Abstract
The C′ position of the C-capping box is the second residue outside of the helix. Statistical analysis of residue distribution at the C′ position in the α-helices' C-capping box showed that different amino acid residues occur with different probabilities, with the strongest preference being for glycine. To understand the physico-chemical basis for this preference, we studied the effects that 17 amino acid substitutions at the C′ position in an α-helix of ubiquitin have on the stability of this protein. We determined the following rank order of amino acid residues at the C′ position with respect to their effect on the stability: Gly>His>Asn>Arg>Lys>Gln>Ala>Phe>Met>Ser>Asp>Glu>Trp>Thr>Pro>Ile>Val. The effect of the amino acid substitutions on the structure also was evaluated by comparing the 1H-15N heteronuclear sequential quantum correlation spectra and showed no significant changes in the structures of the most stable (Gly) and the least stable (Val) variants. The obtained changes in stability highly correlate (r = 0.85) with the statistical distribution of the residues at the C′ position indicating that the measured thermodynamic propensities are unbiased by secondary interactions. We also found that the measured thermodynamic propensities correlate well with the amide hydrogen exchange data on short model peptides (r = 0.85) and the calculated hydration of the peptide backbone (r = 0.88). These results combined with the changes in enthalpy and entropy of unfolding of ubiquitin variants suggest that dehydration of the peptide backbone plays a significant role in defining the thermodynamic propensity scale at the C′ position of the C-capping box in α-helices. This propensity scale is useful for protein secondary structure predictions and protein design.
The first basic structural motif of globular proteins proposed was the α-helix (1). The structure of the α-helix is characterized by hydrogen bonding patterns between amide hydrogen bond donors and carbonyl oxygen acceptors of residues situated four apart in the sequence. This pattern of hydrogen bonding, however, implies that the four initial amide hydrogen bond donors and the four final carbonyl oxygen hydrogen bond acceptors do not have hydrogen bonding partners. The potential effect of this is fraying of the helix ends. In 1988, Richardson and Richardson (2) as well as Presta and Rose (3) analyzed statistical preferences of the amino acid residues at the ends of α-helices and revealed the existence of specific capping interactions at both the N and C termini, which compensate for the unsatisfied hydrogen bonds and thus prevent the ends from fraying. According to Aurora et al. (4) and Harper and Rose (5), amino acid residues in helices and their flanking residues can be labeled as:
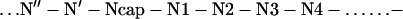 |
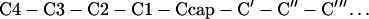 |
where numbered residues have helical backbone dihedral angles (φ = −64 ± 7o; ϕ = −41 ± 7o). Ncap with dihedrals φ = −94 ± 15o and ϕ = 167 ± 5o (5) and Ccap with dihedrals φ ≈−81o and ϕ ≈−2o (6) are boundary residues that belong both to the helix and the adjacent turn. Residues at the C′ position often have positive dihedral angles and often are occupied by a glycine residue (7). What is the physico-chemical basis that defines this preference? One explanation is that Gly introduces flexibility to the peptide backbone, which allows it to adopt a left-handed conformation, i.e., positive φ and ϕ dihedral angles, with a minimal loss of energy (2, 3). Serrano et al. (8) argue that Gly at the C′ position is largely exposed to solvent and substitution of C′ Gly by other amino acid residue lead to dehydration of the peptide backbone, affect the strength of the C′ to C3 hydrogen bond, and decrease stability. To gain deeper insight we have studied the effects amino acid substitutions at the C′ position of the α-helix in ubiquitin have on the stability of this protein. The C-capping box of the α-helix of ubiquitin has a sequence—Ile30Gln31Asp32Lys33Glu34Gly35Ile36Pro37—and shows the characteristics of the αL motif (4) with Gly-35 as a C′ residue, the backbone-backbone hydrogen bond between residues C′ (Gly-35) and C3 (Gln-31) and van der Waals interactions between residues C4 (Ile-30) and C′′ (Ile-36). As expected Gly-35 has positive torsional angles, φ = 81.2o and ϕ = 5.3o (4, 6, 7, 9, 10).
Seventeen amino acid substitutions at the Gly-35 position of ubiquitin were incorporated into the yeast ubiquitin sequence and the effects of these substitutions on ubiquitin stability were measured by differential scanning calorimetry (DSC). In addition, the effect the substitutions had on the ubiquitin structure was evaluated by using NMR spectroscopy. We found that there is a large (up to 30°C, depending on the nature of the amino acid residues) decrease in stability, relative to wild-type (WT) ubiquitin. These large decreases in stability do not seem to be caused by the large structural changes upon substitution as shown by comparison of heteronuclear sequential quantum correlation (HSQC) spectra of ubiquitin variants. Analysis of the changes in the thermodynamic parameters that accompany the decrease in stability and the correlational analysis with the data reported in the literature indicate that the decrease in stability is largely defined by the dehydration of the peptide backbone by the side chain of a residue at the C′ position.
