Abstract
Acidophile bacteria belonging to the Acidithiobacillus genus are pivotal players for the bioleaching of metallic values such as copper. Cell adherence to ores and biofilm formation, mediated by the production of extracellular polymeric substances, strongly favors bioleaching activity. In recent years, the second messenger cyclic diguanylate (c-di-GMP) has emerged as a central regulator for biofilm formation in bacteria. C-di-GMP pathways have been reported in different Acidithiobacillus species; however, c-di-GMP effectors and signal transduction networks are still largely uncharacterized in these extremophile species. Here we investigated Pel exopolysaccharide and its role in biofilm formation by sulfur-oxidizing species Acidithiobacillus thiooxidans. We identified 39 open reading frames (ORFs) encoding proteins involved in c-di-GMP metabolism and signal transduction, including the c-di-GMP effector protein PelD, a structural component of the biosynthesis apparatus for Pel exopolysaccharide production. We found that intracellular c-di-GMP concentrations and transcription levels of pel genes were higher in At. thiooxidans biofilm cells compared to planktonic ones. By developing an At. thiooxidans ΔpelD null-mutant strain we revealed that Pel exopolysaccharide is involved in biofilm structure and development. Further studies are still necessary to understand how Pel biosynthesis is regulated in Acidithiobacillus species, nevertheless these results represent the first characterization of a c-di-GMP effector protein involved in biofilm formation by acidophile species.
Keywords: Acidithiobacillus, biofilm, bioleaching, biomining, c-di-GMP, Pel exopolysaccharide, PelD
1. Introduction
Biomining is an industrial process in which acidophilic leaching microorganisms including bacteria and archaea are used to recover valuable metals such as copper, cobalt and zinc from low-grade sulfidic ores [1,2]. In addition to its advantaging industrial application, bioleaching naturally occurs in any environment where sulfidic minerals are exposed to both water and oxygen, contributing to water contamination through acid mine/rock drainage (AMD/ARD) generation [3,4]. Microbial leaching activity is increased by bacterial attachment on mineral due to the formation of a thin reaction space between ore and cells [5,6] suggesting that the understanding of molecular events involved in biofilm formation by acidophile species may help to improve biomining and mitigate environmental pollution. Due to the focus on the leaching activities of microorganisms, Acidithiobacillus species have been the first and most characterized bioleaching species and were early considered a pivotal player for the biomiming process [7,8]. To date Acidithiobacillus genus encompasses seven gram-negative, acidophilic and chemolithoautotrophic species that can only oxidize reduced inorganic sulfur compounds (RISCs) or both ferrous iron and RISCs [9,10,11,12,13]. All Acidithiobacillus sp. are capable to form biofilms on mineral surfaces [14,15,16]. Moreover, recent ecological studies pointed out that acidophilic bacterial communities from natural environments frequently occur as biofilms in which Acidithiobacillus species are predominant structural members [17,18,19]. Several studies performed with the iron/sulfur oxidizer specie At. ferrooxidans revealed that quorum sensing (QS) communication system mediated by acyl-homoserine lactone molecules modulates biofilm formation [20,21,22]. Nevertheless, since sulfur-oxidizing species At. caldus and At. thioooxidans do not possess any canonical genes for QS [23], it was earlier suggested that biofilm formation should also be regulated by other molecular pathways in the Acidithiobacillus species. Indeed, it has been recently reported that the cyclic diguanylic acid (c-di-GMP) pathway is functional and plays an active role in biofilm formation by different Acidithiobacillus species [24,25].
