Abstract
Aims
Caveolinopathies are a family of genetic disorders arising from alterations of the caveolin-3 (cav-3) gene. The T78M cav-3 variant has been associated with both skeletal and cardiac muscle pathologies but its functional contribution, especially to cardiac diseases, is still controversial. Here, we evaluated the effect of the T78M cav-3 variant on cardiac ion channel function and membrane excitability.
Methods and results
We transfected either the wild type (WT) or T78M cav-3 in caveolin-1 knock-out mouse embryonic fibroblasts and found by immunofluorescence and electron microscopy that both are expressed at the plasma membrane and form caveolae. Two ion channels known to interact and co-immunoprecipitate with the cav-3, hKv1.5 and hHCN4, interact also with T78M cav-3 and reside in lipid rafts. Electrophysiological analysis showed that the T78M cav-3 causes hKv1.5 channels to activate and inactivate at more hyperpolarized potentials and the hHCN4 channels to activate at more depolarized potentials, in a dominant way. In spontaneously beating neonatal cardiomyocytes, the expression of the T78M cav-3 significantly increased action potential peak-to-peak variability without altering neither the mean rate nor the maximum diastolic potential. We also found that in a small cohort of patients with supraventricular arrhythmias, the T78M cav-3 variant is more frequent than in the general population. Finally, in silico analysis of both sinoatrial and atrial cell models confirmed that the T78M-dependent changes are compatible with a pro-arrhythmic effect.
Conclusion
This study demonstrates that the T78M cav-3 induces complex modifications in ion channel function that ultimately alter membrane excitability. The presence of the T78M cav-3 can thus generate a susceptible substrate that, in concert with other structural alterations and/or genetic mutations, may become arrhythmogenic.
Keywords: Ion channels, Caveolin, Electrophysiology, Genetic diseases, Arrhythmia
1. Introduction
Many cellular processes require a concerted and ordered activation of various proteins and signal transduction pathways; to comply with this function, many proteins have been confined to specific membrane microdomains that keep them in close proximity and favour their functional interactions. Caveolae are specialized omega-shaped microdomains localized in the plasma membrane of many cell types.1 In cardiomyocytes, caveolae compartmentalize several ion channels, accessory subunits, and modulatory proteins, whose contributions shape the excitability properties of the plasma membrane. Important structural components of caveolae are caveolins. In humans, three different isoforms of caveolin are expressed: caveolin-1 is ubiquitous, caveolin-2 usually interacts with caveolin-1, while caveolin-3 (cav-3) is the main isoform expressed in muscles.1 Cav-3 binds several proteins including ion channels and modulates their functional properties through this interaction.2 We and others have previously demonstrated that alteration of caveolar domains significantly affects the properties of cardiac HCN4, Kv1.5, Cav1.2, and Nav1.5 channels.3–6
The term caveolinopathies identifies a family of diseases arising from mutations in the cav-3 gene (CAV3). Several variants of CAV3 have been reported in patients with skeletal muscle pathologies such as limb-girdle muscular dystrophy (LMGD), rippling muscle disease (RMD), isolated hyperCKemia and distal myopathy as well as in patients with cardiac pathologies such as long QT syndrome (LQTS), sudden infant death syndrome (SIDS) and hypertrophic cardiomyopathy (HCM).7 The 233C > T variant introducing the amino acid substitution T78M has been identified in patients with RMD, proximal myopathy, LQTS, idiopathic hyperCKemia, and SIDS.8–12 A patient homozygous for the T78M substitution presented both dilated cardiomyophaty, hyperCKemia, and proximal myotonia.10In vitro functional analysis demonstrated that expression of the T78M cav-3 causes changes in the Nav1.5 sodium channel properties, generating a non-inactivating late INa9 and a large reduction of the current density generated by the Kir2.1 potassium channel.13 Even if these findings point to a role of the T78M variant in generating the above pathological phenotypes, recent analysis reported however this same variant in the general (healthy) population at a frequency comparable to that found in LQTS patients.14–16 It thus remains unclear if and how this single variant can generate such different phenotypes. Considering that some cardiac arrhythmias may remain asymptomatic for years, the effect of the T78M substitution in contributing to a pathological phenotype remains unresolved. To shed light on the possible involvement of the T78M cav-3 variant in altering membrane excitability, we analysed (1) the presence of caveolae in cells lacking caveolins but transiently expressing this variant, (2) the interaction and effect of the T78M cav-3 on the properties of heterologously expressed human (h)HCN4 and hKv1.5 channels, and (3) its effect on membrane excitability in spontaneously beating neonatal rat ventricular cardiomyocytes (NRVCs).
2. Methods
For detailed methods, see the Supplementary material online.
All animal procedures were in accordance with the Italian and UE laws (D. Lgs n° 2014/26, 2010/63/UE) and approved by the committee of the Università degli Studi di Milano and by the Italian Minister of Health (protocol number 1197/2015).
2.1 Cell culture and transfection
For electrophysiological recordings and sucrose gradient fractionation experiments, mouse embryonic fibroblasts from cav-1 knock-out mice (3T3 MEF-KO CRL-2753™, ATCC) plated in 35 mm dishes were co-transfected with 1.5 µg of either WT or T78M cav-3, or with 0.75 µg of both, and 1 µg of plasmid containing the ion channel sequence (hHCN4, hKv1.5, hKir2.1) with Fugene HD or ViaFect™ Transfection Reagent (Promega). Neonatal rat (sprague dawley, ENVIGO) ventricular cardiomyocytes were isolated as previously described17 and transfected the day after with the plasmid containing either the WT or the T78M cav-3 using Lipofectamine 2000 (Life Technologies).
For immunofluorescence or electron microscopy, MEF-KO cells were transfected with 1.5 µg of either the WT-EGFP or T78M-EGFP cav-3. For immunofluorescence cells were transfected also with 1 µg of a mixture of pECFP-Mem and pECFP-C1-CAAX plasmids in order to uniformly stain the entire membrane.
For co-immunoprecipitation experiments, MEF-KO cells plated on 60 mm dishes were transiently co-transfected with equimolar quantities of the plasmid containing the ion channel sequence (V5-hKv1.5 or hHCN4) and WT or T78M cav-3-EGFP plasmids with ViaFect™ Transfection Reagent (Promega).
2.2 Electron microscopy and immunofluorescence analysis
24–36 h after transfection, MEF-KO cells expressing either the WT or T78M cav-3-EGFP were selected by flow cytometry (FACSAria II, BD) and subsequently plated on 60 mm dishes, fixed, pelleted, and processed for electron microscopy experiments.
For immunofluorescence analysis WT or T78M cav-3-EGFP and membrane CFP signals were analysed in living cells; images were acquired using a laser confocal microscope (Zeiss LSM710).
2.3 Separation of lipid raft membrane fractions
Discontinuous sucrose gradient experiments were carried out as previously reported (see Supplementary material online and Refs. 3, 18).
2.4 Co-immunoprecipitation (co-IP)
24–36 h after transfection, MEF-KO cells were re-suspended in co-IP buffer and supernatants were immunoprecipitated overnight at 4 °C, using anti-cav-3 antibody (Becton Dickinson). Anti-mouse Dynabeads® (Life Technologies) were used to bind. After elution and denaturation, proteins were run on SDS-PAGE and revealed by western blotting using specific antibodies (see Supplementary material online). All co-IP runs were repeated at least twice.
