Abstract
Antibiotic resistance poses an important threat to global public health and has become a challenge to modern medicine. The occurrence of antibiotic-resistant bacteria in a broad range of foods has led to a growing concern about the impact that food may have as a reservoir of antibiotic resistance genes. Considering Minas Frescal Cheese (MFC)—a typical Brazilian white soft cheese—and its economic and cultural values, in this study, medically relevant antimicrobial-resistance genetic markers (AR genes) were screened, and the occurrence of integrons were evaluated in manufactured MFC using culture-independent approaches. Through a fingerprinting analysis, the tested MFCs were brand-clustered, indicating reproducibility along the production chain. A common core of resistance markers in all brands evaluated and related antimicrobials such as β-lactams, tetracyclines, quinolones, and sulfonamide was detected. Several other markers, including efflux pumps and aminoglycosides-resistance were distributed among brands. Class 1 and 2 integrons were observed, respectively, in 77% and 97% of the samples. The presence of AR genes is of special interest due to their clinical relevance. Taken together, the data may suggest that the production chain of MFC might contribute to the spread of putative drug-resistant bacteria, which could greatly impact human health. Furthermore, detection of class 1 and class 2 integrons in MFC has led to discussions about resistance gene spread in this traditional cheese, providing evidence of potential horizontal transfer of AR genes to human gut microbiota.
Keywords: antibiotic resistance, genetic markers, Minas Frescal Cheese, integron
1. Introduction
As long as antimicrobial-resistant bacteria are being detected worldwide, associated to activities other than health-care and including food chain production, the phenomena will continue to pose a major threat to humans, animals, and environmental health and will become the focus of scientific investigation [1]. For several decades, studies on the dissemination of antibiotic resistance have focused mainly on health-care facilities and clinically relevant bacterial species [2]. However, it has been well documented that antimicrobials widely used in livestock have contributed to the spread of antibiotic resistant bacteria or antimicrobial resistance genes throughout the food production chain up to consumers and surrounding environments. In addition, it is accepted that foodstuff may play an important role in the phenomena since ready to eat food may be cross contaminated with antimicrobial resistant bacteria during processing, becoming an important reservoir and raising questions about its safety [2,3,4,5].
Antimicrobial resistance in foodborne pathogens involves direct risk for consumers’ health, impairs successful antibiotic treatment of infections, and increases the number of hospitalizations [6]. In addition, commensal bacteria found in the food may act as reservoirs for resistant genes which may potentially be transferred to gut microbiota and putative pathogens through horizontal gene transfer [7,8,9].
Resistance-encoding genes can be located on chromosomes or on mobile genetic elements, such as plasmids, transposons, and integrons, making them available for horizontal gene transfer [10]. Integrons are genetic platforms that may integrate gene cassettes (which are promoterless genetic elements) mainly related to antimicrobial resistance; they contain promoter(s) allowing the expression of those inserted cassettes and may be spread and incorporated into transposons and plasmids [11,12].
Dairy products such as milk and cheese have been implicated as potential sources for transmission of antimicrobial-resistant bacteria, with a direct link between the animal microbiota and the human gastrointestinal tract [13]. Minas Frescal Cheese (MFC), a white soft fresh cheese, is one of the most consumed and produced cheeses in Brazil. Due to the low salt content and high moisture, the MFC is an appropriate environment for the growth of putative pathogens. In addition, their manufacture involves technological steps that require the use of equipment without efficient treatments to reduce the microbial load [14,15,16]. Although sanitary legislations consider only classical foodborne pathogens such as coagulase-positive Staphylococcus, Listeria monocytogenes, and coliforms as quality indicators, other drug-resistant putative bacteria (DRB) may occur, posing a health issue [17,18,19].
To date, despite MFC having already been associated with multidrug-resistant bacteria, and considering the major microbial groups related to human health, there are no data on the cheese resistome, or even the clinical resistome including the occurrence of integrons, which would potentially be associated with antimicrobial-resistance genetic marker (AR genes) transfer. Most of the studies evaluating the prevalence of AR genes in cheese samples are based on methodological approaches such as isolation and identification of the resistant bacteria, followed by a molecular analysis of their AR determinants [18,20,21,22,23]. In this regard, a holistic view of cheese-resistome based on culture-independent approaches would retrieve valuable information on its quality, as those drug-resistant associated genes may be harbored by classical pathogens and by putative bacteria (not considered in food sanitary legislations) [20,24].
To contribute to aspects of clinical resistome and microbiological safety of manufactured MFC, this study aimed to perform a screening of clinically relevant antimicrobial-resistance genetic markers and evaluate the occurrence of integrons by culture-independent techniques based on direct analysis of total DNA from cheese samples. In addition, the application of the repetitive extragenic palindromic sequences polymerase chain reaction (rep-PCR) fingerprinting technique to compare the microbial community profile of MFC samples and evaluate its reproducibility was also evaluated.