The second messenger cyclic diguanylate (c-di-GMP) has emerged as a central metabolite that controls several phenotypes in bacteria, including motility and biofilm formation [26,27]. It is well accepted now that high intracellular levels of c-di-GMP repress motility and stimulate biofilm formation [26,27,28,29,30]. Intracellular levels of c-di-GMP are balanced by the antagonist activities of diguanylate cyclases (DGCs) and phosphodiesterases (PDEs). The synthesis of c-di-GMP is performed by GGDEF domain present in DGC enzymes by using two guanosine triphosphate (GTP) molecules as substrate. C-di-GMP degradation is catalyzed by EAL and HD-GYP domains from two different PDE families [26,27]. Several classes of c-di-GMP effectors that differ in their nature as well as in structural basis for c-di-GMP binding have been described [26,27,31]. Although two different classes of c-di-GMP binding riboswitches have been characterized [32], most of c-di-GMP effectors described so far are proteins. These include the PilZ domain of multimeric protein complexes such as bacterial cellulose synthase (BCS) [33], inactive GGDEF domains such as PelD (see further [29]), inactive EAL domains [34], several transcriptional regulators [26,35,36], ATPases involved in protein secretion [37] and cell-cycle regulators with kinase-phosphatase activity [38]. The inner-membrane protein PelD from Pseudomonas aeruginosa was one of the first c-di-GMP effector proteins to be characterized [29]. It is encoded into the seven-gene operon pelABCDEFG, the gene products of which are assembled in a multiprotein membrane complex for Pel biosynthesis [39,40]. The binding of c-di-GMP to the cytoplasmic side of PelD increases the glycosil-transferase activity of Pel biosynthetic machinery [29,39]. Pel apparatus architecture and export mechanism have been now deciphered and the structural composition of Pel exopolysaccharide has been characterized [41,42,43]. In Pseudomonas aeruginosa, Pel exopolysaccharide is involved in cell aggregation and maintenance of biofilm structure [29,41,44].
In the course for the characterization of c-di-GMP pathways in Acidithiobacillus spp. [24,25], comparative genomic studies performed by our group revealed that the complexity of c-di-GMP network differs in Acidithiobacillus species [25]. In addition, it was also pointed out that an open reading frame (ORF) coding for the c-di-GMP effector protein PelD and the corresponding pel-like operon are present only in the sulfur-oxidizing species At. thiooxidans and At. caldus while bcs operon involved in biosynthesis of cellulose, which is also regulated by c-di-GMP, is present on both sulfur/iron and sulfur oxidizer species [25]. Thus, we hypothesized that Pel exopolysaccharide should play a specific role in biofilm formation by sulfur oxidizing Acidithiobacillus species [25]. The purpose of the present work was to challenge this hypothesis. Here we fully characterized a c-di-GMP pathway in At. thiooxidansT type strain ATCC 19377 and we investigated the role of Pel exopolysaccharide on biofilm formation by this extremophile microorganism. By high-performance liquid chromatography (HPLC) and quantitative polymerase chain reaction (qPCR) experiments we demonstrated that intracellular c-di-GMP concentrations and transcription levels of pel genes are increased in At. thiooxidans biofilm cells compared to planktonic ones. In correlation with both results, we demonstrated that Pel exopolysaccharide is involved in biofilm structure by developing an At. thiooxidans ΔpelD null-mutant strain. Finally, this work provides the first evidence that the c-di-GMP pathway and Pel exopolysaccharide are both involved in biofilm formation by acidophilic bacteria.
2. Materials and Methods
2.1. Strains, Plasmids, Primers and Growth Conditions
Strains, plasmids and primers used in this work are described in Tables S1 and S2. At. thiooxidans ATCC 19377T strain was grown at 30 °C in modified Mackintosh (MAC) medium [45] pH 4.5 supplemented with different energetic substrates: 5% w/v elemental sulfur (S°; prills and coupons); 20 mM thiosulfate (Na2S2O3), 10 mM tetrathionate (K2S4O6). Solid medium was obtained by adding 1 mM MgSO4, 8 mg/L Bromocresol Green and 0.88% phytagel (w/v). At. thiooxidansT null-mutant strain ΔpelD was grown in selective media with 100–200 μg/mL kanamycin. Escherichia coli and Salmonella enterica serovar Typhimurium strains were grown at 37 °C in Luria-Bertani (LB) medium (1% Triptone, 0.5% yeast extract, 0.5% NaCl) pH 7.0 and agar (1.5% w/v) was added for solid medium. Selective media for E. coli strains were supplemented with ampicillin (100 μg/mL), trimetroprin (50 μg/mL), chloramphenicol (20 μg/mL) or kanamycin (30 μg/mL). The mating medium for conjugation was made by adding 0.5 mM d-Glucose, 0.05% yeast extract and 50 μM diaminopimelic acid into solid thiosulfate growth medium.