2.5 Electrophysiology
Patch-clamp experiments in the whole-cell configuration were carried out 36–48 h after transfection. MEF-KO cells or cardiomyocytes were enzymatically dissociated and seeded at low density in order to analyse ionic currents from single cells in voltage-clamp mode. Cardiomyocytes were kept at 36 ± 1 °C while MEF-KO cells were kept at room temperature. Action potentials were recorded using current-clamp mode from small spontaneously beating clusters of transfected (fluorescent) cardiomyocytes (3–5 cells) obtained by dissociation of the beating monolayer.
2.6 Genetic analysis
The investigation conforms with the principles outlined in the Declaration of Helsinki. For caveolin-3 gene screening, approval was granted by ethic review board of the University of Milan. Written informed consent was obtained from the patients or parents prior to carrying out genetic analysis on genomic DNA extracted from whole blood or saliva (Puragene Blood Kit, Qiagen). The coding sequence of CAV3 was amplified by PCR using specific primers. Analysis of the amplicons was carried out by DNA sequencing (Bio-Fab Research).
2.7 Computational analysis
The effect of the T78M cav-3 variant on the cellular electrical activity was estimated for both atrial and sinoatrial node (SAN) cell models. Simulations of human atrial action potential were based on the Grandi-Bers model19 and on the Koivumaki model.20 Since a full computational model of human SAN cell is not currently available, the analysis of T78M cav-3 effects on SAN action potentials was based on the Severi-DiFrancesco model of rabbit SAN action potential.21 All numerical simulations and data analysis were performed in Matlab (Mathworks Inc.).
2.8 Statistical analysis
Nested and One-Way ANOVA, followed by Fisher LSD mean comparison, were used to compare multiple groups (see Supplementary material online); Student’s t-test for independent populations was used to compare two groups. Fisher’s exact test was used to compare the frequency of the T78M variant in specific subgroups vs. the general population. Significance level was set to P = 0.05.
3. Results
3.1 T78M cav-3 reaches the plasma membrane, forms caveolae and interacts with ion channels
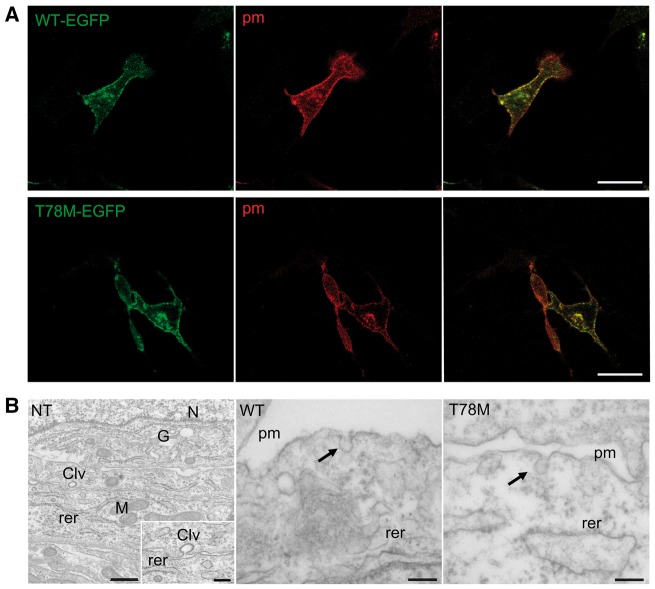
We first analysed the cellular distribution of the WT and T78M cav-3 in MEF-KO cells, which lack endogenous caveolins22 and caveolae (Figure 1B, left), to evaluate whether the T78M cav-3 reaches the plasma membrane or is retained in intracellular compartments, as reported for other cav-3 variants.10 We co-transfected MEF-KO cells with a mix of two plasmids able to target the CFP protein to both caveolar and non-caveolar fractions of the plasma membrane and with either WT (Figure 1A, top) or T78M (Figure 1A, bottom) cav-3 fused to EGFP. Confocal images show that both the WT and the T78M cav-3 are expressed at the plasma membrane.
Figure 1.
WT and T78M cav-3 proteins are expressed at the plasma membrane and form caveolae. (A) Representative confocal images of MEF-KO cells co-transfected with either the WT-EGFP cav-3 (green, top left) or the T78M-EGFP cav-3 (green, bottom left) and the membrane-targeted CFP (pm, red, centre) (exp = 3). Right panels show both signals overlapped (yellow). Scale bar = 20 µm. (B) Representative transmission electron micrographs of thin-sections from non-transfected (NT) MEF-KO cells (left) showing absence of any visible caveola, and from MEF-KO cells transfected with either the WT-EGFP cav-3 (centre) or the T78M-EGFP cav-3 (right) in which typical caveolar structures (arrows) can be seen at the plasma membrane (pm). The inset in the first panel shows a magnified region in which a clathrin-coated vesicle (Clv) is visible. rer, rough endoplasmic reticulum; N, nucleus; G, golgi apparatus; M, mitochondrion; n/exp = 25/3). Scale bar =150 nm.
Subsequently, we evaluated if the T78M cav-3 forms caveolae. In Figure 1B, representative electron microscopy images of MEF-KO cells expressing either the WT (middle panel) or T78M (right panel) cav-3 are shown. While non-transfected MEF-KO cells do not show any visible caveola (Figure 1B, left), caveolae could be detected in cells expressing both cav-3 variants (arrows). A quantification analysis demonstrated that the number of caveolae generated by expressing either the WT or T78M cav-3 were similar (WT: 0.056 ± 0.007 cav/µm n = 25, T78M: 0.053 ± 0.006 cav/µm n = 25, from 3 experiments).
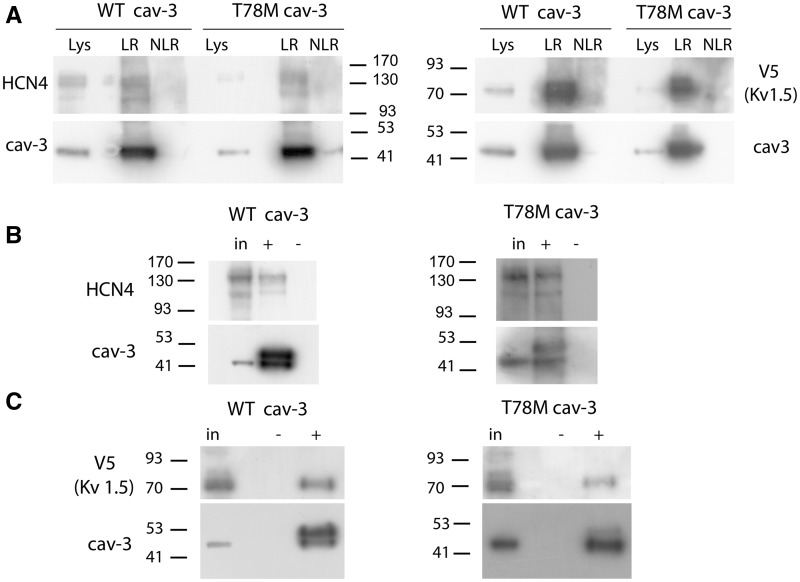
We then evaluated whether the T78M variant is present in lipid rafts when membranes are separated based on their density, as previously reported for the WT cav-3.3 We co-transfected MEF-KO cells with either the WT or T78M cav-3 and either the hKv.1.5 or hHCN4, two ion channels that directly interact with cav-3.4,22–24Figure 2A shows that both the WT and T78M cav-3 reside in the lipid rafts together with hKv1.5 and hHCN4. Furthermore, by co-immunoprecipitation experiments, we evaluated if the T78M variant retains the capacity to interact with these channels. Representative blots in Figure 2 show that both hHCN4 (panel B) and hKv1.5 channels (panel C) co-immunoprecipitate with the WT cav-3 isoform, as previously reported,22,23 and also with T78M cav-3, demonstrating that the point mutation does not disrupt the molecular complex formed by caveolins and these channels.