2. Materials and Methods
2.1. Sample Processing and DNA Extraction
Samples from 35 cheeses from seven different commercially manufactured, trademarked, Minas soft cheeses (A, B, C, D, E, F, and G) were purchased in retail stores in Juiz de Fora (Minas Gerais state, Brazil). To obtain Brazilian sanitary certification, cheeses are made from pasteurized bovine milk. Following acquisition, the samples were kept in an ice box and sent to the laboratory for immediate processing. Total DNA was extracted directly from 5 samples (same batch) of seven different cheese brands. Twenty-five-gram cheese portions were weighted and homogenized in 225 mL of 2% (w/v) sterile sodium citrate (pH 7,5), using a shaker at 250 rpm for 20 min. Aliquots (50 mL) were centrifuged (14,000× g, 10 min), and the supernatant containing the fat layer was discarded. The pellet obtained was resuspended in 10 mL of Tris-EDTA (TE) (10 mM Tris; 1 mM ethylenediaminetetraacetic acid (EDTA); pH 8) and then centrifuged at 14,000× g for 10 min. This step (addition of wash TE buffer and centrifugation) was repeated twice, and the obtained pellet was resuspended in 200 µL TE buffer. DNA was extracted from the cell pellet using the QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany) according to the protocol. DNA concentration was determined by fluorimetry using the QubitTM 2.0 fluorimeter, with the Qubit dsDNA HS Assay kit (Life Technologies, Carlsbad, CA, USA). DNA integrity was assessed by 0.8% agarose gel electrophoresis. The resulting purified DNA was stored at −70 °C until use.
2.2. Repetitive Extragenic Palindromic Sequences Polymerase Chain Reaction
The rep-PCR analysis was performed using the total DNA extracted directly from the cheese samples. The PCR reactions were performed according to Gaber et al. [25], with modifications, using a single primer (GTG)5 (5′-GTGGTGGTGGTGGTG-3′). PCR final concentration contained 1× Master Mix (Promega, Madison, WI, USA), 0.8 μM of the primer, 20 ng of DNA, and ultrapure PCR water (Promega) was added to a final volume of 25 μL. The PCR conditions were: 5 min at 94 °C; 30 cycles of 1 min at 94 °C; 1 min at 50 °C; 2.5 min at 72 °C; and a final extension of 10 min at 72 °C. The PCR products were checked by electrophoresis in 2% (w/v) agarose gel for 2 h at a constant voltage of 100 V in 1× Tris/Acetate/EDTA buffer (TAE). A 100 bp DNA ladder (Ludwig Biotec, Porto Alegre, RS, Brazil) was used as a molecular-size marker. Gels were stained using ethidium bromide, and the images were visualized in ultraviolet light transilluminator (GE Healthcare, Little Chalfont, Bucks, UK). Fingerprints were analyzed using BioNumerics 5.1 (Applied Maths, Sint-Martens-Latem, Belgium). The similarities between the profiles were calculated using the Jaccard coefficient, and the dendrograms were constructed using the Unweighted Pair Group Method with Arithmetic Mean (UPGMA).
2.3. Screening of Antibiotic Resistance Genetic Markers and Integrons
A set of 40 resistance genetic markers of human clinical relevance (clinical resistome), including genetic markers related to efflux pumps and class 1, 2, and 3 integrons, were screened from the metagenomic DNA of the samples by conventional PCR, using specific primers and amplification conditions previously described (Supplementary Table S1). PCR products were analyzed by agarose gel electrophoresis 1.5% (w/v) in 1× Tris/Borate/EDTA (TBE) buffer. DNA bands were visualized after gel staining with ethidium bromide on ultraviolet light transilluminator. Amplicon sizes were estimated by a 100 bp DNA ladder (Ludwig Biotec). Negative controls were performed as reactions without template DNA. As a positive control, amplicons were purified using the QIAquick Gel Extraction Kit (Qiagen), and the DNA fragments obtained were sequenced in an ABI Prism 3730 DNA sequencer (Applied Biosystems, Foster City, CA, USA) and were further blasted against the nucleotide database of the National Center for Biotechnology Information (NCBI). The clustering based on the occurrence of antibiotic resistance markers was performed by a principal component analysis (PCA) using a PAST program (version 3.14).
