Abstract
The aberrant activation of STAT3 occurs in many human cancers and promotes tumor progression. Phosphorylation of a tyrosine at amino acid Y705 is essential for the function of STAT3. Synthesized carbazole derived with fluorophore compound 12 was discovered to target STAT3 phosphorylation. Compound 12 was found to inhibit STAT3-mediated transcription as well as to reduce IL-6 induced STAT3 phosphorylation in cancer cell lines expressing both elevated and low levels of phospho-STAT3 (Y705). Compound 12 potently induced apoptosis in a broad number of TNBC cancer cell lines in vitro and was effective at inhibiting the in vivo growth of human TNBC xenograft tumors (SUM149) without any observed toxicity. Compound 12 also effectively inhibited the growth of human lung tumor xenografts (A549) harboring aberrantly active STAT3. In vitro and in vivo studies showed that the inhibitory effects of 12 on phospho-STAT3 were through up-regulation of the protein–tyrosine phosphatase PTPN6. Our present studies strongly support the continued preclinical evaluation of compound 12 as a potential chemotherapeutic agent for TNBC and cancers with constitutive STAT3 signaling.
Graphical abstract
INTRODUCTION
Current emphasis in cancer drug development has shifted from nonspecific cytotoxic chemotherapies to molecularly targeted reagents.1 The etiology of cancer is multimodal, but great progress has been made in recent times toward the understanding of these diseases. Discovery of oncogenes2 and transcription factors3,4 that play the critical roles in regulating gene expression with the development of cancers has aided in the development of new approaches for the cancer treatment. Signal transducers and activators of transcription 3 (STAT 3),5–7 one of seven known members of the mammalian STAT family, regulates transcription of several genes upon activation of membrane receptors by various cytokines.8 Ligand binding to cytokine receptors triggers activation of cytokine receptor associated kinases such as Janus kinases (JAKs) or receptor tyrosine kinases.8 Activation of STAT3 is achieved by phosphorylation of Y705 residue by these cytokine receptor associated kinases.8 Phosphorylated STAT3 translocates into the nucleus to bind STAT3 responsive elements and regulates expression of target genes.7 Phosphorylation of STAT3 is also regulated by protein tyrosine phosphatases such as tyrosine–protein phosphatase nonreceptor type 6 (PTPN6/SHP-1), tyrosine–protein phosphatase nonreceptor type 11 (PTPN11/SHP-2), and T-cell protein–tyrosine phosphatase (TC-PTP).9
Constitutive STAT3 activation has been detected in a large number of human cancers, and elevated levels of the activated STAT3 are associated with a poor clinical prognosis.10,11 Studies showed that the inhibition of STAT3 expression attenuated the proliferation and survival of STAT3-positive cancer cell lines in vitro and in vivo, while normal tissues were unaffected by loss of STAT3.5,12–14 Furthermore, the causal role of STAT3 in oncogenesis has been confirmed via molecular biology and pharmacology tools in disease-relevant models.15 All of these support STAT3 as a molecular target for cancer drug discovery,9,15–17 however, only one STAT3 inhibitor has been advanced into clinical trials18 because of the physicochemical problems of the studied molecules.16 To address those problems that are an obstacle to the STAT3 inhibitors’ being promoted into clinical trials, further studies have been carried out in the field recently and many advances have been achieved as the results.12,19–22
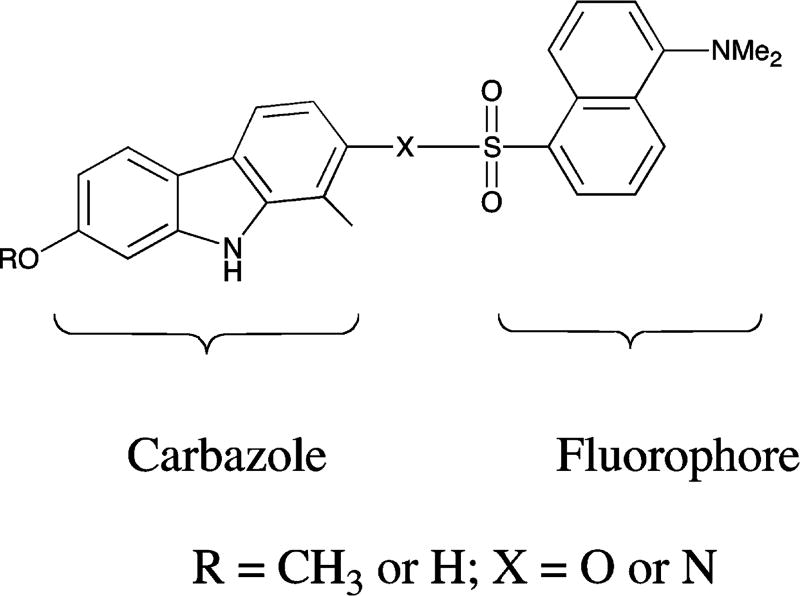
A wide range of synthetic analogues based on natural products have emerged as therapeutically useful antitumor agents.23 Among these natural products, natural and synthetic congeners of the carbazole alkaloids comprise a large group of therapeutically useful antitumor agents in the various stages of clinical development.24–26 Although earlier reports27,28 have demonstrated that some of carbazoles negatively regulate the STAT3-mediated transcription and/or STAT3-DNA binding and phospho-STAT3 with relatively high concentration (30–50 µM), the mechanisms of these regulations still remain unknown. We have been interested in targeting STAT3 pathways.29,30 In an effort to discover targeted therapeutics to treat cancers, we have developed fluorescent small molecules, which could be observed directly as it interacts with receptor-positive cell lines, as potential anticancer agents.31–33 Meanwhile, our previously studies34 showed that the antiproliferative effect on the cancer cell lines of the free carbazole is much less than the corresponding scaffolds with dansyl group. Herein, novel imageable carbazoles (Figure 1) were designed and synthesized, and their biological activities as STAT3 inhibitors were further evaluated.
Figure 1.
Novel carbazoles fluorophore.
RESULTS
Chemistry
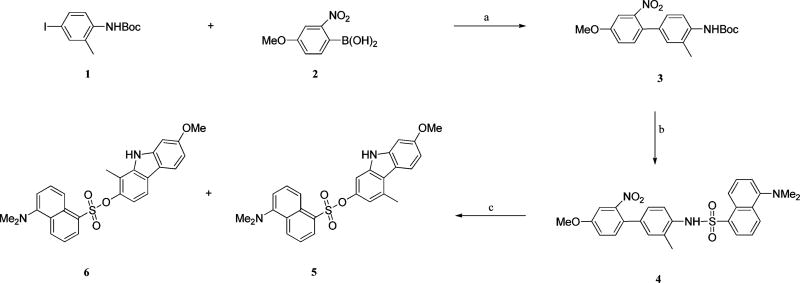
A palladium-catalyzed cross-coupling reaction35 was carried out (Scheme 1) between the commercially available compounds tert-butyl 4-iodo-2-methylphenylcarbamate (1) and 4-methoxy-2-nitrophenylboronic acid (2), providing the intermediate tert-butyl 4′-methoxy-3-methyl-2′-nitrobiphenyl-4-ylcarbamate (3) in a yield of 79%. Deprotection of 3 in the presence of 20% trifluoroacetic acid–dichloromethane and followed by dansylation with dansyl chloride resulted in the formation of the fluorescent compound 4. Treatment of 4 with triphenylphosphine in 1,2-dichlorobenzene under reflux36,37 overnight resulted in the reductive cyclization of the 2-nitrobiphenyl to form a mixture of two carbazole 5 and 6 (ca. 1:1), which were readily separable by chromatography. The structural assignment of the two isomers was based on the 1H NMR coupling patterns of the aryl protons. The 1H NMR spectrum of compound 6 showed two doublets (δ 6.48 and 7.46, J = 8.4 Hz) corresponding to the protons on carbons 3 and 4, respectively, whereas the 1H NMR spectrum of compound 5 showed two singlets (δ 6.61 and 7.52) corresponding to the protons on carbons 1 and 4.
Scheme 1. Synthesis of Compounds 5 and 6a.
aReagents and conditions: (a) Pd(PPh3)4, K2CO3, CH3CN–H2O, reflux, 79%; (b) (i) 20% TFA–DCM, (ii) dansyl chloride, pyridine, 80%; (c) PPh3, 1,2-DCB, reflux under N2, 42% for 5 and 38% for 6.
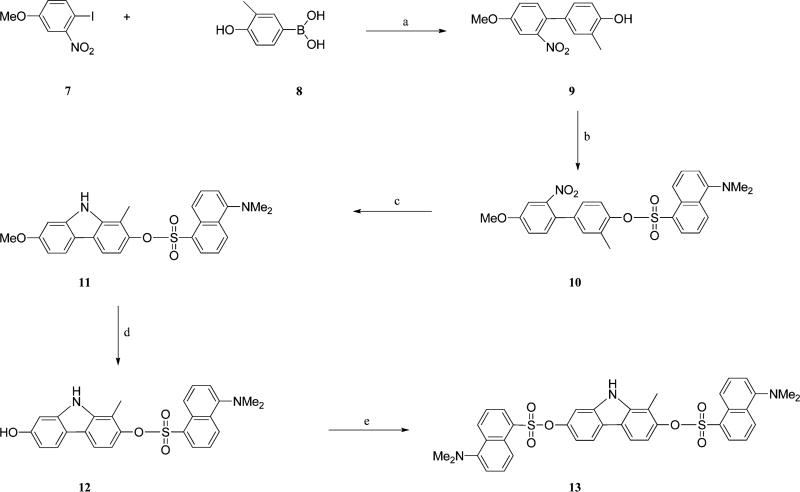
The known compound 1134 was prepared in an improved way as shown in Scheme 2. The commercially available reagents 7 and 8 were coupled under the standard Suzuki reaction condition, providing the biphenyl-4-ol compound 9 (80%). After dansylation (10) and reductive cyclization, two isomers, 7-methoxy-1-methyl-9H-carbazol-2-yl 5-(dimethylamino)-naphthalene-1-sulfonate (11) and 7-methoxy-3-methyl-9H-carbazol-2-yl 5-(dimethylamino)naphthalene-1-sulfonate,34 were formed. Because our previously studies indicated that 11 is around 20 times stronger than the corresponding isomer to the cancer cell lines,34 several conditions were investigated to increase the ratio of 11 to its isomer. We found that the removal of oxygen atmosphere could increase the yield of 11 from 40% to 60% in the presence of triphenylphosphine and 1,2-dichlorobenzene under reflux. The demethylation was carried out in the presence of BBr3 in dichloromethane. Concerned that the dansyl ester is good leaving group, the deprotection was performed at low temperature (−78 °C) and the process of the demethylation was monitored by TLC. The result of TLC indicated that compound 11 was intact at low temperature (−78 °C to −30 °C), even up to 8 h. When the mixture was warmed up to ambient temperature, the demethylation proceeded cleanly and afforded the phenol product 12 with a yield of 81%. Compound 12 was treated with dansyl chloride in anhydrous dichloromethane and triethylamine, resulting in the didansylated compound 13.
