Abstract
Aims
Duchenne muscular dystrophy (DMD) is an inherited devastating muscle disease with severe and often lethal cardiac complications. Emerging evidence suggests that the evolution of the pathology in DMD is accompanied by the accumulation of mitochondria with defective structure and function. Here, we investigate whether defects in the housekeeping autophagic pathway contribute to mitochondrial and metabolic dysfunctions in dystrophic cardiomyopathy.
Methods and results
We employed various biochemical and imaging techniques to assess mitochondrial structure and function as well as to evaluate autophagy, and specific mitochondrial autophagy (mitophagy), in hearts of mdx mice, an animal model of DMD. Our results indicate substantial structural damage of mitochondria and a significant decrease in ATP production in hearts of mdx animals, which developed cardiomyopathy. In these hearts, we also detected enhanced autophagy but paradoxically, mitophagy appeared to be suppressed. In addition, we found decreased levels of several proteins involved in the PINK1/PARKIN mitophagy pathway as well as an insignificant amount of PARKIN protein phosphorylation at the S65 residue upon induction of mitophagy.
Conclusions
Our results suggest faulty mitophagy in dystrophic hearts due to defects in the PINK1/PARKIN pathway.
Keywords: Cardiomyopathy, Autophagy, Mitochondria, Dystrophin, PINK1, PARKIN
1. Introduction
Duchenne Muscular Dystrophy (DMD) is an inherited lethal muscle disease that affects primarily adolescent males. DMD is usually diagnosed in early childhood. The primary defect in DMD is lack of functional cytoskeletal protein dystrophin.1 For a long time, it was considered to be predominantly a skeletal muscle illness clinically associated with progressive debilitating muscle weakness, skeletal deformities, and breathing disorders. In the past, most of DMD patients have indeed succumbed from respiratory failure and only ∼10% have died from cardiac complications. Currently, steroid therapy and assisted ventilators help combat respiratory dysfunctions and prolong the life of DMD patients. Average life expectancy increased from 25 years in 1970 to >40 years in 2011. Statistics also have shown that heart failure has continued to grow as a prominent cause of death. At present, over 40% (and increasing) of DMD patients are dying from heart disease.2,3
On the cellular level, the pathophysiology of cardiac dystrophy is associated with severe oxidative stress and defective intracellular Ca2+ signaling.4–8 Importantly, these cellular pathologies can already be detected in cardiomyocytes isolated from hearts of very young dystrophic (mdx) mice, which did not yet exhibit any clinical signs of the disease. Our recent findings indicate that excessive Ca2+ signaling in young dystrophic hearts largely results from an increased sensitivity of sarcoplasmic reticulum (SR) Ca2+ release channels (ryanodine receptors, RyR2s) to activation by Ca2+.7 Moreover, we identified RyR2 oxidation and ROS-activated CaMKII-dependent phosphorylation as pertinent post-translational modifications and initial causes of RyR2 hypersensitivity. At later stages of the disease, PKA phosphorylation and nitrosation further hypersensitize RyR29,10 contributing to activation of Ca2+-dependent apoptotic and/or necrotic cell death that culminates in the development of fibrosis and reduction in contractility of dystrophic hearts (see ref. 11 for review). It was suggested that RyR2 hypersensitivity eventually increases SR Ca2+ leak and the propensity for arrhythmias, and leads to weak heartbeat and heart failure.
In addition, some mitochondrial abnormalities were identified in hearts of mdx animals at the onset of cardiomyopathy.8,12–14 As disease progressed, the number of mitochondria showing an increased area as well as a loss of normal crista structure gradually increased. Degradation in mitochondrial structure was correlated with a progressive increase in mitochondrial Ca2+ sequestration and mitochondrial depolarization. Overall, it appears that oxidative stress in synergy with intracellular Ca2+ overload increases the propensity for irreversible opening of the mitochondrial permeability transition pore (mPTP) resulting in progressive deterioration of mitochondrial structure and function.
Macroautophagy (commonly referred to as autophagy) is a crucial cellular housekeeping mechanism that enables organisms to maintain cellular homeostasis and normal function by degrading and turning over proteins. To perform this task, autophagy functions together with another protein degradation pathway, the ubiquitin-proteasome system. In addition, autophagy helps to eliminate large protein aggregates and damaged organelles. Loss or inhibition of autophagy results in accumulation of ubiquitinated proteins and development of various pathologies, including cardiomyopathies. In addition to constituent autophagy that ensures quality control of proteins and organelles, autophagy also enables cells to survive various internal and external stresses, such as nutrient deprivation, infection, and inflammation. In heart autophagy is up-regulated under different pathological conditions, such as ischemia/reperfusion, cardiac hypertrophy, and heart failure (reviewed in ref. 15).
Autophagy is a multistep process, which starts with the formation of a double membrane structure called phagophore assembled from various cellular compartments, such as endoplasmic reticulum, Golgi complex, and even mitochondria.16,17 Then the phagophore elongates forming an autophagosome, in which damaged cytoplasmic components are sequestered.18 This is followed by autophagosome fusion with lysosomes and generation of autolysosomes, where the autophagic cargo is degraded by acidic hydrolases.19
Defective mitochondria can be selectively degraded in the process of mitophagy (a.k.a. mitochondrial autophagy). The PINK1/PARKIN-mediated mechanism is one of the most established mitophagy pathways in mammals.20,21 In healthy polarized mitochondria, PTEN-induced putative kinase 1 (PINK1) is continuously degraded by presenilin-associated, rhomboid-like (PARL) protease of the inner mitochondrial membrane (IMM).22,23 The current (and still developing) view is that mitochondrial depolarization results in accumulation of this protein in the outer mitochondrial membrane (OMM).24,25 PINK1 phosphorylates other OMM proteins, including mitofusin 2 (Mfn2), which subsequently recruits the cytosolic protein Parkinson juvenile disease protein 2 (PARKIN).26 PARKIN functions as an E3 ubiqitin ligase.27 It ubiquitinates multiple proteins in OMM leading to the recruitment of SQSTM1 (ubiquitin-binding protein sequestosome 1, a.k.a. p62). SQSTM1, in turn, docks autophagosomal protein LC3 (microtubule-associated protein 1 light chain 3) and triggers mitochondrial autophagy.28 The efficiency of mitophagy is determined by a balance between the availability of mitophagic substrates and the cellular needs for removal of damaged organelles. Reduced autophagy and/or mitophagy may eventually add to accumulation of dysfunctional mitochondria resulting in decreased ATP production and energy starvation of the heart tissue.
The aim of this study was to establish whether and to what extent accumulation of damaged mitochondria and loss of mitochondrial functions in dystrophic hearts result from an impaired autophagic/mitophagic mechanisms. For this, mitochondrial structure and function was assessed and compared in hearts of animals at pre-clinical and clinical stages of the disease. In parallel, autophagy and specific mitochondrial autophagy were examined. Our results indicate substantial structural damage of mitochondria and significant decrease in ATP production in hearts of 12 months and older mdx animals. In these hearts, we also detected enhanced autophagy. At the same time, mitophagy appeared to be suppressed. The faulty mitophagy is likely to be due to a deficiency of the substrates involved in the PINK1/PARKIN pathway. Our results suggest that defective mitophagic mechanisms contribute to a progressive accumulation of damaged mitochondria in dystrophic hearts and deterioration of cardiac functions.
2. Methods
All experiments conformed to the NIH Guide for the Care and Use of Laboratory Animals published by the US National Institute of Health (NIH publication, 8th edition, 2011) and were approved by the Institutional Animal Care and Use Committee of the New Jersey Medical School, Rutgers University, USA. Brief protocol descriptions are below, and all methods are described in detail in Supplementary material online.
2.1. Animals and cell isolation
C57BL10 mice (wild-type, WT) were purchased from Harlan laboratory and C57BL/10ScSn-Dmdmdx/J mice (dystrophin-deficient, mdx) were purchased from the Jackson Laboratory. Mice at the age of 3–4 months (when mdx mice show no sign of the cardiac disease) and 12 months and older (when mdx mice display significant cardiomyopathy) were used in this study. Ventricular myocytes were isolated enzymatically as previously described.7
2.2. Electron microscopy
Hearts were removed, fixed in Formaldehyde, cut in 100 nm sections and imaged with a Phillips CM12 transmission electron microscope at Rutgers Core Imaging Lab.
2.3. ATP production measurements
Luciferin–luciferase assay was used to measure ATP content in heart tissue as previously described.29
2.4. Extraction of mitochondrial fraction
Mouse hearts were removed and homogenized. Mitochondrial fractions were extracted by sucrose gradient centrifugation as previously described.30
2.5. Western blots
Protein extraction and western blotting were performed as in ref. 7
2.6. RNA isolation and real-time reverse transcription PCR
Total RNA was isolated using Tri-Reagent® and Direct-zol RNA MiniPrep kit according to manufacturer’s instructions (Zymo Research, Irvine, CA) and quantified with a Nanodrop ND-1000 Spectrophotometer (Nanodrop Technologies, Wilmington, DE). Real-time PCR was performed using a 7500 Fast Real-Time PCR system (Applied Biosystems) with TaqMan Gene Expression detection assay. Three independent measurements were carried out for each sample.
2.7. In vivo treatment with chloroquine, rapamycin, and DNP (2,4-dinitrophenol)
Mice were intraperitoneally injected 10 mg/kg chloroquine, 10 mg/kg rapamycin, 15 mg/kg DNP, and sacrificed 4, 12, and 12 h later, respectively.
2.8 Confocal imaging of cardiomyocytes
Green (Alexa Fluor 488) and red (MitoTracker® Orange) fluorescence was recorded with an Olympus FluoView-1000 confocal microscope operation in frame scan mode (Olympus).