3. Results
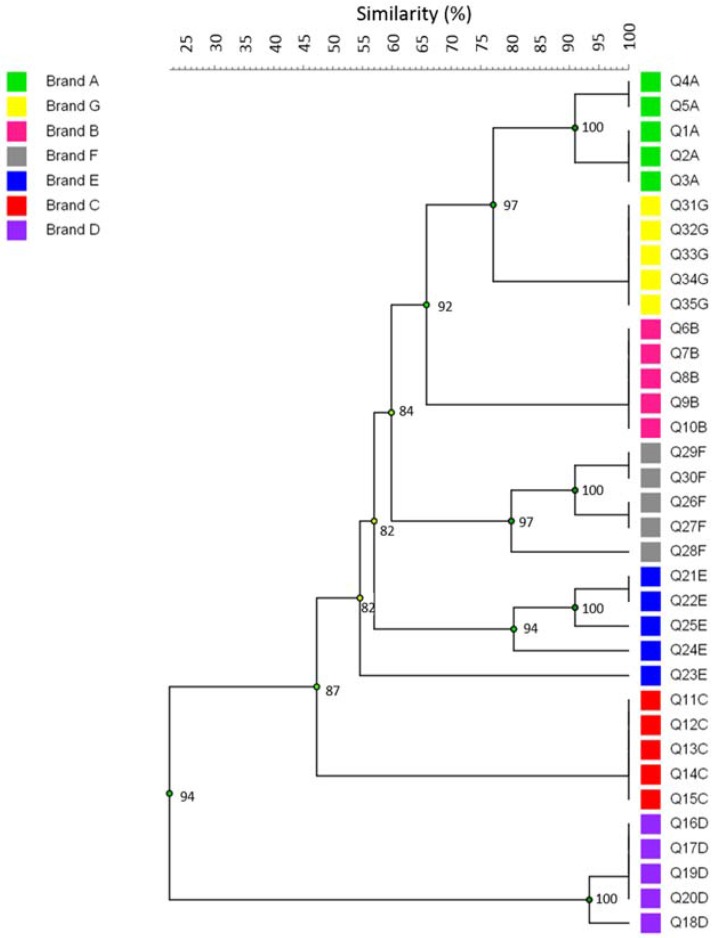
Repetitive extragenic palindromic sequences PCR was considered in this study as a culture-independent method, using the total DNA extracted from the cheese to compare the microbial community profile among samples from the same batch and to provide evidence of its reproducibility. The dendrogram obtained by rep-PCR clustering of cheese samples is presented in Figure 1. In general, considering a coefficient of similarity greater than 80%, the samples were clearly brand-clustered, suggesting standardization along the production chain and that the sampling in this study was reproducible. This result is in agreement with that expected since the products from the same batch are processed under similar conditions. However, one sample from brand E differed from the others, clustering with 55% similarity.
Figure 1.
Clustering of microbial community profile from different brands of Minas Frescal Cheese (MFC). The dendrogram was obtained from repetitive extragenic palindromic sequences polymerase chain reaction (rep-PCR) DNA fingerprint data, using the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) grouping method.
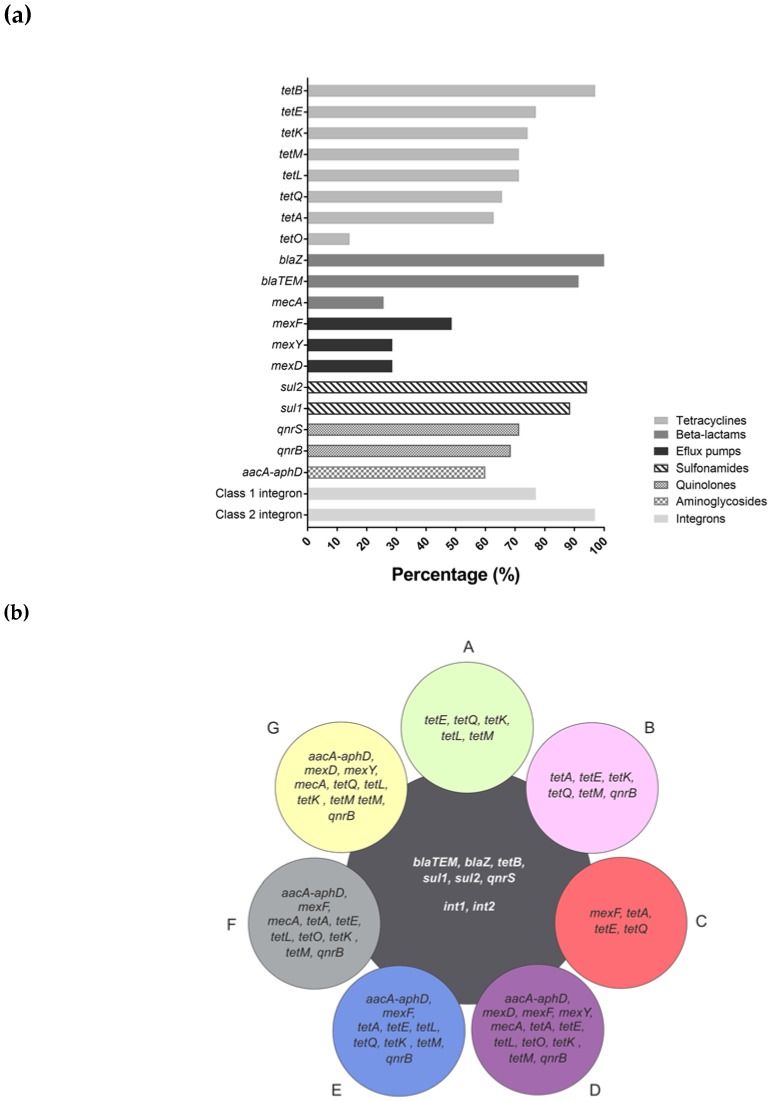
Considering the occurrence of antibiotic resistance markers, Figure 2a shows the percentage of resistance markers obtained in the 35 samples analyzed, according to the pharmacological class. From 40 resistance markers investigated, 14 were detected related to tetracyclines, β-lactams, efflux pumps, sulfonamides, quinolones, and aminoglycosides. On the other hand, resistance markers to macrolide, lincosamide, streptogramin, and erythromycin were not detected in any sample. In particular, the blaZ β-lactamase gene was detected in 100% of the samples and tetB, blaTEM, sul1, and sul2 genes in more than 80%. Despite the high percentage of occurrence, only three resistance markers related to β-lactams were detected, representing 21.4% of the total markers evaluated for this class (14 markers). Resistance markers to tetracyclines were the most prevalent in the analyzed samples. All markers evaluated in this study (eight markers) were detected among the samples, despite the low occurrence of the tetO gene.