Figure 2.
The T78M cav-3 interacts with hHCN4 and hKv1.5 channels and targets them to lipid rafts. (A) Blots showing that hHCN4, hKV1.5, and cav-3 transfected in MEF-KO cells localize into lipid rafts isolated by discontinuous sucrose gradient (n ≥ 2). Lys, lysate; LR lipid raft fractions; NLR non-lipid raft fractions. (B, C) Co-immunoprecipitation (co-IP) experiments from MEF-KO cells co-transfected with either HCN4 (B) or V5-Kv1.5 (C) and WT cav-3 EGFP or T78M cav-3 EGFP (n = 3). An aliquot of the input (in) and of the co-IP eluate (+) were tested by western blot. A negative control (–) was performed by omitting the cav-3 antibody in the IP procedure. The cav-3 signal appear at around 50 kDa because it is a fusion protein with EGFP. The band just above the caveolin signal represents the heavy chain of the mouse IgG used during the immunoprecipitation, since it can be directly recognized by the anti-mouse IgG secondary antibody used in the cav3 western blots (data not shown).
3.2 Effects of the T78M cav-3 on hKv1.5 and hHCN4 channel properties
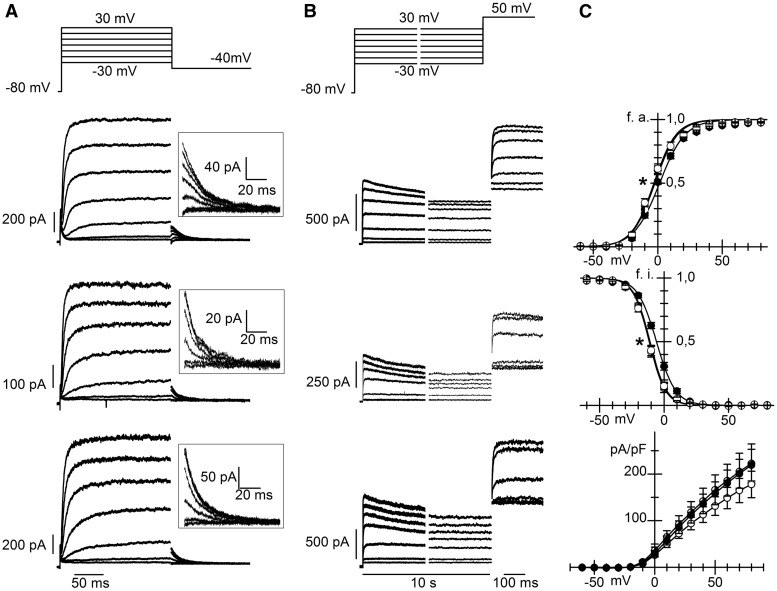
We next evaluated whether T78M cav-3 induces alterations of the ion currents mediated by hKv1.5 or hHCN4 channels. We compared the properties of these channels co-expressed in MEF-KO cells with either the WT or the T78M cav-3. In panels A and B of Figure 3, families of hKv1.5 current traces recorded from MEF-KO cells transfected with the WT (top), the T78M (middle) or both variants (bottom) of cav-3, to mimic heterozygous conditions, are shown. The plot of the mean activation curves (panel C top) shows that hKv1.5 activated at more negative voltages when expressed with the T78M (empty circles) than with the WT cav-3 (filled circles); the shift was 4 mV and was statistically significant. Notably, the expression of equal amounts of WT and T78M cav-3 (WT/T78M, half-filled circles) had an effect similar to that of the T78M cav-3 alone, with a shift of 3.6 mV vs. WT cav-3, suggesting a dominant behaviour.
Figure 3.
The T78M cav-3 affects hKv1.5 channel properties. Representative current traces recorded from MEF-KO cells co-transfected with hKv1.5 and WT cav-3 (top), T78M cav-3 (centre), or both (bottom) during activation (A) and inactivation voltage protocols (B). Insets in A show, on an expanded scale, tail currents recorded at –50 mV used for activation curve analysis. (C) Mean activation curves (top; V1/2 values were: WT 1.41 ± 0.70 mV, n/exp = 27/13; T78M –2.63 ± 1.43*, n/exp = 23/11; WT/T78M –2.21 ± 1.15*, n/exp = 17/5), inactivation curves (centre; WT –5.9 ± 0.9 mV, n/exp = 24/12; T78M –10.9 ± 0.9*, n/exp = 24/11; WT/T78M –11.5 ± 0.6*, n/exp = 18/5) and current density-voltage relations (bottom, WT n/exp = 22/12, T78M n = 26/11, WT/T78M n = 21/5) in the three groups (WT filled circles, T78M open circles, WT/T78M half-filled circles,) *P < 0.05 by nested and One-way ANOVA with Fisher’s test.
A similar negative shift was also evident from the analysis of inactivation curves (Figure 3C, middle); with shifts of 5.0 and 5.6 mV for homomeric T78M and heteromeric WT/T78M vs. WT cav-3, respectively, (P < 0.05). hKv1.5 current densities were similar in all groups over the whole range of voltages tested (Figure 3C, bottom).
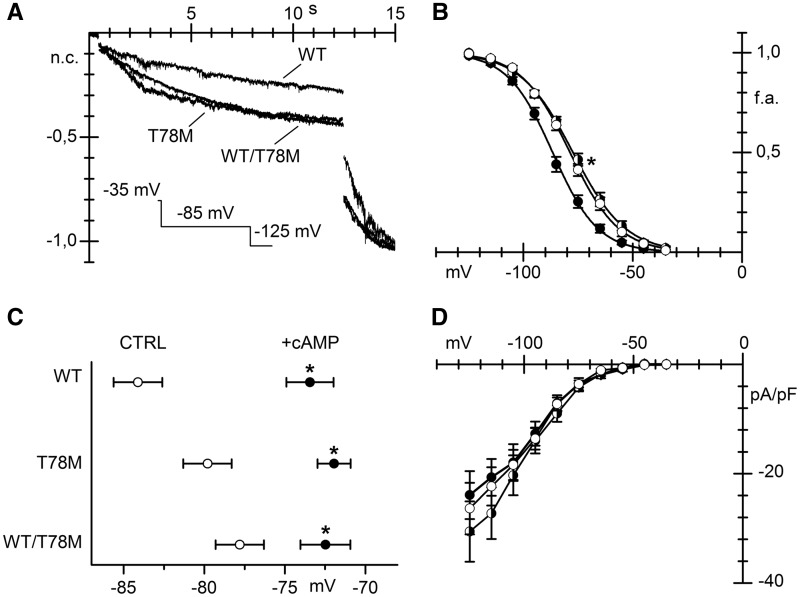
In Figure 4A, normalized hHCN4 current traces, recorded in response to a double step protocol to –85 and –125 mV from MEF KO cells transfected with the WT, the T78M or both (WT/T78M) cav-3 isoforms, are shown. In cells expressing the T78M variant, the fact that at –85 mV the current is larger while at the fully activated voltage of –125 mV the steady state current is similar indicates that hHCN4 activates at more positive potentials than in WT cells. The analysis of the mean activation curves shown in Figure 4B confirmed that co-expression of hHCN4 channels with the T78M cav-3, either alone or together with the WT cav-3, significantly shifted the hHCN4 channel activation to the depolarized direction, relative to co-expression with WT cav-3, by 7.9 and 8.8 mV, respectively. Since a depolarized shift of the hHCN4 current can result from increases intracellular levels of cAMP, we evaluated if saturating concentration of cAMP (10 µM) in the recording pipette shifted the activation curve of hHCN4 also in the presence of T78M cav-3. Figure 4C shows the mean V1/2 recorded in the presence of cAMP (filled circles) and in day-matched control measurements (empty circles) for the hHCN4 co-expressed with the WT, T78M, and WT/T78M cav-3. It is apparent that cAMP significantly shifted the V1/2 curves of hHCN4 channels to more depolarized potentials in all conditions. In the presence of 10 µM cAMP V1/2 values of HCN4 channels were similar. Neither current densities (Figure 4D) nor time constants (not shown) were affected by the cav-3 variant.
