Abstract
An important component of muscle elasticity is the PEVK region of titin, so named because of the preponderance of these amino acids. However, the PEVK region, similar to other elastomeric proteins, is thought to form a random coil and therefore its structure cannot be determined by standard techniques. Here we combine single-molecule electron microscopy and atomic force microscopy to examine the conformations of the human cardiac titin PEVK region. In contrast to a simple random coil, we have found that cardiac PEVK shows a wide range of elastic conformations with end-to-end distances ranging from 9 to 24 nm and persistence lengths from 0.4 to 2.5 nm. Individual PEVK molecules retained their distinctive elastic conformations through many stretch-relaxation cycles, consistent with the view that these PEVK conformers cannot be interconverted by force. The multiple elastic conformations of cardiac PEVK may result from varying degrees of proline isomerization. The single-molecule techniques demonstrated here may help elucidate the conformation of other proteins that lack a well-defined structure.
Elastomeric proteins are predicted to function as random coils and determine the elasticity of muscle and a variety of other tissues (1). In striated muscle fibers the passive elasticity is determined largely by the protein titin, where the PEVK region makes an important contribution (2–4). The advantage of a random coil is that it can be mechanically extended and relaxed without heat dissipation, driven entirely by its entropic elasticity. The mechanical properties of a random coil are determined by the persistence length (a measure of its flexibility) of the polypeptide chain. The entropic elasticity of polymer chains is well characterized by the worm-like-chain (WLC) model of polymer elasticity (5, 6). This model predicts that the force needed to simultaneously extend the ends of n polymer chains of equal contour length, Lc, placed in parallel, to a distance x is of the form
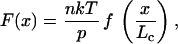 |
where p is the persistence length of the polymer. Hence, measurements of the persistence length of titin in muscle are difficult because they require knowledge of the total number of molecules being stretched (n) in the intact muscle fiber (3, 7). Even when these experiments are done at the single-molecule level, one cannot determine the persistence length if there is any uncertainty about the number of molecules (8). Here we have developed a strategy to generate a fingerprint that will identify unambiguously the stretching of a single molecule containing the PEVK region.
Single-molecule atomic force microscopy (AFM) has become a powerful tool to examine the conformation of proteins under a stretching force (9–15). Force-extension curves provide a characteristic sawtooth pattern that serves as a fingerprint of single molecules in experiments of mechanical unfolding and refolding of proteins containing tandem modules such as the Ig β-sandwiches of titin (12, 13). Although it would seem obvious to engineer a polyPEVK protein for single-molecule AFM experiments, stretching a random coil would give a featureless force-extension curve that would be difficult to distinguish from that observed for a denatured protein fragment, a common occurrence in these experiments. A featureless force-extension curve would also make it difficult to identify single molecules. In this article, we have used protein engineering techniques (12) to assemble a chimeric polyprotein composed of three identical repeats of I27-PEVK, a unit containing the 27th Ig module of human cardiac titin [I27, following the convention of Labeit and Kolmerer (2)] linked to the human cardiac PEVK segment. This experimental design is based on the discovery that the unfolding of tandem protein modules follows their mechanical stability rather than their ordering within the protein (16). This engineered protein, (I27-PEVK)3, combines the uncertain structure of the PEVK with the well characterized I27 module, which can be readily identified by AFM (12). Hence, we use the I27 module as an easily recognizable marker of the boundaries of the PEVK segment.
Materials and Methods
Construction of the (I27-PEVK)3 Polyprotein.