Scheme 2. Synthesis of Compounds 12 and 13a.
aReagents and conditions: (a) Pd(PPh3)4, K2CO3, CH3CN–H2O, reflux, 80%; (b) dansyl chloride, DCM–TEA, 85%; (c) PPh3, 1,2-DCB, reflux under N2, 60%; (d) BBr3, dichloromethane, −78 °C to room temperature, 81%; (e) dansyl chloride, DCM–TEA, 71%.
Biology
Compound 12 Inhibits the STAT3-Mediated Transcription and IL-6-Induced Phosphorylation of STAT3 in Triple-Negative Breast Cancer (TNBC) Cells
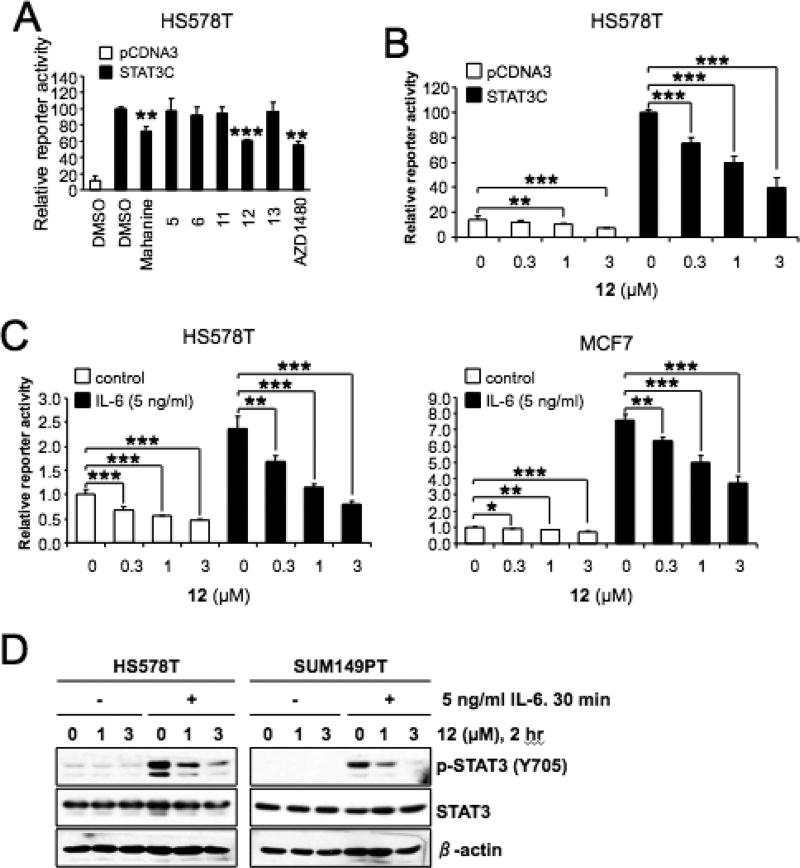
To determine the biological effect of our novel compounds and a natural product carbazole mahanine38,39 on the STAT3 pathway, we performed a typical reporter gene assay. HS578T cells were transfected with the expression vector for a constitutively active STAT3 (STAT3C)40 and the APRE-Luc reporter (a STAT3-responsive reporter).41 The cells were treated with 1 µM of compounds for 24 h, and the luciferase reporter activity was measured (Figure 2A). From this screening, compound 12 was identified as the most potent analogue in reduce the STAT3C-induced reporter gene activity in HS578T cells at 1 µM concentration. The effect of compound 12 on STAT3C-mediated transcription was comparable to that of AZD1480,42 a JAK2 kinase inhibitor (Figure 2A). In HS578T cells transfected with STAT3C expression vector and the APRE-Luc, 12 repressed the STAT3C-dependent transactivation in a dose-dependent manner (Figure 2B). The effect of 12 on the STAT3 pathway was also assessed in the interleukin-6 (IL-6) induction model. HS578T and MCF7 cells, transfected with the APRE-Luc, were pretreated for 1 h with an increasing amount of compound 12 followed by 2 h (for HS578T) or 16 h (for MCF-7) activation with 5 ng/mL of IL-6. Under these conditions, compound 12 significantly reduced the IL-6-induced activation of endogenous STAT3-mediated transcription (Figure 2C). In addition, 12 reduced the IL-6-induced phosphorylation of STAT3 (Y705) in a dose-dependent manner in two separate TNBC cell lines, HS578T and SUM149PT (Figure 2D).
Figure 2.
Repression of STAT3C-mediated transcription by compound 12. (A) Reporter gene assay for STAT3C transactivation in HS578T cells. HS578T cells were transfected with the APRE-Luc and STAT3C expression vector and further treated with 1 µM of compounds for 24 h. (B) HS578T cells were transfected with the APRE-Luc and STAT3C expression vector and further treated with increasing amount of compound 12 for 24 h. (C) HS578T and MCF7 cells were transfected with the APRE-Luc and treated with increasing amount of compound 12 for 1 h. The cells were further treated with 5 ng/mL of IL-6 for 2 h for HS578T or 16 h for MCF7 before reporter gene assay. (A–C) The relative reporter activities are presented as mean ± SD from experiments performed in triplicate. *P < 0.05, **P < 0.01, and ***P < 0.001. (D) Compound 12 inhibits the IL-6-induced phosphorylation of STAT3. HS578T cells were treated with 12 for 2 h as indicated. The IL-6 (5 ng/mL) was added to the culture medium and incubated for 30 min. The cell lysates were analyzed by the indicated antibodies and β-actin was used as a loading control.
Compound 12 Exhibits the Antiproliferative Effects on a Broad Range of Cancer Cell Lines
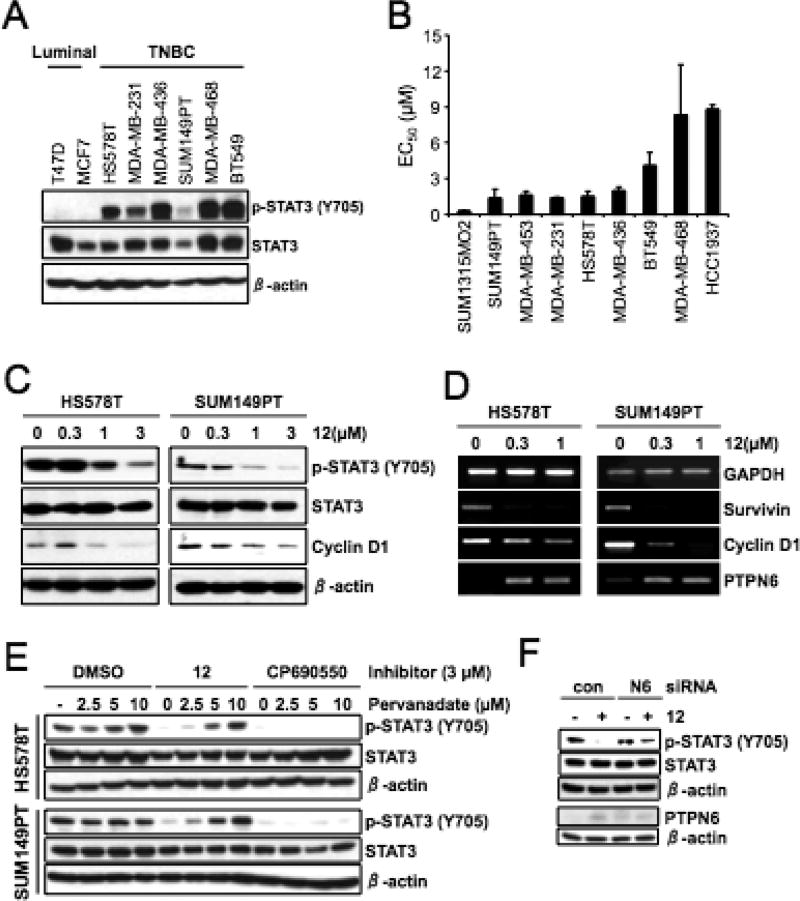
Because activation of STAT3 has been implicated in human cancers, we determined the level of phospho-STAT3 (Y705) in a panel of breast cancer cell lines by Western blot analysis. Interestingly, the level of phospho-STAT3 (Y705) was elevated in all the TNBC cell lines tested as compared to two luminal breast cancer cells, T47D and MCF7 (Figure 3A). We further determined the cytotoxic effects of 12 on the panel of TNBC cell lines. The cells were treated with increasing concentration of 12 for ~72 h, and the viable cells were measured by MTT assay. Interestingly, compound 12 demonstrated antiproliferative affects on a broad range of TNBC cell lines (Figure 3B). Compound 12 also exhibited the antiproliferative effects on additional cancer cell lines, including A431 (a squamous carcinoma cell line), A549 (a lung cancer cell line) and PC-3 (a prostate cancer cell line) with GI50 values 0.16 µM (for A431), 2.5 µM (for A549), and 3 and 7.9 µM (for PC-3), respectively, in MTT assay with ~72 h treatment (data not shown).
Figure 3.
Antiproliferative effects and induction of PTPN6 by 12 in TNBC cells. (A) The elevated expression of phospho-STAT3 (Y705) in TNBC cell lines. The cell lysates from exponentially growing cells were analyzed by indicated antibodies. β-Actin was used as a loading control. (B) EC50 values of 12 were determined by MTT cell viability assay. Data from two independent experiments performed in triplicate are shown as mean ± SEM (C) HS578T and SUM149PT cells were treated with increasing amount of 12 for 24 h and the cell lysates were analyzed by indicated antibodies and β-actin was used as a loading control. (D) RT-PCR analysis of cells treated as in (C), GAPDH was used as a loading control. (E). The cells were treated with compound 12 or CP690550 in the absence or presence of increasing amount of pervanadate for 16 h. The cell lysates were analyzed by indicated antibodies, and β-actin was used as a loading control. (F) The cells were transfected with either control- or PTPN6-siRNA for 3 days and further treated with 12 for 24 h. The cell lysates were subjected to Western blot analysis with indicated antibodies.
