Abstract
The anti-angiogenic and neurogenic pigment epithelium-derived factor (PEDF) demonstrated a potency to control choroidal neovascularization in age-related macular degeneration (AMD) patients. The goal of the present study was the development of an efficient and safe technique to integrate, ex vivo, the PEDF gene into retinal pigment epithelial (RPE) cells for later transplantation to the subretinal space of AMD patients to allow continuous PEDF secretion in the vicinity of the affected macula. Because successful gene therapy approaches require efficient gene delivery and stable gene expression, we used the antibiotic-free pFAR4 mini-plasmid vector to deliver the hyperactive Sleeping Beauty transposon system, which mediates transgene integration into the genome of host cells. In an initial study, lipofection-mediated co-transfection of HeLa cells with the SB100X transposase gene and a reporter marker delivered by pFAR4 showed a 2-fold higher level of genetically modified cells than when using the pT2 vectors. Similarly, with the pFAR4 constructs, electroporation-mediated transfection of primary human RPE cells led to 2.4-fold higher secretion of recombinant PEDF protein, which was still maintained 8 months after transfection. Thus, our results show that the pFAR4 plasmid is a superior vector for the delivery and integration of transgenes into eukaryotic cells.
Keywords: age-related macular degeneration, electroporation, RPE cells, antibiotic-free pFAR4 vector, Sleeping Beauty transposon, ocular gene therapy, transfection, PEDF, VEGF
Introduction
Since the first attempts to treat genetically based diseases using retroviral vectors to deliver transgenes to host cells,1, 2 various non-viral vectors and transgene delivery methods have been developed, including nanoparticle-mediated gene delivery,3 physical methods,4, 5, 6 plasmids,7, 8, 9 and DNA transposons, which are DNA sequences that can move from one location and become integrated into another locus of the genome.10 Even though transposons were discovered in the 1950s, it has been shown only recently that the transposon named Sleeping Beauty (SB), and especially its hyperactive form (SB100X), is able to efficiently and safely integrate transgenes into the host genome, providing sustained transgene expression in quiescent and dividing cells.11, 12
Non-virus-mediated gene delivery often uses plasmid vectors that usually include an antibiotic resistance marker for efficient plasmid manufacturing. However, antibiotic resistance genes present several safety concerns, such as the risk for horizontal gene transfer, which could provide pathogenic bacteria with resistance to antibiotics that are used to treat humans, and risk to patients with severe hypersensitivity to antibiotics.13 In addition, because cell transfection efficiency is inversely correlated with plasmid size,14, 15, 16 several research groups have designed vectors as small as possible and devoid of antibiotic resistance markers8, 17, 18, 19, 20, 21, 22, 23, 24, 25 to increase both transfection efficiency and safety. To produce a small plasmid free of antibiotic resistance marker (pFAR),26 we introduced an amber mutation into the thyA gene of Escherichia coli, resulting in a bacterial strain auxotrophic for thymidine and synthesized de novo a small plasmid (pFAR4) to remove redundant sequences and introduce a suppressor tRNA sequence (the selection marker) expressed from prokaryotic regulatory sequences.26 Introduction of pFAR4 mini-plasmid constructs into the E. coli strain auxotrophic for thymidine suppresses the nonsense mutation, restoring prototrophic growth to the thyA mutant and allowing efficient plasmid production. The reduced size of the pFAR4 vector leads to efficient transfection and expression of transgenes in various tissues, including mouse muscle, skin, and liver as well as transplanted tumor cells.26, 27 To integrate transgenes and support long-term transgene expression in dividing cells, we combined the pFAR4 vector with the hyperactive SB transposon system.
SB is a DNA transposon that belongs to the Tc1/mariner superfamily.28 It is derived from fish transposon sequences that were subjected to site-directed replacements and high-throughput genetic screenings to first “awaken” and subsequently increase its mobility.28, 29 The resulting hyperactive transposase variant, SB100X, is an effective genetic tool for the transposition of any eukaryotic gene flanked by inverted terminal repeats (ITRs).30, 31 The SB100X transposase binds to the ITRs (∼227 bp) and catalyzes transgene transposition from a donor plasmid into the host cells’ genome via a “cut and paste” mechanism.11, 12 The SB100X transposon system displays a nearly random transgene integration profile and does not have preferences for transcriptionally active regions, exhibiting lower genotoxicity than most viral integrative vectors.31, 32, 33, 34, 35
The objectives of combining the pFAR4 vector with the SB100X transposon system were to optimize essential parameters; i.e., efficiency of transfection, transgene expression level in dividing cells (such as HeLa cells), and somatic differentiated cells (such as retinal pigment epithelial [RPE] cells). Our particular interest in RPE cells stems from our objective to develop a gene therapeutic treatment for neovascular age-related macular degeneration (AMD).
AMD is the most common cause of severe vision loss in patients over the age of 60 and the major cause of blindness in industrialized countries.36, 37, 38 There are two distinct types of AMD: a slowly progressing dry (atrophic) form and a rapidly developing wet (neovascular) form, in which choroidal blood vessels grow through Bruch’s membrane into the subretinal space. The symptoms are characterized by the degeneration of RPE cells, alterations in Bruch’s membrane, neural retinal ganglion cell degradation, and the death of photoreceptor cells.
Neovascular AMD (nvAMD) is the result of an imbalance between the retinal anti-angiogenic pigment epithelium-derived factor (PEDF) and the pro-angiogenic vascular endothelial growth factor (VEGF). In pathological states, decreased production of PEDF and/or increased VEGF levels will result in choroidal neovascularization (CNV).39, 40 Current treatments for nvAMD are based on re-establishment of the balance between the PEDF and VEGF proteins, mostly by using biopharmaceuticals that inhibit VEGF, allowing CNV control in 90% of patients and significant vision improvement in 30%–40% of treated patients.41, 42 Effectiveness in responsive patients requires, however, frequent, often monthly, intravitreal injections of short half-life anti-VEGFs, which have been linked to local side effects, such as endophthalmitis, ocular hypertension,43 submacular hemorrhage,44 and rarely occurring thromboembolic events.45 To reduce treatment costs and avoid side effects, an alternative approach for CNV inhibition includes increasing PEDF levels, the natural antagonist of VEGF, by PEDF gene delivery to the retina of AMD patients. Using adenoviral gene vectors, Campochiaro et al.46, 47 reported a significant improvement in 25% of patients but no further follow-up. Our therapeutic approach comprises the subretinal transplantation of genetically modified autologous pigment epithelial cells that continuously secrete the PEDF protein for the life of AMD patients. Initial studies performed using conventional plasmids allowed demonstration, both in vitro and in animal models, of the feasibility of the proposed treatment.48, 49, 50 To implement clinical trials, further improvements are nevertheless required, such as removal of antibiotic resistance markers from the gene vectors.
In this study, we compared pFAR4 constructs and conventional plasmids paired with the SB100X transposon system for transfection efficiency, transposon integration, and transgene expression levels in easy-to-transfect HeLa cells as well as in primary human RPE cells. Here we report that the pFAR4 mini-plasmids mediate higher cell transfection efficiency, superior PEDF expression levels, and sustained, long-term PEDF secretion by RPE cells.
Results
pFAR4 Promotes Higher Transgene Integration Efficiency in HeLa Cells Compared with pT2
The suitability of delivering, integrating, and expressing a transgene in a host cell was preliminary investigated in HeLa cells transfected with a plasmid carrying the transposon and another encoding the SB100X transposase. The transfection and transposition efficiencies were assessed using a colony-forming assay based on the quantification of clones containing the inserted transgene. For this purpose, the neomycin resistance gene expressed from the simian virus 40 (SV40) promoter and carried by the pT2/SV-Neo transposon plasmid51 was subcloned into the pFAR4 mini-plasmid, resulting in the pFAR4-ITRs-SV-Neo construct (Figure 1A). Similarly, the hyperactive SB100X transposase expressed from the human cytomegalovirus (CMV) promoter carried by pCMV-CAT(T7)-SB100X29 was introduced into the pFAR4 mini-plasmid, leading to the pFAR4-CMV-SB100X construct. Thus, the original and the antibiotic-free plasmids contain identical eukaryotic expression cassettes but alternative plasmid backbones (Figure 1A), resulting in a decrease in plasmid size of ∼35%.
Figure 1.
