; Cell Signaling), mouse monoclonal anti-IVL clone SY5 (1:1000, Sigma-Aldrich), and mouse monoclonal anti-β-actin AC-74 (1:10,000; Sigma-Aldrich) were used as primary antibodies. β-actin served as a loading control.
Quantitative Reverse-Transcriptase Polymerase Chain Reaction Assays
RNA extraction and cDNA synthesis were done as described.6, 8, 29 Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) was done with TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA) for NOTCH1 (Hs01062014_m1), NOTCH2 (Hs00225747_m1), NOTCH3 (Hs00166432_m1), NOTCH4 (Hs00270200_m1), JAG2 (Hs00171432_m1), DLL1 (Hs00194509_m1), HES5 (Hs01387463_g1), IVL (Hs00846307_s1), FLG (Hs00863478_g1),and GAPDH (Hs99999905_m1), using the StepOnePlus Real-Time PCR System (Applied Biosystems). The relative level of each mRNA was normalized to GAPDH as an internal control.
RNA-Seq Data Analysis
Raw sequence data with quality scores (GSE58640)32, 33 were downloaded from the NCBI GEO database. The dataset included samples from 10 active EoE patients and 6 healthy control subjects. Sequences for each sample were aligned to the human genome GRCh38.p7 using the STAR aligner (v252b).34 Genomically mapped reads were counted against reference genes as annotated in Gencode (version 25)35 using htseq-count.36 One EoE sample (GSM1415921, EoE_803) was noted to have a low number of mapped reads and was excluded from further analyses. Genes were tested for differential expression between EoE and control subjects using DESeq2,37 yielding fold change, P value, and fdr-adjusted P value for each gene.
Transient Transfection and Dual-Luciferase Assays
Transient transfection of reporter plasmids and luciferase assays were performed as described previously.8 Briefly, 400 ng of 8xCBF1-luc (designated as 8xCSL-luc),38 a Notch-inducible reporter, was transfected per well seeded with 2 × 105 cells 24 hours before transfection in 24-well plates along with 5 ng of phRL-SV40-Renilla luciferase vector (Promega), which was used to calibrate the variation of transfection efficiencies among wells. A total of 40 ng/mL TNF-α was added at 24 hours after transfection and incubated for an additional 72 hours before cell lysis. The mean of firefly luciferase activity was normalized with the cotransfected Renilla luciferase activity. Transfection was carried out at least 3 times, and variation between experiments was not >15%.
Statistical Analysis
Data are presented as mean ± standard error of the mean or mean ± standard deviation and were analyzed by 2-tailed Student t test, Wilcoxon test P < .05 was considered significant. Data were analyzed using the Jmp13 pro ver.13.0.0 software package (SAS Institute, Cary, NC). All authors had access to the study data and reviewed and approved the final manuscript.
Results
Esophageal 3-Dimensional Organoids Display an Explicit Proliferation-Differentiation Gradient
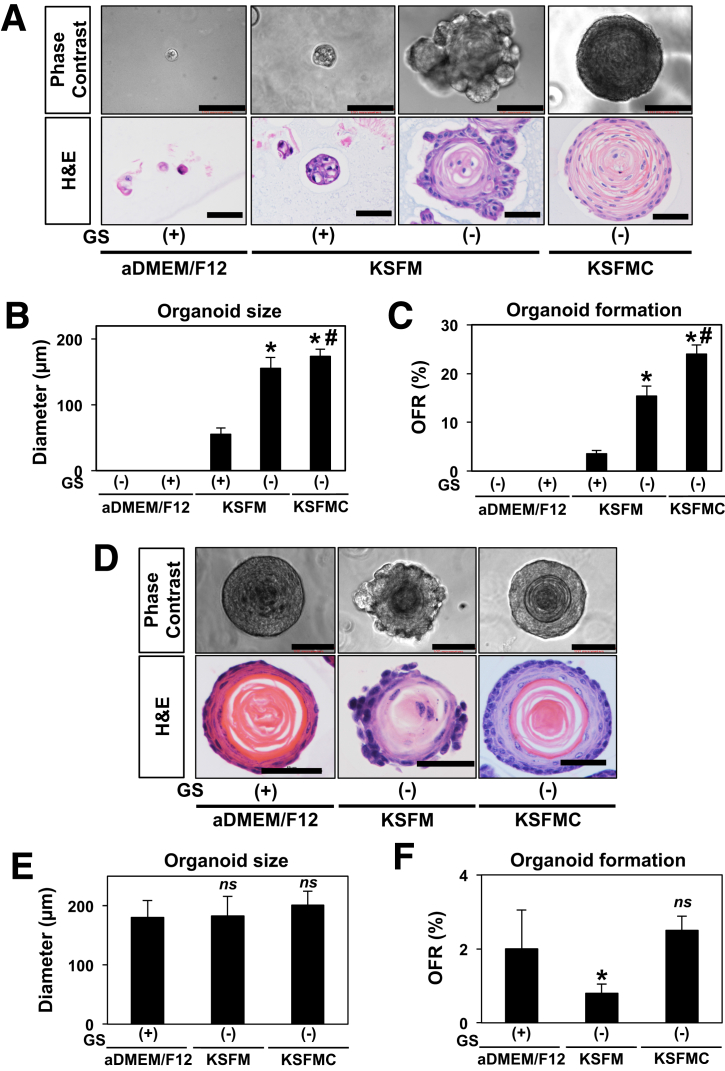
The aDMEM/F12-based media originally described by Sato et al39 to generate 3D organoids from the intestine and other gastrointestinal organs has been successfully used to grow 3D organoids from normal murine esophageal epithelia.2, 27, 31 Our initial attempts to grow human esophageal 3D organoids failed in this medium composition before poor, if any, 3D structure formation was noted in the extensively characterized normal human esophageal cell line EPC2-hTERT25 and in human esophageal cells isolated from endoscopic biopsies from patients with normal esophageal mucosa (n = 4) or EoE (n = 7) (Figure 2A and data not shown). However, on culturing in KSFM, EPC2-hTERT cells gave rise to larger 3D structures with lobulated morphology (Figure 2A and B). Histologic analysis of H&E-stained organoids revealed a keratinized inner cell mass surrounded by less-differentiated basaloid cell layers protruding into the matrix. Interestingly, organoid growth was severely impaired when KSFM was supplemented with growth factors and agents used for the aDMEM/F12 medium (Figure 2A, Table 2). KSFM has low calcium content (0.09 mM) to limit squamous-cell differentiation in EPC2-hTERT monolayer culture where raising Ca2+ to 0.6 mM induces terminal differentiation.8 We suspected that calcium may influence the differentiation gradient in organoids. Addition of CaCl2 to a final concentration of 0.6 mM Ca2+ resulted in the formation of less lobulated 3D structures with a more explicit differentiation gradient (Figure 2A) while also significantly enhancing OFR (Figure 2C). Because higher Ca2+ concentrations (0.9–1.8 mM) did not significantly improve OFR or organoid size compared with KSFMC (data not shown), we have identified KSFM containing 0.6 mM Ca2+ (designated KSFMC) as the optimal medium for 3D organoid formation by EPC2-hTERT cells.
Figure 2.
Optimization of normal human and murine esophageal 3D organoid culture. Esophageal 3D organoid culture was optimized with the normal human esophageal epithelial cell line EPC2-hTERT in A–C and wild-type murine esophageal cells dissociated from epithelial sheets in D–F. A single cell suspension was mixed with Matrigel and cultured for 11 days in indicated media supplemented with EGF and insulin. Unique growth supplements used for murine esophageal 3D organoids27 (ie, Noggin/R-Spondin1-conditioned medium, Wnt3A, A83-01, SB202190, gastrin, and nicotinamide) were added as indicated. KSFM contains 0.09 mM Ca2+ (low Ca2+), whereas KSFMC is supplemented with additional CaCl2 to the final concentration of 0.6 mM Ca2+ (high Ca2+). The aDMEM medium contains 1 mM Ca2+. 3D culture products were photomicrographed under a phase-contrast microscopy and further recovered from Matrigel for H&E staining in A and D. Organoids were defined as 3D structures with a diameter of 50 μm or larger. Note that 3D organoid formation was barely observed in aDMEM/F12 with or without unique supplements. Addition of unique supplements also limited organoid formation in KSFM. 3D organoids grown in KSFM showed a lobular pattern of basaloid cell expansion toward surrounding matrix and had a less explicit differentiation gradient as compared with 3D organoids formed in KSFMC. Average organoid size was determined in B and E. OFR was determined in C and F. Scale bar = 100 μm in phase-contrast images and 50 μm in H&E staining. *P < .0001 vs KSFM with supplements; #P < .05 vs KSFM without supplements; n = 6 in B and n = 8 in C. *P < .05 vs aDMEM/F12; ns, not significant vs aDMEM/F12; n = 6 in E and n = 8 in F. Data represent >2 biologic replicates with similar results. GS, growth supplements.
We further evaluated the influence of culture medium composition on organoid formation by esophageal cells isolated from murine epithelial sheets. aDMEM/F12, KFSM, and KSFMC each gave rise to 3D organoids of comparable size (Figure 2D and E). However, organoids grown in KSFMC and aDMEM/F12 recapitulated better a physiological differentiation gradient as compared with those cultured in KSFM (Figure 2D). Additionally, although organoid formation efficiency was comparable between KSFMC and aDMEM/F12, the latter supplemented either fully27 or partially (with EGF, R-Spondin1, and Noggin only2, 31) (Figure 2E and F, and data not shown); OFR was impaired on culture in KFSM as compared to KSFMC (Figure 2F). Taken together, these findings identify KSFMC as a culture medium that supports 3D organoid formation in esophageal keratinocytes of human and murine origin.
