Abstract
Spinal muscular atrophy (SMA) is a devastating childhood motor neuron disease. SMA is caused by mutations in the survival motor neuron gene (SMN1), leading to reduced levels of SMN protein in the CNS. The actin-binding protein plastin 3 (PLS3) has been reported as a modifier for SMA, making it a potential therapeutic target. Here, we show reduced levels of PLS3 protein in the brain and spinal cord of a mouse model of SMA. Our study also revealed that lentiviral-mediated PLS3 expression restored axonal length in cultured Smn-deficient motor neurons. Delivery of adeno-associated virus serotype 9 (AAV9) harboring Pls3 cDNA via cisterna magna in SMNΔ7 mice, a widely used animal model of SMA, led to high neuronal transduction efficiency. PLS3 treatment allowed a small but significant increase of lifespan by 42%. Although there was no improvement of phenotype, this study has demonstrated the potential use of Pls3 as a target for gene therapy, possibly in combination with other disease modifiers.
Keywords: SMA, plastin 3, gene therapy
Introduction
Spinal muscular atrophy (SMA) is a recessive, monogenic motor neuron disease carried by approximately 1/40 people, and accounting for 1/6,000 to 1/10,000 live births. The most common form, SMA type I, manifests within 6 months of birth as a progressive dystonia, beginning at the lower extremities and progressing inward. Death generally occurs at around 18 months due to respiratory complications, a result of weakened intercostal muscles. The disease is caused by homozygous deletion of survival of motor neuron (SMN1) gene, which encodes a 294-aa protein implicated in RNA splicing (as part of the small nuclear ribonucleoprotein [snRNP] complex), stabilization of the axonal growth cone in neurons, and the formation of neuromuscular junctions. Disease severity correlates with patients’ copy number of SMN2, an inverted set of repeats of SMN1 with a single mutation that results in 90% of its translated product being truncated and non-functional. However, families with identical SMN2 copy numbers may have radically different disease severity implying that additional factors must contribute to the rescue of homozygous SMN1 deletion.
A transcriptome-wide differential expression analysis performed on total RNA from lymphoblastoid cells taken from SMN1-deleted siblings with discordant disease outcomes revealed a significant association between disease severity and Plastin 3 (PLS3) expression.1 Further analysis of blood samples from a diverse population of SMA patients and from healthy controls confirmed this association.1 Stratified analysis of blood samples from SMA type I, II, and III patients demonstrated that postpubertal females only show an inverse correlation between circulating Pls3 and SMA severity, suggesting a role for PLS3 as an age and sex specific modifier of SMA. It has been suggested that this sex-specific effect may be due to PLS3 mapping to Xq23.2
In smn−/− zebrafish model, Pls3 mRNA splicing and protein stability are unaffected, but Pls3 protein levels overall are reduced. Partially restoring PLS3 in these animals rescued presynaptic defects and motor behavior, suggesting that PLS3 is not merely a correlate of SMN expression, but instead plays a significant role in SMA pathology. PLS3 orthologs act as SMN modifier genes in Caenorhabditis elegans and Drosophila, possibly involving pathways of endocytosis and RNA processing.3
Investigations into the function of PLS3 have begun to shed light on its role in protecting against SMA. Removal of PLS3’s ability to bind to or cross-link actin filaments, by deleting one or both of its acting binding domains (ABD1/2), did not fully prevent the molecule from rescuing smn−/− mutant zebrafish. This suggests that PLS3’s role might involve an additional function unrelated to its interaction with actin. Further deletion studies determined that the EF hand motifs and specifically their interaction with Ca2+ ions are essential for PLS3 function in motor neuron development.4 This is supported by the observation in SMA of calcium dysregulation in the growth cone during motor axon outgrowth.5
In the present study, we assess the expression pattern of PLS3 in SMNΔ7, a well-characterized mouse model of SMA,6 as well as evaluate the neuroprotective effect of lentiviral gene transfer of Pls3 in an in vitro model of SMA (purified motor neurons). We provide evidence that PLS3 is important for axonogenesis, since its overexpression rescued the axon length and outgrowth defects associated with Smn depletion in motor neurons derived from SMNΔ7 embryos. AAV9-mediated Pls3 gene delivery in SMNΔ7 through cisterna magna led to efficient gene transfer to motor neurons and marginally extended mouse survival.