Delivery of the Sleeping Beauty Transposon System by the pFAR4 Vector Mediates a Higher Number of Neomycin-Resistant Clones
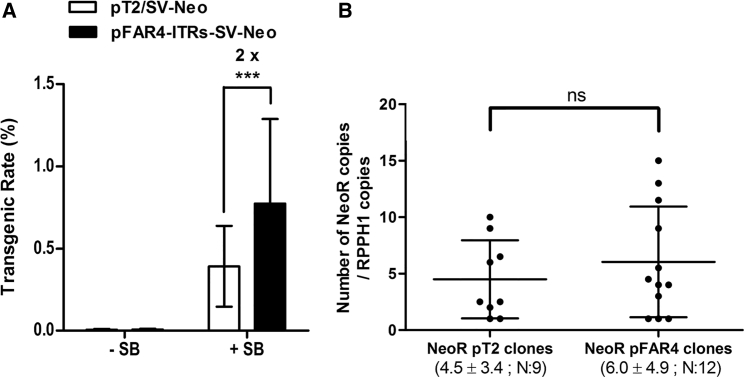
HeLa cells were co-transfected with either pT2/SV-Neo and pCMV-(CAT)T7-SB100X (ratio transposon plasmid:transposase plasmid = 500:50 ng or 50:5 ng) or an equimolar amount of pFAR4-ITRs-SV-Neo and pFAR4-CMV-SB100X (ratio transposon plasmid:transposase plasmid = 328.3:32.5 ng or 32.8:3.3 ng). The total DNA amount was adjusted to 550 ng using pFAR4 empty vector. (A) Both vectors contain identical eukaryotic expression cassettes but alternative plasmid backbones, differing by their size and the selection markers used for their propagation in E. coli. (B) Transgenic rates are the mean number of NeoR colonies after 11 days of growth in selection medium per number of cells seeded ± SD. Data are the average of three independent transfection experiments with replicated cell seedings (n = 23) for the “high” plasmid amount and four independent transfection experiments for the “low” plasmid amount (n = 40). +SB and −SB indicate the presence or absence of the transposase plasmid, respectively. **p < 0.005, ***p < 0.0001, using an unpaired two-tailed t test.
To assess whether the decrease in plasmid size affects the number of transgenic colonies, HeLa cells were co-transfected with either 500 or 50 ng of pT2/SV-Neo combined with 50 or 5 ng of pCMV-CAT(T7)-SB100X, respectively, or with 328.3 or 32.8 ng of pFAR4-ITRs-SV-Neo combined with 32.5 or 3.3 ng of pFAR4-CMV-SB100X, respectively (Figure 1B). A transposase-to-transposon ratio of 1:10 was chosen to avoid the “overproduction inhibition” (OPI) effect that negatively affects SB transposition efficiency in the presence of high transposase levels.52, 53, 54 With higher plasmid amounts, a higher number of neomycin-resistant colonies (∼1.6-fold, p < 0.0001) was obtained when the pFAR4 mini-plasmid was used compared with the pT2 regular plasmid. The effect was even more pronounced, ∼1.8-fold enhancement (p < 0.005), when HeLa cells were transfected with a lower plasmid amount (Figure 1B). In the absence of the SB100X transposase, negligible numbers of neomycin-resistant colonies were obtained, indicating that the higher number of colonies obtained with the pFAR4 vector resulted from transposition events catalyzed by the SB100X transposase.
Because the pFAR4 constructs are significantly smaller than the conventional plasmids, we next investigated the effect of the size of the transposon plasmid on transposition rates by using pFAR4 as the only SB100X delivery vector. Thus, HeLa cells were co-transfected with 5 ng of pFAR4 encoding SB100X and either 50 ng of pFAR4-ITRs-SV-Neo or an equimolar amount of pT2/SV-Neo (76.1 ng). As observed previously, a higher number of neomycin-resistant colonies (a ∼2-fold increase; p < 0.0001) was obtained when the neoR transgene was delivered by pFAR4 compared with the pT2 control plasmid (Figure 2A). These results suggest that the higher number of neomycin-resistant colonies obtained with the pFAR4 constructs is mediated by the smaller size of the pFAR4 transposon mini-plasmid.
Figure 2.
Delivery of the Sleeping Beauty Transposon by pFAR4 Promotes a Higher Number of NeoR Clones
HeLa cells were co-transfected with an equimolar amount of transposon plasmid (pT2/SV-Neo, 76.1 ng; pFAR4-ITRs-SV-Neo, 50 ng) either in the absence (−SB) or presence (+SB) of transposase (5 ng of pFAR4 encoding SB100X). The DNA amount was adjusted to 500 ng using empty pFAR4 plasmid. (A) Eleven days after transfection, NeoR colonies were enumerated. Data represent the mean number of NeoR colonies per number of seeded cells for three independent experiments ± SD; experiments were performed in quadruplicates with various number of cells seeded (n = 40). ***p < 0.0001, using an unpaired two-tailed t test. (B) The number of transposon integration events was determined by qPCR (performed twice in triplicate) and normalized to the RPPH1 gene using genomic DNA prepared from 9 and 12 NeoR clones selected from the three independent transfections of HeLa cells with the pT2 and pFAR4 transposon constructs, respectively. The number of integrated neoR gene copies varied among clones, but the mean (± SD) did not differ statistically with the type of gene vector used (ns, not significantly different, using Mann-Whitney test).
Following the enumeration of colonies after 11 days of selection, neomycin-resistant colonies were propagated in antibiotic-containing medium, and the transposon copy number was determined by qPCR using primers specific to the neomycin resistance gene (Figure 2B). An average of 6.0 ± 1.4 copies (n = 12 clones analyzed) were integrated in the genome of HeLa cells transfected with the neoR gene carried by pFAR4 (pFAR4-ITRs-SV-Neo), whereas 4.5 ± 1.2 copies (n = 9 clones analyzed) were quantified when HeLa cells were transfected with the neoR gene delivered by the pT2 plasmid (pT2/SV-Neo). The statistical analysis of transgene copy number quantified in cells transfected with either pFAR4-ITRs-SV-Neo or pT2/SV-Neo revealed that the difference is not significant. Thus, the pFAR4 plasmid appears to promote a higher number of transgenic cells without increasing the number of transgene copies integrated per cell.
pFAR4 Promotes a Higher Number of Transgenic Cells under Non-selective Conditions
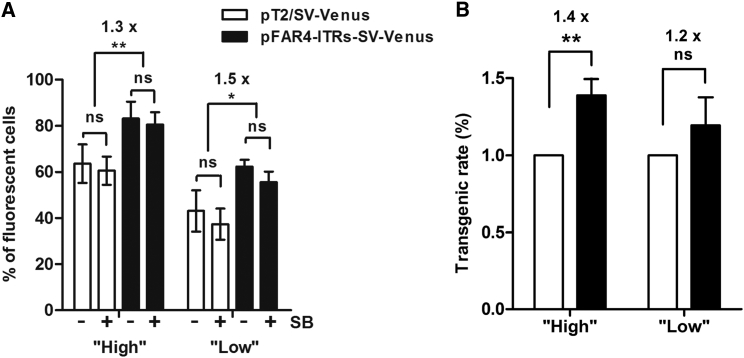
Because one of our goals is to take the pFAR4 mini-plasmid merged with the SB100X transposon system to the clinic, our next step was to assess the combination of both molecular tools in the absence of selection pressure for integration events. For this purpose, the neomycin resistance gene was substituted with a reporter gene encoding Venus, a yellow fluorescent protein, expressed from the SV40 promoter, resulting in two constructs, pFAR4-ITRs-SV-Venus (2,984 bp) and pT2/SV-Venus (4,878 bp). These two plasmid constructs allow assessment of the effect of plasmid size at two main time points reflecting the transfection and transposition events. For this study, HeLa cells were transfected with either pFAR4-ITRs-SV-Venus (300 or 50 ng) or an equimolar amount of pT2/SV-Venus (490 or 82 ng) in the presence or absence of transposase. Two days after transfection, in the presence of 10-fold less pFAR4-CMV-SB100X (30 or 5 ng), a higher percentage of fluorescent cells was observed when the cells were transfected with pFAR4-ITRs-SV-Venus (80.6% for 300 ng and 55.6% for 50 ng) than with an equimolar amount of pT2/SV-Venus (60.6% for 490 ng and 37.4% for 82 ng). Similarly, in the absence of transposase, the percentage of fluorescent cells dropped from 83.2% when the cells were transfected with 300 ng of pFAR4-ITRs-SV-Venus to 63.6% when using 490 ng of pT2/SV-Venus. When the cells were transfected with either 50 ng of pFAR4-ITRs-SV-Venus or 82 ng of pT2/SV-Venus, the percentage of fluorescent cells dropped from 62.3% to 43.1% (Figure 3A). Thus, the difference in size between the pFAR4 and pT2 gene vectors has primarily an effect on transfection efficiency.
Figure 3.