Compound 12 Reduces Phospho-STAT3 by Inducing Protein–Tyrosine Phosphatase PTPN6
Because elevated phospho-STAT3 was detected in various TNBC cell lines, we further treated two TNBC cell lines with compound 12 without extracellular IL-6 stimulation and the level of phospho-STAT3 was determined by Western blot analysis. As shown in Figure 3C, 12 reduced the phospho-STAT3 (Y705) in a dose-dependent manner. We also found that the level of cyclin D1, a transcriptional target of STAT3, was also reduced by compound 12 treatment.
The effect of compound 12 on the mRNA expressions of STAT3 targets was assessed by semiquantitative reverse-transcription PCR. Consistent with Western blot analysis, the level of cyclin D1 mRNA was decreased by compound 12 in two TNBC cells (Figure 3D). Under these conditions, the mRNA expression of survivin, another STAT3 target was also reduced by compound 12. Interestingly the mRNA expression of PTPN6 was induced by compound 12 in a dose-dependent manner (Figure 3D).
To determine the effect of PTPN6 upregulation on STAT3, HS578T, and SUM149PT, cells were treated with compound 12 or CP690550 (a JAK1/2 inhibitor)43 in the presence of a phosphatase inhibitor, pervanadate, for 16 h, and the level of phospho-STAT3 was determined by Western blot analysis. Consistent with the induction of PTPN6 mRNA, the compound 12 mediated reduction of phospho-STAT3 (Y705) was reversed by increasing the amount of pervanadate (Figure 3E). On the contrary, the kinase inhibitor CP690550-mediated inhibition of phospho-STAT3 was not affected by pervanadate.
To further verify the PTPN-dependent dephosphorylation of phospho-STAT3, we performed siRNA-based knockdown of PTPN6 in HS578T cells. HS578T cells were transfected with either control- or PTPN6-siRNA for 3 days and further treated with compound 12 for 24 h, and then the level of proteins were determined by Western blot analysis. Consistent with PTPN6 mRNA expression, treatment of compound 12 induced PTPN6 protein expression in HS578T cells transfected with control-siRNA (Figure 3F). As expected, transfection of PTPN6-siRNA reduced the compound 12 mediated PTPN6 protein expression. Under these conditions, knockdown of PTPN6 reversed the dephosphorylation of STAT3. Taken together, these results suggest that compound 12 inhibits phospho-STAT3 (Y705) by induction of PTPN6 phosphatase in TNBC cells.
Imaging of Compound 12 in Human Breast Cancer Cells
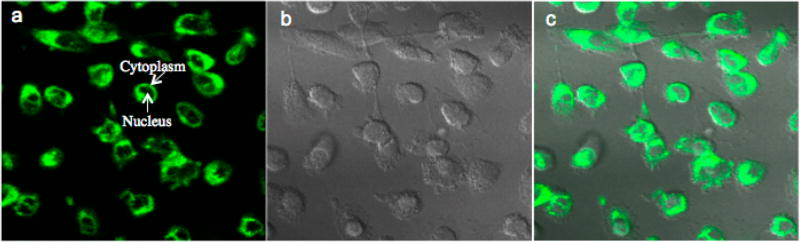
Compound 12 contains a dansyl moiety, which allows it to visibly fluoresce at 552 nm when excited at 370 nm (Figure 4). After treatment of the human breast cancer MDA-MB-231 cells with compound 12, a multiphoton laser was used to excite the compound, and its emission from within the cells was observed by two-photon confocal microscopy (Figure 4). Compound 12 (shown in green) was present within the cytoplasm of the cells but not in the nuclei (Figure 4a).
Figure 4.
Compound 12 fluoresces in the cytoplasm of MDA-MB-231 cells. MDA-MB-231 cells were treated with 10 µM of compound 12 for 1 h. The fluorophore of compound 12 was excited at 725 nm with a multiphoton laser and imaged with a 500–550 nm filter. (a) Compound 12 fluorescence shown in green. (b) Differential interference contrast (DIC) image. (c) Parts a and b are merged.
In Vivo Studies Indicated 12 Significantly Reduces Nonsmall-Cell (A549) Lung Tumor and Human Breast Cancer (SUM 149) Growth in Xenograft Mice
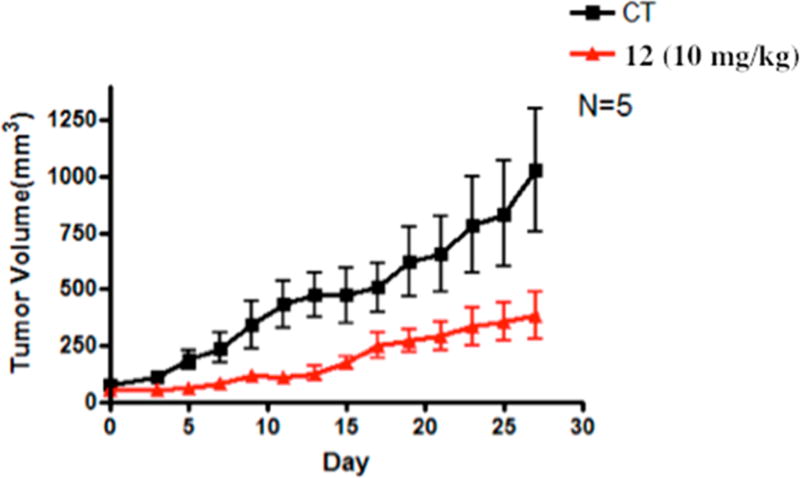
Athymic Balb/c nude mice bearing A549 human nonsmall cell lung tumor that harbor aberrantly active STAT344–46 were dosed via intraperitoneal injection with 10 mg/kg compound 12 at every other day for 27 days (total 13 doses, N = 5, Figure 5). The onset of the antitumor growth effect of compound 12 occurred on day 5 (inhibition rate 68.7%) with the maximum inhibitory effect occurring on day 9 (inhibition rate 73.76%). The duration of inhibition lasted up to 27 days (P < 0.05), with tumor growth inhibition rate between 54.94 to 73.76% during this period.
Figure 5.
Treatment with compound 12 reduces the volume of human nonsmall cell (A549) lung xenograft tumors. Mice bearing A549 tumors were administered 12 via ip 10 mg/kg every other day. Tumor sizes were measured every other day and converted into volumes by L × w × w/2, plotted against days of treatment. Data are present as the mean ± SEM (n = 5, P < 0.05).
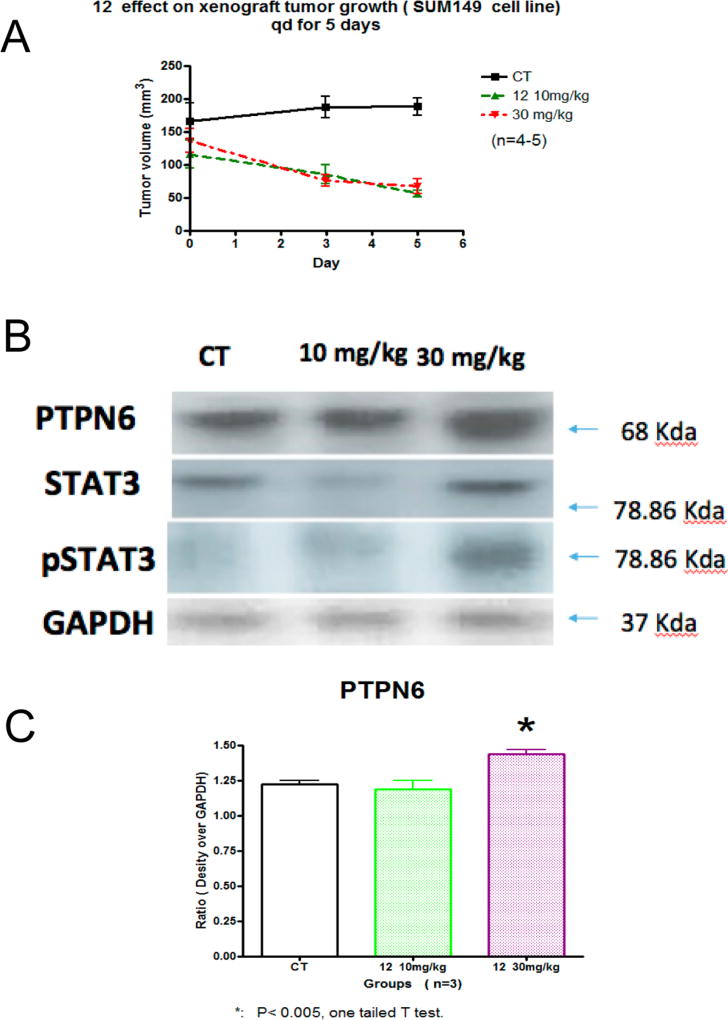
Athymic Balb/c nude mice bearing human TNBC tumor (SUM149) were dosed via intraperitoneal injection with 10 and 30 mg/kg compound 12 at every other day for 5 days (Figure 6A). No significant changes in body weight or obvious signs of toxicity such as loss of appetite, decreased activity, or lethargy were observed during the efficacy study.
Figure 6.
Treatment with compound 12 reduces the volume of human breast cancer (SUM149) xenograft tumors. (A) Mice bearing SUM149 tumors were administered compound 12 via ip 10 and 30 mg/kg every other day. Tumor sizes were measured every other day and converted into volumes by L × w × w/2, plotted against days of treatment. Data are present as the mean ± SEM (n = 5, P < 0.05). (B) Western blot of day 5 for compound 12 treated SUM149 tumors were evaluated for PTPN6, STAT3, and pSTAT3. GAPDH was used as a control. (C) Densitometry of PTPN6 with statistical significance using a one tailed t test (* P < 0.005).
Compound 12 was effective at directly inhibiting the growth of the SUM149 xenograft and resulted in a 57% reduction in tumor volume at day 5 (Figure 6A). Western analysis of the SUM149 tumors treated with compound 12 also revealed an up-regulation of PTPN6 (statistical significant at 30 mg/kg, Figure 6B) and pSTAT3 (Figure 6C). This data strongly supports the potential for use and further preclinical examination of compound 12 in TNBC.