Results
PLS3 Expression in SMA Mouse Model
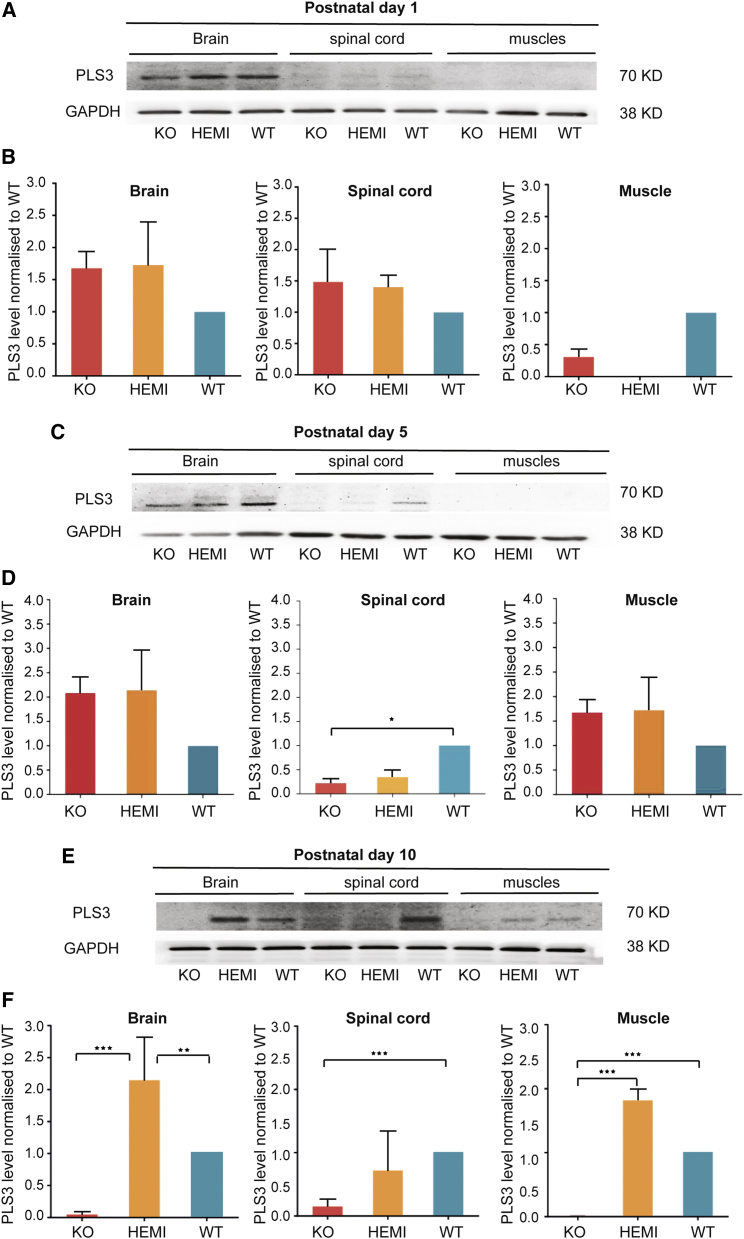
The severity of SMA is determined primarily by levels of SMN protein produced from SMN2 copy of the gene.5, 7 Discordant families have also been described in which siblings have the same genetics but present with different phenotypes, with some siblings affected and others unaffected.8 This suggests that disease modifiers other than SMN may exist. A transcriptome-wide differential expression analysis performed on total RNA from lymphoblastoid cells taken from SMN1-deleted siblings with discordant disease outcomes revealed a significant association between disease severity and PLS3 expression. PLS3, an actin-binding and bundling protein, was identified as a protective modifier of SMA.1 Here, we assessed Pls3 levels within the brain, spinal cord, and muscle of the SMNΔ7 mice (SMN2+/+, SMNΔ7+/+, smn−/−; defined as knockout [KO]) compared to their wild-type (SMN2+/+, SMNΔ7+/+, smn+/+) and carrier (SMN2+/+, SMNΔ7+/+, smn+/−) littermates at postnatal days 1, 5, and 10 (P1, P5, and P10) (n = 3). Proteins were extracted and PLS3 expression measured by western blot. Upon analysis at postnatal day 1, we observed no statistically significant difference between genotypes for PLS3 protein levels in the brain, spinal cord, or in thigh muscle (Figures 1A and 1B). None of the genotypes showed detectable PLS3 protein in muscle and only low detection in the spinal cord (Figures 1A and 1B). This suggests that Smn deficiency does not appear to affect PLS3 levels at P1. There was no discernible difference between protein levels in any of the genotypes at this age, which correlates with the lack of phenotype in P1 pups. Based on the general appearance and measurement of body weight, the KO pups were phenotypically indistinguishable from their WT littermates in the first 48 hr after birth. PLS3 protein levels showed no significant difference in the P5 brains of all genotypes (Figure 1C). However, the current analysis revealed that Pls3 protein levels were significantly lower in P5 spinal cord of KO and carriers when compared to WT (p < 0.05, n = 3) (Figures 1C and 1D). PLS3 was undetectable in muscle of the 3 groups tested (Figures 1C and 1D).
Figure 1.
Plastin 3 Protein Levels in Postnatal Day 1, 5, and 10 SMNΔ7 Mice
Representative images of western blot and fold changes in PLS3 protein levels in brain, spinal cord, and muscle tissues from wild-type (WT), carrier littermates (HEMI), and SMNΔ7 (KO) postnatal day 1 (P1) (A and B), P5 (C and D), and P10 (E and F) neonate mouse pups (n = 3). The membrane was probed with mouse anti-PLS3 antibody; GAPDH was used as the house keeping control (n = 3). Error bars represent SEM. One-way ANOVA *p < 0.05, **p < 0.01, ***p < 0.001.
Western blot analysis performed at postnatal day 10 revealed a significant decline of Pls3 protein levels in the KO brain compared to previous time points (P1 and P5) and the same tissues in carriers and WT (Figures 1E and 1F), suggesting that Smn levels did affect PLS3 expression at P10 in the brain. It is also noteworthy to state the significant reduction of PLS3 protein in spinal cord of carrier pups (Figures 1E and 1F). Surprisingly PLS3 levels were higher in brains of carrier littermates compared to WT (Figures 1E and 1F).