Comparative Analysis of Transfection Efficiency and Transgenic Rate Using the pFAR4 or the pT2 Plasmid to Encode the Venus Gene
HeLa cells were co-transfected with either a high or a low plasmid amount using the following constructs: pFAR4-ITRs-SV-Venus (2,984 bp, 300 or 50 ng) or an equimolar amount of pT2/SV-Venus (4,878 bp, 490 or 82 ng) either without (−SB) or plus SB100X transposase encoded by the pFAR4 plasmid (30 or 5 ng). Using the pGL3 empty vector, the total amount of plasmid was adjusted to 800 or 500 ng for the high or low conditions, respectively. (A) Data represent the mean number of fluorescent cells ± SD, obtained 2 days after six and four independent transfections for the high and low plasmid amounts, respectively. **p < 0.01, *p < 0.05; ns, not statistically different using Mann-Whitney test. (B) To assess transgenic rates, 2 days after transfection, 104 cells were seeded in plates, and the percentage of fluorescent cells was determined by fluorescence-activated cell sorting (FACS) after 5 passages. The transgenic rate in cells transfected with pFAR4-ITRs-SV-Venus was related to that obtained with pT2/SV-Venus, which was set to 1 (± SD). **p < 0.01; ns, not statistically different using Wilcoxon signed-rank test; n = 5 or 3 for the high or low plasmid amount conditions, respectively.
To assess the effect of plasmid size on transgene integration, transfected cells were then propagated for 3 weeks, and fluorescent cells were analyzed by flow cytometry. For both plasmid amounts and in the presence of transposase encoded by pFAR4, the percentage of fluorescent cells varied between 20% and 23% for cells transfected with pT2/SV-Venus and between 27% and 28% for cells transfected with pFAR4-ITRs-SV-Venus. In the absence of the SB100X transposase, the percentage of fluorescent cells was, for all conditions tested, less than 1.3%. When cells were transfected with the Venus gene carried by the pFAR4 plasmid, the number of fluorescent cells was 1.2-fold greater with low and 1.4-fold greater with a high plasmid amount than when the cells were transfected with the pT2/SV-Venus plasmid (Figure 3B). In this latter case, the difference between both plasmid constructs was statistically significant, with p < 0.01.
Thus, under either selective (such as in the presence of antibiotics) or non-selective conditions for transgenic cells, pFAR4 is an efficient vector for the delivery of the SB100X transposon system and the integration of transgenes into the host cell’s genome, a useful feature for the transfection of primary cells or cells sensitive to a high amount of plasmid DNA.
Primary Human RPE Cells a Secrete Higher PEDF Level When Transfected with pFAR4 Constructs
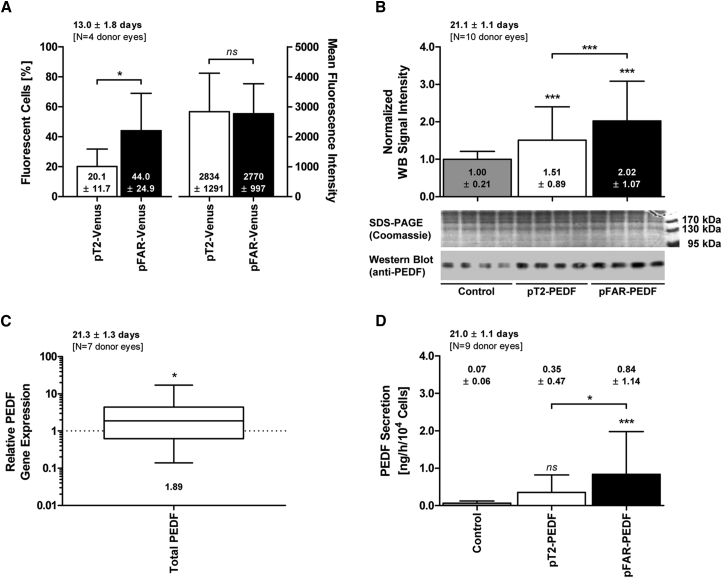
To determine whether the pFAR4 plasmid is an effective vector for the delivery of transgenes to primary somatic cells, we investigated the transfection of RPE cells, which we plan to use as a means to deliver therapeutic molecules to the subretinal space of patients suffering from neovascular AMD. In these experiments, electroporation was used to deliver the plasmids to the cells because lipofection of primary pigment epithelial cells has been shown to be poor.55 In all experiments with RPE cells, SB100X was encoded by the pFAR4 plasmid, and a transposase to transposon ratio of 1 to 16 was used. 13 days after transfection, human RPE cells isolated from 4 donor eyes and transfected with SB100X and the Venus gene carried by the pFAR4 plasmid showed an average of a 2.2-fold greater number of fluorescent cells compared with cells transfected with the transposase gene and the Venus gene encoded by pT2 (p < 0.03). However, the mean fluorescence intensity was similar, indicating that the pFAR4 vector mostly promoted a higher transfection rate (Figure 4A).
Figure 4.
Analysis of Transfection Efficiency, PEDF Expression, and Secretion by Primary Human RPE Cells 13 and 21 Days after Transfection
(A) Eight individual cultures of RPE cells isolated from 4 donor eyes (age, 74.0 ± 6.0 years; gender, 2 males and 2 females; time postmortem, 26.3 ± 3.9 hr; cultivation time before transfection, 48.8 ± 10.0 days) were transfected with 30 ng pFAR4-CMV SB100X SV40 transposase and either 470 ng pT2-ITRs CAGGS Venus (0.11 pmol) or 324 ng pFAR4-ITRs CAGGS Venus (0.11 pmol + 146 ng pFAR4/empty plasmid DNA) or without plasmid DNA as a control. Venus expression for 1 × 105 cells was analyzed by flow cytometry at 13.0 ± 1.8 days after transfection. Statistical analysis using an unpaired two-tailed t test showed a significantly greater number of fluorescent cells when transfected with pFAR4-Venus than when transfected with pT2-Venus (p = 0.0277). However, no difference in mean fluorescence intensity was noted. (B) PEDF secretion was analyzed by western blots of culture medium of 1 × 104 RPE cells isolated from 10 human donor eyes (age, 68.3 ± 14.8 years; gender, 6 males and 4 females; time postmortem, 29.5 ± 19.6 hr; cultivation time before transfection, 39.3 ± 22.3 days) and transfected with equimolar concentrations of pT2-PEDF (470 ng) or pFAR4-PEDF (313 ng + 157 ng pFAR4/empty plasmid DNA) in the presence of 30 ng of SB100X carried by pFAR4 or without plasmid DNA as a control. Culture supernatants of 72 individual control cultures, 116 individual pT2-PEDF cultures, and 114 individual pFAR4-PEDF cultures were analyzed for total PEDF secretion 21.1 ± 1.1 days after transfection. Loading of equal amounts of culture supernatants was proven by SDS-PAGE Coomassie G-250 staining. Using anti-PEDF antibodies, signal intensities obtained with cells transfected with either pFAR4-PEDF or pT2-PEDF were normalized to the signal intensities obtained with control cells electroporated without plasmid DNA. Total PEDF secretion from pT2-PEDF- or pFAR4-PEDF-transfected cells was significantly higher than from cells transfected without plasmid DNA (p ≤ 0.001), and pFAR4-PEDF-transfected cells secreted a significantly higher PEDF level than pT2-PEDF-transfected cells (p ≤ 0.001, one-way ANOVA with Tukey’s multiple comparisons test). (C) Relative PEDF gene expression was analyzed by qPCR for 1 × 104 RPE cells isolated from 7 donor eyes (age, 63.7 ± 15.5 years; gender, 5 males and 2 females; time postmortem, 32.1 ± 23.4 hr; cultivation time before transfection, 37.6 ± 17.8 days), transfected with equimolar concentrations of pT2-PEDF and pFAR4-PEDF (using the same conditions as described above), and cultured for 21.3 ± 1.3 days after transfection. Data for total (endogenous + recombinant) PEDF gene expression are presented as a box and whisker plot (whiskers, minimum to maximum). Total PEDF gene expression in pFAR4-PEDF-transfected cells was related to that obtained with pT2-PEDF-transfected cells, which was set to 1 (dashed line). The difference is statistically significant, p = 0.0406 (one sample t test), exhibiting a 1.89-fold increase. Transfection of RPE cells with pFAR4-PEDF allowed for a significant increase in total PEDF expression (p = 0.0333, unpaired two-tailed t test) compared with the endogenous PEDF level (data not shown). (D) Total PEDF secretion for 1 × 104 cells transfected with equimolar concentrations of pT2-PEDF and pFAR4-PEDF (using the same conditions as described in B) was quantified by ELISA using RPE cells isolated from 9 human donor eyes (age, 67.4 ± 15.4 years; gender, 6 males and 3 females; time postmortem, 30.1 ± 20.7 hours; cultivation time before transfection, 41.6 ± 22.5 days). Culture supernatants of 36 individual control cultures, 32 individual pT2-PEDF cultures, and 32 individual pFAR4-PEDF cultures were analyzed 21.0 ± 1.1 days after transfection. Total PEDF secretion of pT2-PEDF and pFAR4-PEDF transfected cells was compared with non-transfected control cells (p = not significant for pT2-PEDF-transfected cells and p ≤ 0.001 for pFAR4-PEDF-transfected cells), and total PEDF secretion of pT2-PEDF-transfected cells was compared with total PEDF secretion of pFAR4-PEDF-transfected cells (p ≤ 0.05, one-way ANOVA with Tukey’s multiple comparisons test). All data are expressed as mean ± SD.