The strategy used to construct the (I27-PEVK)3 polyprotein was based on directional DNA concatemerization by self-ligation of the sticky ends of the nonpalindromic CTCGGG AvaI restriction site (12). I27-PEVK was made by two separate PCRs on human heart cDNA encoding I27 and PEVK. The first PCR amplified I27 and incorporated a 5′ AvaI site and a 3′ EcoRI site, the second PCR amplified PEVK and incorporated a 5′ EcoRI site and a 3′ AvaI site. The two PCRs products were digested with EcoRI and then ligated together to produce an I27-PEVK construct flanked with the nonpalindromic AvaI sites. After digestion with AvaI and self-ligation, the concatemers were cloned into a custom-made expression vector based on pET-31b(+) vector (Novagen). (I27-PEVK)3 was expressed in the recombination-defective strain BLR (DE3) (Novagen). We were able to harvest full-sized (I27-PEVK)3 without inducing the cells with isopropyl β-d-thiogalactoside (IPTG). There was enough basal expression to detect the protein on a Western blot by using an anti-His tag antibody (Penta-His, Qiagen, Chatsworth, CA) (for unknown reasons, IPTG-induced cells resulted in truncated protein). Then the protein was purified by Ni2+-affinity chromatography and FPLC and kept in PBS/5 mM DTT/0.2 mM EDTA at 4°C.
Electron Microscopy (EM) and Gradient Sedimentation Analysis.
His-bind purified (I27-PEVK)3 and I2712 were sedimented through glycerol gradients (15–40% glycerol in 0.2 M ammonium bicarbonate) at 20°C for 16 h at 38,000 rpm in a Beckman SW-55 rotor, providing an additional purification step before EM. Rotary-shadowed specimens for EM were prepared directly from gradient fractions (17). (PEVK-I27)3 samples were prepared at a high dilution, so that the observed trimers were not likely to be the result of the random proximity of three separate molecules. Electron micrographs were digitized, and dimensions were measured by using National Institutes of Health image, adjusting for the estimated 1-nm-thick metal shell. Two of the three arms of the triangle should correspond to PEVK links, but these were not visible in the electron micrograph. We chose the two shortest arms as the most likely PEVK and used these for statistical analysis.
AFM.
Single-protein AFM has been described in detail (10, 12). In this study we used Si3N4 cantilevers from Digital Instruments, Santa Barbara, CA (with a typical spring constant of ≈100 mN⋅m−1) and Park Scientific, Sunnyvale, CA (with a typical spring constant of ≈12 mN⋅m−1). Calibration of the cantilevers was done in solution by using the equipartition theorem.
Results and Discussion
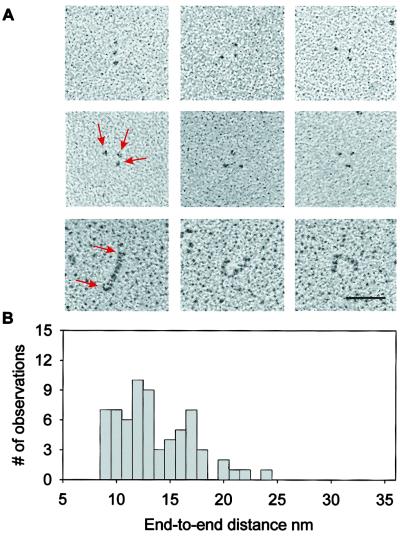
Fig. 1A (Top and Middle) shows rotary-shadowed electron micrographs of the (I27-PEVK)3 polyprotein. The micrographs show groups of three globular domains that appear to be tethered by an invisible string. We interpret these results as evidence of three clearly marked I27 modules separated by two PEVK segments. The PEVK region is 186 aa long (www.embl-heidelberg.de/ExternalInfo/Titin/annotation.html). Hence this region has a predicted contour length of ≈70 nm (0.38 nm/aa × 186 aa). A histogram of the separation between the I27 domains shows a wide distribution with values from 9 to 24 nm, with a suggestion of distinct peaks at 11 and 17 nm. The observed end-to-end length of PEVK is significantly smaller than its contour length, suggesting that the PEVK region in the relaxed state is coiled. Moreover the PEVK region is invisible in the EM micrographs, indicating that PEVK forms a much less compact structure than the folded Ig domain. Bustamante and colleagues (18, 19) investigated the end-to-end distance of DNA molecules adsorbed and equilibrated onto a two-dimensional plane and determined that this distance, R, could be calculated as < R2 > = 4 pL, where p is the persistence length and L is the contour length of a WLC polymer. Assuming that the EM measurements are a good approximation of the expected value of the end-to-end length distribution, we estimate a broad range of persistence lengths for the PEVK segments (0.2–2.3 nm).