Materials and Methods
Mutagenesis and Expression of Ubiquitin Variants.
Site-directed mutagenesis of the yeast ubiquitin gene was carried out by using the QuickChange Site-Directed Mutagenesis kit (Stratagene). The presence of mutations was confirmed by sequencing the entire ubiquitin gene with an Applied Biosystems PRISM 310 Genetic Analyzer. Overexpression of the ubiquitin variants was done in the BL21(DE3) strain of Escherichia coli as described (11). Each variant was purified to apparent homogeneity as described (12). For reasons discussed previously (13), all described ubiquitin variants contained R63K substitution. Overexpression and purification of the 15N-labeled WT and G35V ubiquitin variant was done as described (14). To increase the yield, the protein was expressed as a His-tag fusion and purified by using Ni-column and gel filtration chromatography as described (14). Attachment of His tag does not affect the position of resonances on 1H-15N correlation spectrum (14).
DSC.
DSC experiments were performed on a VP-DSC (Microcal, Northhampton, MA) instrument at a scan rate of 1 deg/min. The protein concentration for ubiquitin mutants varied between 1.0 and 3.0 mg/ml. Detailed procedures of the sample preparation for the DSC experiment were the same as described (12, 15). The partial molar heat capacity of the protein, Cp,pr(T), was obtained from the experimentally measured apparent heat capacity difference between the sample (containing protein solution) and reference (containing corresponding buffer solution) cells, ΔC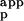 , using the following expression:
, using the following expression:
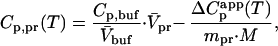 |
1 |
where V̄pr is the partial molar volume of a protein calculated as described (16), Cp,buf and V̄buf are the partial molar heat capacity and the partial molar volume of the aqueous buffer, respectively, mpr is the mass of the protein in the cell, and M is the molar mass of the protein.
Protein concentration was measured spectrophotometrically by using a known extinction coefficient for the WT ubiquitin, ɛ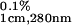 = 0.149 (11–13). This extinction coefficient was used for all but G35W ubiquitin variants because none of those amino acid substitutions introduced residues that absorb in the far UV range. The extinction coefficient for the G35W variant was estimated at ɛ
= 0.149 (11–13). This extinction coefficient was used for all but G35W ubiquitin variants because none of those amino acid substitutions introduced residues that absorb in the far UV range. The extinction coefficient for the G35W variant was estimated at ɛ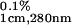 = 0.80 using the methods of Pace et al. (17). Correction for light scattering was taken into account as described (18). Analysis of the heat capacity profiles was done by using nonlinear regression routine nlreg and in-house written scripts (15). Individual curves for a given ubiquitin variant were fit to a two-state transition model with the heat capacities of the native and the unfolded states, the enthalpy of unfolding, ΔHcal, heat capacity of unfolding, ΔCp, and transition temperature, Tm, as independent variables (12). The standard thermodynamic functions under reference conditions were calculated as:
= 0.80 using the methods of Pace et al. (17). Correction for light scattering was taken into account as described (18). Analysis of the heat capacity profiles was done by using nonlinear regression routine nlreg and in-house written scripts (15). Individual curves for a given ubiquitin variant were fit to a two-state transition model with the heat capacities of the native and the unfolded states, the enthalpy of unfolding, ΔHcal, heat capacity of unfolding, ΔCp, and transition temperature, Tm, as independent variables (12). The standard thermodynamic functions under reference conditions were calculated as:
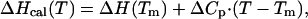 |
2 |
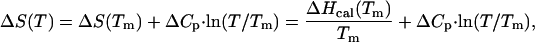 |
3 |
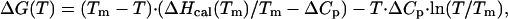 |
4 |
where ΔS(T) and ΔG(T) are the entropy and Gibbs energy functions of a protein, respectively. As a control for the correctness of the experiments we compared the stability of the G35D variant in the background of true yeast ubiquitin. We found the same effect of this substitution on the stability (ΔTm = 9°C; ΔΔG = −5.8 kJ/mol) as in the case of the pseudo-WT protein used for the major part of this study.
NMR Spectroscopy.
1H-15N heteronuclear sequential quantum correlation spectra were acquired on a Bruker spectrometer (DRX-600) in 95% H2O/5% 2H2O as described (14). The solutions were ≈1 mM for the WT-ubiquitin and 0.2 mM G35V-ubiquitin and contained 5 mM acetate buffer, pH 5.0, with added 2H2O to yield a 90% H2O/10% 2H2O solution.
Results
Effects of Amino Acid Substitutions at the C′ Position on the Stability of Ubiquitin.