PEDF secretion by human RPE cells isolated from 10 donor eyes and transfected with SB100X and the human PEDF gene expressed from the CMV promoter and carried by either a pT2 or pFAR4 plasmid was next analyzed by western blot. Three weeks after transfection, RPE cells transfected with pFAR4-PEDF showed an average 100% increase in secreted PEDF (p ≤ 0.001) compared with cells electroporated in the absence of any plasmid, whereas cells transfected with pT2-PEDF showed only an average 50% increase (p ≤ 0.001) (Figure 4B). Three weeks after transfection, PEDF expression was analyzed by quantitative PCR in human RPE cells isolated from seven donor eyes. Compared with endogenous PEDF expression, a significant increase in total (endogenous plus recombinant) PEDF expression was observed, regardless of whether the cells had been transfected with the pFAR4-PEDF or pT2-PEDF plasmids (data not shown). Notably, total PEDF expression was 1.9-fold higher in cells transfected with pFAR4-PEDF than in cells transfected with the PEDF gene carried by pT2 (p < 0.05) (Figure 4C). Finally, secreted PEDF was quantified by ELISA in medium of RPE cells isolated from nine human donor eyes. Three weeks after transfection, cells transfected with pFAR4-PEDF secreted 0.84 ± 1.14 ng of PEDF/hr/104 cells, whereas cells transfected with pT2-PEDF secreted 0.35 ± 0.47 ng of PEDF/hr/104cells. The culture medium of non-transfected cells contained 0.07 ± 0.06 ng of PEDF/hr/104cells (Figure 4D). Thus, transfection of human RPE cells with a pFAR4 encoding PEDF leads to a significant increase in PEDF expression, allowing for a 2.4-fold higher secreted PEDF level than that obtained with pT2-PEDF (p ≤ 0.05) and a 12-fold increase compared with non-transfected control cells (p ≤ 0.001).
Long-term secretion of PEDF by RPE cells was analyzed by immunoblotting for a period of 8 months. PEDF secretion, measured as mean normalized signal intensity, for pT2-PEDF-transfected cells was reduced by 27.8% from 51 to 240 days, whereas, for cells transfected with an equimolar amount of the pFAR4-PEDF plasmid, PEDF secretion increased by 42% (Figure 5A; Table S1). Comparison of secreted PEDF levels by RPE cells transfected with either the pFAR4-PEDF or the pT2-PEDF transposon plasmids showed a statistically significant difference 51 and 103 days after transfection, whereas, at later time points (on days 150, 197, and 240), the difference was no longer statistically significant, even though—referring to the samples of every individual RPE cell isolate—the averaged amounts secreted by the pFAR4-PEDF-transfected cell samples were 1.2- to 5.7-fold higher at 150 days, 1.3- to 18.1-fold higher at 197 days, and 1.2 to 26.7-fold higher at 240 days compared with the averaged amounts secreted by the respective pT2-PEDF-transfected cell samples (Figure 5A; Table S1). Furthermore, the proportion of isolates transfected with pFAR4-PEDF that secreted PEDF at levels superior to the highest level obtained with pT2-PEDF-transfected cells ranged between 25% and 40%.
Figure 5.
Analysis of Long-Term PEDF Expression and Secretion by Primary Human RPE Cells Transfected with SB100X Encoded by pFAR4 and PEDF Carried by Either the pFAR4 or pT2 Plasmid
(A) At the times indicated in the graph, PEDF secretion was analyzed by western blots of culture media of 1 × 104 RPE cells isolated from human donor eyes (numbers as indicated; the number of samples tested is listed in Table S1) transfected with equimolar concentrations of pT2-PEDF or pFAR4-PEDF (7.50 × 1010 plasmid copies) combined with 30 ng of SB100X encoded by the pFAR4 plasmid. The total plasmid amount (500 ng) was adjusted using pFAR4 empty vector. Signal intensities obtained with pT2-PEDF-transfected and pFAR4-PEDF-transfected cells were normalized to the signal intensities obtained with cells electroporated without plasmid DNA. Data are presented as mean ± SD. Statistical analyses were performed to compare level of PEDF secreted by cells transfected with plasmids to that of cells electroporated without plasmid and PEDF values obtained with pFAR4-PEDF-transfected cells versus those of pT2-PEDF cells using one-way ANOVA with Tukey’s multiple comparisons test. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001. See also Table S1. (B) Relative PEDF gene expression was analyzed by qPCR in 1 × 104 RPE cells isolated from 8 human donor eyes (age, 66.3 ± 16.0 years; gender, 5 males and 3 females; time postmortem, 31.1 ± 21.8 hr; cultivation time before transfection, 39.9 ± 23.4 days) and cultured for 180 ± 66 days after transfection with equimolar concentrations of pT2-PEDF and pFAR4-PEDF (as described in A). Data for total (endogenous + recombinant) PEDF gene expression are presented as a box and whisker plot (whiskers, minimum to maximum). PEDF gene expression in pFAR4-PEDF-transfected cells was related to that obtained with pT2-PEDF-transfected cells, which was set to 1 (dashed line). In cells transfected with pFAR4-PEDF, the total PEDF gene expression level was 2.2-fold higher than in cells transfected with pT2-PEDF. This difference is statistically significant, with p = 0.0123, using one-sample t test.
Long-term PEDF gene expression was analyzed in human RPE cells from eight donor eyes cultured for 180 ± 66 days after transfection. For pFAR4-PEDF-transfected cells, a 2.2-fold increase in total (endogenous plus recombinant) PEDF expression was observed compared with the total PEDF expression in pT2-PEDF-transfected cells (Figure 5B). This difference was statistically significant (p = 0.0123).
Discussion
Successful gene therapy requires effective and safe means of modifying a targeted cell or organ affected by a specific gene, whether the gene in question is mutated, silent, or expressed at an insufficient level. During the last two decades, several vectors have been developed to deliver genes to cells both in vivo and in vitro; however, many delivery vectors still have limitations related to efficiency or safety. Efficiency requires not only that the gene is successfully introduced into the cell at therapeutic levels but also that it will express the appropriate therapeutic molecule for the necessary period of time or for life, as may be required. Safety requires that the vector that delivers the gene will have no effect on the host cells and that, if it is required, the integration of the transgene into the host cell’s genome will not disrupt the expression of other genes or prompt the expression of genes that can cause harm to the host cell or the organism. Here we investigated the delivery of the Venus gene and the neomycin resistance gene in HeLa cells and of the PEDF gene in RPE cells using the SB100X transposase to integrate the genes into the host cell’s genome and compared the efficiency of delivery by the pT2 and pFAR4 plasmids. Although retroviral and lentiviral vectors and other transposons show a preference for integration into actively transcribed genes,31, 33, 56 the SB transposon displays a safe, close to random integration pattern in mammalian genomes and is less likely to integrate into transcribed genes or transcriptional regulatory regions.31, 32 The pFAR4 mini-plasmid is not only more effective, but it is also safer because it does not include an antibiotic resistance gene; in fact, both the European Medicines Agency (EMA)57 and the Food and Drug Administration (FDA)58 advise to avoid the use of plasmids that contain antibiotic resistance genes. In this study, it has been shown that pairing the hyperactive SB transposon system (SB100X) and the pFAR4 vector results in greater efficiency, as evidenced by an ∼2-fold increase in neomycin-resistant HeLa cell colonies, a 2.4-fold increase in PEDF secretion into the culture medium of cultured PEDF-transfected primary human RPE cells, as well as higher PEDF expression compared with RPE cells transfected with the pT2 standard plasmid. The increased efficiency of transfection probably results from a faster transit of the small pFAR4-PEDF construct (3,870 bp) through the host cytoplasm to the nucleus compared with the significantly larger pT2-PEDF construct (5,804 bp). In addition, the smaller size of the pFAR4 constructs may favor transposon excision59 because of the shorter distance between the transposon ITRs. Considering that less than 1%–2% of available intracellular transposons are excised from a plasmid60 (M.P. and C.M., unpublished data), the development of molecular tools that mediate more efficient transfection is an important advantage, which is particularly relevant when a limited number of cells is available; for cells, such as primary cells, which are difficult to transfect or when transfected cells are sensitive to high levels of plasmid.61 Therefore, the use of the pFAR4 plasmid paired with the SB100X transposase allows for greater gene expression, a greater transgene product level, and, more importantly, greater safety. Our results showing that an average of six neoR gene copies (range, 1–15) were integrated into the HeLa cell genome compare well with the copy number of chimeric antigen receptor (CAR) transgenes integrated into CD8+T cells (n = 5; range, 3–8) or CD4+T cells (n = 6; range, 3–8) transfected with a SB transposon expressing a CD19-specific CAR.61 Noteworthy is that the smaller size and the higher potency of small DNA vectors (pFAR4 and minicircles) used for the SB delivery did not challenge the nearly close to random integration profile of the SB transposon.61, 62 The superior safety feature of SB, compared with the Tol2 or piggyBac transposons, which tend to integrate into transcription start sites or transcriptional regulatory regions,31 promoted its translation to clinical trials.