2.2. Bioinformatic Analysis
At. thiooxidansT draft genome AFOH01000001 [46] was obtained from National Center for Biotechnology Information (NCBI) Database. Candidate genes for proteins with GGDEF, EAL, HD-GYP and c-di-GMP effectors domains were predicted using the basic local alignment search tool (BLAST) as previously described [25]. Annotation results were visualized with Artemis software [47]. Protein domains were identified using Pfam [48] and Prosite [49]. Transmembrane domain predictions were done by TMHMM Server [50]. The functionality of identified domains was predicted using ClustalO algorithm [51].
2.3. qPCR Experiments
At. thiooxidansT cells were grown in 200 mL of medium with sulfur, thiosulfate or tetrathionate for five days. Planktonic cells were collected by centrifugation at 6000× g for 10 min. Biofilm cells were separated from solid sulfur by incubation with 0.05% Triton X-100 and collected by centrifugation [24]. Total RNA was extracted from both cell sub-populations as previously described, incubated with DNase I at 37 °C for 1 h and purified by phenol-chloroform treatment [24]. The complementary DNA (cDNA) was synthetized from 1 µg of total RNA by using reverse transcriptase and random primers. Then, cDNA was diluted 1/30 with nuclease-free water and used as template for qPCR experiments. Specific primers were designed to analyze transcriptional levels of the pelA, pelD and wcaG genes. 16S rDNA and map were used as housekeeping genes for data normalization [52].
2.4. Nucleotide-Enriched Fraction Extraction and c-di-GMP Analysis
Nucleotide-enriched fractions were extracted from late-exponential growing cells from sulfur, thiosulfate or tetrathionate cultures. Sulfur cultures were separated in two independent cell populations: planktonic and biofilm cells [24]. The extraction was realized by hot lysis and HClO4 treatments [53]. HPLC analysis was performed by a HPLC coupled to photodiode array detector (Waters 1525, 2996) (Waters, Milford, MA, USA) using a 15 cm × 3 mm SUPELCOSIL LC-18-DB C18, 3 μm particle size, Reverse Phase Column (SIGMA, Saint Louis, MO, USA). The liquid chromatography (LC) system consisted of degasser (Waters), binary pump (Waters) and oven (Waters). The mobile phase was methanol (A) and water pH 6.0 (6 mM KH2PO4) (B). Elution conditions were 5 min at 100% B, 15 min linear gradient from 100% B to 20% A and 80% B and finally 10 min with a gradient from 20% A and 80% B to 100% B with a constant flow of 0.4 mL/min. The temperature was set at 30 ± 3 °C. The injection volume was 20 µL. The calibration curve was performed with synthetic c-di-GMP (BIOLOG, Hayward, CA, USA) in a range of 6.9–552 ng (10–800 pmol) for injection. Signal signatures were identified by coincidence of retention times of 12.531 ± 0.295 min and comparison of absorption spectra at 252.4 nm. Data were expressed as pmol c-di-GMP and normalized against cellular wet weight.
2.5. Diguanylate Cyclase Activity
Heterologous complementation assays in Salmonella strain defective in DGC activity were performed as previously described [25]. Briefly, several At. thiooxidansT genes encoding for different proteins with GGDEF domains were amplified from genomic DNA by PCR and cloned into pBAD24 plasmid. Salmonella enterica serovar Typhimurium AdrA1f strain was electrotransformed with pBAD24 recombinant plasmids harboring At. thiooxidansT genes and DGC activity was evaluated by congo red binding assay [25].