Figure 2.
Clinical resistome composition of MFC. (a) Frequency of antimicrobial-resistance genetic markers detection based on their pharmacological properties; (b) Occurrence profile of antimicrobial-resistance markers among the cheese brands (A to G).
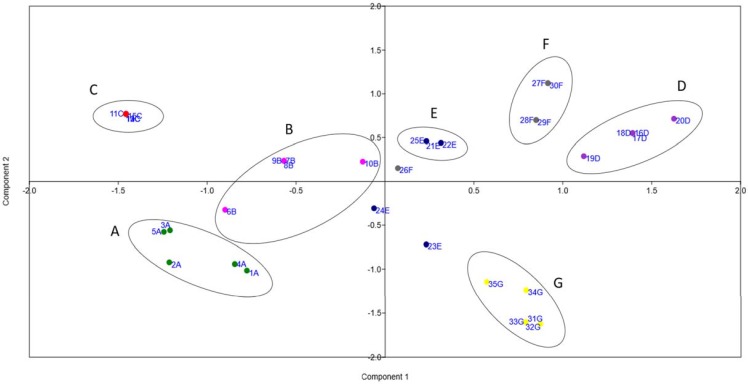
The co-occurrence of different resistance genes was observed in the analyzed brands and each one presented a particular profile (Figure 2b). In general, the frequency of detection and the variety of markers were higher in the brand D samples and lowest in brand C. Nevertheless, a common core of antibiotic resistance genes, related to tetracycline, β-lactam, quinolone, and sulfonamide (tetB, bla-TEM, blaZ, qnrS, sul1, and sul2), were detectable in all brands (Figure 2b). In accordance with the clustering obtained by the rep-PCR fingerprint, the samples were also brand-clustered according to the occurrence profile of resistance markers, as observed in Figure 3.
Figure 3.
Clustering of cheese samples according to the occurrence of antibiotic resistance markers using principal component analysis (PCA). Green: brand A (1A to 5A); pink: brand B (6B to 10B); red: brand C (11C to 15C); purple: brand D (16D to 20D); blue: brand E (21E to 25E); grey: brand F (26F to 30F); yellow: brand G (31G to 35G).
PCR-based detection of integrons was carried out on all the samples, using specific primers for three different intI genes (intl1, intl2 and intl3). Class 1 and 2 integrons were detected, respectively, in 77% and 97% of the samples and also characterized the common core of detectable genes in all brands (Figure 2b). Class 3 integrons were not detected in any sample analyzed.
4. Discussion
In general, microbiota characterization, especially in food microbiology, using culture-dependent approaches may not reveal the overall microbial diversity due to its complexity. On the other hand, culture-independent methods based on molecular biology techniques, such as rep-PCR, have been suggested to globally characterize the microbial population and differentiate cheese samples [26,27].
In this study, a rep-PCR approach as a culture-independent method to compare the microbial community profile of MFC samples and evaluate its reproducibility in the same batches was employed. Of the tested samples, only one from brand E did not follow the 80% similarity criteria within the same batch. This sample clustered with a similarity of 55%, which may represent low quality control during manufacturing, addressing issues related to microbial contamination along food chain production. However, a limitation in this approach was the lack of the assessment of which groups of microorganisms are present or absent in each sample.
Antibiotic resistance genes are widely distributed in several different environments, including food production systems. Food microbiology has also taken advantage of culture-independent, DNA-based molecular methods for the detection and analysis of AR genes directly in food samples, including several cheese types. Devirgiliis et al. [28] investigated the presence of specific antibiotic resistance genes in total DNA extracted from the whole microbiome present in an Italian cheese using molecular approaches. PCR detection of tetracycline and erythromycin resistance genes was performed in DNA from commercial Spanish and Italian cheeses samples by Flórez et al. [20]. In this study, a specific white soft cheese as a model, that is, MFC, which is typically produced in Brazil and consumed fresh was chosen.