Currently, in the United States, ten phase I clinical trials have been launched using the SB transposon. All ten trials are based on the genetic modification of T cells, nine for the treatment of B cell malignancies and one for the treatment of metastatic breast cancer (http://www.abedia.com/wiley/search.php). In Europe, a first-in-man clinical trial pairing the pFAR4 plasmid and the SB100X transposase is expected to be initiated in 2018 (http://www.targetAMD.eu). Here, we have shown that delivery and integration of a transgene are more efficient when the transgene and the transposase are encoded by pFAR4 plasmids. In addition, the safety and biological activity of the protocol have also been shown in animal studies.63 The transgene expression level is not only higher using the pFAR4 and SB100X pair, but it is also maintained over a long period of time. Here we have shown that primary human RPE cells transfected with the pFAR4-PEDF construct secrete sustained levels of PEDF into the cell culture medium for the 8 months the cells were followed, whereas only very low levels were secreted when the cells were transfected using the pT2-PEDF plasmid. It is interesting to note that, during the first 5 months, the difference between PEDF secreted by cells transfected using pFAR4 and pT2 was statistically significant; however, by 5 months, the difference was no longer statistically significant even though the average levels of PEDF were much higher for cells transfected with the pFAR4 plasmid. The discrepancy could be the result of uneven growth of the cells that were isolated from donors of different ages (45 to 84 years), at different postmortem times (18 to 66 hr) and of unknown/different health status. Still, the number of isolates transfected with pFAR4-PEDF that secrete PEDF at levels above the highest value obtained with pT2-PEDF-transfected cells ranged between 25% and 40%.
Thus, our results show that combining the pFAR4 plasmid and the SB transposon system for the delivery and integration of transgenes into the genome of eukaryotic cells using either lipofection or electroporation offers several advantages, such as increased transfection efficiency, decreased safety concerns, sustained gene expression, and reduced gene vector manufacturing costs compared with those of viral vectors. The manufacturing of the pFAR4 mini-plasmid family can be achieved in a growth medium free of thymidine and animal-derived components (C.M., unpublished data), allowing good manufacturing practices-compatible production, making this plasmid family an appealing tool for cell/gene therapy clinical applications.
Materials and Methods
Plasmid Constructs and Manipulation
pFAR4-ITRs is a derivative of the antibiotic-free pFAR4 vector26 that contains the ITR sequence elements extracted from pT2/BH (a gift from P. Hackett, Addgene plasmid 26556; Cambridge MA, USA). pFAR4-ITRs-SV-Neo (3,385 bp), pFAR4-ITRs-CAGGS-Venus (pFAR-Venus, 4,227 bp), and pFAR4-CMV-SB100X (3,094 bp) were constructed using standard cloning procedures and the following donor templates: pT2/SV-Neo (5,155 bp),51 pT2-CAGGS-Venus (pT2-Venus, 6,132 bp),29 and pCMV-CAT(T7)-SB100X (4,752 bp).29 The pT2/SV-Venus (4,878 bp), and pFAR4-ITRs-SV-Venus (2,984 bp) plasmids are pT2/BH and pFAR4-ITRs derivatives, respectively. The two plasmids contain an identical eukaryotic expression cassette composed of the SV40 promoter (amplified from pT2/SV-Neo51) and the Venus sequence (extracted from pT2-CAGGS-Venus29). The pFAR4-ITRs-CMV-PEDF-bovine growth hormone (BGH) (pFAR4-PEDF, 3,870 bp) and pT2/BH-CMV-PEDF-BGH (pT2-PEDF, 5,804 bp) constructs contain a human PEDF cDNA that was generated from ARPE-19 cells (ATCC CRL-2302) and regulatory sequences amplified from pT2-CMV-PEDF/EGFP.64 All pFAR4 derivatives were propagated using the dedicated bacterial strain (TM#47-9a) genetically modified for the production of antibiotic-free plasmids.26 All plasmids were purified using Endofree preparation plasmid kits (Macherey Nagel, Hoerdt, France).
Transfection of HeLa Cells
HeLa cells (ATCC, CCL-2, LGC Standards, Molsheim, France) were cultured in minimal essential medium (MEM) supplemented with GlutaMAX, fetal calf serum (FCS, 10%), streptomycin (100 μg/mL), penicillin (100 U/mL), non-essential amino acids, and pyruvate (1 mM). FCS was purchased from Sigma-Aldrich (Saint-Quentin Fallavier, France), and all other reagents were from Gibco (Life Technologies, Illkirch, France). 3 × 105 cells were transfected in 6-well plates using cationic lipid 2-{3-[bis-(3-amino-propyl)-amino]-propylamino}-N-ditetradecyl carbamoyl methyl-acetamide or di-miristyl aminopropyl aminopropyl (DMAPAP)65 in combination with DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Avanti Polar Lipids). After 24 hr, the transfection mixture was replaced with fresh medium. One day later, various numbers of cells (from 2,000 to 30,000 per well, depending on the transfection conditions) were transferred to selective medium supplemented with Geneticin (G-418, 800 μg/mL, InvivoGen, Toulouse, France). Neomycin-resistant clones were enumerated 11 days later, after staining with crystal violet (0.1% dissolved in water). For each experiment, co-transfection of cells with transposon and transposase plasmids was performed in quadruplicates. When required, the amount of plasmid was adjusted to the same final quantity using empty pFAR4 vector26 or pGL3-basic (Promega, Madison, WI, USA). For the “colony-forming assays,” transgenic rates correspond to transgene-expressing cells normalized to the number of seeded cells.
Isolation and Cultivation of Primary Human RPE Cells
Human eyes from 13 donors (age, 69.4 ± 13.3 years; 7 males and 6 females) were obtained from the Aachen Cornea Bank of the Department of Ophthalmology, University Hospital Rheinisch-Westfaelische Technische Hochschule (RWTH) Aachen. The eyes were removed 29.1 ± 17.0 hr postmortem, after informed consent was obtained, in accord with the Declaration of Helsinki. The procedures for the collection and use of human samples were approved by the institutional ethics committee. For RPE cell isolation, the anterior segment was removed by a circumferential cut approximately 3 mm posterior to the limbus. After careful removal of the vitreous and the retina, the posterior eyecup was filled with 1 mL DMEM/Ham’s F-12 (Biochrom, Berlin, Germany) supplemented with 10% fetal bovine serum (FBS; PAA Laboratories, Pasching, Austria), 80 U/mL penicillin and 80 μg/mL streptomycin (Lonza, Basel, Switzerland), and 2.5 μg/mL amphotericin B (Sigma-Aldrich, Taufkirchen, Germany). The cells were harvested by gently brushing the retinal pigment epithelium with a fire-polished glass spatula. The procedure was repeated once. After the eyecup was rinsed with 1 mL DMEM/Ham’s F-12 supplemented with 10% FBS, 80 U/mL penicillin, 80 μg/mL streptomycin, and 2.5 μg/mL amphotericin B, the cell mixture was plated into 3 wells of a 24-well tissue culture plate. Cultures were maintained at 37°C in a humidified atmosphere of 95% air and 5% CO2 until confluence was reached. The cell culture medium was changed twice a week.
Electroporation of Primary Human RPE Cells and Cultivation of the Transfected Cells
Transfections were performed with the Neon transfection system using the 10 μL kit (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s protocol. The electroporation parameters were as follows: 2 pulses, 1,100 V (pulse voltage), and 20 ms (pulse width). 1 × 105 or 1 × 104 cells in 11 μL resuspension buffer R (Thermo Fisher Scientific) were combined with 2 μL of purified plasmid mixture containing 30 ng of pFAR4-CMV-SB100X transposase plasmid and either 470 ng of pT2-CAGGS-Venus, 470 ng of pT2/BH-CMV-PEDF-BGH, 324 ng of pFAR4-ITRs-CAGGS-Venus, or 313 ng of pFAR4-ITRs-CMV-PEDF-BGH, aiming at transfecting cells with equimolar amount of Venus and PEDF transgenes. Empty pFAR4 vector26 was used to reach the final plasmid amount of 500 ng. For analysis of Venus transgene expression, each experiment comprised 2 transfection controls with electrical field application without plasmid DNA and 2 transfections with electrical field application and plasmid DNA for each of the pT2 and the pFAR4 vector. For analysis of PEDF transgene expression, each experiment comprised 4 transfection controls without electrical field application and without plasmid DNA, 6 transfection controls with electrical field application without plasmid DNA, and 10 transfections with electrical field application and plasmid DNA for each of the pT2 and the pFAR4 vector. Transfected cells were transferred into 24-well tissue culture plates (for 1 × 105 cells) or 48-well tissue culture plates (for 1 × 104 cells) containing 1.0 mL or 0.5 mL of DMEM/Ham’s F-12 supplemented with 10% FBS without antibiotics or antimycotics. Penicillin (80 U/mL), streptomycin (80 μg/mL), and amphotericin B (2.5 μg/mL) were added with the first medium exchange 3 days after electroporation. Cell cultures were either used after 2 or 3 weeks for further analyses or maintained for analysis of long-term PEDF expression and secretion.