2.9. Statistics
Data sets contain results from 3 to 10 mice. N indicates the number of animals used, and n specifies the number of cells studied. Comparisons between two groups of data were made by one-way ANOVA followed by Tukey, Holm–Sidak, or Dunn multiple comparison tests. A P value of <0.05 was considered to be significant. Data are presented as means and standard errors of the means, except data in Figures 3 and 4, which are presented as box plots. Individual measurements are depicted by symbols. Statistical analyses were performed with the Sigma Plot 13 software (Systat Software Inc., San Jose, CA). Image processing and analyses were done with ImageJ software (NIH).
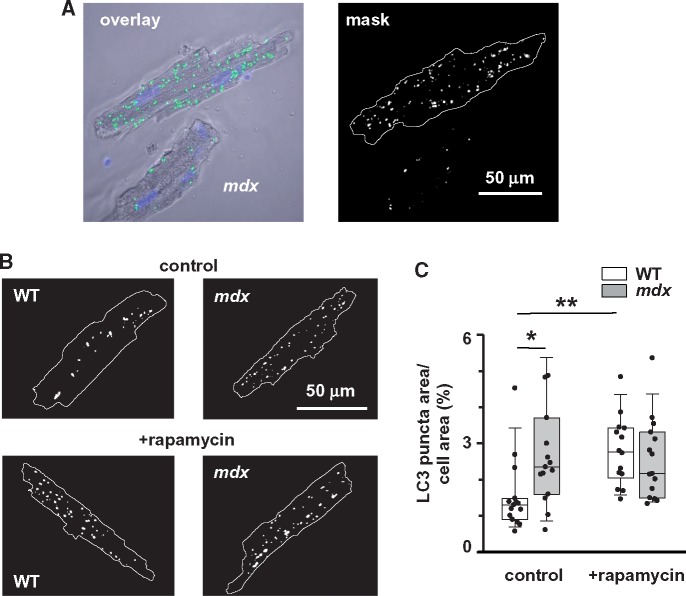
Figure 3.
LC3 puncta are more prominent in resting mdx cardiomyocytes. (A) Left panel shows typical overlay image of cardiomyocyte stained with anti-LC3 antibodies and then with Alexa Fluor 488-congugated secondary antibodies (green puncta on the image) and with nuclear fluorescent label DAPI (blue staining). Right panel shows binary image of the cardiomyocyte, where pixels with the numerical values above the preset threshold are marked in white. (B) Representative binary images of myocytes isolated from mdx and WT animals injected with either vehicle or rapamycin. (C) Pooled data of LC3 puncta area relative to area occupied by a cell in mdx and WT cardiomyocytes isolated from hearts of 12+ months old WT (N = 3, n = 14 and N = 3, n = 19) and mdx (N = 3, n = 15 and N = 3, n = 17) animals with and without rapamycin treatment, respectively. Data obtained from mdx tissue are shown as gray bars whereas white bars illustrate data acquired from WT tissue. (*P < 0.05, **P < 0.01, ANOVA test).
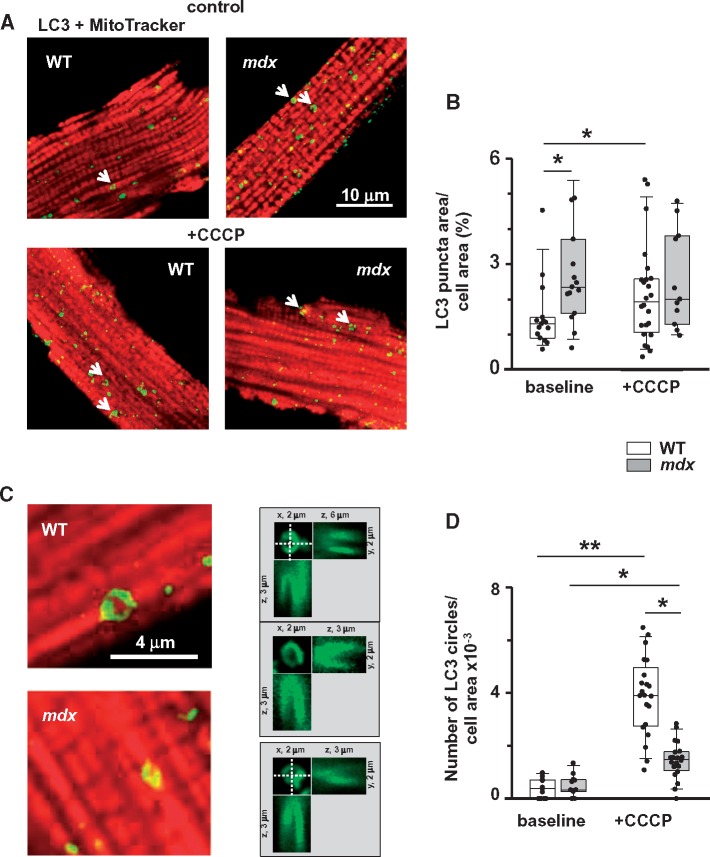
Figure 4.
Mitophagy is compromised in dystrophic cardiomyocytes isolated from hearts of 12+ months old mice (evidence from immunocytochemistry). (A) Representative overlay images of WT and mdx cardiomyocytes incubated with anti-LC3 antibodies and then with Alexa Fluor 488-conjugated secondary antibodies (green puncta on the image) and MitoTracker® Orange (red staining) without (top panels) and with (bottom panels) CCCP treatment. (B) Graph compares area occupied by LC3 puncta per cell area in myocytes with and without treatment with CCCP. Number of cells analysed was for WT (N = 3, n = 15 and N = 3, n = 24) and mdx hearts (N = 3, n = 15 and N = 3, n = 11) without and with incubation with CCCP, respectively. Data indicate enhanced autophagic activity in WT. Arrows on the images point to the ‘circle-like’ LC3 structures, which are likely to be associated with mitochondria. (C) Magnified images of cardiomyocytes containing the mitochondria associated LC3 circles. (D) Graph compares numbers of LC3 circles per cell area in WT (N = 3, n = 11 and N = 3, n = 21) and mdx (N = 3, n = 11 and N = 3, n = 20) myocytes with and without treatment with CCCP, respectively. Only LC3 puncta/circles associated with mitochondrial structures were considered for the analysis. This was ensured by taking additional images within ∼2 μm above and below the initial focal plane. Right panels in (C) show images of three representative mitochondria surrounded by LC3 puncta in three different projections (xy, xz, and yz). Data indicate that mitochondrial uncoupling induced substantially more mitochondria-associated LC3 circles in WT cardiomyocytes. Data obtained from mdx tissue are shown as gray bars whereas white bars illustrate data acquired from WT tissue. (*P < 0.05, **P < 0.01, ANOVA test).
3. Results
To establish the causal link between deterioration of mitochondrial structure/function and compromised autophagy/mitophagy, we examined mitochondrial structure with transmission electron microscopy, determined cellular ATP content with a luciferase assay and established the levels of several major autophagic markers and mitophagic substrates under resting (control) conditions and after forced induction of autophagy or mitophagy. Studies were carried out on cardiomyocytes isolated from 3 to 4 months and 12 months and older WT and mdx mice.
3.1. Mitochondrial structure/function is compromised in dystrophic cardiac myocytes
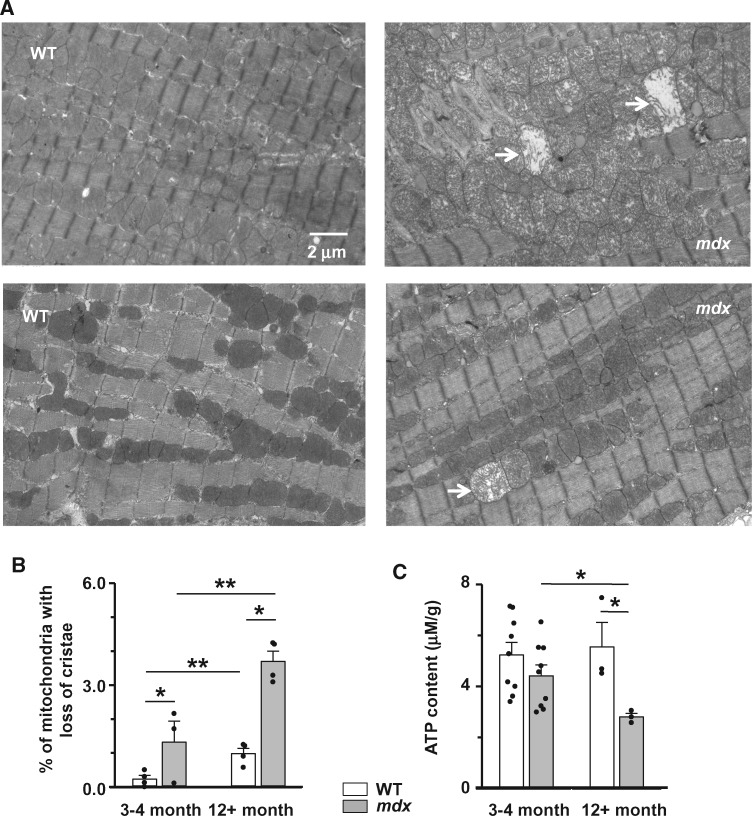
Sections of WT and mdx cardiac muscle were imaged with transmission electron microscopy (Figure 1A). Electron micrographs revealed a significantly larger number of mitochondria with a loss of normal cristae structure in mdx cardiac muscle compared to WT tissue (Figure 1B). Moreover, the number of structurally abnormal mitochondria significantly increased in cardiac tissue of 12 months and older dystrophic animals, which have developed dilated cardiomyopathy.