2.6. Construction and Selection of a At. thiooxidansT ΔpelD Null Mutant Strain
At. thiooxidansT ΔpelD null mutant strain was constructed as described in Castro et al. [25]. First, a ΔpelD suicide plasmid was produced by molecular engineering. Briefly, two different 800 bp DNA fragments carrying 5′ and 3′ extremes of pelD gene were obtained by PCR. Primers harbored specific restriction sites for cloning and fragment production in pGEM-T vector (Promega, Madison, WI, USA). KanR gene was released from plasmid pSKM2 [54] by using restriction enzymes HindIII and XmaI. The plasmid pOT was digested by restriction enzymes SacI and XbaI and then dephosphorylated with alkaline phosphatase. All DNA restriction-fragments were separated by electrophoresis, recovered from agarose gels and quantified. Ligations were performed with T4 DNA ligase at 4 °C overnight and ligation product was transformed into chemocompetent E. coli JM109 cells. Recombinant cells were selected on solid medium supplemented with ampicillin and kanamycin resistance. Suicide plasmid pOT-pelD::kanR was checked by restriction analysis and sequencing (MACROGEN, Seoul, Korea).
For conjugation assays, At. thiooxidansT cells were grown on thiosulfate to reach a 108 cells/mL density. E. coli HB101 strain carrying pOT-pelD::kanR and pR388 [54] plasmids was grown overnight in selective LB medium to inoculate 50 mL of liquid mating medium with corresponding antibiotics. Then E. coli cells were grown overnight at 37 °C and collected. Both At. thiooxidans and E. coli cells fractions were separately washed twice with modified MAC medium (pH 4.5) and collected by centrifugation (6000× g, 10 min). Both cell suspensions were homogeneously mixed (cell ratios 1:1) and 100 μL of this cellular mix was spotted on a sterile polycarbonate filter. Inoculated filters were gently located over a solid mating medium and incubated for 5 days at 30 °C. Then filters were picked off and incubated for 7 days at 30 °C in liquid MAC medium with thiosulfate (20 mM) and kanamycin (200 μg/mL). Finally, cultures were diluted and 100 μL of 10−4, 10−5 and 10−6 dilutions were plated on selective solid MAC medium (200 μg/mL kanamycin) and incubated at 30 °C for several days until the appearance of colonies.
Colonies were first analyzed by PCR with specific kanR primers. They were picked up and re-suspended in 10 μL of MAC medium pH 4.5. The cell suspension was spotted in solid selective MAC medium for growth. A fraction of the grown spot was collected and re-suspended in 100 μL of a 25 mM Tris-HCl (pH 7.5), 3 mM KCl solution. The cell suspension was heated at 100 °C and centrifuged to eliminate cell debris. Thus, the cell lysate was used as a template for the amplification of 16S rDNA, kanR and pelD genes by PCR. Finally, PCR products were run in 1.5% w/v agarose gels in TAE buffer (40 mM Tris, 20 mM acetic acid, and 1 mM EDTA, pH 8.2) and electrophoretic pattern were compared to wild type strain. Positive colonies were finally analyzed by Southern blot analysis using specific pelD and kanR labeled with digoxigenin [25]. Genomic DNA (10 µg) from mutant and wild type strain or control plasmids (1 µg) were digested with SphI or BamHI for pelD or kanR probes, respectively. All DNAs were run in 1% agarose gel using TBE buffer 1× (89 mM Tris, 89 mM boric acid, 2 mM EDTA, pH 7.6). The SSC buffer 10× (1,5 M NaCl, 0.15 M sodium Citrate, pH 7.2) was used to transfer DNA fragments from gel to nylon membrane. Final concentration for probes hybridization was 25 ng/mL and developing was done according to manufacturer’s instructions (Roche, Basel, Switzerland).