Several genetic markers related to antimicrobial resistance have been detected, namely, the tet genes, which are associated with ribosomal protection and tetracycline efflux mechanisms. These were the most prevalent among the processed MFC. Although tetracyclines are well-known for their broad spectrum of activity to Gram-positive and Gram-negative bacteria, the spread of tetracycline-resistant mechanisms limited their use in treating bacterial infections [29]. Worldwide use of tetracycline in livestock production is well-known and leads to the persistence of resistance in the food chain and facilitates its spread among human pathogens. The tetracycline resistance genes have been reported in lactic acid bacteria (LAB) strains isolated from dairy products [28,30] including artisanal and industrial cheeses [20,31].
β-lactams are also clinically relevant antibiotics, for which resistance genes have also been described in bacteria from several foods, including cheeses. The mecA gene related to methicillin resistance, detected in 25.7% of the cheese samples in this study, was previously associated with coagulase-negative Staphylococci in this kind of cheese [18]. Add to that, the blaTEM, related to extended spectrum β-lactamases (ESBL), was detected in 91.4% of the samples. In general, ESBL is mainly encoded by mobile genetic elements such as integrons, transposons, and plasmids, which are easily transferable to other bacteria. Occurrence of blaTEM had already been documented in bacteria isolated from Portuguese soft and semi-soft cheeses [21]. Of especial interest, another β-lactams resistance gene, blaZ, was observed in all tested cheese sample. The data is impressive and may be related to the massive use of penicillin in Brazil, both in animal and human medicine. In cheese samples, blaZ has been reported in isolates of Staphylococcus aureus [32,33,34].
The presence of the aminoglycoside resistance gene (aacA-aphD) in 60% of the cheese samples is also of concern due to its controlled use, although this gene was already associated with coagulase-negative Staphylococci isolated from cheese samples in Turkey [23]. Aminoglycosides are important drugs in human therapy and their use in animal husbandry has been strictly regulated to avoid the selection and spread of aminoglycoside-resistant bacteria [35].
Quinolone resistance genes, qnrB and qnrS, detected at rates of 68.6% and 71.4%, respectively, along with sulfonamide resistance genes, sul1 and sul2, detected at rates of 88.6% and 94.3%, respectively, are also of particular interest; to date, these genetic markers have not been related to cheese. According to the literature, both qnr and sul genes are thought to be integron- and plasmid-born genes and, thus, highly available to horizontal transfer and environmental persistence upon selective pressure [36,37,38].
Bacterial multidrug efflux pumps are antibiotic resistance determinants which may be found in all microorganisms. These transporters can actively extrude a variety of substrates such as antibiotics, heavy metals, dyes, detergents, organic solvents, among others, indicating their importance in bacterial physiology under environmental selective pressure as a defense mechanism. Unspecific efflux pumps that transport several compounds can be associated with bacterial cross-resistance, i.e., simultaneous resistance to different antimicrobial drugs and other xenobiotics [39,40]. Efflux pump genes detected in this study encode Mex-type multidrug resistance efflux pumps (mexD, mexF, and mexY). These Mex systems can export several drugs such as chloramphenicol, fluoroquinolones, and tetracycline and are often described in opportunistic pathogenic bacteria [41,42].
Overall, the screening of AR genes in this study provided information on particularities of MFC processing. Although there is no available information with which to compare the results, taken together, the data may suggest that different manufacturing plants may face different contamination or even cross-contamination failures, leading to a peculiar AR profile in the final product which is available to the consumers. To date, it is not possible to address if the microbial contamination is related to the raw-material used in the dairy industry or related to human handling. However, the MFC resistome would be predictive of regional antimicrobial resistance epidemiology.
In Brazil, despite the lack of statistical information about the use of antibiotics in livestock, the β-lactams and tetracyclines are drugs frequently chosen for the treatment of infections affecting dairy herds [43]. In this study, several resistance-markers related to both treatment and prophylaxis in human and veterinary medicine were detected. According to the literature, food chain contamination including antimicrobial-resistant bacteria may be related to: (i) the use of xenobiotics in dairy, livestock, and agriculture, or (ii) contamination in plant processing or handling [5].
It is well-known that genes encoding antimicrobial resistance are often linked to mobile genetic elements. In this study, class 1 and 2 integrons were detected in 77% and 97% of the cheese samples, respectively, raising issues about AR gene mobilization. According to the literature, class 1, 2, and 3 integrons are historically associated with antimicrobial-resistance gene spread [44]. It has already been suggested that integrons might be associated with resistance to β-lactams, aminoglycosides, quinolones, sulfonamides, macrolides, and tetracyclines [45,46,47].