Figure 1.
Abnormal mitochondrial morphology and impaired ATP synthesis in dystrophic cardiac myocytes. (A) Typical transmission electron micrographs of heart from 12 months and older WT (left panels) and mdx (right panels) mice. Arrows indicate mitochondria with a loss of cristae. (B) Percent of defective mitochondria in cardiac tissue isolated from 3 to 4 months and 12+ months old mdx (gray bars) and WT (white bars) mice. Samples from N = 4 (WT) or N = 3 (mdx) hearts were probed in each experimental cluster. About 10–23 random fields with 50–100 mitochondria were analysed in each experimental group. (C) Total ATP content in hearts from WT (white bars) and mdx (gray bars) mice. Heart samples from 3 to 4 months old (N = 9, mdx and WT) and 12+ (N = 3, both mdx and WT) months old mice were probed. (*P < 0.05, **P < 0.01, ANOVA test).
Mitochondria are the major source of energy in cardiac muscle. Under normal physiological conditions, mitochondria use electron transport through the respiratory chain to generate an electrochemical gradient across the inner mitochondrial membrane. This gradient is used by the ATP-synthase to phosphorylate ADP to ATP. Under pathological conditions (uncoupling and dissipation of mitochondrial membrane potential), ATP-synthase can function in reverse mode, so mitochondria hydrolyse (consume) ATP. Therefore, even a relatively small number of damaged organelles can have a significant impact on mitochondrial ATP production. In agreement with this scenario, the total ATP content is substantially decreased in mdx hearts, especially after clinical onset of the cardiomyopathy (hearts of 12 months and older mice, Figure 1C).
Overall, our data support and extend our previous finding that mitochondrial structure and function significantly worsens during the development of dystrophic cardiomyopathy. In the next several sets of experiments, we will examine whether defective autophagy/mitophagy pathways contribute to the accumulation of damaged mitochondria in dystrophic hearts.
3.2. Specific markers indicate enhanced autophagy in senescent dystrophic hearts
Each step in autophagy requires a concerted effort of numerous proteins and enzymes. Several proteins involved in these steps are widely used as markers of autophagy. LC3 and SQSTM1 (a.k.a. p62) proteins are among them. In mammalian cells LC3 is modified post-translationally to LC3-I, a soluble cytosolic protein. After autophagy is induced, LC3-I is lipidated to form LC3-II that associates with the phagophore membrane. An increase in LC3-II level indicates the increase in number of cellular autophagasomes, which in general but not always is considered to be an indication of enhanced autophagy. SQSTM1 is an ubiquitin and LC3-binding scaffold protein that links ubiquitin-containing protein aggregates to the autophagic structures. It accumulates in the cytoplasm when autophagy is inhibited and decreased levels of SQSTM1 are observed when autophagy is induced.31
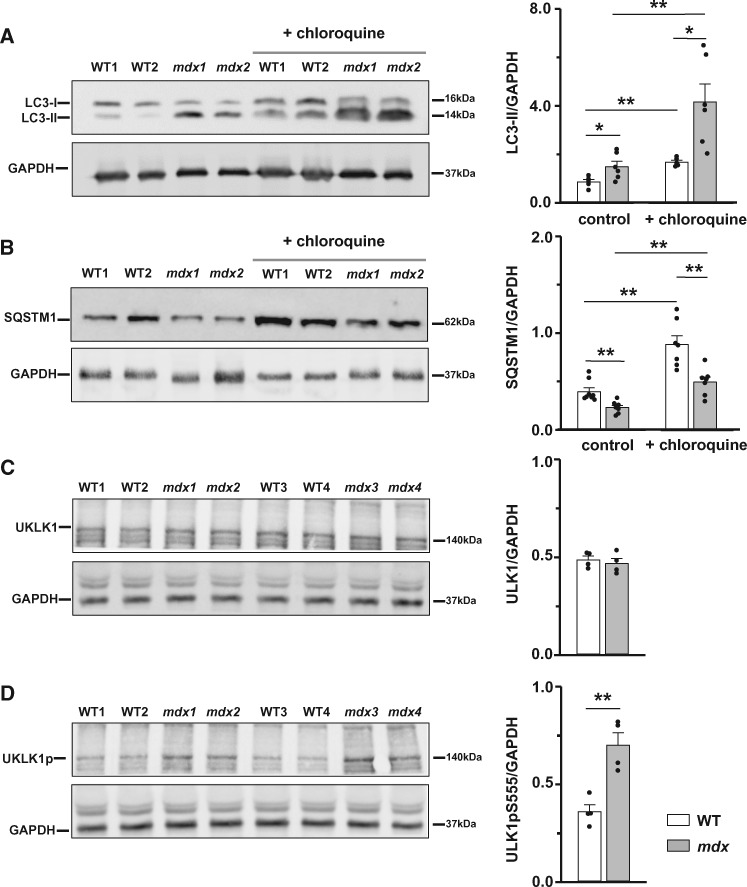
We performed western blots on whole heart homogenates of 12 months and older WT and mdx mice. We detected a significant increase in LC3-II level as well as a decrease in SQSTM1 level in dystrophic cardiac tissue (Figure 2A and B, left panels on the graphs). These results indicate an enhanced basal autophagy in mdx hearts compared to WT cardiac tissue. However, an increase in LC3-II (or increased generation of autophagasoms) may also be due to inhibition or block of downstreams steps of autophagy. To distinguish between these possibilities, we measured LC3-II and SQSTM1 levels in cardiac tissue obtained from animals treated in vivo with the lysosomal inhibitor chloroquine (10 mg/kg) and compared these values with those obtained in control samples obtained from non-treated animals. Block of LC3-II degradation with chloroquine resulted in further accumulation of LC3-II and SQSTM1 levels in both WT and dystrophic tissue (Figure 2A and B, right panels on the graphs). Supplementary material online, Figure S1 shows pairs of data points obtained from the control and treated samples originating from the same gels. The difference in the amount of LC3-II between samples in the presence and absence of chloroquine represents the amount of LC3-II that is delivered to lysosomes for degradation. The graph on Supplementary material online, Figure S1A demonstrates that this difference is significantly larger in mdx hearts, thus indicating an augmented autophagic activity. The difference in SQSTM1 levels was also somewhat larger in dystrophic samples Supplementary material online, Figure S1B.
Figure 2.
Autophagy markers indicate augmented autophagy in mdx hearts. Cardiac tissue was collected from 12 months and older mdx and WT mice under control conditions or after their treatment with chloroquine. Samples were immunoblotted with anti-LC3 (A), anti-SQSTM1 (B), anti-ULK1 (C), anti-phospho-ULK1(Ser555) (D), and anti-GAPDH (all panels) antibodies to determine protein levels of LC3-I, LC3-II, SQSTM1, ULK1, and phosphorylated ULK1. Samples from N = 4–8 senescent hearts were tested in each experimental group. Data obtained from mdx tissue are shown as gray bars whereas white bars illustrate data acquired from WT tissue. (*P < 0.05, **P < 0.01, ANOVA test).
Autophagy can be inducted/enhanced in response to a multiple stress factors, including depletion of ATP. AMP-activated kinase (AMPK) is one of the cellular sensors of ATP. It is believed that AMPK stimulates autophagy in response to energy depletion through phosphorylation and activation of ULK1 protein kinase.32–34 There are multiple phosphorylation sites for AMPK on ULK1. Phosphorylation of S555 of ULK1 is known to contribute to the activation of this kinase and induction of autophagy. Therefore, we compared levels of expression of total ULK1 as well as ULK1 phosphorylated at S555 in mature WT and dystrophic hearts (Figure 2C and D). Increased levels of ULK1-pS555 in dystrophic cardiac tissue strongly suggest enhanced autophagy through AMPK-ULK1 pathway, probably in response to the reduced ATP levels in mature dystrophic hearts (Figure 1C).
3.3. Confocal imaging reveals more LC3 puncta in resting mdx cardiomyocytes
The measurement of LC3 turnover is a reliable indicator of autophagy, however the autophagy can and should also be measured by a combination of other techniques. LC3 protein can be visualized with fluorescence-conjugated antibodies or with fluorescent protein-tagged LC3 probes in live or fixed tissue. In this group of experiments, we visualized LC3 puncta in cardiomyocytes isolated from mdx and WT mice (12 months and older) injected with either vehicle (control conditions) or 10 mg/kg rapamycin (established autophagy inducer). Myocytes were fixed and labeled with primary LC3 antibody followed by Alexa Fluor 488 conjugated secondary antibody. Cells were imaged with confocal microscopy and analysed with an ImageJ software plugin to detect pixels with fluorescent values above the preset threshold. Figure 3A illustrates the original image of a cardiomyocyte (left panel) as well as the binary image (mask, right panel) of the cell where detected LC3 puncta are shown in white. Figure 3B shows representative examples of binary masks obtained from WT and mdx myocytes isolated from hearts of animals treated with the vehicle or rapamycin, respectively. Figure 3C compares the area occupied by LC3 puncta pixels relative to the total cell area under each experimental condition. It should be noted that cardiomyocytes undergo some stress during the cell isolation procedure which, in turn, can influence the autophagy. Nevertheless, the appearance of LC3 puncta was significantly more pronounced in mdx cells under control (no treatment with rapamycin) conditions, confirming the accumulation of autophagic vacuoles in dystrophic hearts initially suggested by experiments with western blotting (Figure 2). Treatment with rapamycin significantly increased LC3 puncta in WT but not in mdx cells. Moreover, the levels of puncta were similar in both types of cells after rapamycin treatment and in mdx cells under control condition. The latter suggest an already maximal activation of autophagy in dystrophic hearts under resting conditions, probably in an attempt to remove cellular components damaged by the disease. The question that arises is: why do so many damaged mitochondria accumulate in the diseased state? In subsequent experiments, we attempted to answer this question by directly looking at mitochondrial autophagy to investigate whether this mechanism is well tuned to remove damaged organelles.