Table 1 summarizes the results of calorimetric studies of 17 variants of ubiquitin with the single-site amino acid substitutions at the C′ position of the C-capping box in ubiquitin. Comparison is done under a fixed set of conditions, 50°C and pH 3.0. This pH was selected because at this pH the transition temperature for the WT protein is 60°C (12) and thus more stable variants will have the unfolding transition completed before reaching the upper limit (110°C) of the operating temperature range of the DSC instrument. The reference temperature was set at 50°C because it is an average temperature of unfolding of all ubiquitin variants studied at pH 3.0, and thus the extrapolation error over the entire set of variants is minimal.
Table 1.
Thermodynamic parameters of unfolding of the position 35 variants of ubiquitin at pH 3.0
| Residue in position 35 | Tm, °C | ΔHcal(Tm), kJ/mol | ΔH(50°C), kJ/mol | ΔS(50°C), J/mol⋅K | ΔG(50°C), kJ/mol | ΔΔG(50°C), kJ/mol | ΔΔH(50°C), kJ/mol | −T⋅ΔΔS(50°C), kJ/mol |
|---|---|---|---|---|---|---|---|---|
| GLY | 59.4 | 205 | 172 | 516 | 5.3 | 0.0 | 0 | 0 |
| HIS | 58.1 | 188 | 160 | 481 | 4.6 | −0.7 | −12 | 11 |
| ASN | 54.1 | 174 | 160 | 488 | 2.4 | −2.9 | −12 | 9 |
| ARG | 53.5 | 157 | 145 | 443 | 1.9 | −3.4 | −27 | 24 |
| LYS | 53.6 | 163 | 150 | 460 | 1.4 | −3.9 | −22 | 18 |
| GLN | 52.1 | 161 | 154 | 473 | 1.2 | −4.1 | −18 | 14 |
| ALA | 50.7 | 145 | 143 | 440 | 0.9 | −4.4 | −29 | 25 |
| PHE | 50.4 | 162 | 161 | 497 | 0.5 | −4.8 | −11 | 6 |
| MET | 50.4 | 153 | 152 | 469 | 0.5 | −4.8 | −20 | 15 |
| SER | 48.7 | 142 | 147 | 456 | −0.3 | −5.6 | −25 | 19 |
| ASP | 49.9 | 154 | 154 | 478 | −0.4 | −5.7 | −18 | 12 |
| GLU | 49.4 | 150 | 152 | 472 | −0.5 | −5.8 | −20 | 14 |
| TRP | 46.0 | 134 | 148 | 464 | −1.9 | −7.2 | −24 | 17 |
| THR | 41.3 | 102 | 132 | 420 | −3.7 | −9.0 | −40 | 31 |
| PRO | 39.6 | 87 | 123 | 393 | −3.9 | −9.2 | −49 | 40 |
| ILE | 36.8 | 90 | 136 | 437 | −5.2 | −10.5 | −36 | 26 |
| VAL | 27.5 | 45 | 124 | 402 | −5.8 | −11.1 | −48 | 37 |
Estimated experimental errors are Tm ± 0.2°C, ΔHcal(Tm) ± 7%.
The WT protein that has Gly at position 35 is the most stable protein. All other 16 aa residues at the C′ position destabilize the ubiquitin molecule to a different degree. The G35V variant is most dramatically destabilized, with a 32°C lower transition temperature. Other strongly destabilizing variants are G35I (ΔTm = −23°C) and G35T (ΔTm = −18°C), which are also β-branched amino acid residues.
Effect of Val Substitution at C′ Position on the Structure of Ubiquitin.