Acute toxicity was evaluated by using the acute toxicity up-and-down procedure,47 where animals were dosed with compound 12 via ip injection and observed for 14 days. The estimated LD50 of 550 mg/kg was determined, with the 95% PL confidence interval between 116.4 and 811 mg/kg.
DISCUSSION AND CONCLUSIONS
Conventional chemotherapeutic agents do not distinguish between healthy and cancer cells. As a newer type of cancer treatment, targeted therapy uses drugs to more precisely identify and attack cancer cells while doing little damage to normal cells. The aberrant activation of STAT3 occurs in the majority of human cancers and its cancer promoting activities have been confirmed involves with cell proliferation, metastasis, angiogenesis, host immune evasion, and resistance to apoptosis.48,49 Given its importance to cancer, STAT3 is considered a molecular target for the development of new cancer therapy.
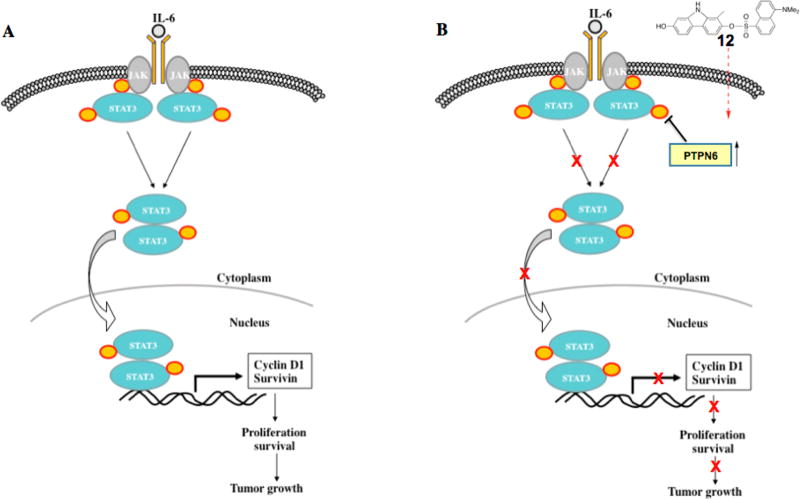
Phosphorylation of a tyrosine at amino acid Y705 is essential for the function of STAT3, and elevated levels of STAT3 phosphorylation have been correlated with tumor invasion, nodal metastasis, and staging.50,51 Phospho-STAT3 has gained much attention,52–54 and although several phospho-STAT3 inhibitors have been reported,18,55–57 only one of them have reached the clinic for several reasons including lacking of specificity and scaffold suitability. Herein we described the discovery and characterization of a novel carbazole compound 12 that strongly inhibits STAT3-mediated transcription as well STAT3 phosphorylation induced by IL-6. IL-6 is the most notable activator of STAT3 pathway and crucial for inducing and maintaining a cancer-promoting inflammatory environment, tumor cell survival through the up-regulation of antiapoptotic genes, and metastasis of tumor cells.5 Recently, it has been demonstrated that TNBC cell lines have autocrine secretion of high levels of inflammatory cytokines including IL-6, IL-8, and CXCL1.58 Consistent with this, we found that TNBC cell lines expressed a high level of phospho-STAT3 without exogenous IL-6 and compound 12 also reduced the activated phospho-STAT3 in these cell lines as well (Figure 7).
Figure 7.
STAT3 pathway (A) and mechanism of compound 12’s STAT3 pathway inhibition (B).
The observed cytoplasmic fluorescence of compound 12 provides visual evidence that the compound enters human breast cancer cells. Further, it provides strong support for molecular target engagement of compound 12 in the cytoplasm and provides a new tool for study of STAT3 mechanistic pathways involved in mediating protein up-regulation via cytoplasmic signaling.
We found that the inhibition of phospho-STAT3 by compound 12 was via the up-regulation of cytoplasmic protein–tyrosine phosphatase PTPN6. Although loss of PTPN6 has been shown to enhance JAK3/STAT3 signaling in ALK-positive anaplastic large-cell lymphoma,59 the roles of PTPN6 in TNBC are not yet understood. One emerging question in our study relates to how compound 12 induces the expression of PTPN6. Because the loss of PTPN6 expression is due to its promoter DNA methylation that is mediated by STAT3/DNMT1,60 we are currently investigating the potential role of compound 12 in the epigenetic regulation of gene expression in TNBC cells via a cytoplasmic control.
Compound 12 was effective at inhibiting the growth of both human TNBC xenograft tumors (SUM149) and human xenograft (A549) tumors that harbor aberrantly active STAT3 without any observed toxicity. Our present studies strongly support the continued preclinical evaluation of compound 12 as a potential chemotherapeutic agent for TNBC and cancers with constitutive STAT3 signaling.
EXPERIMENTAL SECTION
All reagents and solvents were purchased from commercial suppliers and used as received unless noted otherwise. Flash column chromatography separations were done on a Biotage SP1 system monitoring at 254 and 310 nm. NMR spectra were recorded on a Varian 400 spectrometer at 25 °C, operating at 400 MHz for 1H and 100 MHz for 13C NMR. The chemical shifts are expressed in ppm downfield from TMS as an internal standard. Reactions were monitored by thin-layer chromatography (TLC) on silica gel 60 glass slides. The structure of the synthesized compounds follows unequivocally from the mode of synthesis and the m/z values found in their low- and high-resolution mass spectra, TLC, and NMR spectroscopy verified the purity. The HPLC system from Shimadzu consisted of a CBM-20A connector, LC-20AD pumps, an SDP-10AV UV detector, and a SIL-HTA autosampler. Compounds 12 and 13 were monitored at 254 nm by the UV detector. A Symmetry C18 column with 4.6 mm × 250 mm and 5 µm from Waters was used. The mobile phase for 12 consisted of 70% acetonitrile and 30% water containing 0.1% acetic acid or 80% acetonitrile and 20% water containing 0.1% acetic acid. The mobile phase for 13 consisted of 70% acetonitrile and 30% water containing 0.1% acetic acid or 90% acetonitrile and 10% water containing 0.1% acetic acid. The flow rate of mobile phase was 1.0 mL/min. The retention time of 12 was 9.0 or 4.7 min and 13 was 6.4 or 5.1 min. HPLC showed that the purity of both of the compounds is ≥95%.
tert-Butyl 4′-Methoxy-3-methyl-2′-nitrobiphenyl-4-ylcarbamate (3)
4-Methoxy-2-nitrophenylboronic acid 2 (1.07 g, 5.5 mmol) was added to a solution of tert-butyl 4-iodo-2-methylphenylcarbamate 1 (1.66 g, 5.0 mmol) in CH3CN–H2O (20 mL, v/v = 2:1), followed by Pd(PPh3)4 (115 mg, 0.1 mmol) and K2CO3 (2.65 g, 25 mmol). The mixture was refluxed for 16 h. After cooling to room temperature, the mixture was filtered through a Celite pad and the filtrate was concentrated to remove acetonitrile. The residue was diluted with DCM (100 mL) and washed with saturated brine (2 × 30 mL), and the organic phase was dried over Na2SO4 and concentrated. tert-Butyl 4′-methoxy-3-methyl-2′-nitrobiphenyl-4-ylcarbamate (3) (1.42 g, 79%) was afforded after concentration and chromatography (hexane/ethyl acetate = 6:1); mp 136 °C (from ethanol). 1H NMR (400 MHz, CDCl3), δ 7.89 (d, J = 8.4 Hz, 1 H), 7.33–7.29 (m, 2 H), 7.12 (d, J = 2.4 Hz, 1 H), 7.10 (d, J = 2.8 Hz, 1 H), 7.06 (brs, 1 H), 6.31 (brs, 1 H), 3.88 (s, 3 H), 2.26 (s, 3 H), 1.53 (s, 9 H). 13C NMR (100 MHz, CDCl3), δ 158.8, 1559.9, 152.8, 149.5, 136.2, 132.7, 132.1, 129.8, 128.2, 126.5, 120.5, 118.5, 108.9, 80.6, 55.8, 28.3 (3 C), 17.7. Anal. Calcd for C19H22N2O5: C 63.67, H 6.19, N, 7.82. Found: C 63.59, H 6.16, N 7.77.
5-(Dimethylamino)-N-(4′-methoxy-3-methyl-2′-nitrobiphenyl-4-yl)naphthalene-1-sulfonamide (4)
A solution of 3 (1.0 g, 2.79 mmol) in 20% trifluoroacetic acid–dichloromethane (15 mL) was stirred at room temperature for 5 h and concentrated to dryness. The residue was added to EtOH (2 × 3 mL) and concentrated to completely remove the residual trifluoroacetic acid. The product was used for next step without further purification. Dansyl chloride (2.25 g, 8.37 mmol) was added to a solution of the fresh prepared residue in pyridine (15 mL) at 0 °C and warmed up to room temperature. After stirring for 24 h, the mixture was concentrated and chromatographed (hexane/ethyl acetate = 3:1) to afford the 5-(dimethylamino)-N-(4′-methoxy-3-methyl-2′-nitrobiphenyl-4-yl)naphthalene sulfonamide (4) (1.1 g, 80% over two steps). 1H NMR (400 MHz, CDCl3), δ 8.55 (d, J = 8.4 Hz, 1 H), 8.35 (d, J = 8.8 Hz, 1 H), 8.20 (dd, J = 1.2 Hz, 7.6 Hz, 1 H), 7.57 (dd, J = 8.0 Hz, 8.8 Hz, 1 H), 7.48 (t, J = 8.4 Hz, 1 H), 7.30 (d, J = 2.4 Hz, 1 H), 7.20 (m, 2 H), 7.12 (d, J = 9.2 Hz, 1 H), 7.08 (dd, J = 3.2 Hz, 8.4 Hz, 1 H), 6.93 (m, 2 H), 6.54 (s, 1 H), 3.86 (s, 3 H), 2.89 (s, 6 H), 2.00 (s, 3H). 13C NMR (100 MHz, CDCl3), 159.0, 149.4, 134.7, 134.4, 132.6, 131.0, 130.8, 130.2, 129.5, 128.4, 127.6, 126.5, 123.2, 118.6, 108.9, 55.8, 45.4, 17.6. HRMS (TOF-ES+): m/z [M + 1]+ calcd for C26H25N3O5S, 492.1593; found, 492.1660.