3.4. Mitophagy is compromised in dystrophic cardiomyocytes (immunocyto-chemical studies)
We visualized LC3 puncta in isolated cardiomyocytes under control conditions and after treatment of cardiomyocytes with 5 μM of the mitochondrial uncoupler CCCP (established mitophagy inducer) using high magnification confocal imaging. Before or after incubation with the uncoupler, cells were fixed and stained with LC3 antibodies and a mitochondrial marker. Panels on Figure 4A illustrate representative overlay images of WT and mdx cardiomyocytes before and after CCCP treatment. Binary masks were created using images of LC3 fluorescence and analysed, as described earlier. As in experiments shown in Figure 3, the area occupied by LC3 puncta relative to the total cell area was significantly larger in mdx cardiomyocytes under control (no treatment) conditions (Figure 4B). Treatment of cardiomyocytes with CCCP substantially promoted appearance of LC3 puncta in WT but not in mdx cells, suggesting a defective mitochondrial mitophagy in dystrophic cells.
Arrows on the representative images in Figure 4A point toward specific ‘doughnut’ or circle-like LC3 structures that seem to be associated with mitochondria. The size of these circles roughly corresponded to the size of a mitochondrion in cardiomyocytes. Figure 4C shows enlarged overlay images and 2D reconstructions of several circle-like structures. In the next analysis only these structures were taken into account. This was assured by taking additional images within ∼2 μm above and below the initial focal plane. Panels in the middle of Figure 4C show images of three representative mitochondria surrounded by LC3 puncta in three different projections (xy, xz, and yz). The relative number of LC3 circles was similar and relatively low in mdx and WT cells under control conditions without CCCP treatment (Figure 4D), indicating rather low baseline level of mitochondrial mitophagy in both groups of cells. The number of LC3 circles significantly increased after mitochondrial uncoupling. The increase was significantly larger in WT cells also indicating impaired mitophagic mechanisms in dystrophic hearts. Please note that analysis of circle-like LC3 structures is somewhat qualitative as some damaged mitochondria may be targeted with LC3 puncta with shapes other than circle-like. Therefore, experiments utilizing high resolution EM imaging were performed next.
3.5. Mitophagy is compromised in dystrophic cardiomyocytes (EM studies)
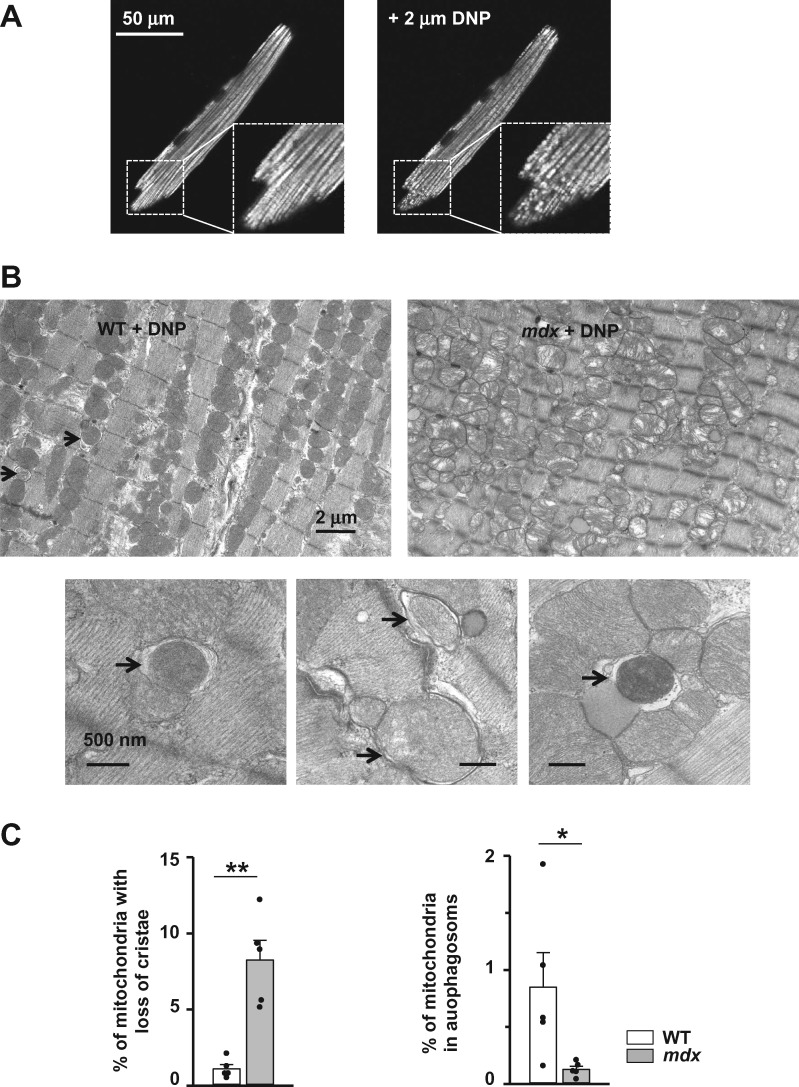
Under resting conditions, autophagic/mitophagic structures are only rarely observed on EM images, both in WT and mdx hearts, rendering their quantification challenging. In an attempt to quantify the mitophagy, we induced this process by injecting animals with 15 mg/kg DNP (2,4-dinitrophenol), a less aggressive mitochondrial uncoupler compared to CCCP. As seen in Figure 5A, incubation of cardiomyocytes isolated from WT mice with 2 μM of DNP resulted in partial mitochondrial depolarization.
Figure 5.
Mitophagy is compromised in dystrophic cardiomyocytes isolated from hearts of 12+ months old mice (evidence from EM studies). (A) Images of TMRE loaded WT cardiomyocyte before (left panel) and after (right panel) DNP treatment. Application of 2 μM DNP resulted in depolarization of some mitochondria (seen as black spots). (B) Typical transmission electron micrographs of heart slices from 12+ months old WT (left panel) and mdx (right panel) mice in vivo treated with DNP. Enlarged images below are taken from WT tissue and show mitochondria included in autophagosomes (indicated by arrows). (C) Left panel shows percent of defective mitochondria in cardiac tissue isolated from mdx and WT mice treated with DNP. Right panel represents percent of mitochondria sequestered in autophagosomes. Samples from N = 5 hearts were analysed in each experimental cluster. About 26–32 random fields with 50–100 mitochondria were analysed in each experimental group. Data obtained from mdx tissue are shown as gray bars whereas white bars illustrate data acquired from WT tissue. (*P < 0.05, **P < 0.01, ANOVA test).
EM images of mdx hearts revealed massive mitochondrial damage after in vivo treatment with DNP (Figure 5B, top right panel and Figure 5C, left panel). The damage of mitochondria in WT cardiac tissue was much less pronounced (Figure 5B, top left panel and Figure 5C, left panel). In contrast to mdx, multiple autophagosomal structures were detected in WT tissue. Moreover, many autophagasomes contained mitochondria (examples are in Figure 5B, bottom). In comparison, the relative number of autophagasomes containing mitochondria, was substantially smaller in dystrophic cardiac tissue (Figure 5C, right panel). Taken together with immunocytochemical studies, these experiments provide additional indications for impaired mitophagy in hearts of dystrophic mice exhibiting cardiomyopathy.
3.6. Mitophagy is compromised in dystrophic cardiomyocytes (biochemical studies)
Results summarized in Figures 4 and 5 strongly suggest defective mitophagy in dystrophic cardiac tissue. In the following experiments, we attempted to gain additional evidence that this is indeed the case.
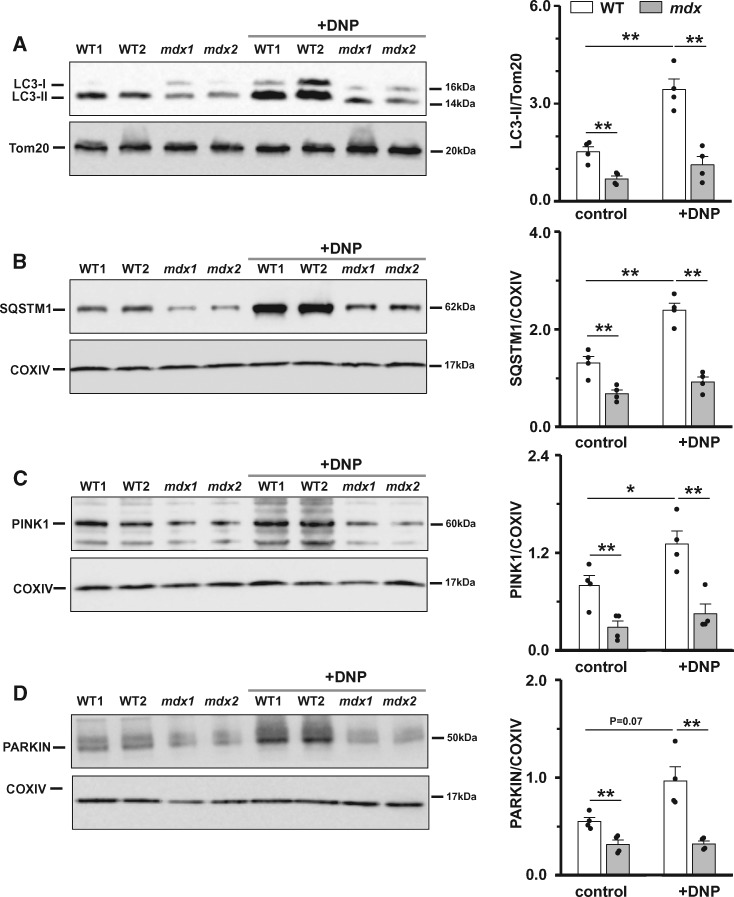
For this, firstly, we checked for the levels of autophagy marker proteins directly associated with mitochondria. Mitochondrial fractionation was performed on heart homogenates. This is a complicated multistep procedure, which in general can influence mitochondrial structure/function. However, currently, it is one of the most useful approaches to assess expression of mitochondria associated proteins.35 Western blots were executed on samples prepared from hearts of control WT and mdx animals, as well as from hearts of animals treated in vivo with the mitochondrial uncoupler DNP. DNP increased the numbers of damaged mitochondria and presumably favoured their removal by autophagy (seen in Figure 5). Figure 6A and B show that level of both lipidated LC3 and SQSTM1 are lower in mitochondrial fractions of mdx mouse heart homogenates under control (non-treated) conditions. Supplementary material online, Figure S2A and B show pairs of data points obtained from control and DNP treated samples obtained from the same gels. The difference in the amount of LC3-II and SQSTM1 between the mitochondrial preparation in the presence and absence of DNP is substantially greater in WT samples. Levels of both LC3-II and SQSTM1 proteins in samples from mdx hearts changed very little after DNP treatment. Overall, the results suggest reduced mitochondrial autophagy in mdx compared to WT mice hearts.