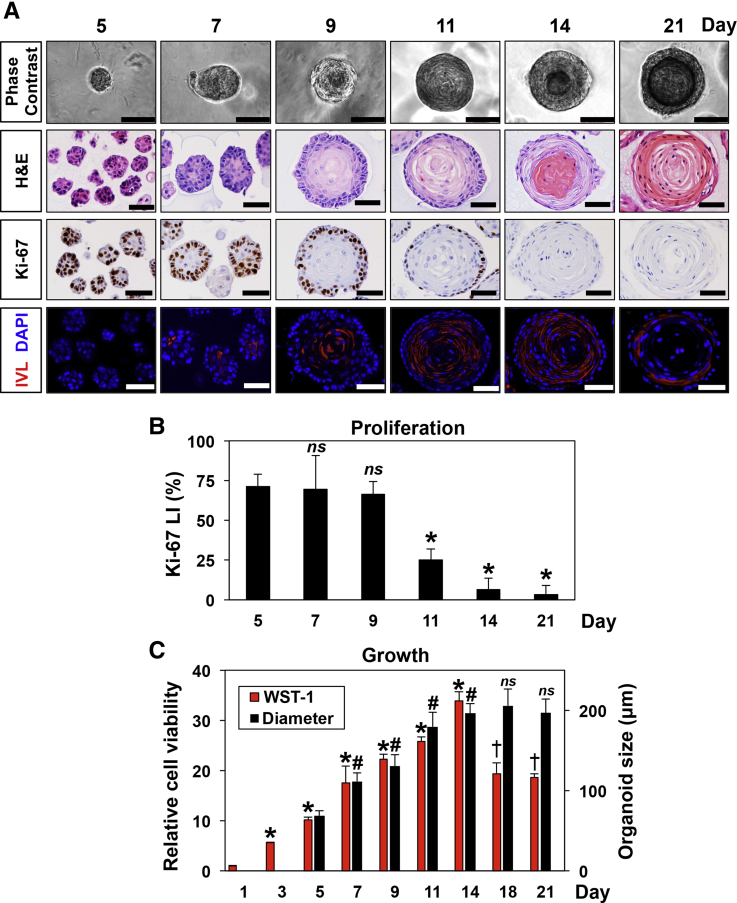
We continued to characterize the growth kinetics of human esophageal 3D organoids in KSFMC medium, using the immortalized normal human esophageal cell line EPC2-hTERT, which has been extensively characterized with respect to proliferation, terminal differentiation, EMT, senescence, apoptosis, motility, and related signaling pathways in response to a variety of physiologically and pathologically relevant stimuli.4, 5, 6, 7, 8, 9, 18, 19, 25, 26, 28, 40, 41, 42 Under monolayer culture conditions, EPC2-hTERT cells are maintained as proliferative and undifferentiated basaloid cells. We initiated organoid formation from a single cell suspension of EPC2-hTERT cells and observed emergence of 3D spherical structures by Day 5 post-plating (Figure 3). These structures then continued to grow until they reached a plateau in size (∼150 μm) between 11 and 14 days (Figure 3A and C) with 3D structures containing 300–600 cells at Day 11, allowing an estimate of a population doubling time of 30.3 ± 1.5 hours (8–9 generations from a single cell). IHC for Ki-67 suggested that cell proliferation is active until Day 9 (Figure 3B) before keratinization becomes prominent at Day 11 (Figure 3A). 3D organoids displayed a proliferation-differentiation gradient with a Ki-67 staining basaloid cell layer and more differentiated cells toward the center of these structures (Figure 3A). Differentiation was corroborated by expression of IVL, an early marker of squamous cell differentiation as documented by IF, which indicated that differentiation begins in a small number cells located within the inner cell mass as early as Day 7 and then increases until Day 14 (Figure 3A). Although both cell number and organoid size contributed to increase through Day 14 (Figure 3C), WST-1 assays suggested that metabolically active cells were decreased by Day 18, indicating that a subset of cells was no longer viable at later time points following terminal differentiation marked by a keratinized core (Figure 3A). When passaged at Day 14, the 3D organoids from EPC2-hTERT cells maintained similar growth kinetics (data not shown). EPC2-hTERT organoids displayed consistent OFR (19.1 ± 2.9%, mean ± standard deviation of 4 biologic replicates) and size (174.8 ± 2.6 μm mean ± standard deviation of 6 biologic replicates) from passage to passage.
Figure 3.
Growth kinetics of human esophageal 3D organoids. Human esophageal 3D organoids were generated with EPC2-hTERT cells in KSFMC medium and analyzed for structures and growth properties at indicated time points. The 3D organoid culture products were photomicrographed under a phase-contrast microscopy and recovered from Matrigel for H&E, IHC for Ki-67, and IF for IVL (A). DAPI-stained cell nuclei. Scale bar = 50 μm. Ki-67 labeling index (LI) was determined at each time point (B). *P < .05 vs Day 5; ns, not significant versus Day 5; n = 8. (C) Average organoid size and viable cells were determined by phase-contrast imaging (A) and WST1 assays, respectively. *P < .05 vs Day 1; and †, vs Day 14 in WST1 assays; n = 3. #P < .05 vs Day 5; and ns, not significant vs Day 14 in organoid size; n = 8. Data represent 2 biologic replicates with similar results.
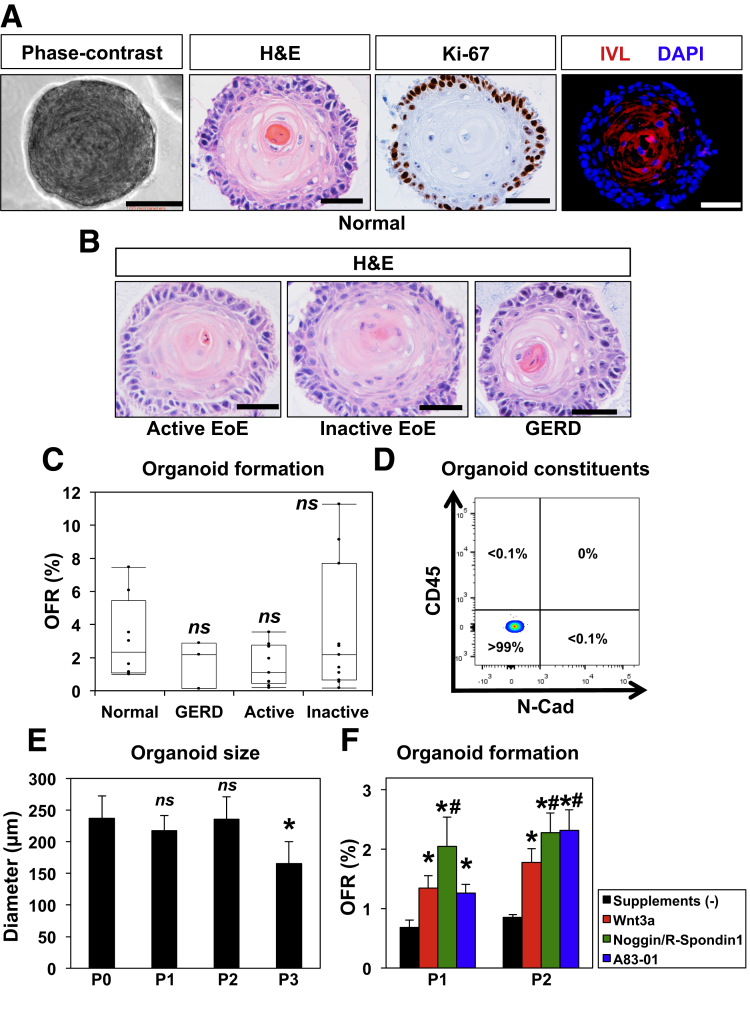
After optimizing organoid culture conditions with EPC2-hTERT cells, we then sought to apply these methods to esophageal organoids derived from patient biopsies (Figure 4). We found that a single biopsy was sufficient to initiate primary esophageal 3D organoids, yielding 105–106 viable cells. Excluding a few cases because of fungal contamination, 3D structures were successfully generated from 34 pediatric patients (100%) (Table 1). We included EPC2-hTERT for quality control purposes in each tissue-derived 3D organoid experiment where outcomes can be influenced by variables including medium components and culture conditions. Esophageal 3D organoids generated from biopsies of normal mucosa were comparable in structure, proliferation, and differentiation (Figure 4A) as compared with those from EPC2-hTERT cells (Figure 3A). Moreover, 3D organoids were generated from patients with GERD and EoE (active and inactive disease) (Table 1) without significant differences in morphology and OFR as compared with 3D organoids from normal mucosa (Figure 4B and C), indicating that esophageal epithelial cells may behave similarly ex vivo despite the presence of reactive epithelial changes (eg, BCH) in the original tissue associated with the in vivo inflammatory milieu of differential disease conditions, such as EoE and GERD.
Figure 4.
Human 3D esophageal organoids from endoscopic biopsies display an explicit proliferation-differentiation gradient ex vivo. Esophageal 3D organoids derived from patient biopsies and normal esophageal mucosa were grown in KSFMC medium. 3D culture products were photomicrographed under a phase-contrast microscopy and further recovered from Matrigel for H&E, IHC for Ki-67, and IF for IVL in A. H&E- and Ki-67-stained panels show from the periphery to the center, basaloid, intermediate (spinous), and keratinized cells with the very center of the organoid showing a terminally differentiated keratinized core. Only basaloid cells are Ki-67 positive. DAPI-stained cell nuclei in IF. Scale bars = 100 μm in phase-contrast image, 50 μm in H&E staining, IHC, and IF. (B and C) Esophageal 3D organoids were generated from patients with normal esophageal mucosa, GERD, active EoE, and inactive EoE. The resulting 3D organoids were analyzed by H&E staining for morphology in B. Scale bar = 50 μm. Organoid formation rate was determined in organoids derived from normal (n = 8), GERD (n = 3), active EoE (n = 11), and inactive EoE (n = 9) patients in C. ns, not significant vs normal as determined with P < .05 being considered statistically significant. (D) Biopsy-derived primary organoids (P01) were recovered from Matrigel and enzymatically dissociated for flow cytometry. Cell surface expression of CD45 and N-cadherin (N-Cad) were evaluated to determine N-Cad-/CD45- epithelial cells and nonepithelial cell (CD45+ or N-Cad+) as shown in a representative dot plot of normal biopsies: N-Cad-/CD45-, 99.9% ± 0.087, N-Cad+/CD45- 0.0078 ± 0.057%, N-Cad-/CD45+ 0.0046 ± 0.039%; mean ± standard error of the mean, <0.05 vs N-Cad-/CD45-, n = 3). Note that nonepithelial cells were detected minimally (<0.1%). (E) Organoid size was determined at the end of each passage (P0-P3). *P < .01 vs P0; ns, not significant vs P0; n = 8. (F) 3D organoids were first grown in KSFMC and then passaged in KSFMC supplemented with or without SB202190, WNT3A, Noggin/R-Spondin1, or A83-01. OFR was determined at the end of passages 2 and 3. *P < .01 vs P2 or 3 and supplements (-); #P < .05 vs P2 or 3 and Wnt3a; n = 6. There was no organoid growth noted in the presence of SB202190 at the end of P2. Data represent ≥2 biologic replicates with similar results.