Figure 1.
EM of individual (I27-PEVK)3 polyproteins. (A) Representative (I27-PEVK)3 (Top and Middle) and I2712 (Bottom) molecules as seen by rotary-shadowing EM. The polyprotein is visible as three small globular particles (I27 modules), apparently connected by an invisible thread (PEVK). In contrast, rotary-shadowed images of I2712 show solid rods with an average length of 58 nm, predicting a folded length of ≈4.8 nm/module. The bar corresponds to 50 nm. (B) A histogram of the average distance between I27 modules, corresponding to the end-to-end distance of PEVK, shows a broad distribution from 9 to 24 nm, with apparent peaks at 11 and 17 nm.
In contrast to the (I27-PEVK)3 polyprotein, rotary-shadowed electron micrographs of an I2712 polyprotein show continuous rod-like proteins (Fig. 1A Bottom) with an average length of 58 nm (not shown), giving an average length of 4.8 nm for the folded I27 module, which is slightly larger than the NMR estimate of 4.4 nm (20). The contrast between the appearance of the I27 polyproteins and that of the I27-PEVK polyprotein (Fig. 1A) supports the view that the PEVK region is largely an unfolded and coiled polypeptide. However, Fig. 1B shows a broad range in the end-to-end distance of the relaxed PEVK segment, implying that PEVK may have multiple conformations with different persistence lengths.
The unfolded conformation of the PEVK segments also was indicated by sedimentation. (I27-PEVK)3 and I2712 sedimented at 2.9 S and 4.2 S, respectively, giving an Smax/S of 2.6 and 2.1. Smax is the S for a compact, unhydrated sphere containing the equivalent mass of protein. Smax/S is equivalent to f/fmin, where f and fmin are the frictional coefficients of the protein and equivalent sphere (21, 22). A value over 2 indicates a highly extended molecule. The Smax/S = 2.1 is consistent with the elongated structure (58 × 2.5 nm) of I2712. The Smax/S = 2.6 of (I27-PEVK)3 indicates an even greater extension. However, the PEVK molecule is coiled and the overall length of the (I27-PEVK)3 seen in the EM pictures of Fig. 1 is shorter than that of I2712. Hence, it is likely that the larger Smax/S ratio results from an increased exposure of the protein to the solvent. The sedimentation data are consistent with the PEVK segments being largely unfolded polypeptide chain, creating a substantial hydrodynamic drag.
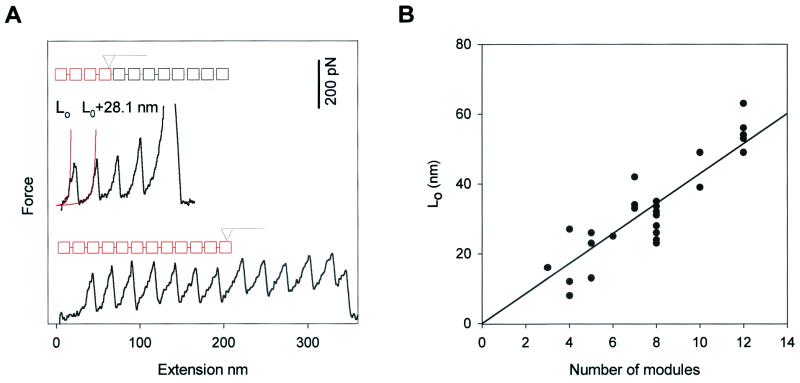
Fig. 2A shows force-extension curves obtained from the I2712 polyprotein. Because the AFM tip picks the protein at random locations, the number of peaks observed serves as a count of the number of modules contained in the segment that was picked up. The number of modules picked up can vary from one to 12. The folded length of a polyprotein can be determined by fitting the WLC model of polymer elasticity (5, 6) to the initial part of the force-extension curve, before any unfolding is observed (Fig. 2A, L0, solid line). The folded length of the polyprotein depends on the number of modules picked up by the AFM tip (n) and the folded length of a single I27 module (4.4 nm) (20). In close agreement with this prediction, a plot of L0 versus n shows a slope of 4.3 nm/module (Fig. 2B, solid line). Hence, both the electron micrographs and the AFM spectrographs give values that agree closely with those determined from the NMR structure of the I27 module.