Figure 4.
The T78M cav-3 alters hHCN4 channel voltage-dependence. (A) Representative current traces elicited by a double hyperpolarizing step protocol to –85 and –125 mV (holding potential –35 mV) recorded from MEF-KO cells expressing hHCN4 and the WT, T78M or both forms of cav-3, as indicated; traces were normalized and overlapped for comparison. (B) Mean hHCN4 activation curves (V1/2 values: WT –86.2 ± 1.1 mV, n/exp = 26/15; T78M –78.3 ± 1.1* mV, n/exp =30/11; WT/T78M –77.4 ± 1.0* mV, n/exp =29/10. *P < 0.05 by nested and One-Way ANOVA with Fisher’s test), obtained from the different groups (WT filled circles, T78M open circles; WT/T78M half-filled circles). (C) Mean V1/2 values from day-matched recordings of hHCN4 currents in control (CTRL, empty circles) or in the presence of 10 µM cAMP in the pipette solution (+cAMP, black circles) in MEF-KO cells transfected with hHCN4 and WT, T78M or both forms of cav-3, as indicated (WT, control: –84.1 ± 1.5 mV n/exp = 6/4, cAMP: –73.4 ± 1.5 mV n/exp = 11/4; T78M, control: –79.8 ± 1.5 mV n/exp = 8/3, cAMP: –71.9 ± 1.0 mV n/exp = 16/3; WT/T78M, control: –77.8 ± 1.5 mV n/exp = 8/3, cAMP: –72.5 ± 1.6 mV n/exp = 8/3). *P < 0.05 by Student’s t-test. (D) Mean current density-voltage relations are shown for comparison (WT, n/exp = 20/10; T78M, n/exp = 14/9; WT/T78M, n = 20/10).
3.3 Effect of the T78M cav-3 on membrane excitability
Taking advantage of the dominant effect of the T78M variant, we transfected either the WT or the T78M cav-3 plasmids in primary cultures of neonatal rat ventricular cardiomyocytes (NRVCs) which endogenously express both cav-3 and cav-1,25 fire spontaneous action potentials and express several types of cardiac ion channels and modulatory proteins. As a readout of the expression of the T78M cav-3 variant in this system we analysed the endogenous If current expressed by NRVCs. In agreement with the hHCN4 data in MEF-KO cells, f-channels activated at significantly more depolarized potentials in cardiomyocytes expressing the T78M than in those expressing the WT cav-3 (see Supplementary material online, FigureS1).
Even though data in the heterologous expression systems showed that the T78M causes gain-of-function changes in both channels studied, the effect of the T78M cav-3 variant on the overall membrane excitability is difficult to predict. It is indeed known that caveolins functionally interact with and modulate many proteins beyond ion channels.26 We thus directly evaluated whether the T78M variant can affect membrane excitability of NRVC.
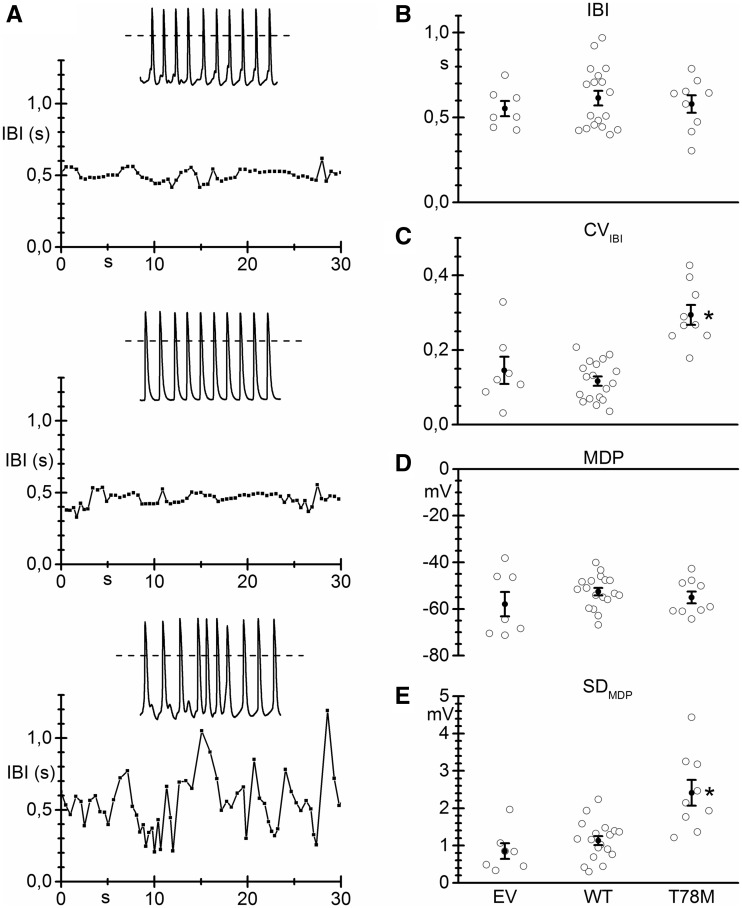
Figure 5A shows representative traces of the time course of the inter-beat interval (IBI) obtained from recordings of spontaneous activity from NRVCs transfected with either the empty vector (top), the WT (middle), or the T78M cav-3 (bottom). The insets represent stretches of original action potential recordings. The presence of the T78M cav-3 increased irregularities in spontaneous firing compared to NRVCs expressing the WT cav-3. Notably, on average the T78M cav-3 expression did not alter the mean IBI (Figure 5B), but caused a significant change only in the inter-beat variability, calculated as the coefficient of variation (CV, Figure 5C).27 Mean maximum diastolic potential (MDP) (Figure 5D) was not affected, while the standard deviation of MDP was significantly higher in the presence of T78M cav-3 (Figure 5E). These data indicate that expression of the T78M variant can indeed cause minor but significant alterations of membrane excitability.
Figure 5.