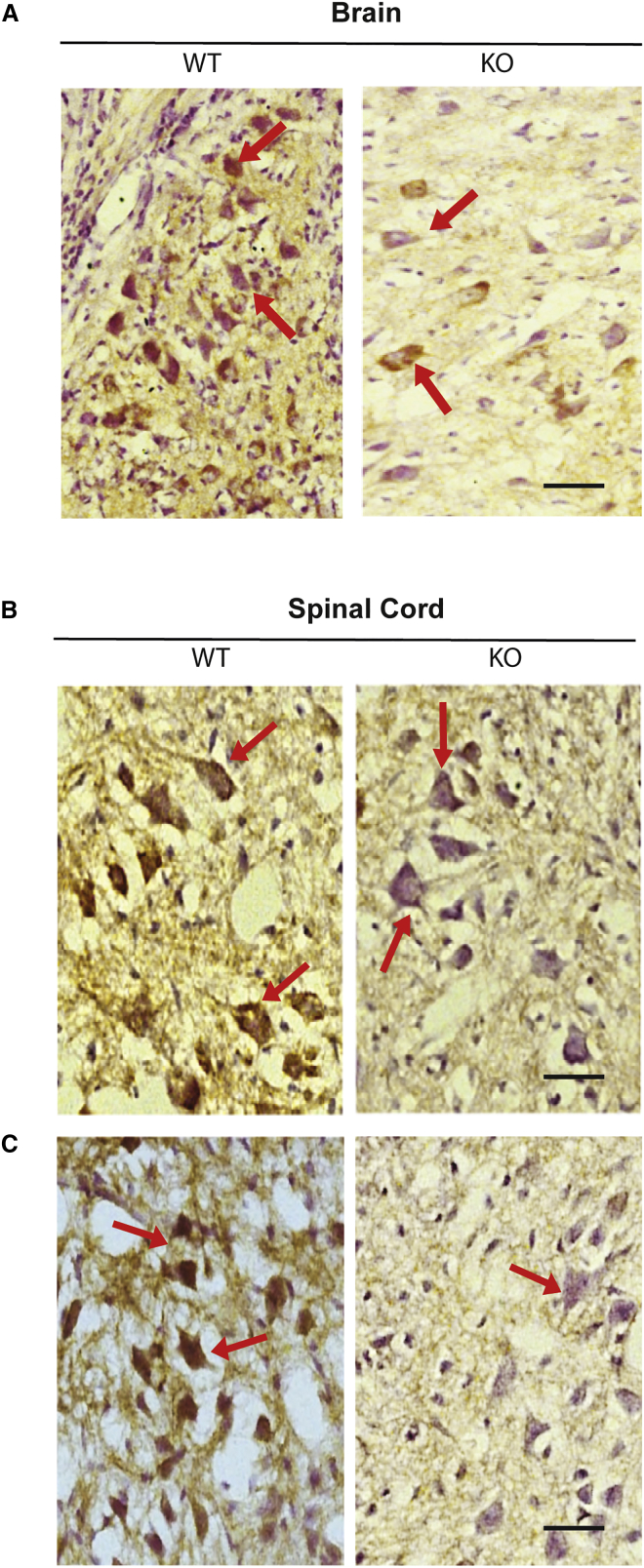
Having established that PLS3 protein is depleted in the brain, spinal cord, and muscle tissues from P10 Smn-deficient mice, we next set out to confirm the western blot findings by carrying out qualitative histological analysis. Consistent with western blotting data (Figures 1E and 1F), immunolabeling of brain and spinal cord sections with anti-PLS3 antibodies revealed reduction in PLS3 staining in P10 KO animals compared to WT group (Figure 2).
Figure 2.
PLS3 Immunoreactivity in the Brain and Spinal Cord Sections of WT and KO P10 Pups
(A) Immunohistochemical staining showing PLS3 expression in the brain from P10 wild-type (WT) and SMNΔ7 mice (KO) (n = 3 per group). Brain sections from WT showing immune reactivity of PLS3 associated with a large number of neurons in the brain cortex (brown) (arrows). SMNΔ7 mice (KO) brain sections showed reduced reactivity of PLS3 associated with neurons (arrows). Scale bar, 50 μm. PLS3 immunoreactivity in the cervical (B) and lumbar (C) spinal cord from P10 wild-type (WT) and SMNΔ7 (KO) mice (n = 3). WT spinal cord sections showing PLS3 immuno-reactivity in large number of neurons in the ventral horn of the spinal cord (brown) (arrows), when compared to sections from SMNΔ7 mice KO section, which showed minimal reactivity of PLS3 associated with neurons (arrows). Scale bar, 50 μm.
PLS3 Overexpression Promotes Motor Neuron Axonal Growth
We report here (Figures 1 and 2) that PLS3 is downregulated in the CNS of Smn-deficient mouse model, suggesting an important role of Pls3 in neurodegeneration associated with SMA. To test whether overexpression of PLS3 would prevent neurodegenerative phenotype, we constructed a lentiviral (LV) harboring human Pls3 cDNA and tested its efficiency in an in vitro model of SMA. We first assured that the generated LV vector can produce PLS3 protein in vitro in a neuronal-like cell line, NSC34 cells. Cells were transduced with LV-PLS3 vector at different MOIs of 5, 10, 20, 40, and 80. The western blot revealed the presence of a 70-kDa band representing the PLS3 protein (Figure 3A). Analysis of the dose response indicated a positive correlation between PLS3 expression and the MOI (Figure 3B). The transduced cells showed no overt signs of toxicity, as indicated by a change in cell morphology, reduced adherence or presence of dead cells, or obvious difference in growth rate even when the viral vector was used at higher MOI.
Figure 3.
Lentiviral-Mediated Pls3 Expression Rescues Axonal Deficit in Smn-Deficient Motor Neurons
(A) Western blot showing NSC34 cells transduced with LV-PLS3 at MOI 5, 10, 20, 40, or 80 and probed with anti-human PLS3 antibody; α-tubulin was the loading control (n = 3). Untransduced (UT) cells were considered as negative control. (B) A dose-response curve of LV-Pls3 at 5 days posttransduction, along with fold change in Pls3 protein levels for each MOI used. (C) Motor neurons isolated from E13 wild-type (WT), carrier (HEMI), and SMNΔ7 (KO) embryos were transduced with LV-Pls3 or LV-GFP at MOI 80. Cells were labeled for Pls3 (blue), tubulin (red), or GFP (green). Scale bars, 20 μm. (D) Impact of LV-Pls3 treatment on axonal length in Smn-deficient motor neurons. The bar chart shows the effect of LV-Pls3 treatment on axonal length in wild-type, carrier (HEMI), and SMN knockout (KO) embryos. One-way ANOVA (*p < 0.05, ***p < 0.001) was considered to be statistically significant. Data generated from four independent experiments (n = 4). Average neurite lengths ± SEM (n = 100 neurites).
To objectively quantify the impact of PLS3 on the neurodegenerative phenotype reported in SMA motor neurons in vitro, we measured neurite length in Smn-deficient motor neurons.9 Enriched spinal motor neurons derived from E13 SMNΔ7 embryos and cultured using the p75 immunopanning method10 were used to assess the impact of LV-PLS3 transduction (MOI 80). Motor neurons were visualized down the microscope and cell included in the analysis if the entire axon length could be seen in a single field. One hundred cells were imaged for each genotype and ImageJ was used to draw a line from the start of the axon at the cell body to the tip of the axon. In the case of axons with branches (Figure 3C), the thickest and longest branch was chosen as the main axon, and other branches were excluded from length calculations.