5-(Dimethylamino)-N-(7-methoxy-3-methyl-9H-carbazol-2-yl)naphthalene-1-sulfonamide (5) and 5-(Dimethylamino)-N-(7-methoxy-1-methyl-9H-carbazol-2-yl)naphthalene-1-sulfonamide (6)
Triphenylphosphine (1.32 g, 5.0 mmol) was added to a solution of 5 (980 mg, 2 mmol) in 1,2-dichlorobenzene (10 mL). The mixture was refluxed under N2 for 48 h, concentrated, and chromatographed (hexane–ethyl acetate–methanol 4:1:0.01) to afford 5-(dimethylamino)-N-(7-methoxy-3-methyl-9H-carbazol-2-yl)-naphthalene-1 sulfonamide (5) (390 mg, 42%) and 5-(dimethylamino)-N-(7-methoxy-1-methyl-9H-carbazol-2-yl)naphthalene-1-sulfonamide (6) (350 mg, 38%).
5: 1H NMR (400 MHz, CDCl3), δ 8.50 (d, J = 8.4 Hz, 1 H), 8.41 (d, J = 8.4 Hz, 1 H), 8.12 (dd, J = 1.2 Hz, 7.2 Hz, 1 H), 7.82 (s, 1 H), 7.74 (d, J = 8.4 Hz, 1 H), 7.53 (dd, J = 7.6 Hz, 8.8 Hz, 1 H), 7.52 (s, 1 H), 7.38 (dd, J = 7.6 Hz, 8.4 Hz, 1 H), 7.18 (d, J = 7.2 Hz, 1 H), 6.82 (d, J = 2.0 Hz, 1 H), 6.78 (dd, J = 7.0 Hz, 8.8 Hz, 1 H), 6.61 (s, 1 H), 3.86 (s, 3 H), 2.88 (s, 6 H), 2.01 (s, 3 H). 13C NMR (100 MHz, CDCl3), δ 159.0, 141.3, 138.2, 134.8, 131.2, 130.5, 130.2, 129.6, 128.3, 123.2, 122.5, 121.5, 120.7, 120.6, 116.4, 115.2, 108.2, 106.1, 94.6, 55.6, 45.4, 17.8. HRMS (TOF-ES+): m/z [M + 1]+ calcd for C26H26N3O3S, 460.1695; found, 460.1720; Anal. Calcd for C26H25N3O3S + 1.2H2O: C 64.90, H 5.74, N, 8.73. Found: C 64.83, H 5.49, N 8.45.
6: 1H NMR (400 MHz, CDCl3), δ 8.52 (d, J = 8.4 Hz, 1 H), 8.41 (d, J = 8.4 Hz, 1 H), 8.00 (dd, J = 1,2 Hz, 7.2 Hz, 1 H), 7.81 (s, 1 H), 7.78 (d, J = 8.8 Hz, 1 H), 7.56 (dd, J = 8.0 Hz, 8.8 Hz, 1 H), 7.46 (d, J = 8.4 Hz, 1 H), 7.37 (dd, J = 7.6 Hz, 8.4 Hz, 1 H), 7.20 (d, J = 7.6 Hz, 1 H), 6.89 (d, J = 2.0 Hz, 1 H), 6.80 (dd, J = 7.0 Hz, 8.8 Hz, 1 H), 6.48 (t, 4.0 Hz, 2 H), 3.87 (s, 3 H), 2.90 (s, 6 H), 2.28 (s, 3 H). 13C NMR (100 MHz, CDCl3), δ 159.1, 155.2, 141.2, 139.1, 134.5, 131.2, 130.5, 130.4, 129.9, 129.7, 128.4, 123.1, 121.1, 118.8, 117.2, 116.8, 115.1, 108.5, 94.7, 55.6, 45.4, 12.1. HRMS (TOF-ES+): m/z [M + 1]+ calcd for C26H26N3O3S, 460.1695; found, 460.1719. Crystallization from ethyl acetate: mp 165–168 °C. Anal. Calcd for C26H25N3O3S + 0.25EtOAc: C 67.34, H 5.65, N, 8.73. Found: C 67.06, H 5.71, N 8.63.
4′-Methoxy-3-methyl-2′-nitrobiphenyl-4-ol (9)
4-Hydroxyphenylboronic acid (8) (1.5 g, 10 mmol) was added to a solution of 4-iodo-3-nitroanisole (7) (3.1 g, 11 mmol) in CH3CN–H2O (30 mL, v/v = 2:1), followed by Pd(PPh3)4 (350 mg, 0.3 mmol) and K2CO3 (5.3 g, 50 mmol), the mixture was refluxed overnight. After cooling to room temperature, the mixture was filtered through a Celite pad. The filtrate was concentrated, diluted with DCM (100 mL), and washed with saturated brine (3 × 30 mL). After concentration and chromatography (hexane/acetone = 6:1) afforded 4′-methoxy-3-methyl-2′-nitrobiphenyl-4-ol (9) (2.1 g, 80%). 1H NMR (400 MHz, CDCl3), δ 7.31 (m, 2 H), 7.12 (dd, J = 2.8 Hz, 8.4 Hz, 1 H), 7.04 (s, 1 H), 6.98 (d, J = 8.0 Hz, 1 H), 6.78 (d, J = 8.0 Hz, 1 H), 3.88 (s, 3 H), 2.26 (s, 3 H). 13C NMR (100 MHz, CDCl3), δ 158.7, 153.7, 132.7, 130.6, 129.5, 128.3, 126.7, 124.1, 118.5, 115.1, 108.8, 55.8, 15.7. MS (TOF-ES+): 260.3 m/z [M + 1]+.
4 ′-Methoxy-3-methyl-2′-nitrobiphenyl-4-yl 5-(Dimethylamino)naphthalene-1-sulfonate (10)
Dansyl chloride (3.14 g, 12 mmol) was added to a solution of 9 (1.5 g, 5.8 mmol) in DCM–TEA (15 mL, v/v, 3:1) with stirring at 0 °C. The mixture was allowed to room temperature and stirred overnight, diluted with DCM (90 mL), and washed with saturated brine (30 mL). After drying over Na2SO4 and concentration, the residue was purified via chromatography (hexane/EtOAc = 5:1) to afforded 4′-methoxy-3-methyl-2′-nitrobiphenyl-4-yl 5-(dimethylamino)naphthalene-1-sulfonate (10) (2.41 g, 85%). 1H NMR (400 MHz, CDCl3), δ 8.67 (d, J = 8.8 Hz, 1 H), 8.49 (d, J = 8.8 Hz, 1 H), 8.19 (dd, J = 1.2 Hz, 7.2 Hz, 1 H), 7.65 (dd, J = 7.6 Hz, 8.8 Hz, 1 H), 7.53 (dd, J = 7.6 Hz, 8.4 Hz, 1 H), 7.33 (d, J = 2.4 Hz, 1 H), 7.25 (m, 2 H), 7.10 (dd, J = 2.8 Hz, 8.8 Hz, 1 H), 7.08 (d, J = 2.4 Hz, 1 H), 6.86 (dd, J = 2.0 Hz, 8.4 Hz, 1 H), 6.66 (d, J = 8.4 Hz, 1 H), 3.87 (s, 3 H), 2.93 (s, 6 H), 2.21 (s, 3 H). 13C NMR (100 MHz, CDCl3), δ 159.2, 149.4, 148.3, 135.9, 132.6, 132.1, 132.0, 131.8, 131.2, 130.8, 130.0, 128.9, 127.4, 126.5, 123.1, 121.8, 118.6, 115.7, 109.0, 55.9, 45.4, 16.6. MS (TOF-ES+): 493.5 m/z [M + 1]+. Anal. Calcd for C26H24N2O6S: C 63.40, H 4.91, N, 5.69. Found: C 63.79, H 4.59, N 5.45.
7-Methoxy-1-methyl-9H-carbazol-2-yl 5-(dimethylamino)-naphthalene-1-sulfonate (11)
A solution of compound 10 (200 mg, 0.41 mmol) and triphenylphosphine (534 mg, 2.03 mmol) in 1,2-DCB (10 mL) was vacuumed and flushed with N2 for three times. The mixture was refluxed for 15 h and concentrated. After chromatography (hexane/ethyl acetate = 5;1) gave 7-methoxy-1-methyl-9H-carbazol-2-yl 5-(dimethylamino)naphthalene-1-sulfonate (11) (117 mg, 60%). NMR data were identical with those of reported.34
7-Hydroxy-1-methyl-9H-carbazol-2-yl 5-(dimethylamino)-naphthalene-1-sulfonate (12)
1.0 M solution of BBr3 (0.22 mL, 0.22 mmol) was added to the solution of compound 11 (48 mg, 0.11 mmol) in anhydrous dichloromethane (5 mL) under N2 with stirring at −78 °C dropwisely. The mixture was warmed up to ambient temperature within 1 h and stirred overnight. The reaction was cooled with ice–water and diluted with dichloromethane (40 mL), and saturated aq NaHCO3 (3.0 mL) was added within 15 min. After of the addition, the mixture was washed with saturated brine aq (20 mL) and the organic phase was dried over Na2SO4 and concentrated, then chromatographed (toluene/acetone = 6:1) to afford 12 (37 mg, 81%, purity >97% based on HPLC analysis, see Supporting Information). 1H NMR (400 MHz, acetone-d6), δ 10.17 (s, 1 H), 8.71 (dt, J = 1.2 Hz, 8.8 Hz, 1H), 8.54 (dt, J = 0.8 Hz, 8.8 Hz, 1 H), 8.38 (brs, 1 H), 8.11(dd, J = 1.2 Hz, 7.2 Hz, 1 H), 7.79 (d, J = 8.4 Hz, 1 H), 7.45 (dd, J = 7.6 Hz, 8.8 Hz, 1 H), 7.61 (dd, J = 7.6 Hz, 8.4 Hz, 1 H), 7.55 (d, J = 8.8 Hz, 1H), 7.36 (dd, J = 0.8 Hz, 7.6 Hz, 1 H), 6.93 (dd, J = 0.4 Hz, 2.0 Hz, 1 H), 6.73 (dd, J = 2.4 Hz, 8.4 Hz, 1 H), 6.34 (d, J = 8.4 Hz, 1 H), 2.92 (s, 6 H), 2.37 (s, 3 H). 13C NMR (100 MHz, acetone-d6), δ 156.7, 152.1, 145.1, 142.4, 139.7, 132.2, 131.8, 130.8, 130.0, 129.8, 128.9, 123.2, 121.7, 120.8, 119.3, 116.3, 115.9, 115.6, 113.7, 112.7, 109.0, 96.7, 44.7, 10.6. HRMS (TOF-ES+): m/z [M + 1]+ calcd for C25H22N2O4S, 447.1379; found, 447.1399. Crystallized from toluene: mp 99–103 °C. Anal. Calcd for C25H22N2O4S +0.25toluene: C 68.42, H 5.15, N, 5.97. Found: C 68.17, H 5.32, N 5.89.