The electrical properties of NRVCs are altered in the presence of T78M cav-3. (A) Time course of the inter-beat interval (IBI) in neonatal cardiomyocytes transfected with the empty vector (EV, top), the WT cav-3 (centre) or the T78M cav-3 (bottom) vectors; insets show 5 s stretches of the original action potential recordings. Scatter plots showing single values (empty circles) and mean values (black circles) of (B) The IBI (EV 0.55 ± 0.04 s, n/exp = 7/2; WT 0.61 ± 0.04 s, n/exp = 18/3; T78M 0.58 ± 0.05 s, n/exp = 9/3), (C) Coefficient of variation of the IBI (CVIBI; EV 0.14 ± 0.04; WT 0.12 ± 0.01; T78M 0.29 ± 0.03), (D) Maximum diastolic potential (MDP; EV –57.9 ± 5.2 mV; WT –52.6 ± 1.6 mV; T78M –55.1 ± 2.5 mV) and of (E) the MDP standard deviation (SDMDP, EV 0.85 ± 0.21; WT 1.13 ± 0.12; T78M 2.41 ± 0.35). *P < 0.05 vs. all other conditions by nested and one-way ANOVA with Fisher’s test.
3.4 Population study and genetic analysis
Although data from the literature indicate that the minor allele frequency (MAF) of the T78M variant is similar in the general population and in cohorts of LQTS patients and SIDS,14,16 we have evaluated the MAF in small cohorts of patients affected by other types of arrhythmias in which HCN4 and Kv1.5 channels are involved.28,29 We screened the CAV3 gene in patients affected by: inappropriate sinus tachycardia (IST, 46 patients), sinus bradycardia (SB, 111 patients) and atrial fibrillation (AF, 16 patients). We also screened DNA samples from 37 stillbirths that did not present mutations in the KCNQ1, KCNH2, and SCN5A genes, the most prevalent LQT-linked genes found in stillbirths30 and from 209 unaffected controls. We found the T78M (CAV3 233C > T) variant in 2 patients with AF, 4 patients with IST and in 2 stillbirths. Frequencies of the T78M variant in these cohorts were thus significantly higher than those found in the bradycardic population (1 patient), in our control population (none) and in the general population, as reported in the ExAC database and in previous works.14,16Table 1 reports the allele frequency of the T78M variant in these cohorts.
Table 1.
Frequencies of the 233C > T Single Nucleotide Polymorphism in the cohorts investigated in this work and in the general population
| Pathology | Allele count/number | Allele frequency |
|---|---|---|
| Inappropriate sinus tachycardia | 4/92* | 4.35% |
| Atrial fibrillation | 2/32* | 6.25% |
| Stillbirth | 2/74* | 2.7% |
| Sinus bradycardia | 1/222 | 0.45% |
| Unaffected controls | 0/418 | 0% |
| General population (ExAC) | 367/120800 | 0.3% |
P < 0.05 by Fisher’s exact test.
3.5 Computational modelling of the T78M effect
Although the above data point to a deleterious effect of the T78M variant, in order to gain insight into its possible role in generating a cellular substrate susceptible to cardiac arrhythmias, we also adopted an in silico approach to verify the effects of the T78M variant by numerical modelling. Since we found the T78M variant at high frequencies in patients with sinus tachycardia as well as in patients with atrial fibrillation, we have used mathematical models of both atrial19 and sinoatrial21 action potentials (Figure 6).
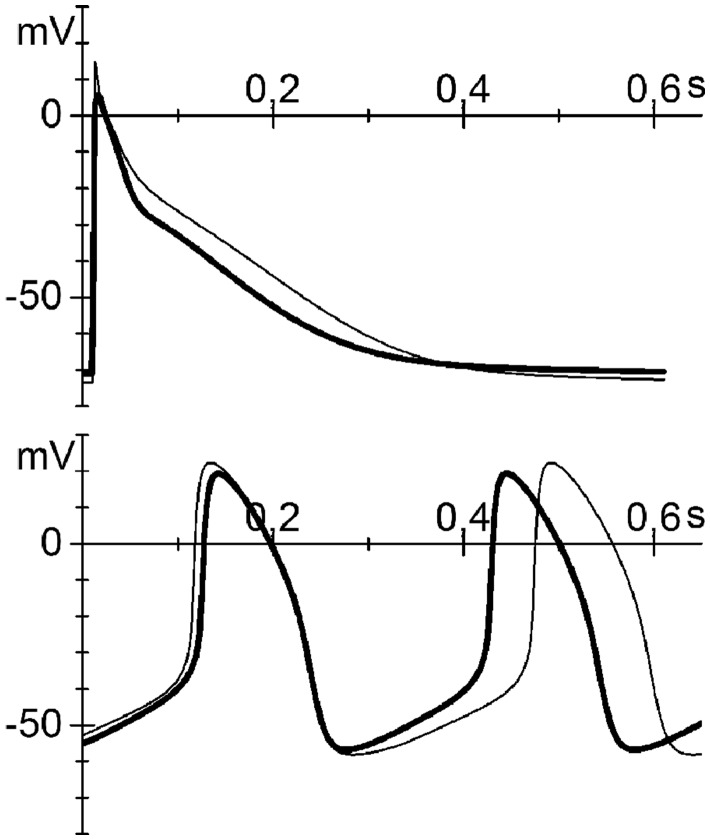
Figure 6.
Mathematical models of both atrial and sinoatrial cells show an arrhythmic contribution of the T78M cav-3. Atrial (top) and sinoatrial (bottom) action potentials generated using the Grandi-Bers human atrial cell model19 and the Severi-DiFrancesco rabbit sinoatrial cell model,20,21 respectively. Thin line, basal conditions (WT); thick line, after insertion of the T78M cav-3-dependent alterations.
In the top panel of Figure 6, the thin line represents the simulated action potential of an atrial cell under control conditions, while the thick line represents the action potential after the introduction of the T78M-related alterations in the If and IKur currents described above and the alterations in the INa current previously reported.9 It has also been previously reported that the T78M mutation is associated with a decrease in the Kir2.1 current amplitude of about 70% at –80 mV.13 According to our numerical reconstruction, this decrease of Kir2.1 current causes a large membrane depolarization incompatible with membrane excitability (see Supplementary material online, FigureS2A). We then checked the effects of the T78M mutation on either native IK1 current in neonatal cardiomyocytes (data not shown) or on hKir2.1 channels expressed in MEF-KO cells (see Supplementary material online, FigureS2B) but we did not observe any alteration. For these reasons, no change in Kir2.1 current was included in our model.
Introducing then only the If and IKur changes described above, the presence of the T78M mutation induced in the atrial action potential a clear shortening of the duration (–11%) at both 50% and 90% of repolarization (APD50 and APD90), a mild depolarization (2.3 mV) of the resting potential and a reduction of the peak potential (–8.4 mV) (Figure 6).
Simulations were also repeated using an alternative computational model of human atrial action potential20 and the results were similar to those of the Grandi-Bers model (APD50 –7%, APD90 –13%, resting potential +0.6 mV; peak +0.1 mV; data not shown).
Simulation of the T78M effects using the Severi-DiFrancesco model of a rabbit SAN cell shows, as expected, an increase in firing rate (18%) due to an increase in the diastolic depolarization rate (Figure 6, bottom), with only minor differences in other action potential parameters (APD90 –6%, Maximum Diastolic Potential +1 mV, upstroke potential –3 mV).