Consistent with previous reports,1, 9 our in vitro study showed a significant reduction in axon length of Smn-deficient motor neurons when compared to cells from either WT or carrier embryos (Figure 3D; p < 0.001). Our studies revealed that overexpression of PLS3 through LV led to a significant rescue (p < 0.001) of average axon length and outgrowth defects associated with Smn downregulation (n = 100) (Figures 3C and 3D). Expression of PLS3 in carrier and wild-type motor neurons also significantly increased axon length (p < 0.05). As expected, the GFP control virus showed no effect on axon length in any genotype. Our data revealed that PLS3 can rescue motor neuron (MN) axon deficits reported in SMNΔ7 primary MNs, providing evidence that PLS3 is important for axonogenesis.
AAV9-Mediated PLS3 Gene Therapy in SMNΔ7 Mouse Model of SMA
We next evaluated the neuroprotective ability of AAV9 encoding PLS3 in a mouse model (SMNΔ7) of SMA. AAV9-Pls3 virus was tested in P1 carrier pups prior efficacy study in SMNΔ7 mice. Four weeks after cisterna magna AAV9-GFP or AAV9-Pls3 vector delivery, animals were culled and spinal cord was collected for transgene assessment. PLS3 expression was assessed into the three different regions: cervical, thoracic, and lumbar spinal cord. The analysis summarized in Figure 4A demonstrates clear PLS3 overexpression in AAV9-Pls3-treated animals compared to GFP controls. A very weak band was seen in the control group injected with AAV9-GFP, indicating cross-species reactivity between the human anti-PLS3 antibody and the endogenous mouse PLS3; however, this was used as the baseline of PLS3 when measuring overexpression.
Figure 4.
AAV9-Mediated Gene Therapy in SMA Mouse Model
(A) Western blot analysis of the spinal cord following cisterna magna delivery of AAV9-Pls3. PLS3 protein levels in three parts of the spinal cord; cervical (C), thoracic (T), and lumbar (L) of carrier (HEMI) pups 4 weeks postdelivery of AAV9-GFP or AAV9-Pls3. The membrane was incubated with anti-PLS3 antibody. (B) Body weight growth in SMNΔ7 injected with AAV9-Pls3 (n = 7), AAV9-GFP (n = 7), or PBS controls (n = 7). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; AAV9-Pls3 versus AAV9-GFP; two-way ANOVA with Dunnett’s multiple comparisons test. (C) Kaplan-Meier survival plot to compare lifespan between all experimental groups of mice (n = 7 per group; overall log-rank test p < 0.05 between all three groups; pairwise comparisons with Bonferroni correction *p < 0.05 between PLS3 and PBS and #p < 0.05 between PLS3 and GFP). (D) AAV9-Pls3 transduction efficiency in spinal motor neurons. Carrier littermate pups were injected at postnatal day 1 with AAV9-Pls3; spinal cords were extracted at 4 weeks postinjection. The cervical and lumbar spinal cord sections were fixed and labeled with calcitonin gene-related peptide (CGRP) antibody as a motor neuronal marker (red), PLS3 antibody as an indicator of transduction (green), and DAPI for visualization of nuclei. Scale bar, 20 μm.
Our in vivo proof-of-concept study assessed the survival of SMNΔ7 mice following delivery of AAV9 vector-encoding PLS3 (AAV9-Pls3) into cerebrospinal fluid (CSF). P1 SMNΔ7 KO pups were injected with 5 μL of AAV9-Pls3 containing 5 × 1010 vg (n = 7) into the cisterna magna. As a control, a second group of SMNΔ7 KO mice was treated with AAV9-GFP (5 × 1010 vg) (n = 7), and a third group of animals injected with PBS (n = 7). Daily assessment of the mice was performed throughout their life to determine whether there was any effect on SMNΔ7 mouse phenotype. We report a significant difference in body weight between AAV9-Pls3-treated group when compared to AAV9-GFP and PBS controls (p < 0.001) (Figure 4B). Kaplan-Meier analysis demonstrated that PLS3-treated mice had longer survival time compared to both untreated and GFP-treated groups (Figure 4C; p < 0.05). Overall, this study revealed a marginal but significant increase in survival of PLS3-treated mice compared to controls.
Finally, transduction efficiency was assessed to ensure that the marginal efficacy was not due to inefficient viral vector delivery to the CNS, in particular to MNs. Cervical and lumbar spinal cord sections were double-labeled for PLS3 and calcitonin gene-related peptide (CGRP), previously used as marker for MNs11 (Figure 4D). PLS3 was detected in motor neurons and neuronal axons, showing that the virus is targeting motor neurons in the spinal cord (Figure 4D). The PLS3-positive MNs were counted and transduction efficiency calculated as the percentage of total CGRP-positive MNs. Not all CGRP-positive motor neurons expressed PLS3, instead quantification revealed around 29% of motor neurons were PLS3 positive.
Discussion
SMA is a debilitating fatal childhood neuromuscular disease, characterized by the degeneration of the anterior horn MNs of the spinal cord due to reduced expression of the SMN protein. In addition to SMN replacement approaches,12, 13 previous studies have reported other disease-modifying genes, which could also be targeted for gene-therapy-mediated treatment for SMA, including PLS3, PTEN, and hnRNP-R.1, 9, 14, 15 Investigation into the function of PLS3 has begun to shed light on its interaction with SMN and its role in neuroprotection against SMA. This study aimed to test whether overexpression of PLS3 by viral-mediated gene therapy could rescue neuronal loss and increase survival of an SMA mouse model and whether it could ultimately be considered for potential clinical therapy.
PLS3 Levels in the SMNΔ7 SMA Mouse Model
Our expression profile study revealed that Pls3 is expressed in neuronal cells in the brainstem and in spinal MNs. Its distribution was as expected in the spinal cord, being expressed in cell bodies of MNs and the white matter where the axons of the neurons are located. This aligns with previous observations that Pls3 co-localizes with Smn in granules throughout motor neuron axons.1 If both proteins are co-localized, they may have the ability to interact. They could influence each other’s stability, which has also been suggested by other groups.3
Our data confirmed differential Pls3 expression between Smn KO mice from the SMNΔ7 line and their wild-type littermates. At birth, there was no difference in Pls3 expression between the genotypes; however, by end-stage of the disease (P10) the KO animals showed significantly reduced or absent Pls3 protein in both CNS and muscles when compared to wild types. Carrier mice also showed lower Pls3 levels in the spinal cord, although not in brain or muscles, suggesting that Smn protein levels are influencing Pls3 expression in some tissues. This supports previous reports of Pls3 downregulation in the brain and spinal cord of other SMA animal models and in human patients.16 This report provides evidence that the SMNΔ7 mouse is a good model for testing whether modulation of PLS3 can be used as a gene therapy for SMA.