SDS-PAGE and Western Blot Analysis
For SDS-PAGE, 15 μL of culture supernatant was mixed with an equal volume of 2× SDS sample buffer66 and heated for 5 min at 95°C. Proteins were separated on a 10% SDS-polyacrylamide gel and transferred onto a 0.45-μm pore size nitrocellulose membrane (Whatman International, Maidstone, UK) using the semi-dry transfer system (Bio-Rad, Hercules, CA), followed by Ponceau S staining to confirm the transfer. SDS-polyacrylamide gels that served as a loading control were stained with GelCode blue stain reagent (Thermo Fisher Scientific) according to the recommendations of the manufacturer. For the detection of total (endogenous + recombinant) PEDF, blots were blocked with 3% BSA/Tris-buffered saline (TBS) for 2 hr at room temperature, incubated for 1 hr at room temperature and overnight at 4°C with anti-PEDF antibodies (rabbit polyclonal, 1:4,000 diluted in 3% BSA/TBS; BioProducts MD, Middletown, MD), followed by incubation for 1 hr at room temperature with horseradish peroxidase-conjugated anti-rabbit antibodies (goat polyclonal, 1:2,000 diluted in 10% milk powder/TBS; Abcam, Cambridge, UK). Protein bands were visualized by chemiluminescence using the LAS-3000 imaging system (FujiFilm, Tokyo, Japan) and evaluated by the open source image processing program ImageJ (W.S. Rasband, NIH, Bethesda, MD, USA; https://imagej.nih.gov/ij/; 1997–2014).
ELISA-Based Quantification of Total PEDF Secretion
Total PEDF secretion was analyzed in culture supernatants of control and PEDF-transfected cells 3 weeks after transfection. Cells were incubated for 24 hr in 0.5 mL of culture medium, from which PEDF was quantified using the ELISAquant kit (BioProducts MD) according to the manufacturer’s protocol and correlated with cell number. Cells were counted using the CASY Cell Counter Model TT (Roche Diagnostics, Mannheim, Germany) after being trypsinized with 0.05% trypsin-0.02% EDTA (PAA Laboratories).
Flow Cytometry
Trypsinized HeLa cells were fixed for 5 min in 1% formaldehyde diluted in PBS and subsequently washed twice in PBS. Fluorescence was quantified using a flow cytometer (Guava easyCyte, Merck Millipore). Fluorescent cells were sorted using either the ARIAIII or FACSJAZZ cell sorters (BD Biosciences, Le Pont de Claix, France). RPE cell monolayers were trypsinized with 0.05% trypsin-0.02% EDTA (PAA Laboratories), washed three times in PBS, suspended in 200 μL PBS, and analyzed using a FACSCalibur (Becton Dickinson, Heidelberg, Germany).
Quantification of Transposon Insertions
Individual NeoR clones obtained after three independent transfections were selected in Geneticin-containing medium for 11 days and passaged, on average, 7 times. Genomic DNA was prepared using the DNeasy blood and tissue kit (QIAGEN, Courtaboeuf, France) according to the manufacturer’s instructions. NeoR gene copy number was determined by qPCR in 384-well plates, each well containing 20 ng of genomic DNA in a final volume of 10 μL, using the TaqMan Gene Expression Master Mix and TaqMan Copy Number Assay made to order (MTO) kits for neomycin resistance gene amplification (Applied Biosystems, Life Technologies, Courtaboeuf, France). Results were normalized by amplifying the RPPH1 gene, which is present as a single copy in the human genome,67 using the TaqMan Copy Number Reference Assay Human RNase P kit (Applied Biosystems). Primers and probes of undisclosed sequences are provided with the kits. Fluorescence measurements were carried out using an ABIPRISM-7900HT sequence detection system (Applied Biosystems) using the following conditions: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles (95°C for 15 s and 60°C for 60 s). Melting curve analysis confirmed the amplification specificity of each primer pair. Data were processed with the ABI-PRISM-7900HT SDS software.
Reverse Transcription and qPCR
Total RNA was isolated using the RNeasy Mini Kit together with the RNase-free DNase set (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol. Reverse transcription was carried out on 0.1 μg total RNA using a reverse transcription system (Promega, Madison, WI). Real-time qPCR reactions were performed on a LightCycler 1.2 instrument using the LightCycler FastStart DNA Master SYBR Green I kit (Roche, Mannheim, Germany) according to the manufacturer’s recommendations. The cDNA samples were run in duplicate using the following primers for detection of the endogenous PEDF gene plus the PEDF gene encoded by the pT2 and the pFAR4 plasmids: GAPDH as the internal control gene (forward [F], 5′-ATC CCA TCA CCA TCT TCC AG-3′; reverse [R], 5′-ATG AGT CCT TCC ACG ATA CC-3′), endogenous PEDF (F, 5′-GCT GGC TTT GAG TGG AAC GA-3′; R, 5′-GTG TCC TGT GGA ATC TGC TG-3′), and endogenous plus recombinant PEDF (F, 5′-CCT GCA GGA GAT GAA GCT GCA-3′; R, 5′-TCC ACC TGA GTC AGC TTG ATG-3′). Reactions were performed with diluted cDNA corresponding to 2 ng of initially used total RNA and a primer concentration of 0.25 μM. Thermal cycler conditions were as follows: initial denaturation at 95°C for 10 min, followed by 50 cycles with denaturation at 95°C for 10 s, annealing at 60°C for 8 s, and elongation at 72°C for 15 s. Melting curve analysis confirmed the amplification specificity of each primer pair. Data were processed with LightCycler software 3.5.3 and evaluated using the comparative CT (2−ΔΔCT) method, which describes relative gene expression.68
Statistical Analysis
Statistical analyses were performed using the tests indicated in the figure legends and GraphPad Prism software version 5.04 (GraphPad, La Jolla, CA).
Author Contributions
Conceptualization, C.M., G.T., S.J., and Z. Izsvák; Investigation, J.P., M.P., M.Q., and N.H.; Resources, C.M., P.W., and Z. Izsvák; Writing – Original Draft, C.M. and S.J.; Writing – Review & Editing, M.P., S.J., N.H., M.Q., J.P., M.K., P.W., Z. Ivics, Z. Izsvák, G.T., D.S., and C.M.; Funding Acquisition, G.T.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
The project has received funding from the European Union’s Seventh Framework Programme for research, technological development and demonstration under grant agreement no. 305134. Anna Dobias, Antje Schiefer (Department of Ophthalmology, University Hospital RWTH Aachen), and Gregg Sealy (Laboratory of Ophthalmology, University of Geneva) are acknowledged for excellent technical support. The Aachen Cornea Bank (Department of Ophthalmology, University Hospital RWTH Aachen) is acknowledged for providing the human donor eyes. P. Hackett (University of Minnesota, USA) is acknowledged for the gift of pT2/BH.
Footnotes
Supplemental Information includes one table and can be found with this article online at https://doi.org/10.1016/j.omtn.2017.12.017.