1-Methyl-9H-carbazole-2,7-diyl Bis(5-(dimethylamino)-naphthalene-1-sulfonate) (13)
Dansyl chloride was added to a solution of 12 (30 mg, 0.067 mmol) in anhydrous dichloromethane (2 mL) in the presence of Et3N (0.5 mL) under N2 with stirring at 0 °C. After stirring for 2 h, the mixture was concentrated and chromatographed (hexane/acetone = 3:1) to give compound 13 (32 mg, 71%, purity >95% based on HPLC analysis, see Supporting Information). 1H NMR (400 MHz, CDCl3), 8.63 (d, J = 8.8 Hz, 1 H), 8.59 (d, J = 8.4 Hz, 1 H), 8.54 (d, J = 8.8 Hz, 2 H), 8.11–8.05 (m, 3 H), 7.71–7.64 (m, 2 H), 7.50–7.38 (m, 3 H), 7.30 (d, J = 8.4 Hz, 1 H), 7.26 (d, J = 7.6 Hz, 2 H), 7.00 (d, J = 2.0 Hz, 1 H), 6.62 (dd, J = 2.0 Hz, 8.4 Hz, 1 H), 6.36 (d, J = 8.8 Hz, 1 H), 2.92 (s, 6 H), 2.91 (s, 6 H), 2.33 (s, 3H). 13C NMR (100 MHz, CDCl3), δ 151.9, 151.8, 147.7, 146.2, 140.0, 139.8, 132.0, 131.9, 131.8, 131.2, 131.1, 130.7, 130.1 (2 C), 129.8, 129.7, 129.0, 128.9, 122.9, 121.8, 120.6, 120.3, 119.6, 119.5, 117.7, 115.6 (2 C), 114.2, 113.9, 113.8, 104.8, 45.4, 11.2. HRMS (TOF-ES+): m/z [M + 1]+ calcd for C37H33N3O6S2, 680.1889; found, 680.1891.
Cell Culture and Reagents
All cell lines, except for SUM1315MO2 and SUM149PT, were obtained from the Tissue Culture Shared Resource of Georgetown University Medical Center. The reagents for cell culture were purchased from Invitrogen (Carlsbad, CA, USA), Lonza (Basel, Switzerland), or Cellgro (Manassas, VA, USA). T47D, HS578T, MDA-MB-436, MDA-MB-453, MDA-MB-468, BT-549, A431, and PC-3 cells were maintained in DMEM containing 10% (v/v) heat-inactivated FBS (HI-FBS; HyClone, Logan, UT, USA) and 100 units/mL penicillin/ streptomycin. MDA-MB-231 and MCF7 cells were maintained in DMEM with 5% HI-FBS and 100 units/mL penicillin/streptomycin. HCC1937 cells were grown in RPMI with 10% HI-FBS, 100 units/mL penicillin/streptomycin, and 1% HEPES. A549 lung cell line (ATCC, Manassas, VA) was cultured in RPMI-1640 with l-glutamine (Mediatech Inc., Herdon, VA), 5% FBS, and 2.5 mM l-glutamine. SUM1315MO2 and SUM149PT were maintained according to manufacturer’s recommendations (Asterand, Detroit, MI, USA). Viable cells were monitored by the Luna automated cell counter (Logos Biosystems, Gyunggi-do, Korea). AZD1480 was purchased from Selleck Chemicals (Houston, TX, USA), and CP690550 was obtained from LC Laboratories (Woburn, MA, USA). Stock solutions of compounds were made at 10 mM concentration in dimethyl sulfoxide (DMSO) and stored at −20 °C in small aliquots. Mahanine was a kind gift from Dr. Samir Bhattacharya and Dr. Bikas C. Pal (Indian Institute of Chemical Biology, Kolkata, India) and dissolved in ethanol. IL-6 was purchased from Sigma-Aldrich (St. Louis, MO, USA).
Multiphoton Laser Imaging
After incubation of slides with a fibronectin/cortactin solution for 30 min, MDA-MB-231 cells were plated onto the slides and incubated overnight. The cells were then treated with a 10 µM solution of 12 or DMSO control and incubated for 1 h before washing with PBS. The slides were fixed with a 4% formaldehyde solution and washed with PBS. The compound was excited at 725 nm with a multiphoton laser and imaged with a 500–550 nm filter.
Plasmid DNAs and Reporter Gene Assay
Transfection and reporter gene assay was performed as described.61 APRE-Luc was described previously51 and kindly provided by Dr. T. Hirano (Osaka University, Osaka, Japan). STAT3C expression DNA was described previously50 and kind gift from Dr. J. Bromberg (Memorial Sloan–Kettering Cancer Center, Weill Cornell Medical College, New York, NY).
Cell Proliferation Assay
MTT (3-(4,5)-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) assay was performed as described previously.62 The half-maximal inhibitory concentration (EC50) and the concentration for 50% of maximal inhibition of cell proliferation (GI50) values were calculated by CompuSyn software V1.0 (ComboSyn, Paramus, NJ).
Western Blot Analysis
Western blot analysis was performed on cell and tissue lysates as described.62 Antibodies used in this study were as follows: phospho-STAT3 (Y705) (no. 9145) and STAT3 (no. 4904) from Cell Signaling (Danvers, MA), Cyclin D1 (sc-717) from Santa Cruz Biotechnologies (Santa Cruz, CA), β-actin (A1978) from Sigma-Aldrich (St. Louis, MO, USA), and GAPDH (no. 3783) from ProsciΨ (Poway, California, USA).
Reverse Transcription-PCR (RT-PCR)
The RT-PCR was performed as described.63 The primers used in this study were synthesized from Sigma Genosys (St. Louise, MO, USA) or Bioneer (Seoul, Korea) with the following sequences: survivin, forward 5′-CCA CTG AGA ACG AGC CAG AC-3′ and reverse 5′-GCA CTT TCT TCG CAG TTT CC-3′; cyclin D1, forward 5′-CCG TCC ATG CGG AAG ATC-3′ and reverse 5′-ATG GCC AGC GGG AAG AC-3′; PTPN6, forward 5′-TCT CTT GCA AGC ATT GGC AAG GTC-3′ and reverse 5′-TGC CAG GAT TCA AAC CCA GAC AGT-3′; GAPDH, forward 5′-GTA TGA CAA CGA ATT TGG CTA CAG-3′ and reverse 5′-AGC ACA GGG TAC TTT ATT GAT GGT-3′.
Transfection of siRNA
To knockdown the PTPN6 expression, transfection of siRNA was performed as described previously.64 In brief, cells in 6-well plates were transfected with 100 nM/well of siRNA by Lipofectamine 2000 reagent (Invitrogen) in serum free media. Four hours after transfection, an equal volume of normal growth media containing 20% FBS was added to each well and the cells were further incubated for 3 days. Transfected cells were further treated with either DMSO or 12 for 24 h before harvest, and then Western blot analysis was performed. Densitometry analysis was performed by ImageJ.65 The siRNAs were purchased from Bioneer (Seoul, Korea) with following sequences: control-siRNA, 5′-GAC GAG CGG CAC GUG CAC AUU-3′; PTPN6-2, 5′-CAG UUC AUU GAA ACC ACU A(dTdT)-3′; and PTPN6-4, 5′-GAG AAC GCU AAG ACC UAC A(dTdT)-3′.
Animals
Balb/c mice and athymic balb/c nude mice were purchased from the NCI. Animals were housed five per cage with microisolater tops and provided food (Purina mice chow) and water ad libitum. The light cycle was regulated automatically (12 h light/dark cycle), and temperature was maintained at 23 ± 1 °C. All animals were allowed to acclimate to this environment for 1 week prior to experimental manipulations. The Georgetown University Animal Care and Use Committee approved all animal studies (protocol no. 11-029) in accordance with the guideline adopted by the National Institutes of Health.
In Vivo Xenograft Study
The stock solution of compound 12 was dissolved in DMSO at 200 mg/mL. The working solution was prepared in a Cremophore and PEG400 mixture (Cremophore 10% and PEG400 3% and 1× PBS 87%) to give a 1 mg/mL solution. Male and female athymic balb/c nude mice (18–22 g) were injected with human A549 lung or SUM149 breast cancer cells (3 × 106 cells in a volume of 0.3 mL) into the subcutaneous tissue of the right auxiliary region of the mice. Three days after the tumor cell inoculation, the mice were randomly sorted into three groups with five mice per group. The tumor-bearing mice were either given an ip injection 10 mg/kg of compound 12 or the vehicle only once a day on every other day. The treatment was initiated when the tumor burden of mice reached around 50 mm3. The tumor size was measured by caliper three times a week to document tumor growth and calculated by the formula: length × width × width/2, and the body weight was measured and recorded. The therapeutic efficacy was evaluated based on body weight loss, tumor growth inhibition [determined by using calipers and calculated with formula: tumor inhibit rate (%) = (tumor volCon − tumor volTre)/tumor volCon].
Statistical Analysis
To compare two groups of interest, the two-tailed Student’s t test was applied for statistical analysis. * indicates P < 0.05, ** indicates P < 0.01, and *** indicates P < 0.001.
Supplementary Material
Acknowledgments
We thank Tissue Culture Shared Resources supported by P30-CA051008-18 (LCCC Cancer Center Support Grant) at Georgetown University Medical Center. We are grateful to Dr. T. Hirano and Dr. J. Bromberg for kind gifts of plasmid DNAs. We also thank Dr. Samir Bhattacharya and Dr. Bikas C. Pal for providing purified mahanine. This work was supported by the Center for Drug Discovery at Georgetown University Medical Center, Susan G. Komen for the Cure (FAS0703858), and the Lombardi Comprehensive Cancer Center, Georgetown University (P30-CA051008).