The molecular mechanisms for PLS3’s involvement in SMA are unclear; there are no structural or sequence similarities between SMN and PLS3 that may suggest they are functionally related, but several theories have been proposed. PLS3 overexpression has been demonstrated to increase levels of F-actin, and it has been suggested that its role in stabilizing actin filaments is crucial to axon development and therefore rescue of SMA pathology. However, the effects of expressing a set of partial Pls3 molecules in zebrafish support a mechanism for PLS3 in SMA beyond that of actin binding.4
Overexpression of PLS3 by Viral Vectors
This study has demonstrated that successful cloning of human Pls3 cDNA into LV or AAV viral vectors was able to significantly overexpress PLS3 protein by at least 5-fold, in vitro and in vivo. In vitro, overexpression of PLS3 does not appear to be toxic to cells. In addition, LV-mediated expression of PLS3 in cells lacking Smn rescued the axonal length deficit. Administration of AAV9-Pls3 to the CNS, via cisterna magna, in postnatal day 1 SMNΔ7 mice, led to significant spinal MN transduction efficiency and extension of SMNΔ7 mouse lifespan. Although PLS3 overexpression induced small but significant extension in SMNΔ7 mouse survival, this gene transfer strategy did not lead to noticeable amelioration of the SMA phenotype. It is worth highlighting that AAV9 administered in P1 pups showed extensive neuronal tropism. Previous studies have shown that astrocytes from SMA-induced pluripotent stem cells and in SMNΔ7 mouse spinal cord exhibit cellular and morphological changes as an indicator of activation before obvious MN loss.17 Our in vivo data and the later reports indicate therefore that early disruption of astrocytes may contribute to SMA disease pathology, which suggests that targeting both MNs and glial cells may be needed for an effective therapy in SMA.
The marginal effect of PLS3 gene transfer in a SMA mouse model could be explained by various factors: (1) SMNΔ7 mouse model displays severe phenotype with short lifespan. The timing of vector application is therefore critical for SMA therapy development. It is likely that AAV-mediated gene transfer would require early start of therapeutic gene expression. However, this was not the case when using single-stranded DNA AAV9 in our study. Future efforts could look at reducing the size of PLS3 to fit the packaging capacity of self-complementary AAV. This could be achieved by identification of the short gene fragment that could lead to full-length protein expression. (2) It is widely reported that motor neurons and other cell types are affected by SMA. Several reports challenge the notion that SMA is solely a disease of motor neurons and demonstrate peripheral organ defects and malfunction in association with SMN deficiency.18, 19, 20 Thus, the choice of route of delivery such as systemic could be important for optimal efficacy. (3) The key feature of SMA is the loss of SMN, which is not addressed by overexpression of PLS3. PLS3 may be able to rescue some SMN functions but not all, such as calcium dysregulation, snRNP assembly, and neuronal development, which has been reported to be implicated in SMA.17 This, along with the previous observation in humans, where PLS3 provides full protection against SMA only in SMN1-deleted individuals carrying three to four SMN2 copies, but not in those with only two SMN2 copies or less, suggests that certain amount of SMN is required in order to benefit from PLS3 overexpression in individuals lacking SMN1.
In summary, evidence has been provided supporting the role of PLS3 in the pathology of SMA, and restoration of PLS3 expression using a LV can rescue the axonal phenotype in Smn-deficient cultured motor neurons. Consistent with recent study,21 we report here that AAV9-mediated PLS3 overexpression can improve the survival of SMNΔ7 mouse model of SMA. The use of PLS3 overexpression to extend SMNΔ7 lifespan demonstrates that PLS3 is a disease modifier of SMA. PLS3 is a potential therapeutic target and can be used in a gene therapy approach but maybe required to be delivered in combination with other disease modulators to give meaningful rescue to SMA and alleviate the disease phenotype.
Materials and Methods
Breeding and Genotyping of Transgenic Mice
Triple mutant mice (FVB.Cg-Tg [SMN2*delta7] 4299Ahmb Tg [SMN2]89Ahmb Smn1tm1Msd/J SMNΔ7 mice) were used for all in vivo studies. SMNΔ7 mice were purchased from The Jackson Laboratory (stock #005025) and were maintained in a controlled facility in a 12-hr dark/12-hr light photocycle (on at 7 a.m./off at 7 p.m.) with free access to food and water. Carrier (SMΔ7+/+; SMN2+/+; Smn+/−) animals were used for breeding and the offspring were genotyped immediately after birth at postnatal day 1 (P1) by PCR amplification of the transgenes according to the protocols provided by The Jackson Laboratory. The mice are lacking the murine Smn gene (Smn−/−),6 but they have two transgenes inserted, a copy of the human SMN gene lacking exon 7 (SMΔ7) and the full-length human SMN2 gene (SMN2+/+). SMNΔ7 copy number was verified as previously described.6 Offspring (SMΔ7+/+; SMN2+/+; Smn−/−) from the same breeding pair were allocated randomly to the experimental groups. Viral vectors (AAV9-PLS3 or AAV9-GFP; 5 × 1010vg) were delivered via cisterna magna in P1 pups. All in vivo experiments were regulated under the UK Home Office Animals (Scientific Procedures) Act 1986.