2.7. Visualization of At. thiooxidansT Biofilms
At. thiooxidansT cells were grown in MAC medium with sulfur (Merck, Darmstadt, Germany) prills and coupons as solid energetic substrate. Sulfur coupons were used for biofilm visualization. Thus, colonized coupons were extracted at different incubation times for 5 days, washed once with aqueous H2SO4 pH 2.0, once with 50 mM Tris-HCl, 1 mM EDTA, pH 7.5 and once with bidistilled water to remove all the remaining planktonic cells. Afterwards cells were fixed overnight with 4% formaldehyde. These coupons were critical point dried, coated with gold and analyzed by Scanning Electron Microscopy (SEM) (LEO 1530VP, LEO Electron Microscopy Inc., Thornwood, NY, USA), as previously described [25].
2.8. Quantification of Extracellular Polymeric Substances
Cells were incubated in 500 mL of MAC medium with sulfur for 5 days. Sulfur-prills colonized with cells were collected and washed twice with 10 mM KH2PO4 pH 4.5 to eliminate any remaining planktonic cells. Then prills were vortexed with 0.05% Triton X-100 in 10 mM KH2PO4 pH 4.5 for 10 min twice to release extracellular polymeric substances (EPS) and attached cells. Sulfur debris and released cells were removed by centrifugation. Supernatant was recollected and EPS were recovered by incubation with ethanol and ultracentrifugation at 4 °C (100,000× g, 1 h). EPS sediment was re-suspended in 300 µL of 20 mM Tris-HCl, 1 mM EDTA pH 7.4 and stored at −20 °C. Carbohydrates determination was done using the Dubois method [55], while protein content was measured using the Bicinchoninic Acid method [56].
3. Results
3.1. Acidithiobacillus thiooxidansT Possesses a Functional c-di-GMP Pathway
The bioinformatical analysis of At. thiooxidansT genome allowed the identification of twenty-five ORFs coding for putative DGCs and PDEs proteins that could be involved in metabolism of c-di-GMP (Figure S1). Twelve of them encode for proteins with both GGDEF/EAL domains (ATHIO_RS1701500, ATHIO_RS1815000, ATHIO_RS0108445, ATHIO_RS1708000, ATHIO_RS0100160, ATHIO_RS0110800, ATHIO_RS179300, ATHIO_RS0102030, ATHIO_RS0113350, ATHIO_RS1813000, ATHIO_RS0104240, ATHIO_RS0114625), 9 for putative DGC with single GGDEF (ATHIO_RS1657500, ATHIO_RS0108455, ATHIO_RS0108485, ATHIO_RS1689500, ATHIO_RS1835500, ATHIO_RS1838000, ATHIO_RS0107955, ATHIO_RS0116120, ATHIO_RS1781000), 3 for putative PDE with single EAL domains (ATHIO_RS0108450, ATHIO_RS1750000, ATHIO_RS0113355) and one for a putative PDE with HD-GYP domain (Figure S2). Moreover, multiple alignment analysis of all these c-di-GMP metabolic domains showed that most of them (15/21 GGDEF domains, 14/15 EAL domains and 1/1 HD-GYP domain) possess all amino acids required for catalytic activity [21]. Different sensor domains such as GAF and PAS have been also predicted inside the amino acid sequence of some of these proteins (17/25) (Figure S2) suggesting that the global intracellular level of c-di-GMP in At. thiooxidansT is modulated by different environmental factors. In addition, fourteen ORFs coding c-di-GMP effector proteins were also identified (Figure S1). Nine of them encode for proteins with PilZ domains including five Type IV pilus assembly proteins (Table S3), one for a PelD-like protein, two for putative transcriptional regulators FleQ, one for an ATPase with MshEN domain and one for a YajQ-like protein. Interestingly, several assigned functions (BlastP Hit) for these putative c-di-GMP effectors were related to biofilm formation such as pilus assembly (ATHIO_RS16400, ATHIO_RS0105675, ATHIO_RS0109125, ATHIO_RS0109755, ATHIO_RS0110790 and ATHIO_RS0114620), motility regulation (ATHIO_RS0108750) and synthesis of exopolysaccharides such as cellulose (ATHIO_RS0101475) and Pel (ATHIO_RS018015). Finally, reverse transcriptase (RT-)PCR experiments using total RNA obtained from planktonic cells grown on sulfur revealed that most of these genes are transcribed (Figure S1).