In conclusion, our data show that the T78M variant is able to reach the plasma membrane and to form caveolae. Nonetheless, the T78M cav-3 modifies the properties of interacting cardiac channels in a way able to produce alterations of the overall electrical activity of the cardiomyocytes.
4. Discussion
4.1 The T78M variant is found in the general population and in patients with rhythm disorders at different frequencies
Among the cav-3 variants, T78M is the one most frequently found in patients with cardiac and muscle diseases.7,12,31 For example, the T78M mutation has been linked to LQTS,8 a pathology that can cause cardiac arrest as a first symptom, and to some cases of SIDS.9 Studies suggesting the association of the T78M variant with one or more pathological conditions, however, do not normally provide evidence for functional alterations,10,11 with the exception of two studies9,13 showing that expression of the T78M cav-3, in heterologous cellular systems, causes pro-arrhythmogenic ion channel modifications. Cronk et al.9 have shown a gain of function of the sodium channel Nav1.5 due to the increase of a non-inactivating late component, while Vaidyanathan and colleagues13 have shown a loss of function of Kir2.1 channels responsible for the IK1 current that stabilizes the resting potential of working cardiomyocytes.
Lately however, genetic analysis has questioned the pathogenic role of the T78M variant; Spadafora et al., reported that T78M cav-3 seems to be a common polymorphism in South Italy,15 while others14,16 found the T78M variant in a cohort of healthy controls with a MAF similar to that found in both LQTS patients and SIDS and in agreement with data from the Exome Sequencing Project. They thus argued against a causative effect of this cav-3 variant in the etiology of those conditions. It is important to underline, however, that the onset of cardiac pathologies occurs mostly late in life and it is usually multifactorial (genetic predisposition, prior diseases, lifestyle); under these conditions, a healthy control appears hard to define.
Here, we report the presence of the T78M variant in patients affected by AF and IST and in stillbirths at a significantly higher frequency (from 2.7 to 6.7%) than in the general population (0.3%). Though an in-depth genetic analysis is still lacking, our findings suggest that the T78M variant, in association with other factors, may contribute to the development of arrhythmic conditions.
4.2 The T78M variants normally target the plasma membrane where they form caveolae and interact with ion channels
The cav-3 protein is essential for proper assembling of membrane caveolae. There are so far few and discordant pieces of evidence concerning the membrane localization of the T78M cav-3 isoform. Immunostaining of T78M cav-3-transfected COS-7 cells showed a distribution pattern similar to that of WT cav-3-transfected cells.10 In another study, however, immunostaining of a muscle biopsy of a patient carrying the T78M variant resulted in a very faint cav-3 signal in most fibres.11 Here, using a cell model without endogenous caveolins, we have shown that the T78M cav-3, like the WT isoform, is expressed at the plasma membrane (Figure 1A). The presence of caveolins in the plasma membrane is a necessary but not sufficient condition to generate the caveolar domain. Therefore, taking advantage of the cellular system lacking caveolae, we analysed the presence of caveolae using electron microscopy in the presence of either the WT or the T78M cav-3. No evident differences in the ability of the system to form caveolae and in the number of caveolae produced in the two conditions were observed. These data elucidate for the first time not only that T78M cav-3 targets the plasma membrane but also that its expression still permits the generation of caveolar structures. Despite this, the functional integrity and molecular composition of the caveolar microdomains cannot be determined based only on the evidence of membrane expression.
The interaction of cav-3 with the proteins residing in the caveolar domain is a determinant of their functionality. To address the functional role of T78M cav-3 in a cardiac context, we focused our attention on two channels, hKv1.5 and hHCN4, which are dysregulated in several cardiac pathologies32–35 and are known to interact with caveolins. Folco and colleagues23 demonstrated that hKv1.5 interacts with cav-3 through the binding of SAP-97, whereas, we have previously shown that mutations in the caveolin binding domain of HCN4 alter both functional properties and trafficking of this channel to the plasma membrane.22 Results obtained with discontinuous sucrose gradient and co-immunoprecipitation experiments show that both channels localize to lipid rafts and are able to interact with both the T78M and WT cav-3 (Figure 2). It is thus likely that also in the presence of the T78M cav-3 the channels reside in caveolae.
4.3 The T78M variant induces functional changes in membrane excitability by affecting ion channel function
While still maintaining an interaction with the channels, T78M cav-3 induces significant alterations of their functional properties. Indeed, electrophysiological analysis revealed that it induces a leftward shift in the activation curves of hKv1.5 channels and a rightward shift in the activation curves of hHCN4 channels in MEF-KO cells. This suggests an increased contribution of both currents during an action potential. In agreement with the proper interaction of channels with T78M cav-3, proteins trafficking to the plasma membrane and thus current densities were not altered.
Despite the previously reported effects of the T78M cav-3 variant on Nav1.5 and on Kir2.1 channels,9,13 and the effects on HCN4 and Kv1.5 channels reported here, the overall action on membrane excitability is difficult to predict. NRVC express all the above ion channels36–38 and modulatory proteins,18 among which cav-1, that may form oligomers with cav-3.25 Moreover, they also possess all the calcium-handling proteins that contribute to membrane excitability and are modulated by caveolae.26 Transfection of the T78M cav-3 variant in NRVCs allowed us to estimate its effect on all these endogenous proteins modulated by cav-3. While the expression of the human WT cav-3 in NRVCs did not affect cell excitability, the expression of the T78M cav-3 caused a clear modification of CVIBI and SDMDP. Notably, neonatal cardiomyocytes show an endogenous If current that, according to our data, was activated in a voltage range significantly more positive in T78M than in the WT cav-3 transfected cells (see Supplementary material online, FigureS1), indicating that alterations in ion channels observed in MEF-KO cells are maintained. Nonetheless, the alterations observed in the presence of T78M cav-3 on membrane excitability do not reproduce the expected effect on a single current, but are the result of the modification of many factors contributing to the integrated electrical activity of cardiomyocytes.
4.4 In silico analysis confirms that the T78M cav-3 alters the action potential shape of atrial and sinoatrial cells
As an alternative approach to evaluate the effect of the T78M on the electrical properties of specific types of cardiomyocytes, we carried out in silico analysis by modelling the alteration in ion currents reported here and in previous works. In this regard, in our analysis we omitted the effect of the T78M mutation on Kir2.1 channels13 because (1) the 70% decrease in IK1 shown by Vaidyanathan et al.13 was not compatible with the generation of action potentials in the atrial models tested (see Supplementary material online, FigureS2A) and (2), we failed to detect any T78M-dependent downregulation of either Kir2-1 channels transfected in MEF-KO cells (see Supplementary material online, FigureS2B) or of endogenous IK1 current in neonatal cardiomyocytes (not shown). We are unable to provide an explanation for this discrepancy. Even in the absence of any modification of the IK1 current, our simulations clearly show that action potentials of both rabbit sinoatrial and human atrial cells are significantly altered. In particular, a reduction of the atrial action potential duration and a more positive activation of HCN channels, such as those we observed, are both involved in the remodelling associated with atrial fibrillation.29,34 Moreover, a gain of function of HCN4 induces a faster rhythm in SAN cells, a condition compatible with phenotypic manifestations of inappropriate sinus tachycardia.28 Although in neonatal cardiomyocytes the overexpression of the T78M cav-3 gives rise to a positive shift of the native If current (see Supplementary material online, FigureS1) we did not see the increase in firing rate, as observed in the SAN model. This discrepancy may occur because NRVCs do not represent a model of SAN cells. Indeed, they have much lower If current density17 than either rabbit3 or mouse SAN.39 Furthermore, they express the IK1 current, which counteracts the If-dependent membrane depolarization. Not surprisingly, therefore, the effect of T78M cav-3 on If has limited impact on NRVC firing rate. Finally, the computational model reproduces a single cell, which includes only known effects on single currents, whereas data from cardiomyocytes were obtained from small clusters of cells in which all the mechanisms, directly or indirectly affected by the mutation, are working.