Supplemental Information
References
- 1.Naldini L. Gene therapy returns to centre stage. Nature. 2015;526:351–360. doi: 10.1038/nature15818. [DOI] [PubMed] [Google Scholar]
- 2.Fischer A., Hacein-Bey-Abina S., Cavazzana-Calvo M. 20 years of gene therapy for SCID. Nat. Immunol. 2010;11:457–460. doi: 10.1038/ni0610-457. [DOI] [PubMed] [Google Scholar]
- 3.Yin H., Kanasty R.L., Eltoukhy A.A., Vegas A.J., Dorkin J.R., Anderson D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014;15:541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 4.Alsaggar M., Liu D. Physical methods for gene transfer. Adv. Genet. 2015;89:1–24. doi: 10.1016/bs.adgen.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Heller R., Heller L.C. Gene electrotransfer clinical trials. Adv. Genet. 2015;89:235–262. doi: 10.1016/bs.adgen.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Young J.L., Dean D.A. Electroporation-mediated gene delivery. Adv. Genet. 2015;89:49–88. doi: 10.1016/bs.adgen.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mairhofer J., Grabherr R. Rational vector design for efficient non-viral gene delivery: challenges facing the use of plasmid DNA. Mol. Biotechnol. 2008;39:97–104. doi: 10.1007/s12033-008-9046-7. [DOI] [PubMed] [Google Scholar]
- 8.Vandermeulen G., Marie C., Scherman D., Préat V. New generation of plasmid backbones devoid of antibiotic resistance marker for gene therapy trials. Mol. Ther. 2011;19:1942–1949. doi: 10.1038/mt.2011.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams J.A., Carnes A.E., Hodgson C.P. Plasmid DNA vaccine vector design: impact on efficacy, safety and upstream production. Biotechnol. Adv. 2009;27:353–370. doi: 10.1016/j.biotechadv.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feschotte C., Pritham E.J. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivics Z., Li M.A., Mátés L., Boeke J.D., Nagy A., Bradley A., Izsvák Z. Transposon-mediated genome manipulation in vertebrates. Nat. Methods. 2009;6:415–422. doi: 10.1038/nmeth.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Izsvák Z., Chuah M.K., Vandendriessche T., Ivics Z. Efficient stable gene transfer into human cells by the Sleeping Beauty transposon vectors. Methods. 2009;49:287–297. doi: 10.1016/j.ymeth.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Solensky R. Hypersensitivity reactions to beta-lactam antibiotics. Clin. Rev. Allergy Immunol. 2003;24:201–220. doi: 10.1385/CRIAI:24:3:201. [DOI] [PubMed] [Google Scholar]
- 14.Bloquel C., Fabre E., Bureau M.F., Scherman D. Plasmid DNA electrotransfer for intracellular and secreted proteins expression: new methodological developments and applications. J. Gene Med. 2004;6(Suppl 1):S11–S23. doi: 10.1002/jgm.508. [DOI] [PubMed] [Google Scholar]
- 15.Kreiss P., Cameron B., Rangara R., Mailhe P., Aguerre-Charriol O., Airiau M., Scherman D., Crouzet J., Pitard B. Plasmid DNA size does not affect the physicochemical properties of lipoplexes but modulates gene transfer efficiency. Nucleic Acids Res. 1999;27:3792–3798. doi: 10.1093/nar/27.19.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chabot S., Orio J., Schmeer M., Schleef M., Golzio M., Teissié J. Minicircle DNA electrotransfer for efficient tissue-targeted gene delivery. Gene Ther. 2013;20:62–68. doi: 10.1038/gt.2011.215. [DOI] [PubMed] [Google Scholar]
- 17.Kreiss P., Cameron B., Darquet A.M., Scherman D., Crouzet J. Production of a new DNA vehicle for gene transfer using site-specific recombination. Appl. Microbiol. Biotechnol. 1998;49:560–567. doi: 10.1007/s002530051213. [DOI] [PubMed] [Google Scholar]
- 18.Chen Z.-Y., He C.-Y., Kay M.A. Improved production and purification of minicircle DNA vector free of plasmid bacterial sequences and capable of persistent transgene expression in vivo. Hum. Gene Ther. 2005;16:126–131. doi: 10.1089/hum.2005.16.126. [DOI] [PubMed] [Google Scholar]
- 19.Cranenburgh R.M., Hanak J.A.J., Williams S.G., Sherratt D.J. Escherichia coli strains that allow antibiotic-free plasmid selection and maintenance by repressor titration. Nucleic Acids Res. 2001;29:E26. doi: 10.1093/nar/29.5.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cranenburgh R.M., Lewis K.S., Hanak J.A.J. Effect of plasmid copy number and lac operator sequence on antibiotic-free plasmid selection by operator-repressor titration in Escherichia coli. J. Mol. Microbiol. Biotechnol. 2004;7:197–203. doi: 10.1159/000079828. [DOI] [PubMed] [Google Scholar]
- 21.Soubrier F., Cameron B., Manse B., Somarriba S., Dubertret C., Jaslin G., Jung G., Caer C.L., Dang D., Mouvault J.M. pCOR: a new design of plasmid vectors for nonviral gene therapy. Gene Ther. 1999;6:1482–1488. doi: 10.1038/sj.gt.3300968. [DOI] [PubMed] [Google Scholar]
- 22.Soubrier F., Laborderie B., Cameron B. Improvement of pCOR plasmid copy number for pharmaceutical applications. Appl. Microbiol. Biotechnol. 2005;66:683–688. doi: 10.1007/s00253-004-1729-9. [DOI] [PubMed] [Google Scholar]
- 23.Pfaffenzeller I., Mairhofer J., Striedner G., Bayer K., Grabherr R. Using ColE1-derived RNA I for suppression of a bacterially encoded gene: implication for a novel plasmid addiction system. Biotechnol. J. 2006;1:675–681. doi: 10.1002/biot.200600017. [DOI] [PubMed] [Google Scholar]
- 24.Mairhofer J., Pfaffenzeller I., Merz D., Grabherr R. A novel antibiotic free plasmid selection system: advances in safe and efficient DNA therapy. Biotechnol. J. 2008;3:83–89. doi: 10.1002/biot.200700141. [DOI] [PubMed] [Google Scholar]
- 25.Luke J., Carnes A.E., Hodgson C.P., Williams J.A. Improved antibiotic-free DNA vaccine vectors utilizing a novel RNA based plasmid selection system. Vaccine. 2009;27:6454–6459. doi: 10.1016/j.vaccine.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marie C., Vandermeulen G., Quiviger M., Richard M., Préat V., Scherman D. pFARs, plasmids free of antibiotic resistance markers, display high-level transgene expression in muscle, skin and tumour cells. J. Gene Med. 2010;12:323–332. doi: 10.1002/jgm.1441. [DOI] [PubMed] [Google Scholar]
- 27.Quiviger M., Arfi A., Mansard D., Delacotte L., Pastor M., Scherman D., Marie C. High and prolonged sulfamidase secretion by the liver of MPS-IIIA mice following hydrodynamic tail vein delivery of antibiotic-free pFAR4 plasmid vector. Gene Ther. 2014;21:1001–1007. doi: 10.1038/gt.2014.75. [DOI] [PubMed] [Google Scholar]
- 28.Ivics Z., Hackett P.B., Plasterk R.H., Izsvák Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 29.Mátés L., Chuah M.K., Belay E., Jerchow B., Manoj N., Acosta-Sanchez A., Grzela D.P., Schmitt A., Becker K., Matrai J. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 30.Hudecek M., Izsvák Z., Johnen S., Renner M., Thumann G., Ivics Z. Going non-viral: the Sleeping Beauty transposon system breaks on through to the clinical side. Crit. Rev. Biochem. Mol. Biol. 2017;52:355–380. doi: 10.1080/10409238.2017.1304354. [DOI] [PubMed] [Google Scholar]
- 31.Narayanavari S.A., Chilkunda S.S., Ivics Z., Izsvák Z. Sleeping Beauty transposition: from biology to applications. Crit. Rev. Biochem. Mol. Biol. 2017;52:18–44. doi: 10.1080/10409238.2016.1237935. [DOI] [PubMed] [Google Scholar]
- 32.Yant S.R., Wu X., Huang Y., Garrison B., Burgess S.M., Kay M.A. High-resolution genome-wide mapping of transposon integration in mammals. Mol. Cell. Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gogol-Döring A., Ammar I., Gupta S., Bunse M., Miskey C., Chen W., Uckert W., Schulz T.F., Izsvák Z., Ivics Z. Genome-wide profiling reveals remarkable parallels between insertion site selection properties of the MLV retrovirus and the piggyBac transposon in primary human CD4(+) T cells. Mol. Ther. 2016;24:592–606. doi: 10.1038/mt.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grabundzija I., Irgang M., Mátés L., Belay E., Matrai J., Gogol-Döring A., Kawakami K., Chen W., Ruiz P., Chuah M.K. Comparative analysis of transposable element vector systems in human cells. Mol. Ther. 2010;18:1200–1209. doi: 10.1038/mt.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hackett P.B., Ekker S.C., Largaespada D.A., McIvor R.S. Sleeping beauty transposon-mediated gene therapy for prolonged expression. Adv. Genet. 2005;54:189–232. doi: 10.1016/S0065-2660(05)54009-4. [DOI] [PubMed] [Google Scholar]