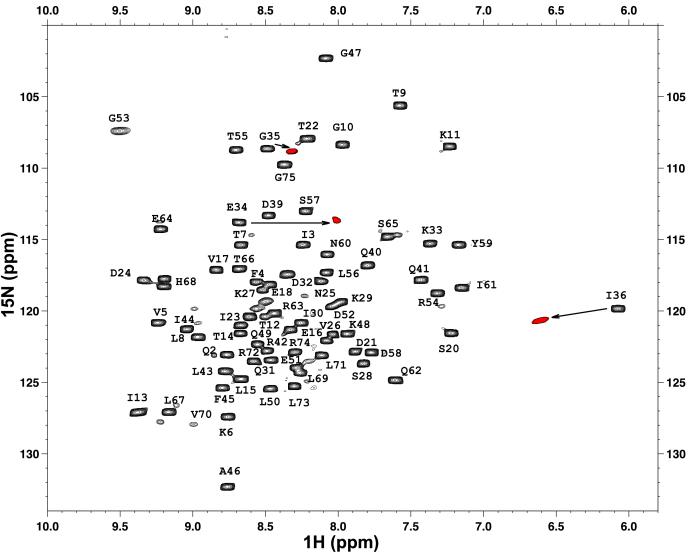
Because the G35V variant showed the lowest stability relative to other position 35-substituted proteins we addressed the question of structural similarity between the WT and G35V variant. We used NMR spectroscopy to characterize the structure of the G35V variant. Because this variant is significantly less stable than the WT, the 1H-15N heteronuclear sequential quantum correlation spectrum, despite having intense cross-peaks, was characterized by a broader resonance dispersion relative to the WT protein. Nevertheless, the overall position of resonances in the G35V variant was very similar to that of the WT. The exceptions were found in the cross-peaks corresponding to the residue at the substitution site (C′) and the residues before (Ccap-E34) and after (C′′–I36). As can be seen from Fig. 1, all three residues in the G35V variant show changes largely along the 1H-axes. The changes appear to be consistent with a small rearrangement of the peptide backbone that alters the relative position of the amide protons (19). The dihedral angles for the C′ position of ubiquitin in the WT protein are φ = 81.2° and ϕ = 5.3°, well outside the allowable positive dihedral angles for any nonglycine residue. It is conceivable that the dihedral angles of the C′ residue in the G35V variant are moving closer to the allowable αL region, leading to the changes in the resonances of the adjacent in-sequence residues. Such changes have been previously observed for T4 lysozyme (20), hen egg lysozyme (19), and rop (21). Overall it is apparent that there is no significant difference between the most stable and the least stable position 35 ubiquitin variant and thus we can assume that the structures of the remaining position 35 ubiquitin variants (with G35P to a lesser degree) are also structurally similar to the WT.
Figure 1.
1H-15N chemical shift correlation spectrum of the WT variant of yeast ubiquitin with the partial assignments of the backbone 15N amide protons (14). The resonances that are significantly different in G35V variant are shown in red with the arrow indicating which resonances have shifted.
Possible Secondary Effect at the C′ Position that Might Bias the Propensity.
The C′ position of the C-capping box in ubiquitin is fully solvent exposed. It was believed for a long time that residues at solvent exposed positions do not have a significant effect on the protein stability (22). This belief was based on the assumption that the only interactions that the surface residue can have are with the solvent, and because in the unfolded state the surface residue interacts with the same solvent, the net change in the interactions between native and unfolded states is zero. However, recently it has been shown that at least one type of interaction, the charge-charge interactions between surface residues, can be an important factor for modulating the protein stability (23). Because some of the C′ position variants of ubiquitin incorporate acidic (Asp, Glu) and basic (His, Lys, Arg) residues instead of uncharged glycine, it is important to estimate the possible effects of incorporating charged residues at the C′ position on the stability of the ubiquitin molecule. We have shown recently that simple electrostatic calculations can reliably predict the effect of substitutions of solvent exposed charged residues in several proteins including ubiquitin (11, 23). We thus performed similar calculations on position 35 of ubiquitin. It appears that substitution of Gly-35 for either acidic or basic residues at this position has a negligible effect on the charge-charge interactions (the expected ΔΔG caused by changes in charge-charge interactions between WT and G35E, G35D, G35R, or G35K is less than 0.3 kJ/mol). Thus under the experimental conditions the side chains of Asp and Glu residues are not expected to have perturbed pKa and will be largely protonated. Taken together, it is unlikely that the charge-charge interactions will bias the intrinsic thermodynamic propensities of the residues at the C′ position of the C-capping box in ubiquitin. This, of course, does not mean that there will not be other electrostatic perturbations upon substitution with the charged residue at this position. One can expect non-negligible interactions of charged side chains at C′ with the helix dipole (24, 25). However, these interactions will exist in all C-capping motifs and thus are considered to be one of the factors defining thermodynamic propensities.
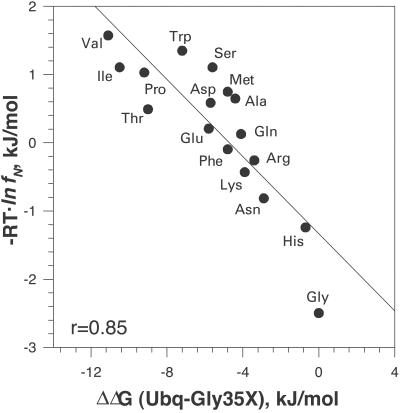
Correlation with the Statistical Distributions.