ABBREVIATIONS USED
- Dansyl
5-(dimethylamino)naphthalene
- TLC
thin layer chromatography
- DCB
dichloronenzene
- DCM
dichloromethane
- TEA
triethylamine
- NMR
nuclear magnetic resonance
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PEG
polyethylene glycol
- GI50
concentration of drug needed to inhibit cell growth by 50%
- EC50
half-maximal effective concentration of drug
- LD50
dose where 50% of test subjects exposed would be killed
- CT
Control
Footnotes
ASSOCIATED CONTENT
1H and 13C NMR spectra of new compounds and HPLC purity analysis of compounds 12 and 13. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare the following competing financial interest(s): Patent applications have been filed by Georgetown University on the behalf of the inventors that are listed as authors in this work.
References
- 1.de Bono JS, Ashworth A. Translating cancer research into targeted therapeutics. Nature. 2010;467:543–549. doi: 10.1038/nature09339. [DOI] [PubMed] [Google Scholar]
- 2.Core CM. Oncogenes and cancer. N. Engl. J. Med. 2008;358:502–511. doi: 10.1056/NEJMra072367. [DOI] [PubMed] [Google Scholar]
- 3.Libermann TA, Zerbini LF. Targeting transcription factors for cancer gene therapy. Current Gene Ther. 2006;6:17–33. doi: 10.2174/156652306775515501. [DOI] [PubMed] [Google Scholar]
- 4.Remond AM, Carroll JS. Defining and target transcription factors for cancer. Genome Biol. 2009;10:311–313. doi: 10.1186/gb-2009-10-7-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu H, Jove R. The STATs of cancer—new molecular targets come of age. Nature Rev. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 6.Darnell JE. Validating Stat3 in cancer therapy. Nature Med. 2005;11:595–596. doi: 10.1038/nm0605-595. [DOI] [PubMed] [Google Scholar]
- 7.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nature Rev. Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bromberg J, Darnell JE., Jr The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19:2468–2473. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- 9.Mankan AK, Greten FR. Inhibiting signal transducer and activator of transcription 3: rationality and rationale design of inhibitors. Expert Opin. Invest. Drugs. 2011;20:1263–1275. doi: 10.1517/13543784.2011.601739. [DOI] [PubMed] [Google Scholar]
- 10.Horiguchi A, Oya M, Shimada T, Uchida A, Marumo K, Murai M. Activation of singal transducer and activator 3 in renal cell carcinoma: a study of incidence and its association with pathological features and clinical outcome. J. Urol. 2002;168:762–765. [PubMed] [Google Scholar]
- 11.Takemoto S, Ushijima K, Kawano K, Yamaguchi T, Terada a, Fujiyoshi N, Nishio S, Tsuda N, Ijichi M, Kakuma T, Kage M, Hori D, Kamura T. Expression of activated signal transducer and activator of transcription-3 predicts poor prognosis in cervical squamous-cell carcinoma. Br. J. Cancer. 2009;101:967–972. doi: 10.1038/sj.bjc.6605212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang XL, Yue PB, Page BDG, Li TS, Zhao W, Namanja AT, Paladino D, Zhao JH, Chen Y, Gunning PT, Turkson J. Orally bioavailable small-molecule inhibitor of transcription factor stat3 regress humane breast and lung cancer xenografts. Proc. Natl. Acad. Sci U. S. A. 2012;109:9623–9628. doi: 10.1073/pnas.1121606109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin L, Amin R, Gallicano GI, Glasgow E, Jogunoori W, Jessup JM, Zasloff M, Marshall JL, Shetty K, Johonson L, Mishra L, He AR. The STAT3 inhibitor NSC 78459 is effective in hepatocellular cancers with disrupted TGF-β signaling. Oncogene. 2009;28:961–972. doi: 10.1038/onc.2008.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sen M, Tosca PJ, Zwayer C, Ryan MJ, Johnson JD, Knostman KA. Lack of toxicity of a STAT3 decoy oligonucleotide. Cancer Chemother. Pharmacol. 2009;63:983–995. doi: 10.1007/s00280-008-0823-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turkson J, Jove R. STAT protein: a novel molecular target for cancer drug discovery. Oncogene. 2000;19:6613–6626. doi: 10.1038/sj.onc.1204086. [DOI] [PubMed] [Google Scholar]
- 16.Debnath B, Xu S, Neamati N. Small molecule inhibitors of signal transducer and activator of transcription 3 (Stat3) protein. J. Med. Chem. 2012;55:6645–6668. doi: 10.1021/jm300207s. [DOI] [PubMed] [Google Scholar]
- 17.Germain D, Frank DA. Targeting the cytoplasmic and nuclear functions of signal transducer and activitors of transcription 3 for cancer therapy. Clin. Cancer Res. 2007;13:5665–5669. doi: 10.1158/1078-0432.CCR-06-2491. [DOI] [PubMed] [Google Scholar]
- 18.Hayakawa F, Sugimoto K, Harada Y, Hashimoto N, Ohi N, Kurahashi S, Naoe T. A novel STAT Inhibitor, OPB-31121, has a significant antitumor effect on leukemia with STAT-addictive oncokinases. Blood Cancer J. 2013;3:e166. doi: 10.1038/bcj.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santos FPS, Kantarjian HM, Jain N, Manshouri T, Thomas DA, Garcia-Manero G, Kennedy D, Estrov Z, Cortes J, Verstovsek S. Phase 2 study of CEP-701, an orally available JAK2 inhibitor, in patients with primary or post-polycythemia vera/essential thrombocythemia myelofibrosis. Blood. 2010;115:1131–1136. doi: 10.1182/blood-2009-10-246363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H, Yang Z, Ding C, Chu L, Zhang Y, Terry K, Liu H, Shen Q, Zhou J. Discovery of O-alkylamino tethered niclosamide derivatives as potent and orally bioavailable anticancer agents. ACS Med. Chem. Lett. 2013;4:180–185. doi: 10.1021/ml3003082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H, Yang Z, Ding C, Chu L, Zhang Y, Terry K, Liu H, Shen Q, Zhou J. Fragment-based drug design and identification of HJC0123, a novel orally bioavailable STAT3 inhibitor for cancer therapy. Eur. J. Med. Chem. 2013;62:498–507. doi: 10.1016/j.ejmech.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyoshi K, Takaishi M, Nakajima K, Ikeda M, Kanda T, Tarutani M, Iiyama T, Asao N, DiGiovanni J, Sano S. Stat3 as a therapeutic target for the treatment of psoriasis: a clinical feasibility study with STA-21, a Stat3 Inhibitor. J. Invest. Dermatol. 2011;131:108–117. doi: 10.1038/jid.2010.255. [DOI] [PubMed] [Google Scholar]
- 23.Newman DJ, Cragg GM. Natural products as sources of new drug discover over the last 25 years. J. Nat. Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 24.Asche C, Demeunynck M. Antitumor carbazoles. Anticancer Agents Med. Chem. 2007;7:246–267. doi: 10.2174/187152007780058678. [DOI] [PubMed] [Google Scholar]
- 25.Long BH, Rose WC, Vyas DM, Matson JA, Forenza S. Discovery of antitumor indolocarbazoles: rebeccamycin, NSC655649, and fluoroindolocarbazoles. Curr. Med. Chem.: Anti-Cancer Agents. 2002;2:255–266. doi: 10.2174/1568011023354218. [DOI] [PubMed] [Google Scholar]
- 26.Prudhomme M. Rebeccamycin analogues as anti-cancer agents. Eur. J. Med. Chem. 2003;38:123–140. doi: 10.1016/s0223-5234(03)00011-4. [DOI] [PubMed] [Google Scholar]
- 27.Arbiser JL, Govindarajan B, Battle TE, Lynch R, Frank DA, Ushio-Fukai M, Perry BN, Stern DF, Bowden GT, Liu A, Klein E, Kolodziejski PJ, Eissa NT, Hossain CF, Nagle DG. Carbazole is a naturally occurring inhibitor of angiogenesis and inflammation isolated from antipsoriatic coal tar. J. Invest. Dermatol. 2006;126:1396–1402. doi: 10.1038/sj.jid.5700276. [DOI] [PubMed] [Google Scholar]
- 28.Saturnino C, Palladino C, Napoli M, Sinicropi MS, Botta A, Sala M, Carcereri de Prati A, Novellino E, Suzuki H. Synthesis and biological evaluation of new N-alkylcarbazole derivatives as STAT3 inhibitors: preliminary study. Eur. J. Med. Chem. 2013;60:112–119. doi: 10.1016/j.ejmech.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 29.Timofeeva OA, Tarasova NI, Zhang X, Chasovskikh S, Cheema AK, Wong H, Brown ML, Dritschilo A. STAT3 suppresses transcription of pro-apoptotic genes in cancer cells with the involvement of its N-terminal domain. Proc. Natl. Acad. Sci U. S. A. 2013;110:1267–1272. doi: 10.1073/pnas.1211805110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Timofeeva OA, Chasovskikh S, Lonskaya I, Tarasova NI, Khavrutskii L, Tarasova S, Zhang X, Korostyshevskiy VR, Cheema A, Zhang L, Dakshanamurthy S, Brown ML, Dritschilo A. Mechanisms of unphosphorylated STAT3 transcription factor binding to DNA. J. Biol. Chem. 2012;287:14192–14200. doi: 10.1074/jbc.M111.323899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kong Y, Jung M, Kang W, Grindrod S, Velena A, Lee SA, Dakshanamurthy S, Yang Y, Miessau M, Zheng C, Dritschilo A, Brown ML. Histone deacetylase cytoplasmic trapping by a novel fluorescent HDAC inhibitor. Mol. Cancer Ther. 2011;10:1591–1599. doi: 10.1158/1535-7163.MCT-10-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kong HS, Tian S, Kong Y, Du G, Zhang L, Jung M, Dritschilo A, Brown ML. Preclinical studies of YK-4-272, an inhibitor of class III histone deacetylases by disruption of nucleocytoplasmic shuttling. Pharm. Res. 2012;29:3373–3383. doi: 10.1007/s11095-012-0832-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walls TH, Grindrod S, Beraud D, Zhang L, Baheti AR, Dakshanamurthy S, Patel MK, Brown ML, Macarthur LH. Synthesis and biological evaluation of a fluorescent analog of phenytoin as a potential inhibitor of neuropathic pain and imaging agent. Bioorg. Med. Chem. 2012;20:5269–5276. doi: 10.1016/j.bmc.2012.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheikh KD, Banerjee PP, Jagadeesh S, Grindord SC, Zhang L, Paige M, Brown ML. Fluorescent epigenetic small molecule induces expression of the tumor suppressor ras-association domain family 1A and inhibits human prostate xenograft. J. Med. Chem. 2010;53:2376–2382. doi: 10.1021/jm9011615. [DOI] [PubMed] [Google Scholar]