OFR for biopsy-derived organoids was smaller than that of EPC2-hTERT (Figure 4C), suggesting heterogeneity of tissue-dissociated cells including dying cells and terminally differentiated cells. Of note, flow cytometry detected few (<0.1%), if any, CD45-positive leukocytes or N-cadherin-positive mesenchymal cells within tissue-derived 3D organoids (Figure 4D and data not shown), even though nonepithelial cells were not excluded from single cell suspensions prepared at the onset of organoid culture in this study. Unlike EPC2-hTERT, biopsy-derived organoids typically exhibited a decrease in size after passage 2 (Figure 4E) and stopped forming organoids after passage 3 (3.8 ± 0.8 on average, n = 10; patients whose organoids were tracked until they no longer formed organoids) (Table 1). When organoids were cultured in the presence of Wnt 3a, Noggin/R-Spondin1 or A83-01, OFR was improved in subsequent passages (Figure 4F); however, these factors were insufficient to extend passage number. Thus, the defined culture conditions may be permissive for the maintenance of keratinocyte progenitors with limited self-renewal capacity.43
Pharmacologic and Genetic Inhibition of Notch Signaling Increases Basaloid Cell Content and Affects the Differentiation Gradient in Esophageal 3-Dimensional Organoids
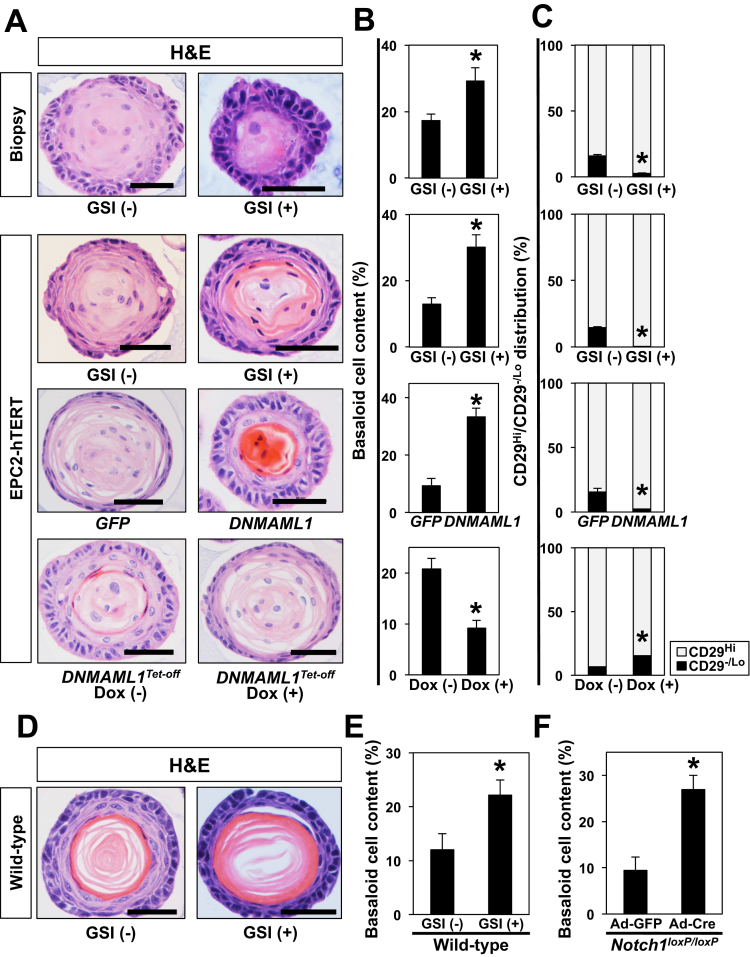
We next explored how the squamous proliferation-differentiation gradient may be regulated in esophageal 3D organoids. Specifically, we interrogated the role of Notch signaling via pharmacologic and genetic inhibition. Notch inhibition is expected to perturb squamous cell differentiation, resulting in accumulation of undifferentiated basal keratinocytes and hyperkeratosis.13 We grew biopsy- and EPC2-hTERT-derived 3D organoids in the presence or absence of the GSI compound E from Day 0. GSI did not affect OFR (data not shown); however, H&E staining of 3D organoids revealed thickening of the outermost basaloid cell layers and concurrent hyperkeratosis in the inner core (Figure 5A and B). Increased basaloid cell content was further corroborated by flow cytometry for CD29 (β1-inetgrin), a basal cell marker (Figure 5C). GSI had a similar effect when treatment was initiated in structurally established 3D organoids at Day 6 (data not shown). These findings were further corroborated in 3D organoids from EPC2-hTERT cells with genetic Notch inhibition (Figure 5A). DNMAML1, a genetic pan-Notch inhibitor, induced a similar phenotype, which was reversed on restoration of Notch activity in the tetracycline-regulatory (Tet-Off) system, allowing doxycycline (DOX)-mediated suppression of ectopically expressed DNMAML1 (Figure 5A). Finally, these findings were recapitulated in murine esophageal organoids on GSI treatment or adenoviral Cre-mediated Notch1 deletion ex vivo (Figure 5D–F). We observed an abrupt terminal differentiation with GSI treatment with increased basaloid cells and no intermediately differentiated cell layers (Figure 5D–F and data not shown). These findings indicate that the differentiation gradient in 3D esophageal organoids is subject to regulation by canonical Notch signaling in human and mice.
Figure 5.
Genetic and pharmacologic inhibition of Notch signaling increases basaloid cell content in esophageal 3D organoids. Esophageal 3D organoids were generated from biopsies of normal esophageal mucosa and EPC2-hTERT and derivatives (A–C) or C57BL/6J murine esophageal epithelial sheets (D–F). Organoids were grown in the presence or absence of 1 μM compound E, a GSI or dimethyl sulfoxide (vehicle), DNMAML1 or GFP (control vector), 1 μg/mL DOX to turn off DNMAML1 expression in cells expressing transgenic DNMAML1 in DNMAML1Tet-Off. H&E-stained 3D organoids in A were evaluated for basaloid cell content under indicated conditions in B. *P < .01 vs GSI (-), GFP, or DOX (-); n = 6. The 3D organoids were enzymatically dissociated and assessed for CD29 expression by flow cytometry in C. Bar diagram (mean ± standard error of the mean) shows the proportion of cells with high CD29 expression (basaloid cells) and low CD29 expression (differentiated cells) in 3D organoids. *P < .01 vs GSI (-), GFP, or DOX (-); n = 3. (D and E) Murine esophageal 3D organoids were grown in the presence or absence of 1 μM compound E (GSI). (F) Single cell suspensions prepared from epithelial sheets isolated from Notch1loxP/loxP mice were incubated with adenovirus expressing either Cre recombinase (Ad-Cre) to induce Notch1 deletion or GFP (Ad-GFP, negative control) before plating in organoid culture. Organoids were stained by H&E in D. Note that GSI-treated organoids display increased basaloid cell content and abrupt terminal differentiation (keratinization), whereas a clear differentiation gradient is present in GSI-untreated control organoids. Basaloid cell content was determined in H&E-stained organoids in E and F. Basaloid cell content is increased on pan-Notch inhibition with GSI or Ad-Cre-mediated genetic deletion of Notch1. *P < .01 vs GSI (-) or GFP. Data represent 2 biologic replicates with similar results.