Fig. 2 shows the correlation of the thermodynamic propensities of different amino acid residues at the C′ position in ubiquitin and statistical analysis of the probabilities of amino acid residues at the C′ position. The latter data were compiled by the Rose group and are based on 1,316 helices from 274 known protein structures (7). The correlation coefficient between the two data sets is 0.85, which can be considered to be quite good, keeping in mind that the residue distribution in the known protein structures is not filtered for the additional interactions that involve the residues at the C′ position of α-helices. We thus can assume at least as a first approximation that the propensities observed in the C′ position of the α-helix in ubiquitin are to a large degree defined by some intrinsic properties. Additional support for this comes from the fact that the thermodynamic propensities obtained at the C′ position of ubiquitin have good correlation (correlation coefficients 0.94 and 0.78) with the effects of amino acid substitutions at the C′ position of two α-helices in barnase (8). It must be noted that barnase double-mutant cycle experiments revealed certain bias due to the secondary interactions of residues at C′ with the residues close in sequence (8, 26). Our data on G35D are in excellent agreement with the effects of similar substitution in T4-lysozyme for which Gray and Matthews (27) reported a decrease in stability of −5.9 kJ/mol at pH 2.0, comparable with −5.7 kJ/mol in ubiquitin (Table 1). The glycine residue in T4 lysozyme is also in the C′ position and has positive dihedrals φ = 76.3° and ϕ = 19.1°.
Figure 2.
Correlation of Gibbs energy change of the C′ ubiquitin variants relative to WT ΔΔG(Ubq-Gly35X) with the normalized probabilities of different residues in the C′ position of an α-helix (7). Normalized probabilities, fN, were converted into Gibbs energy as − R⋅T⋅lnfN, using 298 K as a reference temperature. Linear correlation coefficient is 0.85.
Discussion
We have shown that the amino acid substitutions at the C′ position of α-helix in ubiquitin have rather dramatic effects on the stability of this protein. What are the factors that define this thermodynamic propensity scale? What makes Gly at this position thermodynamically the most favorable residue? Glycine does not have a side chain and its higher flexibility in the unfolded state means a larger loss of entropy upon folding and thus entropically any substitution of Gly must lead to an increase in stability (28). We observe, however, that all residues at position 35 render a protein with lower stability than the Gly-containing WT. We propose that the addition of the side chain at the solvent exposed Gly would reduce the hydration of the protein backbone and the effect of burying the polar peptide backbone would lead to destabilization. Three different facts support this explanation.
Correlation Between the Hydrogen Exchange Rates of Amide Proton and Thermodynamic Propensity at C′ Position.
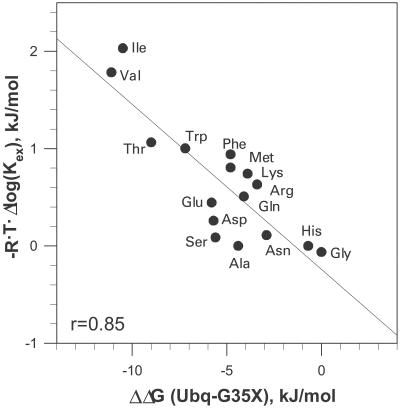
Fig. 3 shows the plot of ΔΔG of the residues at the C′ position in ubiquitin and the ΔΔG obtained from the amide proton exchange rates of different amino acid residues in a short model peptide (29). The original exchange rates were converted into Gibbs energy by using the standard relationship ΔΔG = −R⋅T⋅Δlog(Kex). One needs to keep in mind that the hydrogen exchange in a short peptide occurs very fast and is thus difficult to measure, leading to a relatively large error. Nevertheless, the two data sets clearly correlate (r = 0.85). The difference in the amide exchange rates for different amino acid residues was interpreted in terms of steric blocking effects of the side chain on the accessibility of the backbone amide for the H/D exchange (30). In other words, the difference in amide exchange rates is defined by the accessibility to the solvent. Because this correlates with the thermodynamic propensities of the residues at the C′ position of the α-helix in ubiquitin, we can argue that the same effect, i.e., solvent accessibility of the peptide backbone, contributes to the thermodynamic propensity scale at the C′ position.
Figure 3.
Correlation of Gibbs energy change of the C′ ubiquitin variants relative to WT ΔΔG(Ubq-Gly35X) with the rates of hydrogen exchange of amide protons in short model peptides. The average of base and acid catalysis of the H/D exchange rates relative to Ala, reported by Bai et al. (29) for the left (from the side chain) amide proton, log Kex(X) − log Kex(Ala), was converted to the relative Gibbs energy as ΔΔG = − R⋅T⋅log[Kex(X)/Kex(Ala)] using 298 K as a reference temperature. Linear correlation coefficient is 0.85.