Figure 2.
Characteristic fingerprint of I27 domain unfolding by a stretching force. (A) Stretching of an I2712 polyprotein produces a force-extension curve showing the characteristic sawtooth pattern of unfolding. The force-extension curves show different number of sawteeth, depending on the number of modules picked up by the AFM tip. The red squares represent the modules being picked up in this particular experiment. The solid lines are fits of the data to the WLC model of polymer elasticity. L0 is the contour length of the fully folded polyprotein; upon module unfolding at about ≈200 pN, an additional 28.1 nm will be added to the contour length of the protein. (B) Relationship between L0, determined from the fit to the first sawtooth, and the number of modules picked up by the AFM tip, determined by the number of sawteeth. The solid line is a linear regression of the data with a slope of 4.3 nm/module.
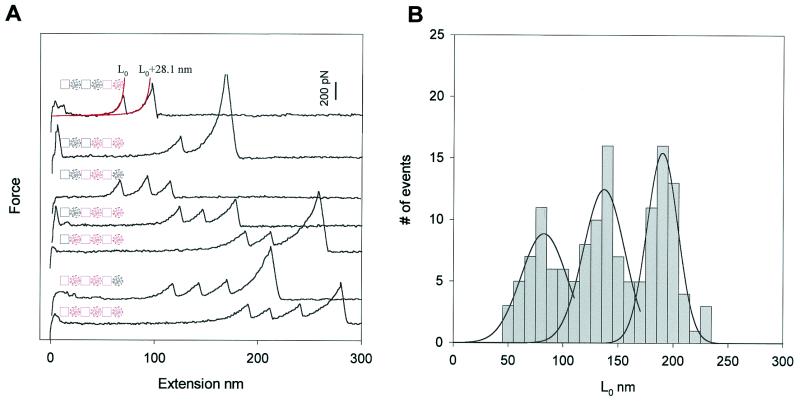
We then used the same techniques to investigate the mechanical nature of the PEVK protein. Fig. 3A shows several force-extension curves for the (I27-PEVK)3 polyprotein. The characteristic fingerprint of the I27 module is that it unfolds at ≈200 pN, extending the contour length of the protein by 28.1 nm (12). In Fig. 3A we show three types of recordings with one (top two traces), two (three middle traces), and three (bottom two traces) I27 unfolding events, excluding the last peak that corresponds to the detachment of the molecule from the AFM tip. In contrast to the I27 polyproteins, the force-extension relationships of the I27-PEVK polyproteins are characterized by a long initial length L0, ranging from 50 to 230 nm, which corresponds to the stretching of one, two, or three PEVK segments. The three traces in the middle of Fig. 3A have two unfolding peaks, but they have very different values of L0, e.g. 68 nm for the first one, 133 nm for the second one, and 211 nm for the third one. Because the polyprotein is constructed in an I27/PEVK alternating pattern, if three I27 unfolding peaks are observed, at least two PEVK segments must have been stretched (see Insets in Fig. 3A). If two I27 unfolding peaks are observed, one, two, or three PEVK segments must have been stretched. If only one I27 module is observed to unfold, there can be either one or two PEVK segments that are stretched but never three, in agreement with our observations. Using this approach, we can be sure that the PEVK segments are stretched and that the mechanical properties of PEVK are represented by the initial part of the force-extension relationships, before any of the I27 modules unfold.
Figure 3.