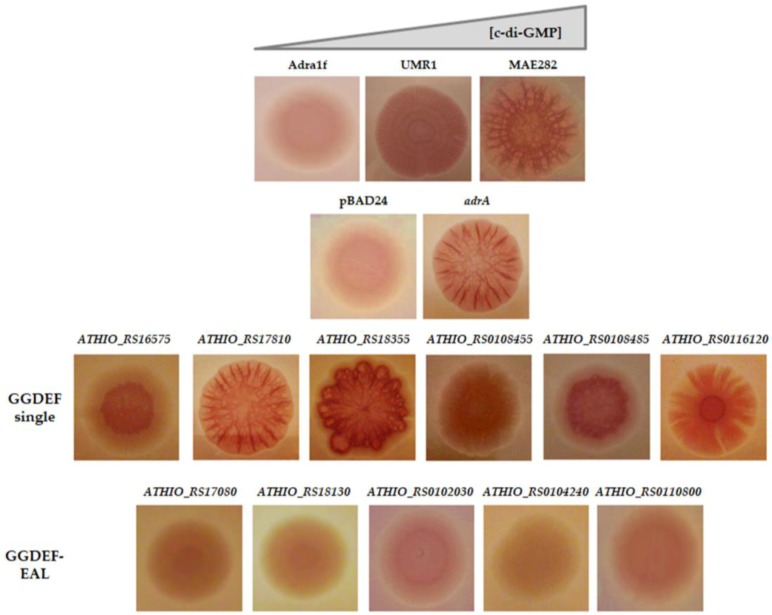
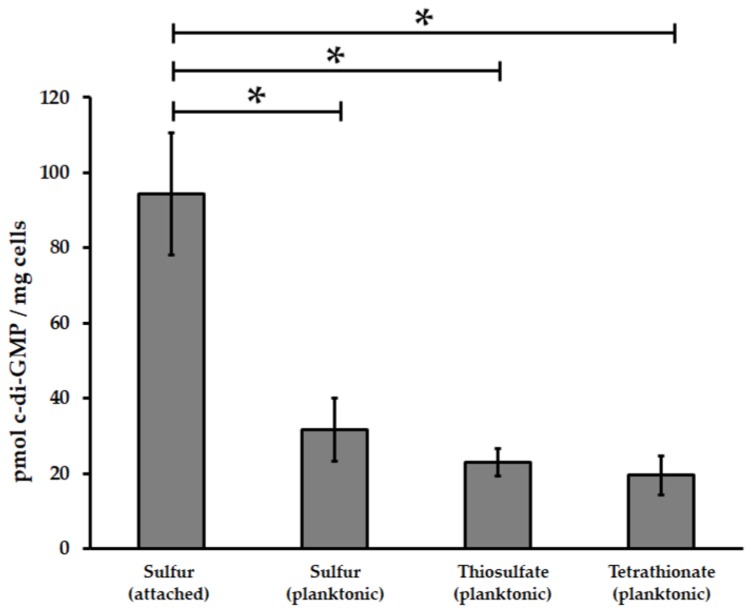
In order to assess DGC activity, 11 GGDEF domains encoding genes from At. thiooxidansT including six putative DGC enzyme with single GGDEF domain and five with both EAL/GGDEF domains (Figure S1) were cloned in Salmonella enterica serovar Typhimurium AdrA1f strain. Congo red phenotypic assays only revealed strong positive DGC activity, based in the DGC activity-induced rdar (rough, dry and red) morphotype of the colony [57], for proteins with single GGDEF domains (Figure 1). In addition, to evaluate the relationship between c-di-GMP pathway and biofilm formation by At. thiooxidansT, intracellular c-di-GMP levels were measured in different cell sub-populations by HLPC. Planktonic populations were obtained from cells grown in thiosulfate, tetrathionate and elemental sulfur, while biofilm cells were obtained from sulfur cultures. In our experimental conditions, intracellular concentrations of c-di-GMP were 3.5-fold higher in five days sulfur-biofilm cells compared to planktonic cells grown in sulfur, thiosulfate or tetrathionate (Figure 2). Both results clearly indicate that c-di-GMP pathway is functional in At. thiooxidansT. In addition, they revealed that attachment of At. thiooxidansT cells to solid energetic substrate is directly related to an increase of intracellular concentration of c-di-GMP.