Figure 6.
Mitophagy is compromised in dystrophic cardiomyocytes (evidence from biochemical studies). Mitochondrial fractions were isolated from hearts of 12+ months old mice under control conditions or after their treatment with DNP. Samples were immunoblotted with anti-LC3, anti-SQSTM1, anti-PINK1, anti-PARKIN, anti-Tom20, and anti-COXIV antibodies to determine mitochondria-associated levels of LC3-II, SQSTM1, PINK1, and PARKIN proteins. Samples from N = 4 hearts were tested in each experiments group. Data obtained from mdx tissue are shown as gray bars whereas white bars illustrate data acquired from WT tissue. (*P < 0.05, **P < 0.01, ANOVA test).
3.7. Defects in mitophagy pathways in dystrophic hearts
The PINK1/PARKIN-mediated mechanism is one of the most established mitophagy pathways in mammals. We therefore first checked for the levels of PINK1 and PARKIN, the two major proteins in this pathway, in mitochondrial fractions prepared from hearts of animals under control conditions and after mitochondrial damage has been induced by in vivo treatment of animals with DNP.
Levels of both PINK1 and PARKIN mitochondria-associated proteins were significantly smaller in hearts from mdx mice compared to WT animals (Figure 6C and D), despite the fact that mitochondria are much more depolarized in mature dystrophic cardiac tissue.14 Moreover, neither level of PINK1 nor PARKIN changed significantly in mdx animals treated with DNP, in contrast to a substantial increase observed in WT mice (see Supplementary material online, Figure S2C and D). These data indicate defective targeting of PARKIN to damaged organelles in dystrophic hearts.
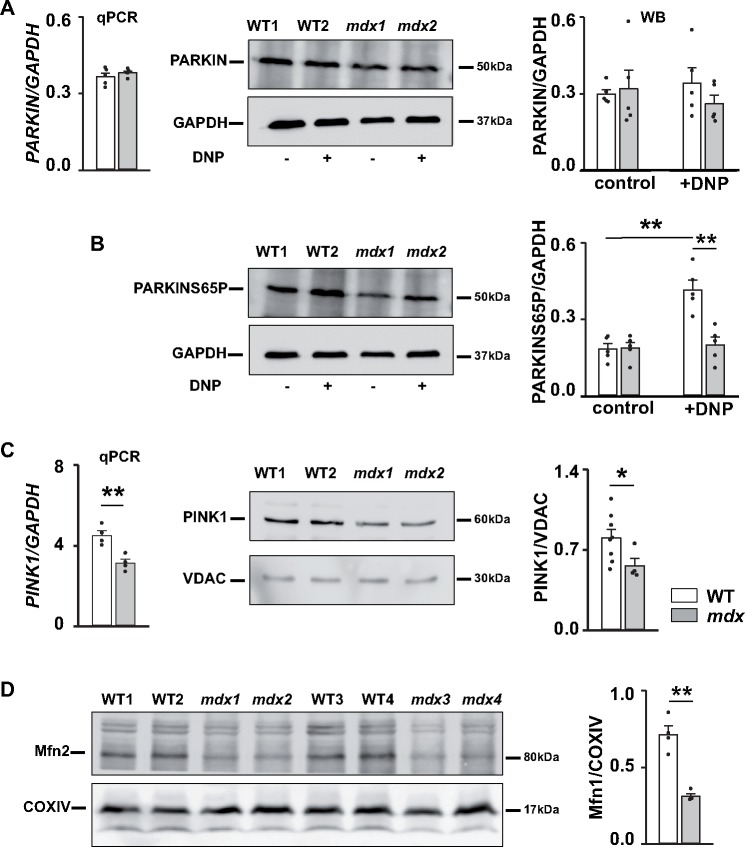
Recently, it has been shown that recruitment of PARKIN to the outer mitochondrial membrane is associated with its phosphorylation at the conserved serine 65 (S65) residue within its ubiquitine-like domain.36–39 Levels of PARKIN and PARKIN phosphorylated at S65 do not seem to be different in whole tissue homogenates from WT and mdx hearts isolated from control (untreated) group of animals (Figure 7A and B and Supplementary material online, Figure S3A and B). However, in vivo treatment of WT mice with DNP drastically increased levels of PARKIN-pS65, whereas treatment of mdx animals did not result in a significant change in PARKIN-S65P (Figure 7B and Supplementary material online, Figure S3B). Similar results were obtained in heart tissue isolated from two animals in vivo treated (injected) with 5 mg/kg CCCP (data not shown). Importantly, the level of PINK1 in whole tissue homogenates was also significantly smaller (Figure 7C), similarly to that previously observed in mitochondrial fractions (Figure 6C). These results present additional evidence of insufficient mitophagy in dystrophic hearts probably due to defects in PINK1/PARKIN-mediated pathway, specifically due to a deficit of PINK1 protein.
Figure 7.
Defects in PINK1/PARKIN-mediated mitophagy in dystrophic hearts. (A, B) Expression of PARKIN. Cardiac tissue was collected from mice treated with either vehicle (control) or DNP. qPCR was used to determine level of gene encoding PARKIN (left panel on A). Samples were also immunoblotted with anti-PARKIN (A), anti-PARKIN-S65P (B), and anti-GAPDH antibodies to determine levels of PARKIN protein and PARKIN protein phosphorylated at S65 residue (summarized at the graphs on the right panels). (C) Expression of PINK1. Cardiac tissue was collected from mice under control conditions. Graph on the left represents levels of expression of gene encoding PINK1. Samples were also immunoblotted with anti-PINK1 and anti-VDAC antibodies to determine protein levels of PINK1 (middle and right panels). (D) Expression of Mfn2 protein, which is involved in mitochondrial mitophagy and mitochondrial dynamics. Overall, samples from N = 3–8 12+ months old mdx and WT hearts were tested. Experiments were carried out on whole tissue homogenates. There were three replicates for each sample during qPCR measurements. Data obtained from mdx tissue are shown as gray bars whereas white bars illustrate data acquired from WT tissue. (*P < 0.05, **P < 0.01, ANOVA test).
Another evidence came from the analysis of the expression of some other proteins, involved in the PINK1/PARKIN pathway. There is a substantial cross talk between mitochondrial dynamics and mitophagy.24,40 In particular, increasing evidence suggest that mitofusin 2 (Mfn2), a protein crucial for mitochondrial fusion, participates in recruiting PARKIN to the outer mitochondrial membrane (OMM) after being phosphorylated by PINK1. As seen on Figure 7D, the level of Mfn2 is significantly reduced in dystrophic hearts. In addition, some other mitochondrial fusion related proteins, such as Mfn1 (mitofusin 1), dynamin-like 120 kDa protein (encoded by the OPA1 gene), and fission related proteins such as Fis1 (mitochondrial fission1 protein) a protein in OMM with which Drp1 (dynamin-related protein 1) protein associates, are also important in maintaining a healthy mitochondrial population. As seen from Supplementary material online, Figure S4 expression of Fis1 and Mfn1 are also significantly reduced in dystrophic cardiac tissue. The deficit of these proteins may also contribute to the lack of proper mitochondrial recycling.
4. Discussion
Most cardiac diseases are associated with mitochondrial dysfunction, at least to some extent. Dystrophic cardiomyopathy is not an exception. We and others reported multiple metabolic and mitochondrial abnormalities in hearts of mdx mice downstream of the primary genetic defect in this disease—lack of dystrophin8,12–14. As dystrophic cardiomyopathy develops (i) the mitochondrial matrix becomes gradually oxidized; (ii) the ability of the organelles to handle increased workload is reduced; (iii) substrate consumption shifts from fatty acids to carbohydrates; (iv) mitochondria become gradually overloaded with Ca2+ and (v) depolarized; and finally (vi) the propensity of irreversible opening of mitochondrial permeability transition pore is increased and the organelles become uncoupled and capable of hydrolysing rather than synthesizing ATP. These functional pathologies are accompanied by a gradual structural damage of the mitochondrial matrix and by the accumulation of impaired organelles in cardiac tissue. A goal of this study was to determine whether defects in housekeeping processes, such as autophagy and more specifically mitophagy, contribute to the progressive accumulation of damaged mitochondria in dystrophic hearts and deterioration of cardiac functions.