Identification and measurement of the elasticity of a PEVK segment. (A) Stretching an (I27-PEVK)3 polyprotein produces a sawtooth pattern only after a long initial spacer, L0. The sawtooth peaks are typical for I27 domain unfolding because they occur at 200 pN and extend the protein by ≈28.1 nm. Events with only one I27 unfolding event show two discrete values of L0: ≈82 nm or ≈135 nm (top two traces). When two or three I27 domains unfold we can measure an even longer value for L0 at ≈190 nm. The discrete values of L0 result from stretching one, two, or three PEVK segments before any I27 module unfolding occurs. The diagrams of the polyprotein accompanying each record show the various combinations of modules picked up by the AFM tip, where red squares and red circles represent I27 modules and PEVK segments being picked up, respectively. (B) Frequency histogram for the initial length L0. The distribution shows three clearly separated peaks (n = 142 recordings). Gaussian fits give distributions that peak at 82, 135, and 190 nm.
Fig. 3B shows a histogram of the initial length L0. The histogram shows three distinct peaks centered at about 82, 135, and 190 nm. The contour length of human cardiac PEVK is ≈70 nm. Thus, this distribution can be explained by assuming that the initial length L0 occurs as approximately integer multiples of about 70 nm. These results strongly suggest that the elastic properties of the initial region of the force-extension curves shown in Fig. 3A are due to the PEVK segments of the (I27-PEVK)3 polyprotein.
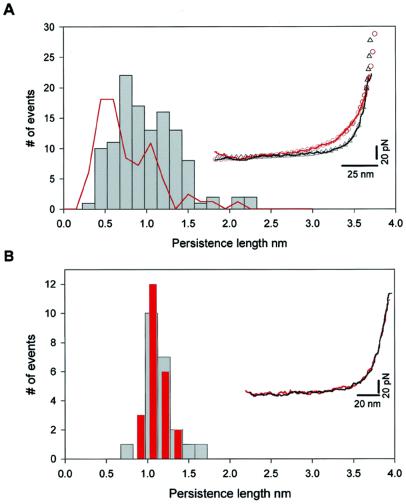
Having determined the contour length of the cardiac PEVK segments, we next measured their persistence length. Stretching a PEVK segment results in a nonlinear monotonic increase in force that can be described by the WLC model of polymer elasticity. The Inset in Fig. 4A shows two examples of force-extension traces where the dotted lines represent nonlinear Leverberg–Marquardt fits to the WLC equation. Within a force resolution of ≈5 pN, the WLC model fits data well and no stable, mechanically resistive structure can be detected, suggesting that the PEVK behaves as a purely entropic spring that has a random coil structure. However, the two force-extension traces do not superimpose, meaning that they have different persistence lengths. The histogram of measured persistence lengths (Fig. 4) shows a broad distribution (p = 0.3–2.3 nm), in reasonable agreement with the EM data (red line). This result provides direct evidence that cardiac PEVK displays multiple conformations with different mechanical flexibility. The small difference in the distributions obtained from the EM and AFM data could be caused by the approximations used to calculate the persistence length of PEVK from its end-to-end distance (19). However, it is remarkable that they agree so closely, validating the single-molecule techniques demonstrated here.
Figure 4.
The PEVK segment of cardiac titin shows multiple mechanical conformations. (A) Frequency histogram of the measured persistence length of the PEVK segment (gray bars). The force-extension relationships are accurately described by the WLC model of polymer elasticity (as shown in the Inset). The close agreement between the WLC model and the data (within ≈5 pN) demonstrates that extension of the PEVK segment does not involve the rupture of hydrogen-bonded structures. The PEVK segment shows a wide range of persistence length values, in agreement with the persistence lengths calculated from the end-to-end distributions observed with EM (red line). (Inset) Two representative PEVK recordings with different persistence lengths. In these two traces only the initial length of a stretched (I27-PEVK)3 polyprotein (before I27 domain unfolding) is shown (solid lines are experimental recordings, open symbols are Levenberg–Marquardt nonlinear fit of WLC model to the individual recordings). The red recording has a persistence length of 0.40 nm, the black recording has a persistence length of 1.08 nm. For comparison, the red recording (with a contour length of 207 nm) was normalized to have the same contour length as the black trace (140 nm). (B) Frequency histogram of the measured persistence length of the PEVK segments of a single (I27-PEVK)3 molecule during repeated stretch and relaxation cycles. The persistence length is narrowly distributed around 1.1 nm and is the same for the stretch (gray bars) and the relaxation (red bars) traces, showing that there is no detectable change in persistence length during these cycles. The scatter is due to the error margin of the fits to the data. (Inset) Two consecutive stretch (black) and relaxation (red) recordings. No hysteresis between stretching and relaxation was observed, indicating that this process is fully reversible.