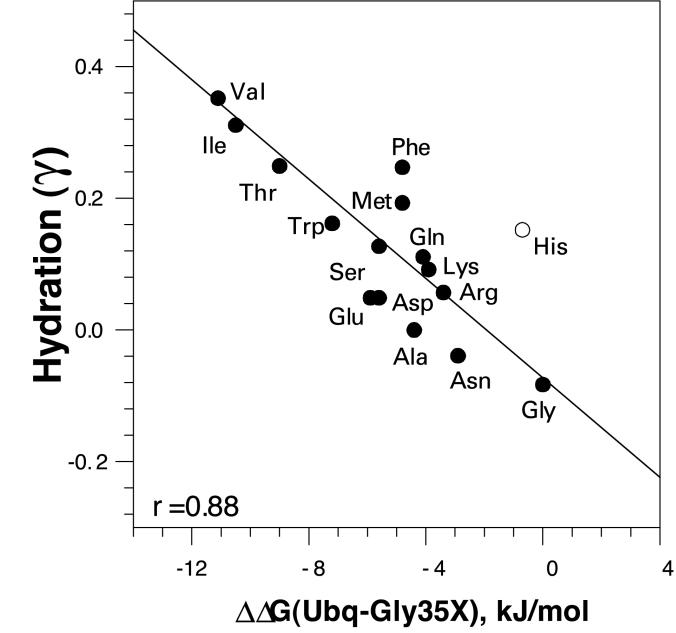
Correlation Between the Backbone Hydration Parameter and the Thermodynamic Propensity at C′ Position.
Energetics of the backbone hydration have been evaluated by Avbelj (31) using a computational approach. The plot of ΔΔG of the residues at the C′ position in ubiquitin and the parameter γ that describes the hydration is shown in Fig. 4. The overall correlation coefficient is 0.85. However, the largest challenge in the calculations that introduces considerable uncertainty is usually the presence of charged amino acid residues. For example, if the histidine residue is omitted from the analysis, the correlation coefficient increases to 0.88. These correlations again can be interpreted as the difference in the hydration of the peptide backbone is caused by the difference in the steric blocking by different side chains, thus contributing to the thermodynamic propensity of the residues at the C′ position of the α-helix.
Figure 4.
Correlation of Gibbs energy change of the C′ ubiquitin variants relative to WT ΔΔG(Ubq-Gly35X) with the parameter γ (31), which represents relative hydration of the peptide backbone. Linear correlation coefficient is 0.88.
The Changes in the Thermodynamic Parameters of Unfolding of the C′ Ubiquitin Variant Are Consistent with the Thermodynamics of Dehydration of the Peptide Backbone.
Thermodynamic parameters of hydration of the peptide backbone have been previously evaluated by using model compound data (32). It was shown that the hydration of the peptide backbone is accompanied by negative enthalpy and negative entropy changes (33). The changes in entropy and enthalpy, however, are such that the Gibbs energy of hydration is negative (33). These characteristic properties are seen in the ΔΔH(50°C) and −TΔΔS(50°C) values of the ubiquitin variants (Table 1). Indeed, all C′ ubiquitin variants have lower enthalpy of unfolding than the WT protein in which Gly in C′ provides the largest backbone exposure. The changes in enthalpy are largest for the least stable C′ variants, all of which have β-branched side chains, and thus are expected to have larger steric blocking effects on the peptide backbone from water (30). The changes in the entropic factor on substitutions at the C′ position follow a similar pattern, i.e., in all cases the changes in the −TΔΔS(50°C) are significant. Again, the increase in the entropic term is the largest for β-branched amino acid residues Thr, Ile, and Val. The loss in enthalpy is nevertheless larger than the gain in entropy leading to an overall decrease in ΔΔG(50°C). This pattern is clearly consistent with the thermodynamic data on the hydration of the peptide backbone and in particular with the enthalpy change upon hydration of the peptide backbone. Analysis of the experimental data on the transfer of different amides into water gives an estimate for ΔHhyd(50°C) = −61 kJ/mol (33), which is consistent with the electrostatic calculations on N-methylacetamide that give a value of −51 kJ/mol (34). Both of these estimates are higher than the result of the calculations done on a more realistic model, an Ala-based model peptide in extended conformation, −33 kJ/mol (34). For comparison, the observed averaged change in ΔΔH(50°C) upon substitution of Gly to any other residue is −26 ± 12 kJ/mol (Table 1). It is important to note the rather small spread in the ΔΔH(50°C) values for nonglycine residues. This observation is consistent with the estimates for the peptide backbone burial in different amino acid residues relative to Gly. Calculations of Creamer et al. (35) show that any residue other than Gly have a very similar amount of surface area (ASA) exposed (average ASA = 25 ± 2 Å2 with the spread from 20 to 29 Å2). In contrast the Gly residues has 65 Å2 of backbone ASA exposed (35). If indeed hydration enthalpy can be scaled to the surface area, the predicted decrease in the enthalpy caused by the difference in hydration upon burial of peptide backbone in Gly and non-Gly residues will be
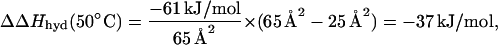 |
which corresponds reasonably well to the experimentally observed changes in ΔΔH(50°C). It is remarkable that although changes in the enthalpies of hydration as calculated from the surface area changes for the peptide backbone averaged over all variants closely predict the average changes in the enthalpies of unfolding of the ubiquitin variants, the correlation between ΔΔH(50°C) and ΔASA for the peptide backbone for individual variants, even excluding glycine, is poor (r = 0.55, data not shown). This finding is possibly caused by two factors: (i) surface area of peptide backbone is very small and thus is prone to the relatively large errors because of the small variations in the conformation, and (ii) the enthalpy of hydration does not scale well with the surface area when the changes in the ASA are very small (see ref. 34 for the theoretical evidence of this effect). The decrease in the entropy of unfolding of ubiquitin variants is also consistent with the idea of significant contribution from the dehydration of polar peptide backbone, although by default the other factors such as side-chain configurational entropy also will be in play.