4.5 Study limitations
The results of the genetic analysis performed on arrhythmic patients is limited by the sample size and an extended analysis would be necessary to possibly correlate the T78M cav-3 variant with the pathological conditions studied.
Another limitation of the present study is that our experiments were all based on transient transfection of either the WT or mutated cav-3, an approach that does not allow to control levels of expression. An alternative and more sophisticated approach to study the effect of the T78M variant would be to generate induced pluripotent stem cells from patients and differentiate them into functional cardiomyocytes. This approach is at this moment unfeasible in our lab because of the lack of patient-derived cells to reprogram.
5. Conclusions
Our data provide the first evidence that the mutant T78M cav-3 normally reaches the plasma membrane and generates caveolae, yet it modifies at the same time the functional properties of the interacting pacemaker hHCN4 channels and the atrial-specific hKv1.5 channels, both important for the physiological activity of sinoatrial and atrial cells, respectively. Moreover, we show that the presence of the T78M cav-3 can significantly alter the overall electrical activity both in vitro in spontaneously beating cardiomyocytes and in silico in atrial and sinoatrial cell models. These data, together with the observation that the T78M variant is found more frequently in patients with rhythm disorders, suggest that the presence of the T78M variant, through alteration of the functional contribution of multiple ion channels can, under specific conditions, predispose to initiation of life-threatening arrhythmias.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Conflict of interest: none declared.
Funding
This work was supported by Fondazione CARIPLO [2014-1090 to A.Ba., 2014-0728 to D.D. and 2014-0822 to M.Ba.].
Supplementary Material
References
- 1. Cohen AW, Hnasko R, Schubert W, Lisanti MP.. Role of caveolae and caveolins in health and disease. Physiol Rev 2004;84:1341–1379. [DOI] [PubMed] [Google Scholar]
- 2. Balijepalli RC, Kamp TJ.. Caveolae, ion channels and cardiac arrhythmias. Prog Biophys Mol Biol 2008;98:149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barbuti A, Gravante B, Riolfo M, Milanesi R, Terragni B, DiFrancesco D.. Localization of pacemaker channels in lipid rafts regulates channel kinetics. Circ Res 2004;94:1325–1331. [DOI] [PubMed] [Google Scholar]
- 4. Martens JR, Sakamoto N, Sullivan SA, Grobaski TD, Tamkun MM.. Isoform-specific localization of voltage-gated K+ channels to distinct lipid raft populations. Targeting of Kv1.5 to caveolae. J Biol Chem 2001;276:8409–8414. [DOI] [PubMed] [Google Scholar]
- 5. Balijepalli RC, Foell JD, Hall DD, Hell JW, Kamp TJ.. Localization of cardiac L-type Ca(2+) channels to a caveolar macromolecular signaling complex is required for beta(2)-adrenergic regulation. Proc Natl Acad Sci USA 2006;103:7500–7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yarbrough TL, Lu T, Lee HC, Shibata EF.. Localization of cardiac sodium channels in caveolin-rich membrane domains: regulation of sodium current amplitude. Circ Res 2002;90:443–449. [DOI] [PubMed] [Google Scholar]
- 7. Gazzerro E, Sotgia F, Bruno C, Lisanti MP, Minetti C.. Caveolinopathies: from the biology of caveolin-3 to human diseases. Eur J Hum Genet 2010;18:137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vatta M, Ackerman MJ, Ye B, Makielski JC, Ughanze EE, Taylor EW, Tester DJ, Balijepalli RC, Foell JD, Li Z, Kamp TJ, Towbin JA.. Mutant caveolin-3 induces persistent late sodium current and is associated with long-QT syndrome. Circulation 2006;114:2104–2112. [DOI] [PubMed] [Google Scholar]
- 9. Cronk LB, Ye B, Kaku T, Tester DJ, Vatta M, Makielski JC, Ackerman MJ.. Novel mechanism for sudden infant death syndrome: persistent late sodium current secondary to mutations in caveolin-3. Heart Rhythm 2007;4:161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Traverso M, Gazzerro E, Assereto S, Sotgia F, Biancheri R, Stringara S, Giberti L, Pedemonte M, Wang X, Scapolan S, Pasquini E, Donati MA, Zara F, Lisanti MP, Bruno C, Minetti C.. Caveolin-3 T78M and T78K missense mutations lead to different phenotypes in vivo and in vitro. Lab Invest 2008;88:275–283. [DOI] [PubMed] [Google Scholar]
- 11. Ricci G, Scionti I, Ali G, Volpi L, Zampa V, Fanin M, Angelini C, Politano L, Tupler R, Siciliano G.. Rippling muscle disease and facioscapulohumeral dystrophy-like phenotype in a patient carrying a heterozygous CAV3 T78M mutation and a D4Z4 partial deletion: Further evidence for “double trouble” overlapping syndromes. Neuromuscul Disord 2012;22:534–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hedley PL, Kanters JK, Dembic M, Jespersen T, Skibsbye L, Aidt FH, Eschen O, Graff C, Behr ER, Schlamowitz S, Corfield V, McKenna WJ, Christiansen M.. The role of CAV3 in long-QT syndrome: clinical and functional assessment of a caveolin-3/Kv11.1 double heterozygote versus caveolin-3 single heterozygote. Circ Cardiovasc Genet 2013;6:452–461. [DOI] [PubMed] [Google Scholar]
- 13. Vaidyanathan R, Vega AL, Song C, Zhou Q, Tan BH, Berger S, Makielski JC, Eckhardt LL.. The interaction of caveolin 3 protein with the potassium inward rectifier channel Kir2.1: physiology and pathology related to long qt syndrome 9 (LQT9). J Biol Chem 2013;288:17472–17480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Refsgaard L, Holst AG, Sadjadieh G, Haunso S, Nielsen JB, Olesen MS.. High prevalence of genetic variants previously associated with LQT syndrome in new exome data. Eur J Hum Genet 2012;20:905–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Spadafora P, Liguori M, Andreoli V, Quattrone A, Gambardella A.. CAV3 T78M mutation as polymorphic variant in South Italy. Neuromuscul Disord 2012;22:669–670. [DOI] [PubMed] [Google Scholar]
- 16. Andreasen C, Refsgaard L, Nielsen JB, Sajadieh A, Winkel BG, Tfelt-Hansen J, Haunso S, Holst AG, Svendsen JH, Olesen MS.. Mutations in genes encoding cardiac ion channels previously associated with sudden infant death syndrome (SIDS) are present with high frequency in new exome data. Can J Cardiol 2013;29:1104–1109. [DOI] [PubMed] [Google Scholar]