We also probed the dissipative properties of the (I27-PEVK)3 polyprotein. The Inset in Fig. 4B shows a single stretch-relaxation cycle of the PEVK segments. Under these conditions, the force-extension curve was fully reversible because the forward (in black) and backward (in red) traces could be superimposed, in contrast to the hysteresis observed during Ig domains unfolding (8–10, 12). The persistence length measured from the same molecule during many consecutive stretch-relaxation cycles surprisingly shows a much narrower distribution (from 0.8 to 1.7 nm), in contrast to the wide distribution of persistence lengths measured on different molecules (Fig. 4A). This result indicates that individual PEVK molecules retain their distinctive elastic conformations through many stretch-relaxation cycles, and these distinctive conformations cannot be interconverted by force.
Previous studies of PEVK have provided insights into the mechanical properties of this segment (3, 7, 8, 23–25). However, the ensemble-averaged elasticity in intact myofibrils (3, 7) and uncertainty of the number of molecules in single-molecule experiments (8) may have prevented the detection of the PEVK's multiple conformations. The broad range of mechanical conformations of cardiac PEVK observed in our studies demonstrate the utility of a “fingerprint” in single-molecule experiments and provide a general approach to study the mechanical properties and conformations of other proteins.
It is well known that including l-prolyl residues into a polypeptide chain has significant structural effects. Polyproline chains have two distinct secondary structure conformations named polyproline type I and polyproline type II (PPII) helices. Cooperative transitions between these two conformations are caused by proline isomerization where a change from trans to cis leads to a transition from type II to type I helix (26). Isomerization from trans to cis results in significantly different end-to-end lengths, suggesting different persistence lengths (27, 28). Recent CD and two-dimensional NMR studies have revealed the existence of short PPII helix conformations in a 28-aa-long PEVK module from human fetal titin (29, 30). Because the energy difference between trans- and cis-prolyl residues is only 1–2 kcal/mol (31), if a fraction of PEVK PPII helix changes conformation caused by the trans-cis transition, this could readily explain our data. Because the cis conformation is shorter than the trans, one might expect that stretching the PEVK molecule would force all of the cis l-prolyl residues into the longer trans form. However, the activation energy barrier from cis to trans is large, 21 kcal/mol (32), and the gain in length is small (1.0 Å). We estimated the most probable force required for a mechanically driven transition from cis to trans according to (33, 34),
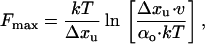 |
where v is the loading rate (200 pN/s), Δxu is the distance between cis and the transition state, and αo is the isomerization rate from cis to trans in the absence of force. Using a value for αo of 2 × 10−3 s−1 (35), we calculate that the cis to trans transition will occur at ≈600 pN and therefore is unlikely to happen under our experimental conditions. This may explain why we observe a broad range of persistence lengths in different PEVK molecules (different amounts of cis and trans l-prolyl residues) that cannot be interconverted by force.
In addition to the cis and trans forms of the l-prolyl residue, Schimmel and Flory (36) noted that the two allowed conformations of the trans prolyl residue can significantly alter the compactness of a polypeptide chain. For example, a polyalanine chain containing 10% of isolated l-prolyl residues was shown to change its end-to-end length by a factor of ≈1.6 when all of the trans prolyl residues changed between these rotamers. Furthermore, because there is no change in contour length of the polypeptide between these two conformers, they cannot be interconverted by an applied force. Hence, PEVK chains with different proportions of the two conformers of the trans prolyl residue also could explain our results.