Intracellular Ca2+ overload and excessive generation of reactive oxygen species (ROS) by NOX2 (NADPH oxidase type 2) are among very early pathophysiological features of dystrophic cardiomyopathy.11 It has been shown that in the heart these conditions not only stimulate cellular pathophysiological pathways, such as apoptosis and necrosis, but also initiate autophagy as a cardioprotective response mechanism under increasing stress.41,42 In addition, progressive mitochondrial damage and mitochondrial uncoupling leads to a severe decrease in ATP production by these organelles, a metabolic signal that further promotes autophagy.43 In agreement with these expectations our experiments revealed enhanced autophagy in dystrophic hearts of 12 months and older animals compared to WT hearts. This conclusion is based both on changes in major autophagic markers (LC3-II and SQSTM1) as well as on the increased appearance of LC3 puncta under control conditions (Figures 2 and 3, respectively). To ensure that changes in autophagic markers were due to alterations in the induction of autophagy but not due to changes in marker synthesis/degradation, experiments were repeated in the presence of the lysosomal inhibitor chloroquine.
Yet another indication of enhanced autophagy in dystrophic hearts come from experiments with ULK-1. ULK-1 is a conserved substrate of AMP-activated protein kinase, which is considered to be one of the cellular energy sensors. Upon activation, AMPK phosphorylates ULK-1 at several residues thus initiating/enhancing autophagy. Our studies revealed an increased level of ULK-1 protein, phosphorylated at S555 residue, in dystrophic tissue. This likely happened in response to activation of AMPK by ATP depletion and enhanced autophagy.
Previous findings in skeletal muscle from young mdx mice show a substantial inhibition of autophagic flux.44–46 Stimulation of autophagy with non-specific autophagy-reactivating treatment ameliorated skeletal muscle pathology, at least to some extent.44 Treatment of mice with agonists of AMPK 46 or simvastatin (a cholesterol lowering drug47) and inhibition of NOX2-Src kinase pathways45 has also been shown to enhance autophagy and improve skeletal muscle function in dystrophy. However, the suppression of autophagy appears to be muscle type dependent.48 This may provide a plausible explanation for the difference in initiation of autophagy in dystrophic skeletal muscle and cardiac tissues.
It also should be noted that in our experiments we mostly assessed basal (not induced) autophagy by following the formation of autophagasomes. It is possible, however, that when autophagy is induced by starvation or mTOR inhibition by rapamycin, the induced autophagic flux can be reduced despite the enhanced basal autophagy, as it has been reported for skeletal muscle of a myotonic dystrophy type I and an amyotropic lateral sclerosis mouse model.49,50 We may not rule out this possibility for dystrophic hearts, as in our experiments in vivo administration of rapamycin to mdx mice did not promote further autophagosome formation. However, as we stated earlier, it can also be because autophagy is already maximally activated in mdx hearts. Interestingly, a recent report by Bibee et al.51 showed some increase in autophagy in mdx hearts after treatment with rapamycin. Possible reasons for the difference can include (but not limited to) the age of animals studied (17 months old in ref. 51 vs. 12 months old in our study) as well as the method by which rapamycin has been delivered to the animals in addition to length of treatment (injection of rapamycin nanoparticles for 4 weeks in ref. 51 vs. single injection of the drug in solvent following 12 hours of waiting time in our experiments).
If autophagy is up-regulated in dystrophic hearts then one would expect that mitochondrial autophagy is also enhanced. However, experiments in Figure 4 indicated otherwise. Treatment of cardiomyocytes with mitochondrial uncoupler CCCP substantially promoted appearance of LC3 puncta in WT but not in mdx cells, indicating insufficient mitochondrial autophagy mechanism supposed to remove depolarized organelles in dystrophic cells. Mitophagy deficiency in mdx hearts was also revealed by experiments with ‘in vivo’ uncoupling of the organelles with DNP (Figure 5). Electron micrographs of cardiac heart tissue obtained from animals treated with the mitochondrial uncoupler DNP revealed much more damaged mitochondria in dystrophic tissue compared with WT. However, the number of autophagosomes (including autophagosomes containing mitochondria) was much larger in WT tissue compared with mdx, where autophagosomes were extremely scarce. Altogether these data clearly indicate a severe deficit of mitophagy in dystrophic heart, despite the enhanced autophagy.
The PINK1/PARKIN-mediated pathway is currently one of the best-known mitophagy pathways. PINK1 was initially found as a highly conserved Parkinson’s disease susceptible protein. In neurons PINK1 has been shown to have a protective function against oxidative stress-induced cell death. In particular, PINK1 has been reported to protect neurons from various mitochondrial dysfunctions, reduce mitochondrial cytochrome C release and mitigate apoptotic cell death (reviewed in refs 21,52). PINK1 detects mitochondrial damage and recruits PARKIN to initiate mitophagy and remove compromised organelles. The role of PINK1 protein in cardiac pathology is much less defined. A severe cardiac hypertrophy was reported in PINK1 knockout mice.53 A substantial decrease in PINK1 protein levels has been found in heart samples from patients with end-stage HF.53 Loss of PINK1 increases the hearts vulnerability to ischemia-reperfusion.54 Overall, it seems that PINK1 exerts some cardioprotective effect.
It was suggested that PINK1/PARKIN-mediated mitophagy plays significant role in cardiac pathology, as a potent inducible cardiac stress-induced response. In damaged mitochondria PINK1 accumulated in OMM, phosphorylates Mfn2, which in turn recruits cytosolic protein PARKIN. PARKIN ubiquitinates several mitochondrial proteins thus labeling mitochondria for recognition by autophagosomes.25,28 With western blotting, we determined that levels of PINK1 and PARKIN proteins were significantly decreased in mitochondrial fractions prepared from hearts of 12 months and older dystrophic mice compared to WT animals (Figure 6). Moreover, levels of PINK1 and Mfn2 were also greatly reduced in tissue homogenates (Figure 7). It is possible that a deficit of PINK1 and Mfn2 limit the targeting of PARKIN to depolarized organelles and restricts PINK1/PARKIN driven mitophagy.
New emerging evidence strongly suggests that post-translational modifications of PINK1, Mfn2, and PARKIN are critical regulators of their functions at different stages of mitophagy.38,39 In particular, the recruitment of PARKIN to the OMM of depolarized mitochondria is associated with its phosphorylation at the S65 residue in the ubiquitin-like domain via PINK1/Mfn2-mediated phosphorylation. Recently developed antibodies against PARKIN-S65P55 provided an opportunity to estimate levels of PARKIN translocation to OMM. Results in Figure 7 illustrate that the level of phosphorylation of PARKIN at S65 significantly increased in WT cardiac tissue after induction of mitophagy by in vivo treatment with the mitochondrial uncoupler DNP, while changes in levels of PARKIN-S65P were negligible in dystrophic tissue.
Overall, findings by several groups, including our laboratory, revealed multiple mitochondrial dysfunctions in dystrophic cardiomyopathy. Severe oxidative stress and cellular Ca2+ overload seem to precede this devastating pathology. Our present studies demonstrate a deficiency of mitophagy in DMD. Moreover, it is likely to be associated with defects in the PINK1/PARKIN mitophagic pathway. Whether it is only due to insufficient expression of key proteins of the pathway or also due to modification of mitophagy regulatory pathways has yet to be determined by future work. However, the results presented here suggest that impaired mitophagy adds to inability to degrade enough impaired organelles and contributes to the progressive accumulation of damaged mitochondria and deterioration of cardiac functions in dystrophic hearts. Our results also suggest that specific interference with mitophagic pathways can be potentially beneficial for treatment of DMD.
Supplementary Material
Acknowledgements
This work was supported by NIH (HL093342 to N.S.). We are grateful to Drs. D. Fraidenraich, L. Gasper, C. Suzuki, A.Thomas, L.H. Xie, and J. Zhou for the insightful discussion, Dr. R. Janicek for help with data analysis and representation, and Ms. L. Liu for technical assistance.
Conflict of interest: none declared.