Western Blotting
Brain, spinal cord, and muscle tissues from SMNΔ7 mice were homogenized in radioimmunoprecipitation assay (RIPA) buffer (5% Tris-HCl [pH 7.]), 1% NP-40, 0.5% Sodium deoxycholate, 0.01% SDS, 150 mM NaCl, 0.2 mM EDTA) supplemented with 1% protease inhibitor cocktail (Sigma-Aldrich, S8830). Subsequently, total protein concentrations were determined using the BCA Protein Assay Kit (Pierce, 23225) according to the manufacturer’ protocol. Denatured proteins samples (20 μg protein/sample) were separated by electrophoresis on 12% SDS-PAGE and transferred onto a polyvinylidene difluoride (PVDF) membrane (Bio-Rad). The membranes were probed with the following primary antibodies: polyclonal rabbit anti-mouse PLS3 ARP56623_P050 PLS3 antibody-middle region (Aviva, 1/500), mouse anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Sigma-Aldrich, 1/1,000). Secondary antibodies were goat anti-rabbit immunoglobulin (Ig) (DakoCytomation, P0214, 1/1,000) and goat anti-mouse IgG (DakoCytomation, P0448, 1/10,000). Signals were detected using chemiluminescence reagent (Super Signal West Pico, Pierce) according to standard protocols. Densitometry of the signals was measured using Chemilmager software. Values were normalized to the average of the reference proteins (GAPDH) from the same mouse. At least three replicates were performed for each sample.
Immunohistochemistry
Animals were terminally anesthetized with pentobarbital and then transcardially perfused with PBS followed by 4% paraformaldehyde in PBS. Tissues were then postfixed for 24 to 48 hr in paraformaldehyde at 4°C. Brains and spinal cords were then cryoprotected in 30% sucrose for at least 24 hr at 4°C and mounted in optimal cutting temperature compound (OCT) (Fisher Scientific, 14-373-65). Tissue sections of 20-μm thickness were prepared on a sliding cryostat microtome (Leica) and collected onto gelatin-coated microscope slides. Immunohistochemistry was then performed with rabbit antibody to PLS3 (1:50; Abcam) followed by Alexa Fluor 568 goat anti-rabbit IgG (1:1000; Invitrogen A11036).
Primary Motor Neuron Culture
Primary motor neurons were purified from lumbar spinal cords of embryonic day 13 (E13) mouse embryos and cultured at 37°C with 5% CO2 at 95% humidity. In brief, 10 mm coverslips were coated with 5% poly-DL-ornithine hydrobromide (PORN) (Sigma, P8638) overnight, washed with Hank’s Balanced Salt Solution (HBSS) (Sigma, H8264) then coated with 2.5μg/mL Laminin (Invitrogen, 23017-015) in incubator at 37°C with 5% CO2 at 95% humidity. Embryos were collected at E13 after sacrificing pregnant mice by cervical dislocation. Dissection buffer Beta-Mercaptoenthanol—10 nM concentration 1:1,000 (β-ME) (Invitrogen, 313500) in HBSS, was used to place the extracted spinal cords. The cells were suspended for 15 min at 37°C with 0.1% trypsin, Worthington 3× crystallized, dissolved in HBSS with NaOH added to adjust the pH and filtered through a 0.22 μm sterile filter, (Worthington, TLR3-3707). Trypsin inhibitor—Sigma, 500 mg dissolved in 49 mL HBSS and 1ml of 1 M HEPES (Sigma, T-6522) and filtered through a 0.22 μm sterile filter then was used to deactivate trypsin.
In order to select motor neurons, P75 panning antibody—p75 NGF receptor antibody (Abcam, ab8877) monoclonal antibody against extracellular domain of p75 was used at a concentration of 1:5,000 in 10 mM Tris (pH 9.5) to precoat the plate before placing the cell suspension in order to specifically select motor neurons. The primary motor neurons were detached by depolarizing solution (30mM KCl [Sigma, P9333] + 0.8% NaCl [Sigma-Aldrich, S7653]), while the unattached cells were washed off with (Neurobasal medium [NB medium] GIBCO [Invitrogen], with 0.2 μM β-ME). The primary motor neurons were plated on the precoated coverslips at concentration of 2,000–5,000 cells/coverslip in GIBCO NB supplemented with 2% of B27 (GIBCO, 12587-010), 1% glutamax (GIBCO, 35050), 2% (v/v) horse serum donor (Linaris, SHD3250YK), 100 U/mL of penicillin and 100 μg/mL streptomycin, 0.2 μM β-ME, and 0.5 mM L-glutamine (GIBCO, 35050) and incubated for 24 hr before transduction. Cells were incubated at 37°C in a humidified incubator with 5% CO2.
Construction and Production of AAV-PLS3 Vectors
A single-stranded DNA AAV plasmid AAV expressing PLS3 was generated by extracting the PLS3 gene from pcDNA3.1-PLS3-V5/His6 vector and cloning into pAAV cytomegalovirus (CMV) multiple cloning site (MCS). First, PLS3 was PCR amplified using the following primers: PLS3-EcoRI-forward, GAT GCC GAATTC CAG CGT GCC ACC ATG GAT GAG ATG GCT ACC ACT, and PLS3-XbaI-reverse, AGCAA TCTAGA TTA CAC TCT CTT CAT TCC C. The PCR product was treated with restriction enzymes EcoRI and XbaI and subcloned into EcoRI-XbaI digested pAAV CMV MCS vector. High-titer AAV9 vectors were then prepared using a three-plasmid transient co-transfection system involving a plasmid encoding the Rep2Cap9 sequence (pAAV2/9), a helper plasmid (pHelper, Stratagene) and the vector genomes (AAV-GFP or AAV-PLS3) as previously described.22, 23 AAV9 titers were evaluated by quantitative PCR assays using primers directed against GFP and linearized AAV-GFP as a standard curve.22, 23
In Vivo Studies
All mouse experiments were approved by the University of Sheffield Ethical Review Sub-Committee and the UK Animal Procedures Committee (London, UK) and performed according to the Animal (Scientific Procedures) Act 1986, under the Project License 40/3739. SMNΔ7 mice were purchased from The Jackson Laboratory (stock #005025) and were maintained in a controlled facility in a 12-hr dark/12-hr light photocycle (on at 7 a.m./off at 7 p.m.) with free access to food and water. Carrier (SMΔ7+/+; SMN2; Smn+/−) animals were used for breeding, and the offspring were genotyped immediately after birth at postnatal day 1 (P1) by PCR amplification of the transgenes according to the protocols provided by The Jackson Laboratory. SMNΔ7 copy number was verified as previously described.6 Offspring (SMNΔ7+/+; SMN2; Smn−/−) from the same breeding pair were allocated randomly to the experimental groups. Viral delivery was achieved as previously reported.23 In brief, AAV9-PLS3 (n = 7) or AAV9-GFP (n = 7) was administered via cisterna magna in postnatal day 1 pups. 5 × 1010 vg of viral vector solution was injected to each animal using a 33-gauge Hamilton syringe (ESS Lab). The animals were left to recover before being rolled in sawdust from their cage and returned to the cage with their mother.