What forces during protein assembly could cause such a variation in proline isomers from one molecule to another that result in PEVK molecules of very different flexibility? Our engineered proteins were made in bacteria and are typically used several days after purification. One might expect that the isomerization tendency will be the same for each proline and therefore average out about the same in each molecule. However, one must look at the protein as a whole. If a PPII helix begins to form, it is likely that it will favor a trans-proline rotamer, in a cooperative folding reaction. Hence, different PEVK molecules could acquire stable, but complex, combinations of PPII and coiled conformations.
We propose that cardiac PEVK behaves as an entropic spring that has a coiled structure overall with PPII helix and unordered structure coexisting inside the coil. Because the PPII helical structures are predominantly dictated by steric effect, the elasticity of PEVK remains entropic. The conformation and mechanical properties of PEVK depend on the available conformers of the l-prolyl residue in the PEVK sequence. Because these conformers can be switched enzymatically, it is possible that the elasticity of PEVK, in vivo, can be regulated by yet unknown signaling mechanisms.
Acknowledgments
We thank J. Kerkvliet for help in polyprotein engineering and P. E. Marszalek for helpful discussions. This work was supported by National Institute of Health grants to J.M.F.
Abbreviations
- WLC
worm-like-chain
- AFM
atomic force microscopy
- EM
electron microscopy
- PPII
polyproline type II
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Tatham A S, Shewry P R. Trends Biochem Sci. 2000;25:567–571. doi: 10.1016/s0968-0004(00)01670-4. [DOI] [PubMed] [Google Scholar]
- 2.Labeit S, Kolmerer B. Science. 1995;270:293–296. doi: 10.1126/science.270.5234.293. [DOI] [PubMed] [Google Scholar]
- 3.Linke W A, Ivemeyer M, Mundel P, Stockmeier M R, Kolmerer B. Proc Natl Acad Sci USA. 1998;95:8052–8057. doi: 10.1073/pnas.95.14.8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Linke W A, Granzier H. Biophys J. 1998;75:2613–2614. doi: 10.1016/S0006-3495(98)77706-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marko J F, Siggia E D. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 6.Bustamante C, Marko J F, Siggia E D, Smith S B. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 7.Trombitas K, Greaser M, Labeit S, Jin J P, Kellermayer M, Helmes M, Granzier H. J Cell Biol. 1998;140:853–859. doi: 10.1083/jcb.140.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kellermayer M, Smith S, Granzier H, Bustamante C. Science. 1997;276:1112–1116. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- 9.Rief M, Gautel M, Oesterhelt F, Fernandez J M, Gaub H E. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 10.Marszalek P E, Lu H, Li H B, Carrion M, Oberhauser A F, Schulten K, Fernandez J M. Nature (London) 1999;402:100–103. doi: 10.1038/47083. [DOI] [PubMed] [Google Scholar]
- 11.Yang G L, Cecconi C, Baase W A, Vetter I R, Breyer W A, Haack J A, Matthews B W, Dahlquist F W, Bustamante C. Proc Natl Acad Sci USA. 2000;97:139–144. doi: 10.1073/pnas.97.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carrion-Vazquez M, Oberhauser A F, Fowler S B, Marszalek P E, Broedel S E, Clarke J, Fernandez J M. Proc Natl Acad Sci USA. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher T E, Marszale P E, Fernandez J M. Nat Struct Biol. 2000;7:719–724. doi: 10.1038/78936. [DOI] [PubMed] [Google Scholar]