Time for primary review: 46 days
References
- 1. Ervasti JM, Campbell KP.. A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J Cell Biol 1993;122:809–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Finsterer J, Stöllberger C.. The heart in human dystrophinopathies. Cardiology 2003;99:1–19. [DOI] [PubMed] [Google Scholar]
- 3. Kieny P, Chollet S, Delalande P, Le Fort M, Magot A, Pereon Y, Perrouin Verbe B.. Evolution of life expectancy of patients with Duchenne muscular dystrophy at AFM Yolaine de Kepper centre between 1981 and 2011. Ann Phys Rehab Med 2013;56:443–454. [DOI] [PubMed] [Google Scholar]
- 4. Yasuda S, Townsend D, Michele DE, Favre EG, Day SM, Metzger JM.. Dystrophic heart failure blocked by membrane sealant poloxamer. Nat Cell Biol 2005;436:1025–1029. [DOI] [PubMed] [Google Scholar]
- 5. Williams IA, Allen DG.. Intracellular calcium handling in ventricular myocytes from mdx mice. Am J Physiol Heart Circ Physiol 2006;292:H846–H855. [DOI] [PubMed] [Google Scholar]
- 6. Jung DW, Baysal K, Brierley GP.. The sodium-calcium antiport of heart mitochondria is not electroneutral. J Biol Chem 1995;270:672–678. [DOI] [PubMed] [Google Scholar]
- 7. Kyrychenko S, Polakova E, Kang C, Pocsai K, Ullrich ND, Niggli E, Shirokova N.. Hierarchical accumulation of RyR post-translational modifications drives disease progression in dystrophic cardiomyopathy. Cardiovasc Res 2013;97:666–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jung C, Martins AS, Niggli E, Shirokova N.. Dystrophic cardiomyopathy: amplification of cellular damage by Ca2+ signalling and reactive oxygen species-generating pathways. Cardiovasc Res 2008;77:766–773. [DOI] [PubMed] [Google Scholar]
- 9. Fauconnier J, Thireau J, Reiken S, Cassan C, Richard S, Matecki S, Marks AR, Lacampagne A.. Leaky RyR2 trigger ventricular arrhythmias in Duchenne muscular dystrophy. Proc Natl Acad Sci U S A 2010;107:1559–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sarma S, Li N, van Oort RJ, Reynolds C, Skapura DG, Wehrens XHT.. Genetic inhibition of PKA phosphorylation of RyR2 prevents dystrophic cardiomyopathy. Proc Natl Acad Sci U S A 2010;107:13165–13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shirokova N, Niggli E.. Cardiac phenotype of Duchenne Muscular Dystrophy: insights from cellular studies. J Mol Cell Cardiol 2013;58:217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Khairallah M, Khairallah R, Young ME, Dyck JRB, Petrof BJ, Rosiers Des C.. Metabolic and signaling alterations in dystrophin-deficient hearts precede overt cardiomyopathy. J Mol Cell Cardiol 2007;43:119–129. [DOI] [PubMed] [Google Scholar]
- 13. Viola HM, Davies SMK, Filipovska A, Hool LC.. L-type Ca2+ channel contributes to alterations in mitochondrial calcium handling in the mdx ventricular myocyte. Am J Physiol Heart Circ Physiol 2013;304:H767–H775. [DOI] [PubMed] [Google Scholar]
- 14. Kyrychenko V, Poláková E, Janíček R, Shirokova N.. Mitochondrial dysfunctions during progression of dystrophic cardiomyopathy. Cell Calcium 2015;58:186–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nishida K, Otsu K.. Autophagy during cardiac remodeling. J Mol Cell Cardiol 2016;95:11–18. [DOI] [PubMed] [Google Scholar]
- 16. Tooze SA, Yoshimori T.. The origin of the autophagosomal membrane. Nat Cell Biol 2010;12:831–835. [DOI] [PubMed] [Google Scholar]
- 17. Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J.. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 2010;141:656–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xie Z, Klionsky DJ.. Autophagosome formation: core machinery and adaptations. Nat Cell Biol 2007;9:1102–1109. [DOI] [PubMed] [Google Scholar]
- 19. He C, Klionsky DJ.. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 2009;43:67–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Scarffe LA, Stevens DA, Dawson VL, Dawson TM.. Parkin and PINK1: much more than mitophagy. Trends Neurosci 2014;37:315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pickrell AM, Youle RJ.. The roles of PINK1, Parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron 2015;85:257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Narendra DP, Jin SM, Tanaka A, Suen D-F, Gautier CA, Shen J, Cookson MR, Youle RJ, Green DR.. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 2010;8:e1000298.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ.. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol 2010;191:933–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dorn GW II, Vega RB, Kelly DP.. Mitochondrial biogenesis and dynamics in the developing and diseased heart. Genes Dev 2015;29:1981–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dorn GWII. Parkin-dependent mitophagy in the heart. J Mol Cell Cardiol 2016;95:42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen Y, Dorn GW.. PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science 2013;340:471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Seirafi M, Kozlov G, Gehring K.. Parkin structure and function. FEBS J 2015;282:2076–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saito T, Sadoshima J.. Molecular mechanisms of mitochondrial autophagy/mitophagy in the heart. Circ Res 2015;116:1477–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chida J, Yamane K, Takei T, Kido H.. An efficient extraction method for quantitation of adenosine triphosphate in mammalian tissues and cells. Anal Chim Acta 2012;727:8–12. [DOI] [PubMed] [Google Scholar]
- 30. Huber LA, Pfaller K, Vietor I.. Organelle proteomics: implications for subcellular fractionation in proteomics. Circ Res 2003;92:962–968. [DOI] [PubMed] [Google Scholar]
- 31. Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, Ahn HJ, Ait-Mohamed O, Ait-Si-Ali S, Akematsu T, Akira S, Al-Younes HM, Al-Zeer MA, Albert ML, Albin RL, Alegre-Abarrategui J, Aleo MF, Alirezaei M, Almasan A, Almonte-Becerril M, Amano A, Amaravadi R, Amarnath S, Amer AO, Andrieu-Abadie N, Anantharam V, Ann DK, Anoopkumar-Dukie S, Aoki H, Apostolova N, Arancia G, Aris JP, Asanuma K, Asare NYO, Ashida H, Askanas V, Askew DS, Auberger P, Baba M, Backues SK, Baehrecke EH, Bahr BA, Bai X-Y, Bailly Y, Baiocchi R, Baldini G, Balduini W, Ballabio A, Bamber BA, Bampton ETW, Bánhegyi G, Bartholomew CR, Bassham DC, Bast RC, Batoko H, Bay B-H, Beau I, Béchet DM, Begley TJ, Behl C, Behrends C, Bekri S, Bellaire B, Bendall LJ, Benetti L, Berliocchi L, Bernardi H, Bernassola F, Besteiro S, Bhatia-Kissova I, Bi X, Biard-Piechaczyk M, Blum JS, Boise LH, Bonaldo P, Boone DL, Bornhauser BC, Bortoluci KR, Bossis I, Bost F, Bourquin J-P, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady NR, Brancolini C, Brech A, Brenman JE, Brennand A, Bresnick EH, Brest P, Bridges D, Bristol ML, Brookes PS, Brown EJ, Brumell JH, Brunetti-Pierri N, Brunk UT, Bulman DE, Bultman SJ, Bultynck G, Burbulla LF, Bursch W, Butchar JP, Buzgariu W, Bydlowski SP, Cadwell K, Cahová M, Cai D, Cai J, Cai Q, Calabretta B, Calvo-Garrido J, Camougrand N, Campanella M, Campos-Salinas J, Candi E, Cao L, Caplan AB, Carding SR, Cardoso SM, Carew JS, Carlin CR, Carmignac V, Carneiro LAM, Carra S, Caruso RA, Casari G, Casas C, Castino R, Cebollero E, Cecconi F, Celli J, Chaachouay H, Chae H-J, Chai C-Y, Chan DC, Chan EY, Chang RC-C, Che C-M, Chen C-C, Chen G-C, Chen G-Q, Chen M, Chen Q, Chen SS-L, Chen WLi, Chen X, Chen X, Chen X, Chen Y-G, Chen Y, Chen Y, Chen Y-J, Chen Z, Cheng A, Cheng CHK, Cheng Y, Cheong H, Cheong J-H, Cherry S, Chess-Williams R, Cheung ZH, Chevet E, Chiang H-L, Chiarelli R, Chiba T, Chin L-S, Chiou S-H, Chisari FV, Cho CH, Cho D-H, Choi AMK, Choi DSeok, Choi KS, Choi ME, Chouaib S, Choubey D, Choubey V, Chu CT, Chuang T-H, Chueh S-H, Chun T, Chwae Y-J, Chye M-L, Ciarcia R, Ciriolo MR, Clague MJ, Clark RSB, Clarke PGH, Clarke R, Codogno P, Coller HA, Colombo MI, Comincini S, Condello M, Condorelli F, Cookson MR, Coombs GH, Coppens I, Corbalan R, Cossart P, Costelli P, Costes S, Coto-Montes A, Couve E, Coxon FP, Cregg JM, Crespo JL, Cronjé MJ, Cuervo AM, Cullen JJ, Czaja MJ, D’Amelio M, Darfeuille-Michaud A, Davids LM, Davies FE, De Felici M, de Groot JF, de Haan CAM, De Martino L, De Milito A, De Tata V, Debnath J, Degterev A, Dehay B, Delbridge LMD, Demarchi F, Deng YZ, Dengjel J, Dent P, Denton D, Deretic V, Desai SD, Devenish RJ, Di Gioacchino M, Di Paolo G, Di Pietro C, Díaz-Araya G, Díaz-Laviada I, Diaz-Meco MT, Diaz-Nido J, Dikic I, Dinesh-Kumar SP, Ding W-X, Distelhorst CW, Diwan A, Djavaheri-Mergny M, Dokudovskaya S, Dong Z, Dorsey FC, Dosenko V, Dowling JJ, Doxsey S, Dreux M, Drew ME, Duan Q, Duchosal MA, Duff K, Dugail I, Durbeej M, Duszenko M, Edelstein CL, Edinger AL, Egea G, Eichinger L, Eissa NT, Ekmekcioglu S, El-Deiry WS, Elazar Z, Elgendy M, Ellerby LM, Eng KE, Engelbrecht A-M, Engelender S, Erenpreisa J, Escalante R, Esclatine A, Eskelinen E-L, Espert L, Espina