- 36.Michalska-Małecka K., Kabiesz A., Nowak M., Śpiewak D. Age related macular degeneration - Challenge for future: Pathogenesis and new perspectives for the treatment. Eur. Geriatr. Med. 2015;6:69–75. [Google Scholar]
- 37.Mehta S. Age-related macular degeneration. Prim. Care. 2015;42:377–391. doi: 10.1016/j.pop.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Velez-Montoya R., Oliver S.C., Olson J.L., Fine S.L., Quiroz-Mercado H., Mandava N. Current knowledge and trends in age-related macular degeneration: genetics, epidemiology, and prevention. Retina. 2014;34:423–441. doi: 10.1097/IAE.0000000000000036. [DOI] [PubMed] [Google Scholar]
- 39.Kolomeyer A.M., Sugino I.K., Zarbin M.A. Characterization of conditioned media collected from aged versus young human eye cups. Invest. Ophthalmol. Vis. Sci. 2011;52:5963–5972. doi: 10.1167/iovs.10-6440. [DOI] [PubMed] [Google Scholar]
- 40.Kwak N., Okamoto N., Wood J.M., Campochiaro P.A. VEGF is major stimulator in model of choroidal neovascularization. Invest. Ophthalmol. Vis. Sci. 2000;41:3158–3164. [PubMed] [Google Scholar]
- 41.Pieramici D.J., Avery R.L. Ranibizumab: treatment in patients with neovascular age-related macular degeneration. Expert Opin. Biol. Ther. 2006;6:1237–1245. doi: 10.1517/14712598.6.11.1237. [DOI] [PubMed] [Google Scholar]
- 42.Rosenfeld P.J., Rich R.M., Lalwani G.A. Ranibizumab: Phase III clinical trial results. Ophthalmol. Clin. North Am. 2006;19:361–372. doi: 10.1016/j.ohc.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 43.Adelman R.A., Zheng Q., Mayer H.R. Persistent ocular hypertension following intravitreal bevacizumab and ranibizumab injections. J. Ocul. Pharmacol. Ther. 2010;26:105–110. doi: 10.1089/jop.2009.0076. [DOI] [PubMed] [Google Scholar]
- 44.Krishnan R., Goverdhan S., Lochhead J. Submacular haemorrhage after intravitreal bevacizumab compared with intravitreal ranibizumab in large occult choroidal neovascularization. Clin. Experiment. Ophthalmol. 2009;37:384–388. doi: 10.1111/j.1442-9071.2009.02043.x. [DOI] [PubMed] [Google Scholar]
- 45.Carneiro A.M., Barthelmes D., Falcão M.S., Mendonça L.S., Fonseca S.L., Gonçalves R.M., Faria-Correia F., Falcão-Reis F.M. Arterial thromboembolic events in patients with exudative age-related macular degeneration treated with intravitreal bevacizumab or ranibizumab. Ophthalmologica. 2011;225:211–221. doi: 10.1159/000323943. [DOI] [PubMed] [Google Scholar]
- 46.Campochiaro P.A., Nguyen Q.D., Shah S.M., Klein M.L., Holz E., Frank R.N., Saperstein D.A., Gupta A., Stout J.T., Macko J. Adenoviral vector-delivered pigment epithelium-derived factor for neovascular age-related macular degeneration: results of a phase I clinical trial. Hum. Gene Ther. 2006;17:167–176. doi: 10.1089/hum.2006.17.167. [DOI] [PubMed] [Google Scholar]
- 47.Campochiaro P.A. Gene transfer for neovascular age-related macular degeneration. Hum. Gene Ther. 2011;22:523–529. doi: 10.1089/hum.2011.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnen S., Kazanskaya O., Armogan N., Stickelmann C., Stöcker M., Walter P., Thumann G. Endogenic regulation of proliferation and zinc transporters by pigment epithelial cells nonvirally transfected with PEDF. Invest. Ophthalmol. Vis. Sci. 2011;52:5400–5407. doi: 10.1167/iovs.10-6178. [DOI] [PubMed] [Google Scholar]
- 49.Kuerten D., Johnen S., Harmening N., Souteyrand G., Walter P., Thumann G. Transplantation of PEDF-transfected pigment epithelial cells inhibits corneal neovascularization in a rabbit model. Graefes Arch. Clin. Exp. Ophthalmol. 2015;253:1061–1069. doi: 10.1007/s00417-015-2954-x. [DOI] [PubMed] [Google Scholar]
- 50.Johnen S., Djalali-Talab Y., Kazanskaya O., Möller T., Harmening N., Kropp M., Izsvák Z., Walter P., Thumann G. Antiangiogenic and Neurogenic Activities of Sleeping Beauty-Mediated PEDF-Transfected RPE Cells In Vitro and In Vivo. BioMed Res. Int. 2015;2015:863845. doi: 10.1155/2015/863845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cui Z., Geurts A.M., Liu G., Kaufman C.D., Hackett P.B. Structure-function analysis of the inverted terminal repeats of the sleeping beauty transposon. J. Mol. Biol. 2002;318:1221–1235. doi: 10.1016/s0022-2836(02)00237-1. [DOI] [PubMed] [Google Scholar]
- 52.Geurts A.M., Yang Y., Clark K.J., Liu G., Cui Z., Dupuy A.J., Bell J.B., Largaespada D.A., Hackett P.B. Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol. Ther. 2003;8:108–117. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 53.Hackett P.B. Integrating DNA vectors for gene therapy. Mol. Ther. 2007;15:10–12. doi: 10.1038/sj.mt.6300065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mikkelsen J.G., Yant S.R., Meuse L., Huang Z., Xu H., Kay M.A. Helper-Independent Sleeping Beauty transposon-transposase vectors for efficient nonviral gene delivery and persistent gene expression in vivo. Mol. Ther. 2003;8:654–665. doi: 10.1016/s1525-0016(03)00216-8. [DOI] [PubMed] [Google Scholar]
- 55.Thumann G., Stöcker M., Maltusch C., Salz A.K., Barth S., Walter P., Johnen S. High efficiency non-viral transfection of retinal and iris pigment epithelial cells with pigment epithelium-derived factor. Gene Ther. 2010;17:181–189. doi: 10.1038/gt.2009.124. [DOI] [PubMed] [Google Scholar]
- 56.Wilson M.H., Coates C.J., George A.L., Jr. PiggyBac transposon-mediated gene transfer in human cells. Mol. Ther. 2007;15:139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- 57.European Medicines Agency (2001). Note for Guidance on the Quality, Preclinical and Clinical Aspects of Gene Transfer Medicinal Products. https://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2009/10/WC500003977.pdf.
- 58.Food and Drug Administration (1998). Guidance for human somatic cell therapy and gene therapy. http://www.fda.gov/downloads/BiologicsBloodVaccines/GuidanceComplianceRegulatoryInformation/Guidances/CellularandGeneTherapy/ucm081670.pdf
- 59.Izsvák Z., Ivics Z., Plasterk R.H. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J. Mol. Biol. 2000;302:93–102. doi: 10.1006/jmbi.2000.4047. [DOI] [PubMed] [Google Scholar]
- 60.Bell J.B., Aronovich E.L., Schreifels J.M., Beadnell T.C., Hackett P.B. Duration of expression and activity of Sleeping Beauty transposase in mouse liver following hydrodynamic DNA delivery. Mol. Ther. 2010;18:1796–1802. doi: 10.1038/mt.2010.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Monjezi R., Miskey C., Gogishvili T., Schleef M., Schmeer M., Einsele H., Ivics Z., Hudecek M. Enhanced CAR T-cell engineering using non-viral Sleeping Beauty transposition from minicircle vectors. Leukemia. 2017;31:186–194. doi: 10.1038/leu.2016.180. [DOI] [PubMed] [Google Scholar]
- 62.Thumann G., Harmening N., Prat-Souteyrand C., Marie C., Pastor M., Sebe A., Miskey C., Hurst L.D., Diarra S., Kropp M. Engineering of PEDF-Expressing Primary Pigment Epithelial Cells by the SB Transposon System Delivered by pFAR4 Plasmids. Mol. Ther. Nucleic Acids. 2017;6:302–314. doi: 10.1016/j.omtn.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garcia-Garcia L., Recalde S., Hernandez M., Bezunartea J., Rodriguez-Madoz J., Johnen S., Diarra S., Marie C., Izsvák Z., Ivics Z. Long-term PEDF release in rat iris and retinal epithelial cells after Sleeping Beauty Transposon-mediated gene delivery. Mol. Ther. Nucleic Acids. 2017;9:1–11. doi: 10.1016/j.omtn.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnen S., Izsvák Z., Stöcker M., Harmening N., Salz A.K., Walter P., Thumann G. Sleeping Beauty transposon-mediated transfection of retinal and iris pigment epithelial cells. Invest. Ophthalmol. Vis. Sci. 2012;53:4787–4796. doi: 10.1167/iovs.12-9951. [DOI] [PubMed] [Google Scholar]
- 65.Byk, G., Scherman, D., Schwartz, B., and Dubertret, C. (2001). Lipopolyamines as transfection agents and pharmaceutical uses thereof. US patent 6171612 B1, filed November 8, 1996, and published January 9, 2001.
- 66.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 67.Baer M., Nilsen T.W., Costigan C., Altman S. Structure and transcription of a human gene for H1 RNA, the RNA component of human RNase P. Nucleic Acids Res. 1990;18:97–103. doi: 10.1093/nar/18.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.