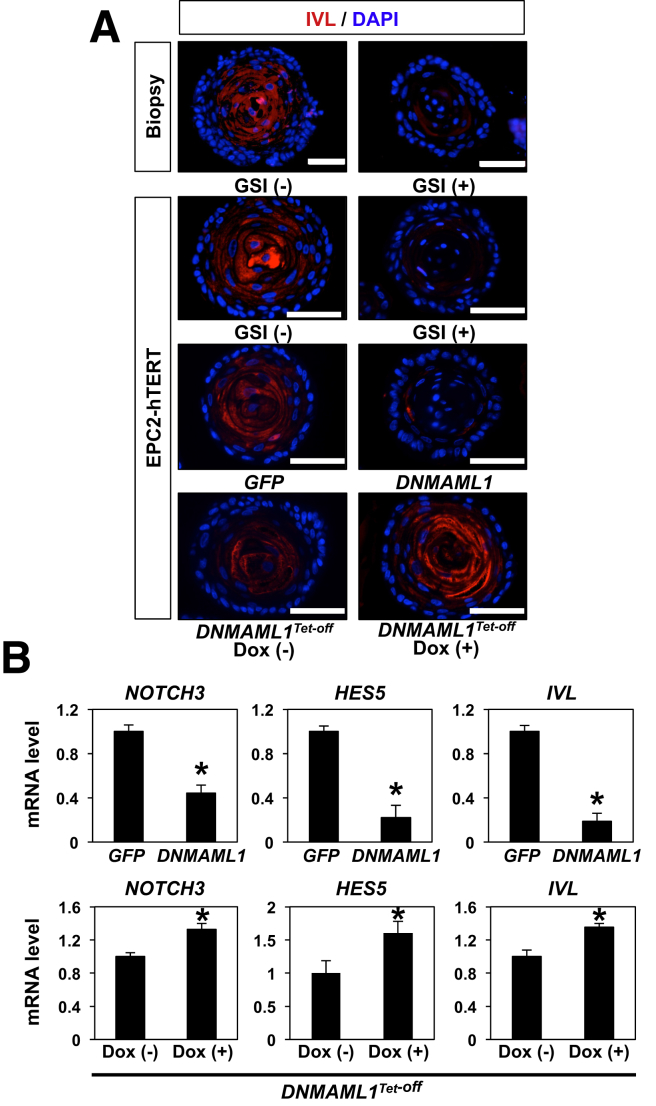
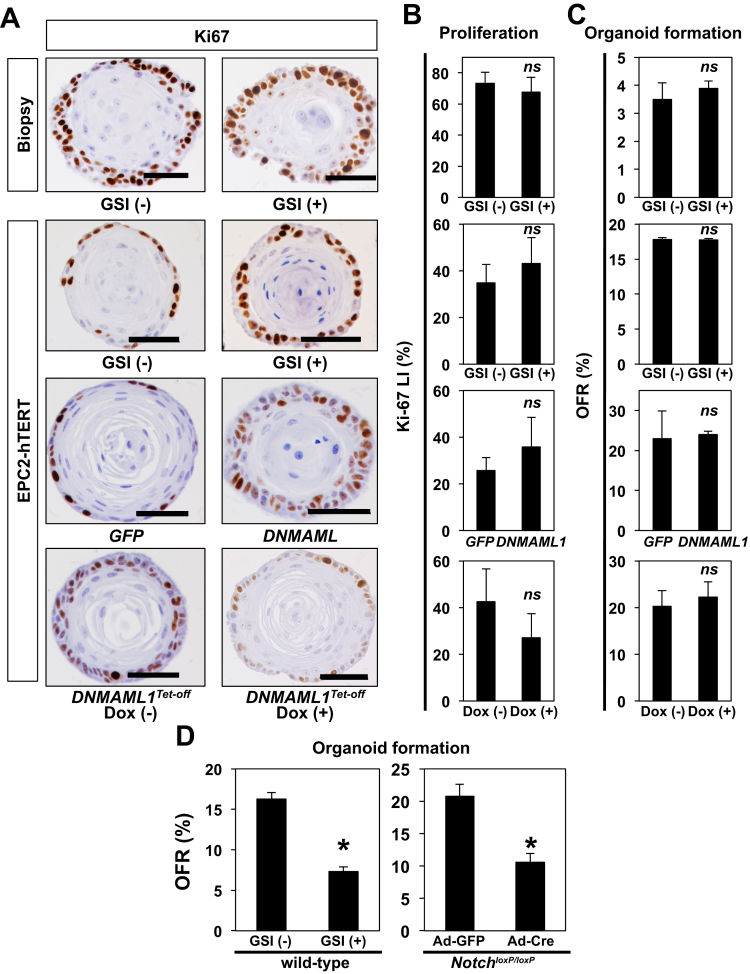
Notch1 regulates squamous cell differentiation via Notch3 in the esophageal epithelium as validated in K14Cre;DNMAML1loxP-stop-loxP mice and EPC2-hTERT cells in monolayer and 3D organotypic cultures, the latter an independent form of human tissue engineering.8 We examined IVL expression by IF in human esophageal 3D organoids on pharmacologic and genetic Notch inhibition. Both GSI and DNMAML1 decreased expression of IVL (Figure 6A). IVL expression was restored on DOX-mediated suppression of DNMAML1 in the Tet-off system (Figure 6A). Additionally, qRT-PCR assays showed that Notch inhibition via DNMAML1 decreased mRNA for IVL, NOTCH3, and HES5, all transcriptional targets for the activated form of Notch1 and reciprocal upregulation on DOX-mediated suppression of DNMAML1 in the Tet-Off system (Figure 6B). Of note, Notch inhibition did not affect Ki-67 expression significantly in esophageal 3D organoids (Figure 7), suggesting that basaloid-cell accumulation may be accounted for by delayed commitment to terminal differentiation, but not increased proliferation. Moreover, Notch inhibition suppressed OFR in mice, but not human (Figure 7), in agreement with our recent findings in neoplastic murine esophageal 3D organoids,44 suggesting a potential species difference in the role of Notch signaling in esophageal epithelial cells.
Figure 6.
Pan-Notch inhibition results in suppression of IVL and Notch target genes in human esophageal 3D organoids. Esophageal 3D organoids in Figure 5 were analyzed for IVL expression by IF (A). Organoids were analyzed by qRT-PCR for mRNA expression of NOTCH3, HES5, and IVL. GAPDH served as an internal control *P < .01 vs GFP in B; *P < .01 vs DOX (-) in B; n = 3. Data represent 2 biologic replicates with similar results.
Figure 7.
Pan-Notch inhibition decreases OFR in murine, but not human, esophageal 3D organoids without affecting basaloid cell proliferation in both human and mice. Human esophageal 3D organoids in Figure 5 were analyzed for Ki-67 expression by IHC under indicated conditions (A and B). Ki-67 labeling index (LI) in A was determined in B. *P < .01 vs GSI (-), GFP, or DOX (-); n = 6. OFR for human (C) and murine (D) esophageal 3D organoids under indicated conditions in Figure 5 was determined. Note that OFR was decreased on pan-Notch inhibition with GSI or Ad-Cre-mediated genetic deletion of Notch1 in (D). ns, not significant vs GSI (-), GFP, or DOX (-); n = 3 in (C). *P < .01 vs GSI (-) or GFP; n = 6 in (D). Data represent 2 biologic replicates with similar results.
Eosinophilic Esophagitis–Relevant Cytokines Inhibit Notch-Dependent Squamous-Cell Differentiation, Promoting Basaloid Cell Accumulation in 3-Dimensional Organoids
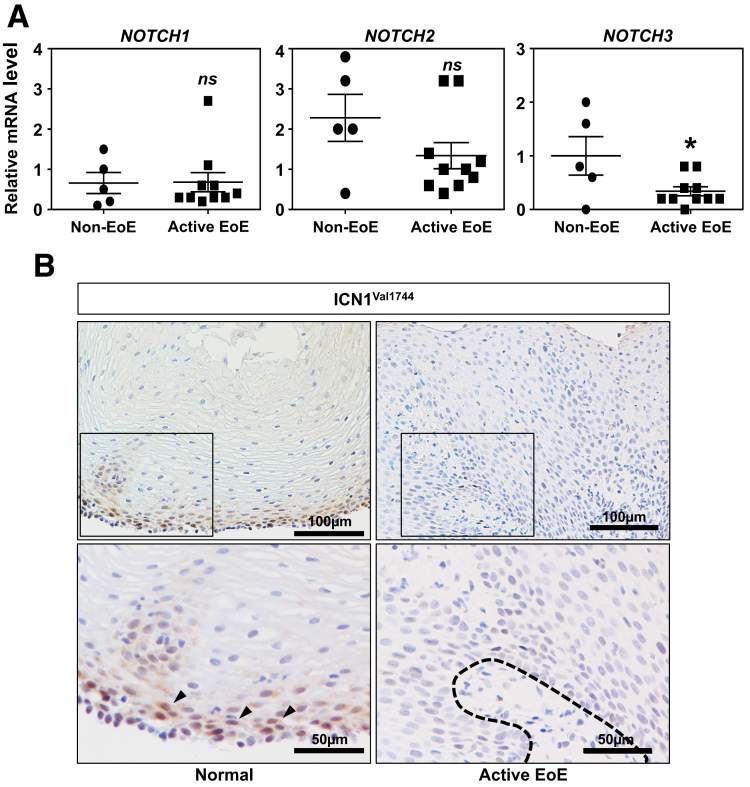
Reactive epithelial changes represented by BCH and spongiosis are common histopathologic features in esophageal pathologies including EoE.14 Perturbed squamous cell differentiation has been implicated in epithelial barrier defects,17 essential in the pathogenesis of EoE.16, 45, 46 To gain insights as to how squamous cell differentiation is affected in EoE, we mined RNA sequencing data (GSE58640) on well-annotated EoE cases32, 33 to find a 40% downregulation of NOTCH3 mRNA expression in the active EoE population compared with non-EoE control (P = .0006). We validated independently this result by qRT-PCR using esophageal biopsy specimens from a pediatric cohort at the The Children’s Hospital of Philadelphia (Figure 8A). This change in NOTCH3 was unique among all Notch receptor paralogs because NOTCH1 and NOTCH2 showed no difference and NOTCH4 was not detected in any of the biopsies (data not shown). NOTCH3 modulates esophageal cell fate toward squamous cell differentiation.8 Although NOTCH3 protein was not detectable, our preliminary IHC analysis suggested that esophageal biopsies from active EoE patients (n = 5) display downregulation of ICN1Val1744, the activated form of NOTCH1 that is expressed in the basal compartment of normal squamous epithelia in non-EoE normal biopsies (n = 4) (Figure 8B and data not shown). We hypothesized that EoE-relevant cytokines influence Notch signaling to alter the differentiation gradient in esophageal epithelium.
Figure 8.
Notch3 mRNA and ICN1Val1744 protein are downregulated in active EoE in esophageal biopsies. (A) qRT-PCR was performed to evaluate the mRNA levels of Notch receptor paralogs (Notch1–4) in esophageal biopsies from non-EoE (n = 5) and active EoE (n = 10) patients. ns, not significant vs non-EoE; *P < .05 vs non-EoE. P value was determined by 2-tailed Student t test. Note that Notch4 mRNA was not detectable by PCR. (B) IHC for ICN1Val1744 was performed on biopsies from patients with non-EoE normal esophageal mucosa (n = 5) and active EoE with BCH (n = 4). Arrows indicate nuclear localization of ICN.
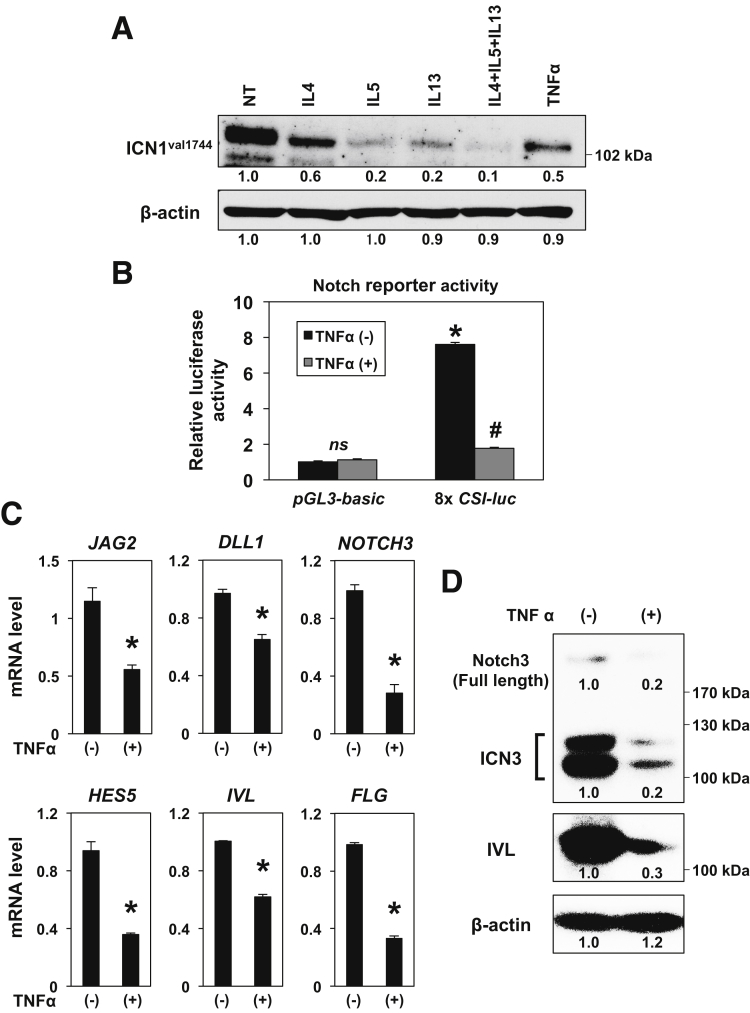
To determine the effect of EoE-relevant cytokines on Notch signaling, we carried out immunoblot analysis for ICN1Val1744 in EPC2-hTERT cells stimulated with or without cytokines, either alone or in combination. We included TNF-α, which has been implicated in broader disease contexts including GERD and radiation esophagitis. All of these cytokines suppressed ICN1Val1744 expression (Figure 9A). We next performed transfection assays using the 8xCSL-luc Notch reporter and found that TNF-α suppresses Notch-dependent transcriptional activity in EPC2-hTERT cells (Figure 9B). Moreover, TNF-α decreased in EPC2-hTERT 3D organoids mRNA expression for Notch ligands (JAG2, DLL1) and ICN1Val1744 targets and squamous cell differentiation markers (NOTCH3, HES5, IVL, and FLG) (Figure 9C). This was further corroborated by immunoblotting for NOTCH3 and IVL in EPC2-hTERT 3D organoids (Figure 9D).