Thus it appears that the decrease in the stability of the C′ ubiquitin variants is largely enthalpic, which is consistent with the changes in thermodynamic properties upon dehydration of the polar peptide backbone. Obviously the dehydration of the peptide backbone is not the only parameter that contributes to the observed differences in ΔΔG values. Other factors such as burial of the Cβ atom (e.g., ref. 36), difference in the intrinsic configuration entropy (e.g., ref. 37), and side-chain charge or side-chain dipole interactions with the helix dipole (e.g., ref. 38) will contribute as well. Nevertheless, it seems that the major factor in the thermodynamic propensity at the C′ position is defined by the dehydration of the peptide backbone. This is not the first time that the specific interactions of the peptide backbone with the solvent have been implicated in the modulation of protein stability. Interactions between water and peptide backbone were shown to be important determinants of helix propensities (39). The osmophobic effect is also believed to affect protein stability (40), primarily because of the unfavorable solvation of the peptide backbone by the osmolytes.
Concluding Remarks.
In this article we have established the propensity scale for the C′ position of the C-capping box of α-helices. Analysis of thermodynamic parameters and their correlation with some structural properties suggests that dehydration of the peptide backbone plays a significant role in defining these propensities, thus underlying the fundamental importance of water for helix formation. In addition, these results can be directly used in protein engineering and design: this propensity scale provides a tool for modulation of protein stability by amino acid substitution at the C′ position of α-helices.
Acknowledgments
We thank Chris Falzone for the NMR experiments and George Rose for thoughtful comments and Marimar Lopez for discussions of the manuscript. This work was supported by National Institutes of Health Grant GM54537 (to G.I.M.).
Abbreviations
- DSC
differential scanning calorimetry
- WT
wild type
- ASA
amount of surface area.
References
- 1.Pauling L, Corey R B, Branson H R. Proc Natl Acad Sci USA. 1951;37:205–210. doi: 10.1073/pnas.37.4.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richardson J S, Richardson D C. Science. 1988;240:1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- 3.Presta L G, Rose G D. Science. 1988;240:1632–1641. doi: 10.1126/science.2837824. [DOI] [PubMed] [Google Scholar]
- 4.Aurora R, Srinivasan R, Rose G D. Science. 1994;264:1126–1130. doi: 10.1126/science.8178170. [DOI] [PubMed] [Google Scholar]
- 5.Harper E T, Rose G D. Biochemistry. 1993;32:7605–7609. doi: 10.1021/bi00081a001. [DOI] [PubMed] [Google Scholar]
- 6.Preissner R, Bork P. Biochem Biophys Res Commun. 1991;180:660–665. doi: 10.1016/s0006-291x(05)81116-7. [DOI] [PubMed] [Google Scholar]
- 7.Aurora R, Rose G D. Protein Sci. 1998;7:21–38. doi: 10.1002/pro.5560070103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serrano L, Sancho J, Hirshberg M, Fersht A R. J Mol Biol. 1992;227:544–559. doi: 10.1016/0022-2836(92)90906-z. [DOI] [PubMed] [Google Scholar]
- 9.Milner-White E J. J Mol Biol. 1988;199:503–511. doi: 10.1016/0022-2836(88)90621-3. [DOI] [PubMed] [Google Scholar]
- 10.Bork P, Preissner R. Biochem Biophys Res Commun. 1991;180:666–672. doi: 10.1016/s0006-291x(05)81117-9. [DOI] [PubMed] [Google Scholar]
- 11.Loladze V V, Ibarra-Molero B, Sanchez-Ruiz J M, Makhatadze G I. Biochemistry. 1999;38:16419–16423. doi: 10.1021/bi992271w. [DOI] [PubMed] [Google Scholar]
- 12.Thomas S T, Makhatadze G I. Biochemistry. 2000;39:10275–10283. doi: 10.1021/bi0000418. [DOI] [PubMed] [Google Scholar]