- 17. Avitabile D, Crespi A, Brioschi C, Parente V, Toietta G, Devanna P, Baruscotti M, Truffa S, Scavone A, Rusconi F, Biondi A, D'alessandra Y, Vigna E, DiFrancesco D, Pesce M, Capogrossi MC, Barbuti A.. Human cord blood CD34+ progenitor cells acquire functional cardiac properties through a cell fusion process. Am J Physiol Heart Circ Physiol 2011;300:H1875–H1884. [DOI] [PubMed] [Google Scholar]
- 18. Rybin VO, Xu X, Lisanti MP, Steinberg SF.. Differential targeting of beta -adrenergic receptor subtypes and adenylyl cyclase to cardiomyocyte caveolae. A mechanism to functionally regulate the cAMP signaling pathway. J Biol Chem 2000;275:41447–41457. [DOI] [PubMed] [Google Scholar]
- 19. Grandi E, Pandit SV, Voigt N, Workman AJ, Dobrev D, Jalife J, Bers DM.. Human atrial action potential and Ca2+ model: sinus rhythm and chronic atrial fibrillation. Circ Res 2011;109:1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Koivumaki JT, Korhonen T, Tavi P, Impact of sarcoplasmic reticulum calcium release on calcium dynamics and action potential morphology in human atrial myocytes: a computational study. PLoS Comput Biol 2011;7:e1001067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Severi S, Fantini M, Charawi LA, DiFrancesco D.. An updated computational model of rabbit sinoatrial action potential to investigate the mechanisms of heart rate modulation. J Physiol (Lond) 2012;590:4483–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barbuti A, Scavone A, Mazzocchi N, Terragni B, Baruscotti M, Difrancesco D.. A caveolin-binding domain in the HCN4 channels mediates functional interaction with caveolin proteins. J Mol Cell Cardiol 2012;53:187–195. [DOI] [PubMed] [Google Scholar]
- 23. Folco EJ, Liu GX, Koren G.. Caveolin-3 and SAP97 form a scaffolding protein complex that regulates the voltage-gated potassium channel Kv1.5. Am J Physiol Heart Circ Physiol 2004;287:H681–H690. [DOI] [PubMed] [Google Scholar]
- 24. Barbuti A, Terragni B, Brioschi C, DiFrancesco D.. Localization of f-channels to caveolae mediates specific beta2-adrenergic receptor modulation of rate in sinoatrial myocytes. J Mol Cell Cardiol 2007;42:71–78. [DOI] [PubMed] [Google Scholar]
- 25. Lal H, Verma SK, Feng H, Golden HB, Gerilechaogetu F, Nizamutdinov D, Foster DM, Glaser SS, Dostal DE.. Caveolin and beta1-integrin coordinate angiotensinogen expression in cardiac myocytes. Int J Cardiol 2013;168:436–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harvey RD, Calaghan SC.. Caveolae create local signalling domains through their distinct protein content, lipid profile and morphology. J Mol Cell Cardiol 2012;52:366–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Clay JR, DeHaan RL.. Fluctuations in interbeat interval in rhythmic heart-cell clusters. Role of membrane voltage noise. Biophys J 1979;28:377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baruscotti M, Bucchi A, Milanesi R, Paina M, Barbuti A, Gnecchi-Ruscone T, Bianco E, Vitali-Serdoz L, Cappato R, DiFrancesco D.. A gain-of-function mutation in the cardiac pacemaker HCN4 channel increasing cAMP sensitivity is associated with familial Inappropriate Sinus Tachycardia. Eur Heart J 2017;38:280–288. [DOI] [PubMed] [Google Scholar]
- 29. Heijman J, Voigt N, Nattel S, Dobrev D.. Cellular and molecular electrophysiology of atrial fibrillation initiation, maintenance, and progression. Circ Res 2014;114:1483–1499. [DOI] [PubMed] [Google Scholar]
- 30. Crotti L, Tester DJ, White WM, Bartos DC, Insolia R, Besana A, Kunic JD, Will ML, Velasco EJ, Bair JJ, Ghidoni A, Cetin I, Van Dyke DL, Wick MJ, Brost B, Delisle BP, Facchinetti F, George AL, Schwartz PJ, Ackerman MJ.. Long QT syndrome-associated mutations in intrauterine fetal death. JAMA 2013;309:1473–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gazzerro E, Bonetto A, Minetti C.. Caveolinopathies: translational implications of caveolin-3 in skeletal and cardiac muscle disorders. Handb Clin Neurol 2011;101:135–142. [DOI] [PubMed] [Google Scholar]
- 32. Christophersen IE, Olesen MS, Liang B, Andersen MN, Larsen AP, Nielsen JB, Haunso S, Olesen SP, Tveit A, Svendsen JH, Schmitt N.. Genetic variation in KCNA5: impact on the atrial-specific potassium current IKur in patients with lone atrial fibrillation. Eur Heart J 2013;34:1517–1525. [DOI] [PubMed] [Google Scholar]
- 33. Milanesi R, Baruscotti M, Gnecchi-Ruscone T, DiFrancesco D.. Familial sinus bradycardia associated with a mutation in the cardiac pacemaker channel. N Engl J Med 2006;354:151–157. [DOI] [PubMed] [Google Scholar]
- 34. Stillitano F, Lonardo G, Giunti G, Del Lungo M, Coppini R, Spinelli V, Sartiani L, Poggesi C, Mugelli A, Cerbai E.. Chronic atrial fibrillation alters the functional properties of If in the human atrium. J Cardiovasc Electrophysiol 2013;24:1391–1400. [DOI] [PubMed] [Google Scholar]
- 35. Macri V, Mahida SN, Zhang ML, Sinner MF, Dolmatova EV, Tucker NR, McLellan M, Shea MA, Milan DJ, Lunetta KL, Benjamin EJ, Ellinor PT.. A novel trafficking-defective HCN4 mutation is associated with early-onset atrial fibrillation. Heart Rhythm 2014;11:1055–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guo W, Kada K, Kamiya K, Toyama J.. IGF-I regulates K(+)-channel expression of cultured neonatal rat ventricular myocytes. Am J Physiol 1997;272:H2599–H2606. [DOI] [PubMed] [Google Scholar]
- 37. He Y, Pan Q, Li J, Chen H, Zhou Q, Hong K, Brugada R, Perez GJ, Brugada P, Chen YH.. Kir2.3 knock-down decreases IK1 current in neonatal rat cardiomyocytes. FEBS Lett 2008;582:2338–2342. [DOI] [PubMed] [Google Scholar]
- 38. Kaufmann SG, Westenbroek RE, Zechner C, Maass AH, Bischoff S, Muck J, Wischmeyer E, Scheuer T, Maier SK.. Functional protein expression of multiple sodium channel alpha- and beta-subunit isoforms in neonatal cardiomyocytes. J Mol Cell Cardiol 2010;48:261–269. [DOI] [PubMed] [Google Scholar]
- 39. Baruscotti M, Bucchi A, Viscomi C, Mandelli G, Consalez G, Gnecchi-Rusconi T, Montano N, Casali KR, Micheloni S, Barbuti A, DiFrancesco D.. Deep bradycardia and heart block caused by inducible cardiac-specific knockout of the pacemaker channel gene Hcn4. Proc Natl Acad Sci USA 2011;108:1705–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.