Figure 9.
TNF-α suppresses Notch-dependent transcription, Notch ligands, and Notch1 target genes in human esophageal epithelial cells. Effects of cytokine stimulation were determined in EPC2-hTERT cells in monolayer culture (A and B) and 3D organoids (C and D). (A) Cells were stimulated for 24 hours with or without indicated cytokines either alone or in combination and subjected to immunoblotting for ICN1Val1744. NT, no treatment. β-actin served as a loading control. To measure Notch-dependent transcriptional activity, cells were transfected with 8xCSL-luc reporter construct and then treated with or without 40 ng/mL of TNF-α for 48 hours before cells were lysed for luciferase assays in B. pGL3-Basic was used to determine the background luciferase activity. ns, not significant vs pGL3-Basic and TNF-α (-); *P < .0001 vs pGL3-Basic and TNF-α (-); #P < .0001 vs 8xCSL-luc and TNF-α (-); n = 4. Established 3D organoids were treated with 5 ng/mL TNF-α for 72 hours from Day 5 and subjected to qRT-PCR (C) or immunoblotting (D) for indicated molecules. β-actin served as an internal control. *P < .01 vs TNF-α (-); n = 3 in C. (D) ICN3 denotes the intracellular domain of NOTCH3. (A and D) Numbers under each band represent the signal intensity relative to NT set as 1. Data represent 2 biologic replicates with similar results.
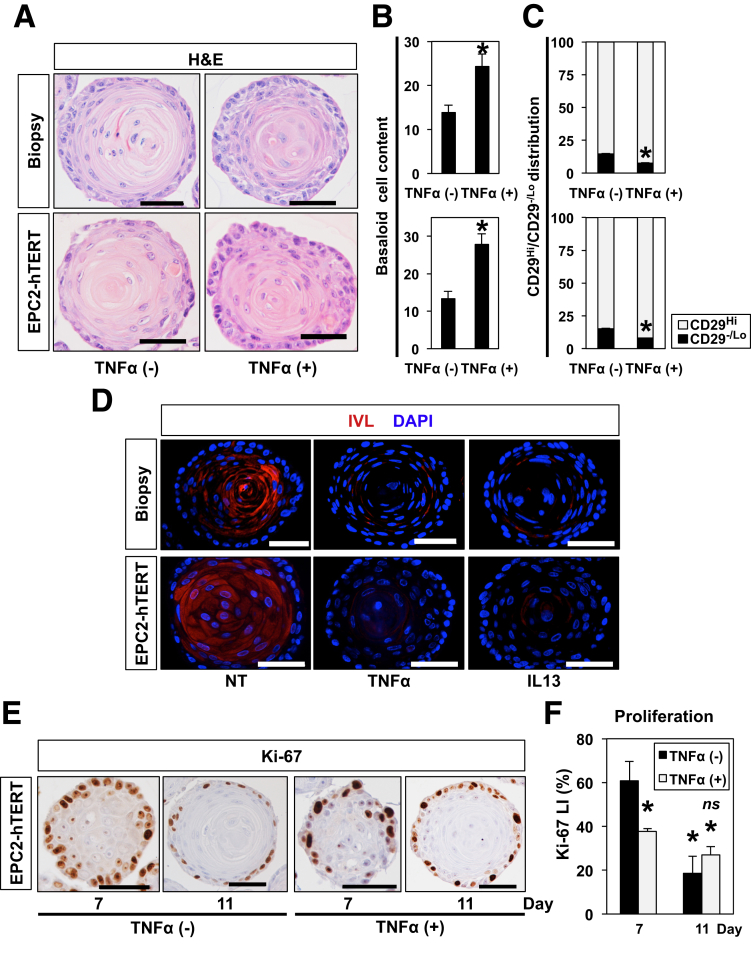
Because TNF-α induces an expansion of the basaloid cell population in murine esophageal 3D organoids,41 we further tested the effects of TNF-α stimulation in human esophageal 3D organoids generated from normal biopsies. When established 3D organoids were stimulated with TNF-α, the resulting 3D organoids displayed augmented basaloid cell content (Figure 10A and B) with flow cytometry demonstrating that TNF-α stimulation increased the CD29 high basaloid cell content consistent with the BCH-like phenotype (Figure 10C). We confirmed further by IF that decreased IVL expression in human esophageal 3D organoids generated with biopsies from normal mucosal and EPC2-hTERT cells (Figure 10D). Additionally, similar changes were detected in human esophageal 3D organoids stimulated with IL13 (Figure 10D), another EoE-relevant cytokine that not only promotes basal cell expansion but suppresses squamous-cell differentiation in the context of EoE.5, 41
Figure 10.
TNF-α increases basaloid cell content with a concurrent suppression of Notch target genes in human esophageal 3D organoids. Esophageal 3D organoids were grown for 11 days from normal esophageal mucosa or EPC2-hTERT cells in the presence or absence of 1 ng/mL TNF-α54 or 1 ng/mL IL13 (D). Resulting 3D organoids were analyzed by H&E staining for morphology in A from which basaloid cell content was determined in B. Scale bars = 50 μm. *P < .0001 vs TNF-α (-); n = 6. (C) 3D organoids were enzymatically dissociated and assessed for CD29 expression. Bar diagram shows the proportion of cells with high CD29 expression (basaloid cells) and low CD29 expression (differentiated cells) in 3D organoids. *P < .0001 vs TNF-α (-); n = 3. (D) Organoids were analyzed for IVL expression by IF. NT, no treatment. (E) EPC2-hTERT 3D organoids grown in the presence or absence of 5 ng/mL TNF-α were harvested at Days 7 and 11. Cell proliferation was assessed by IHC for Ki-67 in from which Ki-67 labeling index (LI) was determined in F. Scale bars = 50 μm. *P < .0001 vs TNF-α (-) at Day 7; ns, not significant vs TNF-α (-) at Day 11; n = 6. Representative images are shown. Scales bars = 50 μm. Data represent 2 biologic replicates with similar results.
Induction of differentiation in 3D organoids was mirrored by a decrease in Ki-67 expression (Figure 3A and B). When evaluated in the presence of TNF-α stimulation, Ki67 downregulation was accelerated on treating with TNF-α from Day 5 (Figure 10E and F), indicating that the increase in basaloid cell content was not accounted for by increased proliferation, but rather an impaired differentiation. Thus, the observed changes induced by EoE-relevant cytokines may represent a phenocopy of Notch inhibition in human and murine esophageal 3D organoids (Figure 5, Figure 6, Figure 7).
In aggregate, these data from our innovative approaches indicate that Notch signaling may be suppressed to affect squamous cell differentiation as a reactive epithelial change under EoE-related inflammatory conditions and possibly other conditions including acid reflux in GERD.
Discussion
Major accomplishments of this study on esophageal 3D organoids established ex vivo include (1) development of a highly reliable protocol that can be commonly applicable for human and mice, (2) understanding physiological functions and regulatory mechanisms via pharmacologic and genetic manipulations47 and characterization of normal and diseased esophageal conditions, and (3) modeling epithelial pathologies associated with esophagitis to gain substantial novel molecular insights about the mechanism underlying reactive esophageal epithelial changes in disease conditions.
We found that human esophageal 3D organoids may not necessarily grow in culture conditions that were previously optimized for murine esophageal 3D organoids,27 revealing potential species differences. The relatively simple formula of KSFMC supported the growth of both human and murine esophageal organoids with comparable growth and structural properties. Among factors and agents originally used for murine esophageal 3D organoids,27 R-Spondin1 and Noggin, Wnt3A, or A83-01 improved OFR in passaged organoids in KSFMC; however, SB202190, a p38 mitogen-activated protein kinase inhibitor, impaired organoid formation by human cells. Oxidative stress may induce apoptosis via p38 mitogen-activated protein kinase.48 We did not use N2 and B27 supplements that provide redundant insulin, transferrin, and selenium, which are present in KSFM; however, B27 also contains antioxidants, which we did not supplement with in KSFMC. Given the inhibitory effect of SB202190, human esophageal cells may be sensitive to p38 mitogen-activated protein kinase–mediated processes, raising the possibility that antioxidants may alleviate cellular stress and improve cell survival. Our organoid protocol shows a high success rate from endoscopic biopsies; however, OFR was usually higher at P1 compared with P0, suggesting that tissue processing may affect cell viability. Alternatively, cells isolated from biopsies likely contain stromal cells, such as fibroblasts and immune cells, thereby underestimating the number of epithelial cells seeded for P0. Although nonepithelial cells were barely detectable within normal tissue-derived 3D organoids, this study does not exclude the possibility that more nonepithelial cells may be present in organoids from biopsies with disease conditions (eg, EoE) to influence organoid formation. Additionally, tissue-derived cells are possibly primed by factors in the tissue microenvironment (eg, inflammatory cytokines). Moreover, organoid characteristics are likely influenced by variables, such as species, gene polymorphism, gender, and age. Future studies may require fluorescence-activated cell sorting before organoid formation. Unbiased molecular analyses (eg, RNA-sequencing), comparing EPC2-hTERT and tissue-derived organoids grown in the presence or absence of disease-relevant cytokines, eosinophils, and other cell types, will better define molecular characteristics of 3D organoids in both physiological and disease contexts.