- 13.Makhatadze G I, Lopez M M, Richardson J M, 3rd, Thomas S T. Protein Sci. 1998;7:689–697. doi: 10.1002/pro.5560070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loladze V V, Ermolenko D N, Makhatadze G I. Protein Sci. 2001;10:1343–1352. doi: 10.1110/ps.370101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makhatadze G I. In: Current Protocols in Protein Chemistry. Coligan J E, Dunn B M, Ploegh H L, Speicher D W, Wingfield P T, editors. Vol. 2. New York: Wiley; 1998. pp. 7.9.1–7.9.14. [Google Scholar]
- 16.Makhatadze G I, Medvedkin V N, Privalov P L. Biopolymers. 1990;30:1001–1010. doi: 10.1002/bip.360301102. [DOI] [PubMed] [Google Scholar]
- 17.Pace C N, Vajdos F, Fee L, Grimsley G, Gray T. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winder A F, Gent W L. Biopolymers. 1971;10:1243–1251. doi: 10.1002/bip.360100713. [DOI] [PubMed] [Google Scholar]
- 19.Masumoto K, Ueda T, Motoshima H, Imoto T. Protein Eng. 2000;13:691–695. doi: 10.1093/protein/13.10.691. [DOI] [PubMed] [Google Scholar]
- 20.Nicholson H, Soderlind E, Tronrud D E, Matthews B W. J Mol Biol. 1989;210:181–193. doi: 10.1016/0022-2836(89)90299-4. [DOI] [PubMed] [Google Scholar]
- 21.Predki P F, Agrawal V, Brunger A T, Regan L. Nat Struct Biol. 1996;3:54–58. doi: 10.1038/nsb0196-54. [DOI] [PubMed] [Google Scholar]
- 22.Jaenicke R. Prog Biophys Mol Biol. 1999;71:155–241. doi: 10.1016/s0079-6107(98)00032-7. [DOI] [PubMed] [Google Scholar]
- 23.Sanchez-Ruiz J M, Makhatadze G I. Trends Biotechnol. 2001;19:132–1354. doi: 10.1016/s0167-7799(00)01548-1. [DOI] [PubMed] [Google Scholar]
- 24.Matthews B W. Annu Rev Biochem. 1993;62:139–160. doi: 10.1146/annurev.bi.62.070193.001035. [DOI] [PubMed] [Google Scholar]
- 25.Rohl C A, Baldwin R L. Methods Enzymol. 1998;295:1–26. doi: 10.1016/s0076-6879(98)95032-7. [DOI] [PubMed] [Google Scholar]
- 26.Sancho J, Serrano L, Fersht A R. Biochemistry. 1992;31:2253–2258. doi: 10.1021/bi00123a006. [DOI] [PubMed] [Google Scholar]
- 27.Gray T M, Matthews B W. J Biol Chem. 1987;262:16858–16864. doi: 10.2210/pdb1l16/pdb. [DOI] [PubMed] [Google Scholar]
- 28.Matthews B W, Nicholson H, Becktel W J. Proc Natl Acad Sci USA. 1987;84:6663–6667. doi: 10.1073/pnas.84.19.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bai Y, Milne J S, Mayne L, Englander S W. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bai Y, Englander S W. Proteins. 1994;18:262–266. doi: 10.1002/prot.340180307. [DOI] [PubMed] [Google Scholar]
- 31.Avbelj F. J Mol Biol. 2000;300:1335–1359. doi: 10.1006/jmbi.2000.3901. [DOI] [PubMed] [Google Scholar]
- 32.Makhatadze G I, Privalov P L. J Mol Biol. 1993;232:639–659. doi: 10.1006/jmbi.1993.1416. [DOI] [PubMed] [Google Scholar]
- 33.Makhatadze G I, Privalov P L. Adv Protein Chem. 1995;47:307–425. doi: 10.1016/s0065-3233(08)60548-3. [DOI] [PubMed] [Google Scholar]
- 34.Avbelj F, Luo P, Baldwin R L. Proc Natl Acad Sci USA. 2000;97:10786–10791. doi: 10.1073/pnas.200343197. . (First Published September 12, 2000; 10.1073/pnas.200343197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Creamer T P, Srinivasan R, Rose G D. Biochemistry. 1997;36:2832–2835. doi: 10.1021/bi962819o. [DOI] [PubMed] [Google Scholar]
- 36.Blaber M, Zhang X J, Matthews B W. Science. 1993;260:1637–1640. doi: 10.1126/science.8503008. [DOI] [PubMed] [Google Scholar]
- 37.Creamer T P, Rose G D. Proteins. 1994;19:85–97. doi: 10.1002/prot.340190202. [DOI] [PubMed] [Google Scholar]
- 38.Marqusee S, Baldwin R L. Proc Natl Acad Sci USA. 1987;84:8898–8902. doi: 10.1073/pnas.84.24.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo P, Baldwin R L. Proc Natl Acad Sci USA. 1999;96:4930–4935. doi: 10.1073/pnas.96.9.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, Bolen D W. Biochemistry. 1995;34:12884–12891. doi: 10.1021/bi00039a051. [DOI] [PubMed] [Google Scholar]