Design and Production of LV
A LV expressing PLS3 was generated by extracting the PLS3 gene from pcDNA3.1-PLS3-V5/His6 Vector, provided by Prof. Brunhilde Wirth, (University Hospital of Cologne, Germany) and cloning into the LV backbone (SIN-PGK-cPPT-WHV; kindly provided by Dr. Nicole Deglon). Double digestion for all the samples with BamHI and XhoI restriction enzymes confirmed the presence of the insert. In addition, sequencing of the insert was achieved using primers targeted to PGK promoter: PGK forward, 5′- GGGAATTCCGATAATCAAC-3′ and PGK reverse, 5′-CCTTCGCTTTCTGGGCTCA-3′. The sequencing confirmed the correct orientation of the insert.
LV production was achieved as previously described.24 In brief, 3 × 106 HEK293T cells were transfected after 24 hr of plating with a mix of four plasmids: pMD.2G: plasmid encoding the vesicular stomatitis virus G (VSV-G) envelope, pCMVDR8.92; packaging plasmid encoding all the viral genes needed in trans, SIN-W-PGK; transfer plasmid containing (PLS3) and pRSV-Rev; plasmid encoding the rev protein of human immunodeficiency virus (HIV)-1. Viral production was performed as previously described.24 At 72 hr posttransfection, the viral supernatant was harvested, filtered through 0.45 μm filter and concentrated by ultracentrifugation for 90 min at 19,000 rpm and at 4°C. The viral pellet was suspended in 1.8 mL PBS supplemented with 1% BSA (Sigma), divided into 50 μL aliquots and stored at −80°C. Finally, LV titration was performed by Enzyme Linked Immunosorbent Assay (ELISA) which targets HIV-1 p24 antigen (ZMC No. 0801111, ZeptoMetrix Corporation).
In Vitro Transduction and Immunostaining of Primary Motor Neurons
Primary motor neurons were isolated at E13 from SMA (Smn+/+; SMN2+/+; Smn−/−) and carrier littermate (Smn+/+; SMN2+/+; Smn+/−) embryos. Twenty-four hours after plating, MNs were transduced with LV-PLS3 or LV-GFP virus at an MOI of 5, 10, 20, 40, and 80 for immunocytochemistry. Transduction efficiency was assessed by counting the GFP-positive cells under the fluorescence microscope after transduction with LV-GFP and was found to be 70%–80% at an MOI of 40. Plates were incubated at 37°C and 5% CO2 for 7 days and immunostained with anti-Tubulin and anti-Pls3. Double staining with anti-tubulin 1:500 and anti-Pls3 1:500 was performed overnight. Then the secondary antibodies were then added at 1:200 for 2 hr at RT: goat anti-rabbit A568 Alexa red (Pls3), goat anti-mouse A350 blue (Tubulin). Images were taken by confocal microscope. The axon length was measured by ImageJ plugin NeuronJ.
Statistical Analysis
Statistical analysis was performed with GraphPad Prism 6. Statistical significance was assessed with one- or two-way ANOVA. A value of p < 0.05 was considered to be statistically significant.
Author Contributions
A.A. conducted the experiments; E.K. produced lentivectors and AAV; I.C. and K.E.L. assisted with mouse work; K.I. and K.N. helped with cell line and motor neuron cultures. M.A. supervised the work. A.A. and M.A. designed the experiments and wrote the paper.
Conflicts of Interest
The authors have declared that no conflicts of interest exist.
Acknowledgments
We wish to thank Prof. Brunhilde Wirth and Dr. Nicole Deglon for kindly sharing the pcDNA3.1-PLS3-V5/His6 and the lentiviral vector backbone (SIN-PGK-cPPT-WHV), respectively. This work was supported by the European Research Council grant (294745 GTNCTV). M.A. is also supported by an MRC DPFS Award (129016). A.A. was supported by scholarship from King Abdulaziz University. E.K. was sponsored by Eve Davis Studentship and ERC Advanced Award no. 294745 GTNCTV. M.A., I.C., and K.E.L. were sponsored by ERC Advanced Award no. 294745 GTNCTV.