Unlike murine esophageal 3D organoids passaged at least 10 times,2 self-renewal in human esophageal 3D organoids may be limited under our culture conditions. Organoids may hit the Hayflick limit for replicative senescence because we estimate doubling of 8–9 generations occurs per passage. Such a premise was corroborated in 3D organoids generated with telomerase-immortalized EPC2-hTERT cells that were indefinitely passaged. In addition to “culture stress,” a plausible explanation is depletion of a unique subset of stem-like cells that are capable of initiating organoid formation. Because several self-renewal factors tested in KSFMC did not rescue the growth arrest of biopsy-derived esophageal 3D organoids, additional niche factors need to be explored. Within 3D organoids, cell proliferation declines as differentiation becomes more overt, indicating that differential biologic processes may occur at different time points. Future studies should determine the window for exposure to specific medium components.
We identified Ca2+ as a key media component factor with regard to promoting optimal recapitulation of the esophageal proliferation-differentiation gradient ex vivo. Added into KSFM via CaCl2, Ca2+ may facilitate cell-cell contact via adhesion molecules, such as E-cadherin, to activate Notch signaling and squamous cell differentiation. Our study validates single cell-derived esophageal 3D organoids as an excellent platform to study cell fate regulation in a physiologically relevant context. Data from Notch inhibition confirmed the physiological role of Notch signaling in squamous cell differentiation, although Notch activity seems to be dispensable for proliferation. Restorable DNMAML1 expression in the Tet-Off system precluded the possibility that unique cell clones refractory to Notch-dependent differentiation were selected before organoid formation. Moreover, our data complemented previous studies using an independent 3D culture system7, 8, 23, 28 that is suitable to study cell lines or primary culture, but not cells directly isolated from biopsies, because this method requires a large number of cells (>106 per well) grown in monolayer culture in advance. EGF may inhibit Notch to limit squamous cell differentiation.49 Thus, Notch-mediated generation of the differentiation gradient in esophageal 3D organoids can also be influenced by EGF present in KSFM. Given the growth properties of organoids, it is plausible that decreased direct exposure of the inner cell mass to EGF may permit Notch activation and terminal differentiation while leaving the proliferative outmost basaloid cell layer undifferentiated.
Our study provides proof of principle that organoids can be used to model epithelial pathologies related to esophagitis. EoE patient-derived esophageal 3D organoids displayed normal tissue architecture in the absence of inflammatory stimuli or potentially influence of a disease-associated unique microbiome that may be suppressed in the presence of antibiotics in KSFMC, confirming for the first time the “reactive” nature of BCH. TNF-α and other cytokines inhibited Notch signaling as a potential explanation of reactive epithelial changes, a novel premise. We demonstrated that Notch-mediated squamous cell differentiation is suppressed by EoE-relevant cytokines, providing novel mechanistic insight into the functional interplay between the inflammatory milieu and epithelial tissue architecture in the esophagus. Inhibition of the Notch activity by TNF-α may imply a common mechanism underlying BCH associated with esophagitis beyond EoE; however, further studies are needed to determine as to how these cytokines inhibit Notch signaling in esophageal epithelium and how inclusion of broader cytokines and other cell types in the 3D organoid system may better recapitulate differential disease conditions. Given the modulatory role of Notch3 in squamous cell differentiation as a direct transcriptional target for the activated form of Notch1, our finding of Notch3 downregulation in active EoE biopsies warrants further investigation of the role of Notch signaling in esophageal epithelial pathobiology, especially in the context of barrier defects associated with aberrant squamous cell differentiation. It should be noted that Notch3 may limit EMT in normal esophageal epithelial cells.28 EMT has been documented in the esophageal epithelium in patients with EoE.50 We have previously identified TNF-α as an essential inducer of EMT in the context of EoE.19 Thus, the cell fate regulation of squamous cell differentiation and EMT may be influenced by inflammatory cytokines via Notch3.
Interestingly, Notch-mutant mice display atopic dermatitis-like disease in the skin with epithelial induction of Tslp expression, BCH, spongiosis, hyperkeratosis, and dermal eosinophilia with concurrent upregulation of IL4 and IL13,13 all relevant to EoE. Because genetic abnormality in esophageal Tslp expression may promote T helper 2 cell responses in EoE,15 future applications of patient-derived organoids include testing for Tslp expression to predict disease susceptibility. Additionally, 3D organoids can be used to explore novel therapy targeting Notch, other signaling pathways, and other epithelial biologic processes, such as autophagy as implicated in the pathogenesis of EoE30, 51 and other esophageal diseases.31, 52, 53
Lack of remarkable differences in organoid structures from diseased mucosa compared with those from normal mucosa suggested that reactive epithelial changes may be normalized ex vivo in the absence of an inflammatory milieu. There were no apparent immune cells or other cell types detected in organoids generated from esophagitis biopsies, suggesting a selection of KSFMC for epithelial cells. Although further optimization is required, coculture experiments in esophageal 3D organoids with nonepithelial cell types including immune cells or fibroblasts can be performed to gain insights into their interplays in the pathogenesis of EoE.29 Because esophageal fibroblasts produce cytokines in response to epithelial conditioned media,42 epithelial-stroma crosstalk may be explored in the 3D organoid platform. Such studies are currently underway, revealing the important role of BCH in fibroblast activation and extracellular matrix tissue remodeling. Finally, building a biopsy-derived organoid library will have a high potential to gain insights into the genetic and epigenetic alterations or intrinsic cellular sensitivity to inflammatory cytokines and pharmacologic agents that may be amenable for translation in personalized medicine.
Acknowledgements
The authors thank Mr. Ben Rhoades and the staff of the Molecular Pathology and Imaging Core, Host-Microbial Analytic and Repository Core, Cell Culture/iPS Core, and Flow Cytometry and Cell Sorting Facilities at the University of Pennsylvania for technical support. They also thank members of the Rustgi laboratory for helpful discussions. Prasanna M. Chandramouleeswaran is currently in the Graduate program in Cell Biology, Physiology, and Metabolism at the University of Pennsylvania.
Footnotes
Author contributions Yuta Kasagi, Prasanna M. Chandramouleeswaran, Amanda B. Muir, and Hiroshi Nakagawa wrote the manuscript. Hiroshi Nakagawa and Amanda B. Muir conceived ideas, analyzed, and interpreted data. Yuta Kasagi, Prasanna M. Chandramouleeswaran, Koji Tanaka, Medha Sharma, John W. Tobias, Joshua Wang, Veronique Giroux, Kathryn E. Hamilton, and Kelly A. Whelan collected, analyzed, and interpreted data. Andres J. Klein-Szanto performed pathologic analyses. Alain J. Benitez, Gary W. Falk, Maureen DeMarshall, and Amanda B. Muir procured clinical samples and analyzed data. Veronique Giroux, Kathryn E. Hamilton, Kelly A. Whelan, Jonathan M. Spergel, Amanda B. Muir, Hiroshi Nakagawa, and Anil K. Rustgi obtained funding. Kelly A. Whelan and Jonathan M. Spergel edited the manuscript. Amanda B. Muir and Hiroshi Nakagawa directed the study.
Conflicts of interest The authors disclose no conflicts.
Funding This study was supported by the following National Institutes of Health Grants: R01DK114436 (P.M.C., A.B.M., H.N.), P01CA098101 (P.M.C., K.A.W., K.T., A.J.K.-S., V.G., H.N., A.K.R.), P30ES013508University of Pennsylvania Center of Excellence in Environmental Toxicology (H.N., A.K.R.), K01DK103953 (K.A.W.), F32CA174176 (K.A.W.), K01DK100485 (K.E.H.), K08DK106444 (A.B.M.), F32DK100088 (A.B.M.), Molecular Pathology and Imaging Core, Host-Microbial Analytic and Repository Core and Cell Culture and iPS Core Facilities at the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases P30-DK050306Center of Molecular Studies in Digestive and Liver Diseases. Additional support was provided by NASPGHAN Foundation/Takeda Pharmaceutical Products Inc Research Innovation Award (H.N.); U54CA163004 (H.N., M.D., G.W.F. and A.K.R.); Joint Penn-CHOP Center in Digestive, Liver and Pancreatic Medicine at the University of Pennsylvania Perelman School of Medicine (K.A.W., H.N.); and the American Cancer Society (RP-10-033-01-CCE, A.K.R.). Koji Tanaka is a recipient of the Japan Society for the Promotion of Science Postdoctoral Fellowship. Veronique Giroux is a recipient of the Fonds de Recherche du Québec – Santé Postdoctoral Fellowship (P-Giroux-27692 and P-Giroux-31601). Jonathan M. Spergel is funded by Stuart Starr Endowed Chair, Food Allergy Research Education. Jonathan M. Spergel and Amanda B. Muir are funded in part by the Consortium of Eosinophilic Gastrointestinal Disease Researchers (CEGIR). CEGIR (U54 AI117804) is part of the Rare Diseases Clinical Research Network, an initiative of the Office of Rare Diseases Research, National Center for Advancing Translational Sciences, and is funded through collaboration between National Institute of Allergy and Infectious Diseases, National Institute of Diabetes and Digestive and Kidney Diseases, and National Center for Advancing Translational Sciences. CEGIR is also supported by patient advocacy groups including American Partnership for Eosinophilic Disorders, Campaign Urging Research for Eosinophilic Disorders, and Eosinophilic Family Coalition.