Overall, the high prevalence of antibiotic resistance genes in MFC samples raises issues about its clinical resistome composition and the threat of the cheese ecosystem as a potential reservoir of resistance genes. In addition, the presence of class 1 and 2 integrons may lead to the risk of AR gene mobilization to human intestinal microbiota and putative pathogens. A larger scale investigation with metagenomic studies would clarify correlations between the bacterial community of MFC and the resistance markers detected in this study.
Acknowledgments
We gratefully acknowledge the Post-Graduate Program in Biological Sciences (PPGCBIO/UFJF) and CAPES for the scholarship. This study was supported by grants from Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4425/9/2/106/s1, Table S1: Primers used in this study.
Author Contributions
C.G.D. and V.L.d.S. conceived and designed the experiments; A.C.L.d.P. and J.M.d.A.C. performed the experiments; A.C.L.d.P., J.D.M., and A.C.d.A. analyzed the data; A.C.L.d.P. wrote the manuscript. All authors read and approved the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Roca I., Akova M., Baquero F., Carlet J., Cavaleri M., Coenen S., Cohen J., Findlay D., Gyssens I., Heure O.E., et al. The global threat of antimicrobial resistance: Science for intervention. New Microbes New Infect. 2015;6:22–29. doi: 10.1016/j.nmni.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landers T.F., Cohen B., Wittum T.E., Larson E.L. A Review of antibiotic use in food animals: Perspective, policy, and potential. Public Health Rep. 2012;127:4–22. doi: 10.1177/003335491212700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Founou L.L., Founou R.C., Essack S.Y. Antibiotic resistance in the food chain: A developing country-perspective. Front. Microbiol. 2016;7:1–19. doi: 10.3389/fmicb.2016.01881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bengtsson-Palme J. Antibiotic resistance in the food supply chain: Where can sequencing and metagenomics aid risk assessment? Curr. Opin. Food Sci. 2017;14:66–71. doi: 10.1016/j.cofs.2017.01.010. [DOI] [Google Scholar]
- 5.Verraes C., van Boxstael S., van Meervenne E., van Coillie E., Butaye P., Catry B., Schaetzen M.A., van Huffel X., Imberechts H., Dierick K., et al. Antimicrobial resistance in the food chain: A review. Int. J. Environ. Res. Public Health. 2013;10:2643–2669. doi: 10.3390/ijerph10072643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doyle M.E. Multidrug-resistant pathogens in the food supply. Foodborne Pathog. Dis. 2015;12:261–279. doi: 10.1089/fpd.2014.1865. [DOI] [PubMed] [Google Scholar]
- 7.Huddleston J.R. Horizontal gene transfer in the human gastrointestinal tract: Potential spread of antibiotic resistance genes. Infect. Drug Resist. 2014;7:167–176. doi: 10.2147/IDR.S48820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devirgiliis C., Barile S., Perozzi G. Antibiotic resistance determinants in the interplay between food and gut microbiota. Genes Nutr. 2011;6:275–284. doi: 10.1007/s12263-011-0226-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rolain J.M. Food and human gut as reservoirs of transferable antibiotic resistance encoding genes. Front. Microbiol. 2013;4:1–10. doi: 10.3389/fmicb.2013.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Hoek A.H.A.M., Mevius D., Guerra B., Mullany P., Roberts A.P., Aarts H.J.M. Acquired antibiotic resistance genes: An overview. Front. Microbiol. 2011;2:1–27. doi: 10.3389/fmicb.2011.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillings M.R. Integrons: Past, present, and future. Microbiol. Mol. Biol. Rev. 2014;78:257–277. doi: 10.1128/MMBR.00056-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng Y., Bao X., Ji L., Chen L., Liu J., Miao J., Chen D., Bian H., Li Y., Yu G. Resistance integrons: Class 1, 2 and 3 integrons. Ann. Clin. Microbiol. Antimicrob. 2015;14:45. doi: 10.1186/s12941-015-0100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathur S., Singh R. Antibiotic resistance in food lactic acid bacteria—A review. Int. J. Food Microbiol. 2005;105:281–295. doi: 10.1016/j.ijfoodmicro.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Gomes B.C., Franco B.D.G.D.M., De Martinis E.C.P. Microbiological food safety issues in Brazil: Bacterial pathogens. Foodborne Pathog. Dis. 2013;10:197–205. doi: 10.1089/fpd.2012.1222. [DOI] [PubMed] [Google Scholar]