References
- 1.Oprea G.E., Kröber S., McWhorter M.L., Rossoll W., Müller S., Krawczak M., Bassell G.J., Beattie C.E., Wirth B. Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science. 2008;320:524–527. doi: 10.1126/science.1155085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stratigopoulos G., Lanzano P., Deng L., Guo J., Kaufmann P., Darras B., Finkel R., Tawil R., McDermott M.P., Martens W. Association of plastin 3 expression with disease severity in spinal muscular atrophy only in postpubertal females. Arch. Neurol. 2010;67:1252–1256. doi: 10.1001/archneurol.2010.239. [DOI] [PubMed] [Google Scholar]
- 3.Dimitriadi M., Sleigh J.N., Walker A., Chang H.C., Sen A., Kalloo G., Harris J., Barsby T., Walsh M.B., Satterlee J.S. Conserved genes act as modifiers of invertebrate SMN loss of function defects. PLoS Genet. 2010;6:e1001172. doi: 10.1371/journal.pgen.1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyon A.N., Pineda R.H., Hao T., Kudryashova E., Kudryashov D.S., Beattie C.E. Calcium binding is essential for plastin 3 function in Smn-deficient motoneurons. Hum. Mol. Genet. 2014;23:1990–2004. doi: 10.1093/hmg/ddt595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGovern V.L., Gavrilina T.O., Beattie C.E., Burghes A.H. Embryonic motor axon development in the severe SMA mouse. Hum. Mol. Genet. 2008;17:2900–2909. doi: 10.1093/hmg/ddn189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le T.T., Pham L.T., Butchbach M.E., Zhang H.L., Monani U.R., Coovert D.D., Gavrilina T.O., Xing L., Bassell G.J., Burghes A.H. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 7.McAndrew P.E., Parsons D.W., Simard L.R., Rochette C., Ray P.N., Mendell J.R., Prior T.W., Burghes A.H. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am. J. Hum. Genet. 1997;60:1411–1422. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernal S., Also-Rallo E., Martínez-Hernández R., Alías L., Rodríguez-Alvarez F.J., Millán J.M., Hernández-Chico C., Baiget M., Tizzano E.F. Plastin 3 expression in discordant spinal muscular atrophy (SMA) siblings. Neuromuscul. Disord. 2011;21:413–419. doi: 10.1016/j.nmd.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Rossoll W., Jablonka S., Andreassi C., Kröning A.K., Karle K., Monani U.R., Sendtner M. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of β-actin mRNA in growth cones of motoneurons. J. Cell Biol. 2003;163:801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiese S., Herrmann T., Drepper C., Jablonka S., Funk N., Klausmeyer A., Rogers M.L., Rush R., Sendtner M. Isolation and enrichment of embryonic mouse motoneurons from the lumbar spinal cord of individual mouse embryos. Nat. Protoc. 2010;5:31–38. doi: 10.1038/nprot.2009.193. [DOI] [PubMed] [Google Scholar]
- 11.Azzouz M., Ralph G.S., Storkebaum E., Walmsley L.E., Mitrophanous K.A., Kingsman S.M., Carmeliet P., Mazarakis N.D. VEGF delivery with retrogradely transported lentivector prolongs survival in a mouse ALS model. Nature. 2004;429:413–417. doi: 10.1038/nature02544. [DOI] [PubMed] [Google Scholar]
- 12.Foust K.D., Wang X., McGovern V.L., Braun L., Bevan A.K., Haidet A.M., Le T.T., Morales P.R., Rich M.M., Burghes A.H., Kaspar B.K. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat. Biotechnol. 2010;28:271–274. doi: 10.1038/nbt.1610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Valori C.F., Ning K., Wyles M., Mead R.J., Grierson A.J., Shaw P.J., Azzouz M. Systemic delivery of scAAV9 expressing SMN prolongs survival in a model of spinal muscular atrophy. Sci Transl Med. 2010;2:35ra42. doi: 10.1126/scitranslmed.3000830. [DOI] [PubMed] [Google Scholar]
- 14.Ning K., Drepper C., Valori C.F., Ahsan M., Wyles M., Higginbottom A., Herrmann T., Shaw P., Azzouz M., Sendtner M. PTEN depletion rescues axonal growth defect and improves survival in SMN-deficient motor neurons. Hum. Mol. Genet. 2010;19:3159–3168. doi: 10.1093/hmg/ddq226. [DOI] [PubMed] [Google Scholar]
- 15.Little D., Valori C.F., Mutsaers C.A., Bennett E.J., Wyles M., Sharrack B., Shaw P.J., Gillingwater T.H., Azzouz M., Ning K. PTEN depletion decreases disease severity and modestly prolongs survival in a mouse model of spinal muscular atrophy. Mol. Ther. 2015;23:270–277. doi: 10.1038/mt.2014.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hao T., Wolman M., Granato M., Beattie C.E. Survival motor neuron affects plastin 3 protein levels leading to motor defects. J. Neurosci. 2012;32:5074–5084. doi: 10.1523/JNEUROSCI.5808-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGivern J.V., Patitucci T.N., Nord J.A., Barabas M.A., Stucky C.L., Ebert A.D. Spinal muscular atrophy astrocytes exhibit abnormal calcium regulation and reduced growth factor production. Glia. 2013;61:1418–1428. doi: 10.1002/glia.22522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamilton G., Gillingwater T.H. Spinal muscular atrophy: going beyond the motor neuron. Trends Mol. Med. 2013;19:40–50. doi: 10.1016/j.molmed.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Ottesen E.W., Howell M.D., Singh N.N., Seo J., Whitley E.M., Singh R.N. Severe impairment of male reproductive organ development in a low SMN expressing mouse model of spinal muscular atrophy. Sci. Rep. 2016;6:20193. doi: 10.1038/srep20193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szunyogova E., Zhou H., Maxwell G.K., Powis R.A., Francesco M., Gillingwater T.H., Parson S.H. Survival Motor Neuron (SMN) protein is required for normal mouse liver development. Sci. Rep. 2016;6:34635. doi: 10.1038/srep34635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaifer K.A., Villalón E., Osman E.Y., Glascock J.J., Arnold L.L., Cornelison D.D.W., Lorson C.L. Plastin-3 extends survival and reduces severity in mouse models of spinal muscular atrophy. JCI Insight. 2017;2:e89970. doi: 10.1172/jci.insight.89970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulcahy P.J., Binny C., Muszynski B., Karyka E., Azzouz M. Adeno-associated vectors for gene delivery to the nervous system. In: Bo X., Verhaagen J., editors. Gene Delivery and Therapy for Neurological Disorders. Springer Science+Business Media; 2015. pp. 1–22. [Google Scholar]
- 23.Lukashchuk V., Lewis K.E., Coldicott I., Grierson A.J., Azzouz M. AAV9-mediated central nervous system-targeted gene delivery via cisterna magna route in mice. Mol. Ther. Methods Clin. Dev. 2016;3:15055. doi: 10.1038/mtm.2015.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Déglon N., Tseng J.L., Bensadoun J.C., Zurn A.D., Arsenijevic Y., Pereira de Almeida L., Zufferey R., Trono D., Aebischer P. Self-inactivating lentiviral vectors with enhanced transgene expression as potential gene transfer system in Parkinson’s disease. Hum. Gene Ther. 2000;11:179–190. doi: 10.1089/10430340050016256. [DOI] [PubMed] [Google Scholar]