V, Fan H, Fan J, Fan Q-W, Fan Z, Fang S, Fang Y, Fanto M, Fanzani A, Farkas T, Farré J-C, Faure M, Fechheimer M, Feng CG, Feng J, Feng Q, Feng Y, Fésüs L, Feuer R, Figueiredo-Pereira ME, Fimia GM, Fingar DC, Finkbeiner S, Finkel T, Finley KD, Fiorito F, Fisher EA, Fisher PB, Flajolet M, Florez-McClure ML, Florio S, Fon EA, Fornai F, Fortunato F, Fotedar R, Fowler DH, Fox HS, Franco R, Frankel LB, Fransen M, Fuentes JM, Fueyo J, Fujii J, Fujisaki K, Fujita E, Fukuda M, Furukawa RH, Gaestel M, Gailly P, Gajewska M, Galliot B, Galy V, Ganesh S, Ganetzky B, Ganley IG, Gao F-B, Gao GF, Gao J, Garcia L, Garcia-Manero G, Garcia-Marcos M, Garmyn M, Gartel AL, Gatti E, Gautel M, Gawriluk TR, Gegg ME, Geng J, Germain M, Gestwicki JE, Gewirtz DA, Ghavami S, Ghosh P, Giammarioli AM, Giatromanolaki AN, Gibson SB, Gilkerson RW, Ginger ML, Ginsberg HN, Golab J, Goligorsky MS, Golstein P, Gomez-Manzano C, Goncu E, Gongora C, Gonzalez CD, Gonzalez R, González-Estévez C, González-Polo RA, Gonzalez-Rey E, Gorbunov NV, Gorski S, Goruppi S, Gottlieb RA, Gozuacik D, Granato GE, Grant GD, Green KN, Gregorc A, Gros F, Grose C, Grunt TW, Gual P, Guan J-L, Guan K-L, Guichard SM, Gukovskaya AS, Gukovsky I, Gunst J, Gustafsson AB, Halayko AJ, Hale AN, Halonen SK, Hamasaki M, Han F, Han T, Hancock MK, Hansen M, Harada H, Harada M, Hardt SE, Harper JW, Harris AL, Harris J, Harris SD, Hashimoto M, Haspel JA, Hayashi S-i, Hazelhurst LA, He C, He Y-W, Hébert M-J, Heidenreich KA, Helfrich MH, Helgason GV, Henske EP, Herman B, Herman PK, Hetz C, Hilfiker S, Hill JA, Hocking LJ, Hofman P, Hofmann TG, Höhfeld J, Holyoake TL, Hong M-H, Hood DA, Hotamisligil GS, Houwerzijl EJ, Høyer-Hansen M, Hu B, Hu C-AA, Hu H-M, Hua Y, Huang C, Huang J, Huang S, Huang W-P, Huber TB, Huh W-K, Hung T-H, Hupp TR, Hur GM, Hurley JB, Hussain SNA, Hussey PJ, Hwang JJ, Hwang S, Ichihara A, Ilkhanizadeh S, Inoki K, Into T, Iovane V, Iovanna JL, Ip NY, Isaka Y, Ishida H, Isidoro C, Isobe K-i, Iwasaki A, Izquierdo M, Izumi Y, Jaakkola PM, Jäättelä M, Jackson GR, Jackson WT, Janji B, Jendrach M, Jeon J-H, Jeung E-B, Jiang H, Jiang H, Jiang JX, Jiang M, Jiang Q, Jiang X, Jiang X, Jiménez A, Jin M, Jin S, Joe CO, Johansen T, Johnson DE, Johnson GVW, Jones NL, Joseph B, Joseph SK, Joubert AM, Juhász G, Juillerat-Jeanneret L, Jung CH, Jung Y-K, Kaarniranta K, Kaasik A, Kabuta T, Kadowaki M, Kagedal K, Kamada Y, Kaminskyy VO, Kampinga HH, Kanamori H, Kang C, Kang KB, Kang KI, Kang R, Kang Y-A, Kanki T, Kanneganti T-D, Kanno H, Kanthasamy AG, Kanthasamy A, Karantza V, Kaushal GP, Kaushik S, Kawazoe Y, Ke P-Y, Kehrl JH, Kelekar A, Kerkhoff C, Kessel DH, Khalil H, Kiel JAKW, Kiger AA, Kihara A, Kim DR, Kim D-H, Kim D-H, Kim E-K, Kim H-R, Kim J-S, Kim JH, Kim JC, Kim JK, Kim PK, Kim SW, Kim Y-S, Kim Y, Kimchi A, Kimmelman AC, King JS, Kinsella TJ, Kirkin V, Kirshenbaum LA, Kitamoto K, Kitazato K, Klein L, Klimecki WT, Klucken J, Knecht E, Ko BCB, Koch JC, Koga H, Koh J-Y, Koh YH, Koike M, Komatsu M, Kominami E, Kong HJ, Kong W-J, Korolchuk VI, Kotake Y, Koukourakis MI, Kouri Flores JB, Kovács AL, Kraft C, Krainc D, Krämer H, Kretz-Remy C, Krichevsky AM, Kroemer G, Krüger R, Krut O, Ktistakis NT, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012;8:445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ.. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011;331:456–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roach PJ. AMPK -> ULK1 -> Autophagy. Mol Cell Biol 2011;31:3082–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim J, Kundu M, Viollet B, Guan K-L.. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011;13:132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Picard M, Taivassalo T, Ritchie D, Wright KJ, Thomas MM, Romestaing C, Hepple RT, Polymenis M.. Mitochondrial structure and function are disrupted by standard isolation methods. PLoS One 2011;6:e18317.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kazlauskaite A, Kondapalli C, Gourlay R, Campbell DG, Ritorto MS, Hofmann K, Alessi DR, Knebel A, Trost M, Muqit MMK.. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser 65. Biochem J 2014;460:127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, Youle RJ.. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol 2014;205:143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ordureau A, Heo J-M, Duda DM, Paulo JA, Olszewski JL, Yanishevski D, Rinehart J, Schulman BA, Harper JW.. Defining roles of PARKIN and ubiquitin phosphorylation by PINK1 in mitochondrial quality control using a ubiquitin replacement strategy. Proc Natl Acad Sci U S A 2015;112:6637–6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Durcan TM, Fon EA.. The three ‘P’s of mitophagy: PARKIN, PINK1, and post-translational modifications. Genes Dev 2015;29:989–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen H, Chan DC.. Mitochondrial dynamics-fusion, fission, movement, and mitophagy-in neurodegenerative diseases. Hum Mol Genet 2009;18:R169–R176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z.. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 2007;26:1749–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Høyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T, Bianchi K, Fehrenbacher N, Elling F, Rizzuto R, Mathiasen IS, Jäättelä M.. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-β, and Bcl-2. Mol Cell 2007;25:193–205. [DOI] [PubMed] [Google Scholar]
- 43. Kroemer G, Mariño G, Levine B.. Autophagy and the integrated stress response. Mol Cell 2010;40:280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. De Palma C, Morisi F, Cheli S, Pambianco S, Cappello V, Vezzoli M, Rovere-Querini P, Moggio M, Ripolone M, Francolini M, Sandri M, Clementi E.. Autophagy as a new therapeutic target in Duchenne muscular dystrophy. Cell Death Dis 2012;3:e418.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pal R, Palmieri M, Loehr JA, Li S, Abo-Zahrah R, Monroe TO, Thakur PB, Sardiello M, Rodney GG.. Src-dependent impairment of autophagy by oxidative stress in a mouse model of Duchenne muscular dystrophy. Nat Commun 2014;5:4425–4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pauly M, Daussin F, Burelle Y, Li T, Godin R, Fauconnier J, Koechlin-Ramonatxo C, Hugon G, Lacampagne A, Coisy-Quivy M, Liang F, Hussain S, Matecki S, Petrof BJ.. AMPK activation stimulates autophagy and ameliorates muscular dystrophy in the mdx mouse diaphragm. Am J Pathol 2012;181:583–592. [DOI] [PubMed] [Google Scholar]
- 47. Whitehead NP, Kim MJ, Bible KL, Adams ME, Froehner SC.. A new therapeutic effect of simvastatin revealed by functional improvement in muscular dystrophy. Proc Natl Acad Sci U S A 2015;112:12864–12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Spitali P, Grumati P, Hiller M, Chrisam M, Aartsma-Rus A, Bonaldo P.. Autophagy is impaired in the tibialis anterior of dystrophin null mice. PLoS Curr 2013. doi: 10.1371/currents.md.e1226cefa851a2f079bbc406c0a21e80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xiao Y, Ma C, Yi J, Wu S, Luo G, Xu X, Lin PH, Sun J, Zhou J.. Suppressed autophagy flux in skeletal muscle of an amyotrophic lateral sclerosis mouse model during disease progression. Physiol Rep 2015;3:e12271–e12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brockhoff M, Rion N, Chojnowska K, Wiktorowicz T, Eickhorst C, Erne B, Frank S, Angelini C, Furling D, Ruegg MA, Sinnreich M, Castets P.. Targeting deregulated AMPK/mTORC1 pathways improves muscle function in myotonic dystrophy type I. J Clin Invest 2017;127:549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bibee KP, Cheng YJ, Ching JK, Marsh JN, Li AJ, Keeling RM, Connolly AM, Golumbek PT, Myerson JW, Hu G, Chen J, Shannon WD, Lanza GM, Weihl CC, Wickline SA.. Rapamycin nanoparticles target defective autophagy in muscular dystrophy to enhance both strength and cardiac function. FASEB J 2014;28:2047–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Grenier K, McLelland G-L, Fon EA.. Parkin- and PINK1-dependent mitophagy in neurons: will the real pathway please stand up? Front Neurol 2013;4:100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Billia F, Hauck L, Konecny F, Rao V, Shen J, Mak TW.. PTEN-inducible kinase 1 (PINK1)/Park6 is indispensable for normal heart function. Proc Natl Acad Sci U S A 2011;108:9572.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Siddall HK, Yellon DM, Ong S-B, Mukherjee UA, Burke N, Hall AR, Angelova PR, Ludtmann MHR, Deas E, Davidson SM, Mocanu MM, Hausenloy DJ, Kukreja R.. Loss of PINK1 increases the heart’s vulnerability to ischemia-reperfusion injury. PLoS One 2013;8:e62400.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, Burchell L, Walden H, Macartney TJ, Deak M, Knebel A, Alessi DR, Muqit MMK.. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol 2012;2:120080–120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.