References
- 1.Jones P.H., Watt F.M. Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell. 1993;73:713–724. doi: 10.1016/0092-8674(93)90251-k. [DOI] [PubMed] [Google Scholar]
- 2.Giroux V., Lento A.A., Islam M., Pitarresi J.R., Kharbanda A., Hamilton K.E., Whelan K.A., Long A., Rhoades B., Tang Q., Nakagawa H., Lengner C.J., Bass A.J., Wileyto E.P., Klein-Szanto A.J., Wang T.C., Rustgi A.K. Long-lived keratin 15+ esophageal progenitor cells contribute to homeostasis and regeneration. J Clin Invest. 2017;127:2378–2391. doi: 10.1172/JCI88941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalabis J., Oyama K., Okawa T., Nakagawa H., Michaylira C.Z., Stairs D.B., Figueiredo J.L., Mahmood U., Diehl J.A., Herlyn M., Rustgi A.K. A subpopulation of mouse esophageal basal cells has properties of stem cells with the capacity for self-renewal and lineage specification. J Clin Invest. 2008;118:3860–3869. doi: 10.1172/JCI35012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andl C.D., Mizushima T., Nakagawa H., Oyama K., Harada H., Chruma K., Herlyn M., Rustgi A.K. Epidermal growth factor receptor mediates increased cell proliferation, migration, and aggregation in esophageal keratinocytes in vitro and in vivo. J Biol Chem. 2003;278:1824–1830. doi: 10.1074/jbc.M209148200. [DOI] [PubMed] [Google Scholar]
- 5.Jiang M., Ku W.Y., Zhou Z., Dellon E.S., Falk G.W., Nakagawa H., Wang M.L., Liu K., Wang J., Katzka D.A., Peters J.H., Lan X., Que J. BMP-driven NRF2 activation in esophageal basal cell differentiation and eosinophilic esophagitis. J Clin Invest. 2015;125:1557–1568. doi: 10.1172/JCI78850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kagawa S., Natsuizaka M., Whelan K.A., Facompre N., Naganuma S., Ohashi S., Kinugasa H., Egloff A.M., Basu D., Gimotty P.A., Klein-Szanto A.J., Bass A.J., Wong K.K., Diehl J.A., Rustgi A.K., Nakagawa H. Cellular senescence checkpoint function determines differential Notch1-dependent oncogenic and tumor-suppressor activities. Oncogene. 2015;34:2347–2359. doi: 10.1038/onc.2014.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naganuma S., Whelan K.A., Natsuizaka M., Kagawa S., Kinugasa H., Chang S., Subramanian H., Rhoades B., Ohashi S., Itoh H., Herlyn M., Diehl J.A., Gimotty P.A., Klein-Szanto A.J., Nakagawa H. Notch receptor inhibition reveals the importance of cyclin D1 and Wnt signaling in invasive esophageal squamous cell carcinoma. Am J Cancer Res. 2012;2:459–475. [PMC free article] [PubMed] [Google Scholar]
- 8.Ohashi S., Natsuizaka M., Yashiro-Ohtani Y., Kalman R.A., Nakagawa M., Wu L., Klein-Szanto A.J., Herlyn M., Diehl J.A., Katz J.P., Pear W.S., Seykora J.T., Nakagawa H. NOTCH1 and NOTCH3 coordinate esophageal squamous differentiation through a CSL-dependent transcriptional network. Gastroenterology. 2010;139:2113–2123. doi: 10.1053/j.gastro.2010.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harada H., Nakagawa H., Takaoka M., Lee J., Herlyn M., Diehl J.A., Rustgi A.K. Cleavage of MCM2 licensing protein fosters senescence in human keratinocytes. Cell Cycle. 2008;7:3534–3538. doi: 10.4161/cc.7.22.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griswold D.E., Webb E.F., Badger A.M., Gorycki P.D., Levandoski P.A., Barnette M.A., Grous M., Christensen S., Torphy T.J. SB 207499 (Ariflo), a second generation phosphodiesterase 4 inhibitor, reduces tumor necrosis factor alpha and interleukin-4 production in vivo. J Pharmacol Exp Ther. 1998;287:705–711. [PubMed] [Google Scholar]
- 11.Rangarajan A., Talora C., Okuyama R., Nicolas M., Mammucari C., Oh H., Aster J.C., Krishna S., Metzger D., Chambon P., Miele L., Aguet M., Radtke F., Dotto G.P. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001;20:3427–3436. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pant A.R., Graham S.M., Allen S.J., Harikul S., Sabchareon A., Cuevas L., Hart C.A. Lactobacillus GG and acute diarrhoea in young children in the tropics. J Trop Pediatr. 1996;42:162–165. doi: 10.1093/tropej/42.3.162. [DOI] [PubMed] [Google Scholar]
- 13.Dumortier A., Durham A.D., Di Piazza M., Vauclair S., Koch U., Ferrand G., Ferrero I., Demehri S., Song L.L., Farr A.G., Leonard W.J., Kopan R., Miele L., Hohl D., Finke D., Radtke F. Atopic dermatitis-like disease and associated lethal myeloproliferative disorder arise from loss of Notch signaling in the murine skin. PLoS One. 2010;5:e9258. doi: 10.1371/journal.pone.0009258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins M.H., Martin L.J., Alexander E.S., Boyd J.T., Sheridan R., He H., Pentiuk S., Putnam P.E., Abonia J.P., Mukkada V.A., Franciosi J.P., Rothenberg M.E. Newly developed and validated eosinophilic esophagitis histology scoring system and evidence that it outperforms peak eosinophil count for disease diagnosis and monitoring. Dis Esophagus. 2017;30:1–8. doi: 10.1111/dote.12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothenberg M.E., Spergel J.M., Sherrill J.D., Annaiah K., Martin L.J., Cianferoni A., Gober L., Kim C., Glessner J., Frackelton E., Thomas K., Blanchard C., Liacouras C., Verma R., Aceves S., Collins M.H., Brown-Whitehorn T., Putnam P.E., Franciosi J.P., Chiavacci R.M., Grant S.F., Abonia J.P., Sleiman P.M., Hakonarson H. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Genet. 2010;42:289–291. doi: 10.1038/ng.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sherrill J.D., Kc K., Wu D., Djukic Z., Caldwell J.M., Stucke E.M., Kemme K.A., Costello M.S., Mingler M.K., Blanchard C., Collins M.H., Abonia J.P., Putnam P.E., Dellon E.S., Orlando R.C., Hogan S.P., Rothenberg M.E. Desmoglein-1 regulates esophageal epithelial barrier function and immune responses in eosinophilic esophagitis. Mucosal Immunol. 2014;7:718–729. doi: 10.1038/mi.2013.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rochman M., Travers J., Miracle C.E., Bedard M.C., Wen T., Azouz N.P., Caldwell J.M., Kc K., Sherrill J.D., Davis B.P., Rymer J.K., Kaufman K.M., Aronow B.J., Rothenberg M.E. Profound loss of esophageal tissue differentiation in patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2017;140:738–749. doi: 10.1016/j.jaci.2016.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohashi S., Natsuizaka M., Wong G.S., Michaylira C.Z., Grugan K.D., Stairs D.B., Kalabis J., Vega M.E., Kalman R.A., Nakagawa M., Klein-Szanto A.J., Herlyn M., Diehl J.A., Rustgi A.K., Nakagawa H. Epidermal growth factor receptor and mutant p53 expand an esophageal cellular subpopulation capable of epithelial-to-mesenchymal transition through ZEB transcription factors. Cancer Res. 2010;70:4174–4184. doi: 10.1158/0008-5472.CAN-09-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muir A.B., Lim D.M., Benitez A.J., Modayur Chandramouleeswaran P., Lee A.J., Ruchelli E.D., Spergel J.M., Wang M.L. Esophageal epithelial and mesenchymal cross-talk leads to features of epithelial to mesenchymal transition in vitro. Exp Cell Res. 2013;319:850–859. doi: 10.1016/j.yexcr.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nadatani Y., Huo X., Zhang X., Yu C., Cheng E., Zhang Q., Dunbar K.B., Theiss A., Pham T.H., Wang D.H., Watanabe T., Fujiwara Y., Arakawa T., Spechler S.J., Souza R.F. NOD-Like receptor protein 3 inflammasome priming and activation in Barrett's epithelial cells. Cell Mol Gastroenterol Hepatol. 2016;2:439–453. doi: 10.1016/j.jcmgh.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lancaster M.A., Knoblich J.A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 22.Liacouras C.A., Furuta G.T., Hirano I., Atkins D., Attwood S.E., Bonis P.A., Burks A.W., Chehade M., Collins M.H., Dellon E.S., Dohil R., Falk G.W., Gonsalves N., Gupta S.K., Katzka D.A., Lucendo A.J., Markowitz J.E., Noel R.J., Odze R.D., Putnam P.E., Richter J.E., Romero Y., Ruchelli E., Sampson H.A., Schoepfer A., Shaheen N.J., Sicherer S.H., Spechler S., Spergel J.M., Straumann A., Wershil B.K., Rothenberg M.E., Aceves S.S. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128:3–20. doi: 10.1016/j.jaci.2011.02.040. quiz 21–22. [DOI] [PubMed] [Google Scholar]
- 23.Kalabis J., Wong G.S., Vega M.E., Natsuizaka M., Robertson E.S., Herlyn M., Nakagawa H., Rustgi A.K. Isolation and characterization of mouse and human esophageal epithelial cells in 3D organotypic culture. Nat Protoc. 2012;7:235–246. doi: 10.1038/nprot.2011.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X., Klein R., Tian X., Cheng H.T., Kopan R., Shen J. Notch activation induces apoptosis in neural progenitor cells through a p53-dependent pathway. Dev Biol. 2004;269:81–94. doi: 10.1016/j.ydbio.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 25.Harada H., Nakagawa H., Oyama K., Takaoka M., Andl C.D., Jacobmeier B., von Werder A., Enders G.H., Opitz O.G., Rustgi A.K. Telomerase induces immortalization of human esophageal keratinocytes without p16INK4a inactivation. Mol Cancer Res. 2003;1:729–738. [PubMed] [Google Scholar]