- 15.Carvalho J.D.G., Viotto W.H., Kuaye A.Y. The quality of Minas Frescal cheese produced by different technological processes. Food Control. 2007;18:262–267. doi: 10.1016/j.foodcont.2005.10.005. [DOI] [Google Scholar]
- 16.Brito J.R.F., Santos E.M.P., Arcuri E.F., Lange C.C., Brito M.A.V.P., Souza G.N., Cerqueira M.M.P.O., Beltran J.M.S., Call J.E., Liu Y., et al. Retail survey of Brazilian milk and Minas Frescal cheese and a contaminated dairy plant to establish prevalence, relatedness, and sources of Listeria monocytogenes isolates. Appl. Environ. Microbiol. 2008;74:4954–4961. doi: 10.1128/AEM.01828-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Visotto R.G., Oliveira M.A., Prado S.O.T., Bergamini A.M.M. Queijo Minas Frescal: Perfil higiênico-sanitário e avaliação da rotulagem. Rev. Inst. Adolfo Lutz. 2011;70:8–15. [Google Scholar]
- 18.Fontes C.O., Silva V.L., de Paiva M.R.B., Garcia R.A., Resende J.A., Ferreira-Machado A.B., Diniz C.G. Prevalence, antimicrobial resistance, and virulence characteristics of mecA-encoding coagulase-negative Staphylococci isolated from soft cheese in Brazil. J. Food Sci. 2013;78:594–599. doi: 10.1111/1750-3841.12088. [DOI] [PubMed] [Google Scholar]
- 19.Nunes C.S.R., Souza C.P., Pereira K.S., Del Aguila E.M., Paschoalin V.M.F. Identification and molecular phylogeny of coagulase-negative staphylococci isolates from Minas Frescal cheese in southeastern Brazil: Superantigenic toxin production and antibiotic resistance. J. Dairy Sci. 2016;99:2641–2653. doi: 10.3168/jds.2015-9693. [DOI] [PubMed] [Google Scholar]
- 20.Belén Flórez A., Alegría Á., Rossi F., Delgado S., Felis G.E., Torriani S., Mayo B. Molecular identification and quantification of tetracycline and erythromycin resistance genes in Spanish and Italian retail cheeses. BioMed Res. Int. 2014 doi: 10.1155/2014/746859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amador P., Fernandes R., Prudêncio C., Brito L. Resistance to β-lactams in bacteria isolated from different types of Portuguese cheese. Int. J. Mol. Sci. 2009;10:1538–1551. doi: 10.3390/ijms10041538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furlaneto-Maia L., Rocha K.R., Henrique F.C., Giazzi A., Furlaneto M.C. Antimicrobial resistance in Enterococcus sp isolated from soft cheese in Southern Brazil. Adv. Microbiol. 2014;4:175–181. doi: 10.4236/aim.2014.43023. [DOI] [Google Scholar]
- 23.Kürekci C. Short communication: Prevalence, antimicrobial resistance, and resistant traits of coagulase-negative staphylococci isolated from cheese samples in Turkey. J. Dairy Sci. 2016;99:2675–2679. doi: 10.3168/jds.2015-10725. [DOI] [PubMed] [Google Scholar]
- 24.Perry J.A., Wright G.D. The antibiotic resistance “mobilome”: Searching for the link between environment and clinic. Front. Microbiol. 2013;4:138. doi: 10.3389/fmicb.2013.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaber A., Hassan M.M., Dessoky E.-D.S., Attia A.O. In vitro antimicrobial comparison of Taif and Egyptian Pomegranate Peels and seeds extracts. J. Appl. Biol. Biotechnol. 2015;3:12–17. doi: 10.7324/JABB.2015.3203. [DOI] [Google Scholar]
- 26.Perin L.M., Sardaro M.L. S, Nero L.A., Neviani E., Gatti M. Bacterial ecology of artisanal Minas cheeses assessed by culture-dependent and -independent methods. Food Microbiol. 2017;65:160–169. doi: 10.1016/j.fm.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 27.Perin L.M., Bello B.D., Belviso S., Zeppa G., Carvalho A.F., Cocolin F., Nero L.A. Microbiota of Minas cheese as influenced by the nisin producer Lactococcus lactis subsp. lactis GLc05. Int. J. Food Microbiol. 2015;214:159–167. doi: 10.1016/j.ijfoodmicro.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 28.Devirgiliis C., Caravelli A., Coppola D., Barile S., Perozzi G. Antibiotic resistance and microbial composition along the manufacturing process of Mozzarella di Bufala Campana. Int. J. Food Microbiol. 2008;128:378–384. doi: 10.1016/j.ijfoodmicro.2008.09.021. [DOI] [PubMed] [Google Scholar]