- 14.Bustamante C, Macosko J C, Wuite G J. Nat Rev Mol Cell Biol. 2000;1:130–136. doi: 10.1038/35040072. [DOI] [PubMed] [Google Scholar]
- 15.Clausen-Schaumann H, Seitz M, Krautbauer R, Gaub H E. Curr Opin Chem Biol. 2000;4:524–530. doi: 10.1016/s1367-5931(00)00126-5. [DOI] [PubMed] [Google Scholar]
- 16.Li H B, Oberhauser A F, Fowler S B, Clarke J, Fernandez J M. Proc Natl Acad Sci USA. 2000;97:6527–6531. doi: 10.1073/pnas.120048697. . (First Published May 23, 2000; 10.1073/pnas.120048697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fowler W E, Ecrickson H P. J Mol Biol. 1979;134:241–249. doi: 10.1016/0022-2836(79)90034-2. [DOI] [PubMed] [Google Scholar]
- 18.Rivetti C, Guthold M, Bustamante C. J Mol Biol. 1996;264:919–932. doi: 10.1006/jmbi.1996.0687. [DOI] [PubMed] [Google Scholar]
- 19.Rivetti C, Walker C, Bustamante C. J Mol Biol. 1998;280:41–59. doi: 10.1006/jmbi.1998.1830. [DOI] [PubMed] [Google Scholar]
- 20.Improta S, Politou A S, Pastore A. Structure (London) 1996;4:323–337. doi: 10.1016/s0969-2126(96)00036-6. [DOI] [PubMed] [Google Scholar]
- 21.Tanford C. Physical Chemistry of Macromolecules. New York: Wiley; 1961. [Google Scholar]
- 22.Garcia de la Torre J, Bloomfield V A. Q Rev Biophys. 1981;14:81–139. doi: 10.1017/s0033583500002080. [DOI] [PubMed] [Google Scholar]
- 23.Tskhovrebova L, Trinick J, Sleep J A, Simmons R M. Nature (London) 1997;387:308–312. doi: 10.1038/387308a0. [DOI] [PubMed] [Google Scholar]
- 24.Tskhovrebova L, Trinick J. J Mol Biol. 1997;265:100–106. doi: 10.1006/jmbi.1996.0717. [DOI] [PubMed] [Google Scholar]
- 25.Greaser M L, Wang S M, Berri M, Mozdziak P, Kumazawa Y. Adv Exp Med Biol. 2000;481:67–88. doi: 10.1007/978-1-4615-4267-4_4. [DOI] [PubMed] [Google Scholar]
- 26.Engel J. Biopolymers. 1966;4:945–948. [Google Scholar]
- 27.Schimmel P L, Flory P J. Proc Natl Acad Sci USA. 1967;58:52–59. doi: 10.1073/pnas.58.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tonelli A E. J Mol Biol. 1974;86:627–635. doi: 10.1016/0022-2836(74)90185-5. [DOI] [PubMed] [Google Scholar]
- 29.Gutierrez-Cruz G, Heerden A, Wang K. J Biol Chem. 2001;276:7442–7449. doi: 10.1074/jbc.M008851200. [DOI] [PubMed] [Google Scholar]
- 30.Ma K, Kan L, Wang K. Biochemistry. 2001;40:3427–3438. doi: 10.1021/bi0022792. [DOI] [PubMed] [Google Scholar]
- 31.Wuthrich K, Grathwohl G. FEBS Lett. 1974;43:337–340. doi: 10.1016/0014-5793(74)80674-5. [DOI] [PubMed] [Google Scholar]
- 32.Pauling L. The Nature of the Chemical Bond. 3rd Ed. Ithaca, NY: Cornell Univ. Press; 1960. [Google Scholar]
- 33.Evans E, Ritchie K. Biophys J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Izrailev S, Stepaniants S, Balsera M, Oono Y, Schulten K. Biophys J. 1997;72:1568–1581. doi: 10.1016/S0006-3495(97)78804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brandts J F, Halvorson H R, Brennan M. Biochemistry. 1975;14:4953–4963. doi: 10.1021/bi00693a026. [DOI] [PubMed] [Google Scholar]
- 36.Schimmel P L, Flory P J. J Mol Biol. 1968;34:105–120. doi: 10.1016/0022-2836(68)90237-4. [DOI] [PubMed] [Google Scholar]