- 26.Chandramouleeswaran P.M., Shen D., Lee A.J., Benitez A., Dods K., Gambanga F., Wilkins B.J., Merves J., Noah Y., Toltzis S., Yearley J.H., Spergel J.M., Nakagawa H., Malefyt R., Muir A.B., Wang M.L. Preferential secretion of thymic stromal lymphopoietin (TSLP) by terminally differentiated esophageal epithelial cells: relevance to eosinophilic esophagitis (EoE) PLoS One. 2016;11:e0150968. doi: 10.1371/journal.pone.0150968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeWard A.D., Cramer J., Lagasse E. Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population. Cell Rep. 2014;9:701–711. doi: 10.1016/j.celrep.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohashi S., Natsuizaka M., Naganuma S., Kagawa S., Kimura S., Itoh H., Kalman R.A., Nakagawa M., Darling D.S., Basu D., Gimotty P.A., Klein-Szanto A.J., Diehl J.A., Herlyn M., Nakagawa H. A NOTCH3-mediated squamous cell differentiation program limits expansion of EMT-competent cells that express the ZEB transcription factors. Cancer Res. 2011;71:6836–6847. doi: 10.1158/0008-5472.CAN-11-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muir A.B., Dods K., Henry S.J., Benitez A.J., Lee D., Whelan K.A., DeMarshall M., Hammer D.A., Falk G., Wells R.G., Spergel J., Nakagawa H., Wang M.L. Eosinophilic Esophagitis-associated chemical and mechanical microenvironment shapes esophageal fibroblast behavior. J Pediatr Gastroenterol Nutr. 2016;63:200–209. doi: 10.1097/MPG.0000000000001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whelan K.A., Merves J.F., Giroux V., Tanaka K., Guo A., Chandramouleeswaran P.M., Benitez A.J., Dods K., Que J., Masterson J.C., Fernando S.D., Godwin B.C., Klein-Szanto A.J., Chikwava K., Ruchelli E.D., Hamilton K.E., Muir A.B., Wang M.L., Furuta G.T., Falk G.W., Spergel J.M., Nakagawa H. Autophagy mediates epithelial cytoprotection in eosinophilic oesophagitis. Gut. 2017;66:1197–1207. doi: 10.1136/gutjnl-2015-310341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whelan K.A., Chandramouleeswaran P.M., Tanaka K., Natsuizaka M., Guha M., Srinivasan S., Darling D.S., Kita Y., Natsugoe S., Winkler J.D., Klein-Szanto A.J., Amaravadi R.K., Avadhani N.G., Rustgi A.K., Nakagawa H. Autophagy supports generation of cells with high CD44 expression via modulation of oxidative stress and Parkin-mediated mitochondrial clearance. Oncogene. 2017;36:4843–4858. doi: 10.1038/onc.2017.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherrill J.D., Kiran K.C., Blanchard C., Stucke E.M., Kemme K.A., Collins M.H., Abonia J.P., Putnam P.E., Mukkada V.A., Kaul A., Kocoshis S.A., Kushner J.P., Plassard A.J., Karns R.A., Dexheimer P.J., Aronow B.J., Rothenberg M.E. Analysis and expansion of the eosinophilic esophagitis transcriptome by RNA sequencing. Genes Immun. 2014;15:361–369. doi: 10.1038/gene.2014.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kottyan L.C., Davis B.P., Sherrill J.D., Liu K., Rochman M., Kaufman K., Weirauch M.T., Vaughn S., Lazaro S., Rupert A.M., Kohram M., Stucke E.M., Kemme K.A., Magnusen A., He H., Dexheimer P., Chehade M., Wood R.A., Pesek R.D., Vickery B.P., Fleischer D.M., Lindbad R., Sampson H.A., Mukkada V.A., Putnam P.E., Abonia J.P., Martin L.J., Harley J.B., Rothenberg M.E. Genome-wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat Genet. 2014;46:895–900. doi: 10.1038/ng.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mudge J.M., Harrow J. Creating reference gene annotation for the mouse C57BL6/J genome assembly. Mamm Genome. 2015;26:366–378. doi: 10.1007/s00335-015-9583-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anders S., Pyl P.T., Huber W. HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeffries S., Capobianco A.J. Neoplastic transformation by Notch requires nuclear localization. Mol Cell Biol. 2000;20:3928–3941. doi: 10.1128/mcb.20.11.3928-3941.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sato T., Stange D.E., Ferrante M., Vries R.G., Van Es J.H., Van den Brink S., Van Houdt W.J., Pronk A., Van Gorp J., Siersema P.D., Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 40.Kinugasa H., Whelan K.A., Tanaka K., Natsuizaka M., Long A., Guo A., Chang S., Kagawa S., Srinivasan S., Guha M., Yamamoto K., St Clair D.K., Avadhani N.G., Diehl J.A., Nakagawa H. Mitochondrial SOD2 regulates epithelial-mesenchymal transition and cell populations defined by differential CD44 expression. Oncogene. 2015;34:5229–5239. doi: 10.1038/onc.2014.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whelan K.A., Merves J.F., Giroux V., Tanaka K., Guo A., Chandramouleeswaran P.M., Benitez A.J., Dods K., Que J., Masterson J.C., Fernando S.D., Godwin B.C., Klein-Szanto A.J., Chikwava K., Ruchelli E.D., Hamilton K.E., Muir A.B., Wang M.L., Furuta G.T., Falk G.W., Spergel J.M., Nakagawa H. Autophagy mediates epithelial cytoprotection in eosinophilic oesophagitis. Gut. 2017;66:1197–1207. doi: 10.1136/gutjnl-2015-310341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muir A.B., Dods K., Noah Y., Toltzis S., Chandramouleeswaran P.M., Lee A., Benitez A., Bedenbaugh A., Falk G.W., Wells R.G., Nakagawa H., Wang M.L. Esophageal epithelial cells acquire functional characteristics of activated myofibroblasts after undergoing an epithelial to mesenchymal transition. Exp Cell Res. 2015;330:102–110. doi: 10.1016/j.yexcr.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dover R., Potten C.S. Heterogeneity and cell cycle analyses from time-lapse studies of human keratinocytes in vitro. J Cell Sci. 1988;89(Pt 3):359–364. doi: 10.1242/jcs.89.3.359. [DOI] [PubMed] [Google Scholar]
- 44.Natsuizaka M., Whelan K.A., Kagawa S., Tanaka K., Giroux V., Chandramouleeswaran P.M., Long A., Sahu V., Darling D.S., Que J., Yang Y., Katz J.P., Wileyto E.P., Basu D., Kita Y., Natsugoe S., Naganuma S., Klein-Szanto A.J., Diehl J.A., Bass A.J., Wong K.K., Rustgi A.K., Nakagawa H. Interplay between Notch1 and Notch3 promotes EMT and tumor initiation in squamous cell carcinoma. Nat Commun. 2017;8:1758. doi: 10.1038/s41467-017-01500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simon D., Radonjic-Hosli S., Straumann A., Yousefi S., Simon H.U. Active eosinophilic esophagitis is characterized by epithelial barrier defects and eosinophil extracellular trap formation. Allergy. 2015;70:443–452. doi: 10.1111/all.12570. [DOI] [PubMed] [Google Scholar]
- 46.Katzka D.A., Ravi K., Geno D.M., Smyrk T.C., Iyer P.G., Alexander J.A., Mabary J.E., Camilleri M., Vaezi M.F. Endoscopic mucosal impedance measurements correlate with eosinophilia and dilation of intercellular spaces in patients with eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2015;13:1242–1248. doi: 10.1016/j.cgh.2014.12.032. [DOI] [PubMed] [Google Scholar]
- 47.Saver JL, Jovin TG, Smith WS, Albers GW, Baron JC, Boltze J, Broderick JP, Davis LA, Demchuk AM, DeSena S, Fiehler J, Gorelick PB, Hacke W, Holt B, Jahan R, Jing H, Khatri P, Kidwell CS, Lees KR, Lev MH, Liebeskind DS, Luby M, Lyden P, Megerian JT, Mocco J, Muir KW, Rowley HA, Ruedy RM, Savitz SI, Sipelis VJ, Shimp SK 3rd, Wechsler LR, Wintermark M, Wu O, Yavagal DR, Yoo AJ, Consortium SV. Stroke treatment academic industry roundtable: research priorities in the assessment of neurothrombectomy devices. Stroke 2013;44:3596–3601 [DOI] [PMC free article] [PubMed]
- 48.Zarubin T., Han J. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 2005;15:11–18. doi: 10.1038/sj.cr.7290257. [DOI] [PubMed] [Google Scholar]
- 49.Kolev V., Mandinova A., Guinea-Viniegra J., Hu B., Lefort K., Lambertini C., Neel V., Dummer R., Wagner E.F., Dotto G.P. EGFR signalling as a negative regulator of Notch1 gene transcription and function in proliferating keratinocytes and cancer. Nat Cell Biol. 2008;10:902–911. doi: 10.1038/ncb1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kagalwalla A.F., Akhtar N., Woodruff S.A., Rea B.A., Masterson J.C., Mukkada V., Parashette K.R., Du J., Fillon S., Protheroe C.A., Lee J.J., Amsden K., Melin-Aldana H., Capocelli K.E., Furuta G.T., Ackerman S.J. Eosinophilic esophagitis: epithelial mesenchymal transition contributes to esophageal remodeling and reverses with treatment. J Allergy Clin Immunol. 2012;129:1387–1396. doi: 10.1016/j.jaci.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Merves J.F., Whelan K.A., Benitez A.J., Muir A.B., Furuta G.T., Wang M.L., Falk G.W., Spergel J.M., Nakagawa H. ATG7 gene expression as a novel tissue biomarker in eosinophilic esophagitis. Am J Gastroenterol. 2016;111:151–153. doi: 10.1038/ajg.2015.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kong J., Whelan K.A., Laczko D., Dang B., Caro Monroig A., Soroush A., Falcone J., Amaravadi R.K., Rustgi A.K., Ginsberg G.G., Falk G.W., Nakagawa H., Lynch J.P. Autophagy levels are elevated in barrett's esophagus and promote cell survival from acid and oxidative stress. Mol Carcinog. 2016;55:1526–1541. doi: 10.1002/mc.22406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roesly H.B., Khan M.R., Chen H.D., Hill K.A., Narendran N., Watts G.S., Chen X., Dvorak K. The decreased expression of Beclin-1 correlates with progression to esophageal adenocarcinoma: the role of deoxycholic acid. Am J Physiol Gastrointest Liver Physiol. 2012;302:G864–G872. doi: 10.1152/ajpgi.00340.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meurs K.M., Stern J.A., Atkins C.E., Adin D., Aona B., Condit J., DeFrancesco T., Reina-Doreste Y., Keene B.W., Tou S., Ward J., Woodruff K. Angiotensin-converting enzyme activity and inhibition in dogs with cardiac disease and an angiotensin-converting enzyme polymorphism. J Renin Angiotensin Aldosterone Syst. 2017;18 doi: 10.1177/1470320317737184. 1470320317737184. [DOI] [PMC free article] [PubMed] [Google Scholar]