- 29.Grossman T.H. Tetracycline antibiotics and resistance. Cold Spring Harb. Perspect. Med. 2016;6 doi: 10.1101/cshperspect.a025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huys G., D’Haene K., Collard J.-M., Swings J. Prevalence and molecular characterization of tetracycline resistance in Enterococcus isolates from food. Appl. Environ. Microbiol. 2004;70:1555–141032. doi: 10.1128/AEM.70.3.1555-1562.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Devirgiliis C., Zinno P., Stirpe M., Barile S., Perozzi G. Functional screening of antibiotic resistance genes from a representative metagenomic library of food fermenting microbiota. BioMed Res. Int. 2014:1–9. doi: 10.1155/2014/290967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carfora V., Caprioli A., Marri N., Sagrafoli D., Boselli C., Giacinti G., Giangolini G., Sorbara L., Dottarelli S., Battisti A., et al. Enterotoxin genes, enterotoxin production, and methicillin resistance in Staphylococcus aureus isolated from milk and dairy products in Central Italy. Int. Dairy J. 2015;42:12–15. doi: 10.1016/j.idairyj.2014.10.009. [DOI] [Google Scholar]
- 33.Jamali H., Paydar M., Radmehr B., Ismail S., Dadrasnia A. Prevalence and antimicrobial resistance of Staphylococcus aureus isolated from raw milk and dairy products. Food Control. 2006;54:383–388. doi: 10.1016/j.foodcont.2015.02.013. [DOI] [Google Scholar]
- 34.Spanu V., Spanu C., Virdis S., Cossu F., Scarano C., De Santis E.P.L. Virulence factors and genetic variability of Staphylococcus aureus strains isolated from raw sheep’s milk cheese. Int. J. Food Microbiol. 2012;153:53–57. doi: 10.1016/j.ijfoodmicro.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 35.Jaimee G., Halami P.M. Emerging resistance to aminoglycosides in lactic acid bacteria of food origin—An impending menace. App. Microbiol. Biotechnol. 2016;100:1137–1151. doi: 10.1007/s00253-015-7184-y. [DOI] [PubMed] [Google Scholar]
- 36.Rodríguez-Martínez J.M., Cano M.E., Velasco C., Martínez-Martínez L., Pascual Á. Plasmid-mediated quinolone resistance: An update. J. Infect. Chemother. 2011;17:149–182. doi: 10.1007/s10156-010-0120-2. [DOI] [PubMed] [Google Scholar]
- 37.Phuong Hoa P.T., Nonaka L., Hung Viet P., Suzuki S. Detection of the sul1, sul2, and sul3 genes in sulfonamide-resistant bacteria from wastewater and shrimp ponds of north Vietnam. Sci. Total Environ. 2008;405:377–384. doi: 10.1016/j.scitotenv.2008.06.023. [DOI] [PubMed] [Google Scholar]
- 38.Sköld O. Sulfonamide resistance: Mechanisms and trends. Drug Resist. Updat. 2000;3:155–160. doi: 10.1054/drup.2000.0146. [DOI] [PubMed] [Google Scholar]
- 39.Piddock L.J.V. Multidrug-resistance efflux pumps—Not just for resistance. Nat. Rev. Microbiol. 2006;4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 40.Blanco P., Hernando-Amado S., Reales-Calderon J.A., Corona F., Lira F., Alcalde-Rico M., Bernardini A., Sanchez M.B., Martinez J.L. Bacterial multidrug efflux pumps: Much more than antibiotic resistance determinants. Microorganisms. 2016;4 doi: 10.3390/microorganisms4010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kievit T., Parkins M., Gillis R., Srikumar R., Ceri H., Poole K., Iglewski B.H., Storey D. Multidrug efflux pumps: Expression patterns and contribution to antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 2001;45:1761–1770. doi: 10.1128/AAC.45.6.1761-1770.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ueda O., Wexler H.M., Hirai K., Shibata Y., Yoshimura F., Fujimura S. Sixteen homologs of the Mex-type multidrug resistance efflux pump in Bacteroides fragilis. Antimicrob. Agents Chemother. 2005;49:2807–2815. doi: 10.1128/AAC.49.7.2807-2815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galvani J.W.C., Tondo E.C., Brandelli A. Commonly screened antibiotics in raw milk from dairy plants under State Inspection in Rio Grande do Sul, Brazil. Braz. J. Vet. Res. Anim. Sci. 2017;57:147–153. doi: 10.11606/issn.1678-4456.bjvras.2017.118547. [DOI] [Google Scholar]
- 44.Cambray G., Guerout A.-M., Mazel D. Integrons. Ann. Rev. Genet. 2010;44:141–166. doi: 10.1146/annurev-genet-102209-163504. [DOI] [PubMed] [Google Scholar]
- 45.Mazel D. Integrons: Agents of bacterial evolution. Nat. Rev. Microbiol. 2006;4:608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 46.Partridge S.R., Tsafnat G., Coiera E., Iredell J.R. Gene cassettes and cassette arrays in mobile resistance integrons: Review article. FEMS Microbiol. Rev. 2009;33:757–784. doi: 10.1111/j.1574-6976.2009.00175.x. [DOI] [PubMed] [Google Scholar]
- 47.Ahmed A.M., Shimamoto T., Shimamoto T. Characterization of integrons and resistance genes in multidrug-resistant Salmonella enterica isolated from meat and dairy products in Egypt. Int. J. Food Microbiol. 2014;189:39–44. doi: 10.1016/j.ijfoodmicro.2014.07.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.