- 35.Miyaura N, Suzuki A. Palladium-catalyzed cross-coupling reaction of organoboron compounds. Chem. Rev. 1995;95:2457–2483. [Google Scholar]
- 36.Freeman AW, Urvoy M, Criswell ME. Triphenylphosphine-mediated reductive cyclization of 2-nitrobiphenyls: a practical and convenient synthesis of carbazoles. J. Org. Chem. 2005;70:5014–5019. doi: 10.1021/jo0503299. [DOI] [PubMed] [Google Scholar]
- 37.Naffziger MR, Ashburn BO, Perkins JR, Carter RG. Diels–Alder approach for the construction of halogenated o-nitro biaryl templates and application to the total synthesis of the anti-HIV agent siamenol. J. Org. Chem. 2007;72:9857–9865. doi: 10.1021/jo070740r. [DOI] [PubMed] [Google Scholar]
- 38.Nakahara K, Trakoontivakorn G, Alzoreky NS, Onishi-Kameyama M, Yoshida M. Antimutagenicity of some edible Thai plants, and a bioactive carbazole alkioid, mahanine, isolated from Micromelum minutum. J. Agric. Food Chem. 2002;50:4798–4802. doi: 10.1021/jf025564w. [DOI] [PubMed] [Google Scholar]
- 39.Roy MK, Thalang VN, Trakoontivakorn G, Nakahara K. Mahanine, a carbazole alkaloid from Micromelum minutum, inhibits cell growth and induces apotosis through a mitochondrial dependent pathway. Br. J. Pharmacol. 2005;145:145–155. doi: 10.1038/sj.bjp.0706137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bromberg JF, Wrzeszcznska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 41.Nakajima K, Yamanaka Y, Nakae K, Kojima H, Ichiba M, Kiuchi N, Fukada T, Hibi M, Hirano T. A central role for Stat3 in IL-6-induced regulation of growth and differentiation in M1 leukemia cells. EMBO J. 1996;15:3651–3658. [PMC free article] [PubMed] [Google Scholar]
- 42.Hedvat M, Huszar D, Herrmann A, Gozgit JM, Schroeder A, Sheehy A, Buettner R, Proia D, Kowolik CM, Hix H, Armstrong B, Bebernitz G, Weng S, Wang L, Ye M, McEachern K, Morosini D, Bell K, Alimzhanov M, Ioannidis S, McCoon P, Cao ZA, Yu H, Jove R, Zinda M. The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer Cell. 2009;16:487–497. doi: 10.1016/j.ccr.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manshouri T, Quintás-Cardama A, Nussenzveig RH, Gaikwad A, Estrov Z, Prchal J, Cortes JE, Kantarjian HM, Verstovsek S. The JAK kinase inhibitor CP-690, 550 suppresses the growth of human polycythemia vera cells carrying the JAK2V617F mutation. Cancer Sci. 2008;99:1265–1273. doi: 10.1111/j.1349-7006.2008.00817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao M, Gao FH, Wang JY, Liu F, Yuan HH, Zhang WY, Jiang B. JAK2/STAT3 signaling pathway activation mediates tumor angiogenesis by upregulation of VEGF and bFGF in non-small-cell lung cancer. Lung Cancer. 2011;73:366–374. doi: 10.1016/j.lungcan.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 45.Yin ZJ, Jin FG, Liu TG, Fu EQ, Xie YH, Sun RL. Overexpression of STAT3 potentiates growth, survival, and radio-resistance of non-small-cell lung cancer (NSCLC) cells. J. Surg. Res. 2011;171:675–683. doi: 10.1016/j.jss.2010.03.053. [DOI] [PubMed] [Google Scholar]
- 46.Song L, Turkson J, Karras JG, Jove R, Haura EB. Activation of STAT3 by receptor tyrosine kinases and cytokines regulates survival in human non-small-cell carcinoma cells. Oncogene. 2003;22:4150–4165. doi: 10.1038/sj.onc.1206479. [DOI] [PubMed] [Google Scholar]
- 47.OECD. OECD Guides for Testing of Chemicals. Organization for Economic Cooperation and Development; Washington, DC: 2001. Acute oral toxicity-up and down procedure. http://ntp.niehs.nih.gov/iccvam/suppdocs/feddocs/oecd/oecd_gl425-508.pdf. [Google Scholar]
- 48.Real P, Sierra A, De JA, Segovia J, Lopez-Vega J, Fernandez- Luna J. Resistance to chemotherapy via Stat3-dependent overexpression of Bcl-2 in metastatic breast cancer cells. Oncogene. 2002;21:7611–7618. doi: 10.1038/sj.onc.1206004. [DOI] [PubMed] [Google Scholar]
- 49.Wang T, Niu G, Kortylewiski M, Burdelya L, Shain K, Zhang S, Bhattacharya R, Gabrilovin D, Heller R, Coppola D, Dalton W, Jove R, Pardoll D, Yu H. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nature Med. 2004;10:48–54. doi: 10.1038/nm976. [DOI] [PubMed] [Google Scholar]
- 50.Kusaba T, Nakayama T, Yamazumi K, Yakata Y, Yoshizaki A, Nagayasu T, Sekine I. Expression of p-STAT3 in human colorectal adenocarcinoma and adenoma; correlation with clinicopathological factors. J. Clin. Pathol. 2005;58:833–838. doi: 10.1136/jcp.2004.023416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma XT, Wang S, Ye YJ, Du RY, Cui ZR, Somsouk M. Constitutive activation of Stat3 signaling pathway in human colorectal carcinoma. World J. Gastroenterol. 2004;10:1569–1573. doi: 10.3748/wjg.v10.i11.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim E, Kim M, Woo D-H, Shin Y, Shin J, Chang N, Oh YT, Kim H, Rheey J, Nakano I, Lee C, Joo KM, Rich JN, Nam D-H, Lee J. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23:839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berishaj M, Gao SP, Ahmed S, Leslie K, Al-Ahmadie H, Gerald WL, Bornmann W, Bromberg JF. Stat3 is tyrosine-phosphorylated through the interleukin-6/glycoprotein 130/Janus kinase pathway in breast cancer. Breast Cancer Res. 2007;9:R32. doi: 10.1186/bcr1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang P, Zhao Y, Zhu X, Sedwick D, Zhang X, Wang Z. Cross-talk between phospho-STAT3 and PLCγ1 plays a critical role in colorectal tumorigenesis. Mol. Cancer Res. 2011;9:1418–1428. doi: 10.1158/1541-7786.MCR-11-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin L, Deangelis S, Foust E, Fuchs J, Li C, Li P-K, Schwartz EB, Lesinski GB, Benson D, Lü J, Hoyt D, Lin J. A novel small molecule inhibits STAT3 phosphorylation and DNA binding activity and exhibits potent growth suppressive activity in human cancer cells. Mol. Cancer. 2010;9:217. doi: 10.1186/1476-4598-9-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu A, Liu Y, Jin Z, Hu Q, Lin L, Jou D, Yang J, Xu Z, Wang H, Li C, Lin J. XZH-5 inhibits STAT3 phosphorylation and enhances the cytotoxicity of chemotherapeutic drugs in human breast and pancreatic cancer cells. PLoS One. 2012;7:e46624. doi: 10.1371/journal.pone.0046624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gu F-M, Li Q-L, Gao Q, Jiang J-H, Huang X-Y, Pan JF, Fan J, Zhou J. Sorafenib inhibits growth and metastasis of hepatocellular carcinoma by blocking STAT3. World J. Gastroenterol. 2011;17:3922–3932. doi: 10.3748/wjg.v17.i34.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hartman ZC, Poage GM, den Hollander P, Tsimelzon A, Hill J, Panupinthu N, Zhang Y, Mazumdar A, Hilsenbeck SG, Mills GB, Brown PH. Growth of triple-negative breast cancer cells relies upon coordinate autocrine expression of the proinflammatory cytokines IL-6 and IL-8. Cancer Res. 2013;73:3470–3480. doi: 10.1158/0008-5472.CAN-12-4524-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han Y, Amin HM, Franko B, Frantz C, Shi X, Lai R. Loss of SHP1 enhances JAK3/STAT3 signaling and decreases proteosome degradation of JAK3 and NPM-ALK in ALK+ anaplastic large-cell lymphoma. Blood. 2006;108:2796–2803. doi: 10.1182/blood-2006-04-017434. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Q, Wang HY, Marzec M, Raghunath PN, Nagasawa T, Wasik MA. STAT3- and DNA methylatransferase 1-mediated epigenetic silencing of SHP-1 tyrosine phosphatase tumor suppressor gene in malignant T lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 2005;102:6948–6953. doi: 10.1073/pnas.0501959102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kang HJ, Yi YW, Kim HJ, Hong YB, Seong YS, Bae I. BRCA1 negatively regulated IGF-1 expression through an estrogen-responsive element-like site. Cell Death Dis. 2012;3:e336. doi: 10.1038/cddis.2012.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yi YW, Hong W, Kang HJ, Kim HJ, Zhao W, Wang A, Seong YS, Bae I. Inhibition of the PI3K/AKT pathway potentiates cytotoxicity of EGFR kinase inhibitors in triple-negative breast cancer cells. J. Cell. Mol. Med. 2013;17:648–656. doi: 10.1111/jcmm.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bea I, Rih JK, Kim HJ, Kang HJ, Haddad B, Kirilyuk A, Fan S, Avantaggiati ML, Rosen EM. BRCA1 regulates gene expression for orderly mitotic progression. Cell Cycle. 2005;4:1641–1666. doi: 10.4161/cc.4.11.2152. [DOI] [PubMed] [Google Scholar]
- 64.Yi YW, Kang HJ, Kim HJ, Kong Y, Brown ML, Bae I. Targeting mutant p53 by a SIRT1 activator YK-3-237 inhibits the proliferation of triple-negative breast cancer cells. Oncotarget. 2013;4:984–994. doi: 10.18632/oncotarget.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schneider CA, Rasband WS, Eliceiri KW. NIH image to imageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.