Figure 1.
Heterologous complementation in Salmonella enterica serovar Typhimurium strain AdrA1f of diguanylate cyclase activity from single GGDEF and GGDEF/EAL encoding genes of Acidithiobacillus thiooxidansT. The rdar (rough, dry and red) morphotype [57], which is induced by diguanylate cyclase (DGC) activity was analyzed on congo red agar plates and compared to wild type (UMR1), DGC null-mutant (AdrA1f), DGC complemented (padrA), negative control (pBAD24 without insert) and a phosphodiesterase null-mutant (MAE 282) strains.
Figure 2.
Comparative analysis of c-di-GMP levels from biofilm and planktonic At. thiooxidansT cells grown in different energetic substrates. The nucleotide-enriched extracts were analyzed by high-performance liquid chromatography (HPLC) coupled to photodiode array detector. Values represent the average of tree independent experiments ± standard deviation. Significant differences made by a one-way analysis of variance (ANOVA) test (p < 0.05) are noted (*).
3.2. Pel Genes Are Overexpressed in At. thiooxidansT Attached Cells
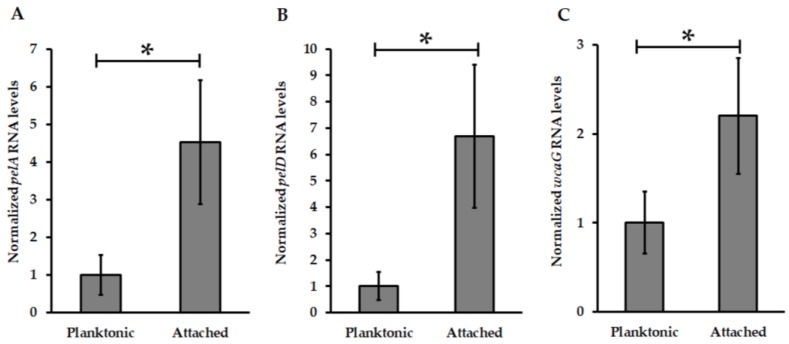
Among the 14 putative c-di-GMP effectors proteins identified in the available draft genome of At. thiooxidansT, a pelD orthologous gene was found. Moreover, the analysis of genomic context revealed that this pelD-like is located inside a putative pel operon which is present in other At. thiooxidans strains [58]. Interestingly, pel operon structure from At. thiooxidansT is similar to pel operon from At. caldus including for the presence downstream pelG of an additional wcaG gene coding for an enzyme with uridine diphosphate (UDP)-glucose-4-epimerase activity (Figure S3). As previously noted [25,58,59], pel-like operon is harbored by Acidithiobacillus species that can oxidize only RISC suggesting a specific role in biofilm formation by At. caldus and At. thiooxidans. To determine the role of Pel biosynthesis apparatus, transcription levels of pelA, pelD and wgcA genes encoding for the deacetylase PelA, the c-di-GMP effector protein PelD and an UDP-Glucose-4-epimerase respectively were measured by qPCR experiments using total RNA obtained from planktonic and biofilm cells of At. thiooxidansT. Compared to planktonic cells, transcription levels of pelA, pelD and wcaG genes were increased in At. thiooxidans biofilm cells 4.5-, 6.7- and 2.2-fold, respectively (Figure 3).
Figure 3.
Transcription levels analysis of Pel biosynthesis machinery encoding genes from At. thiooxidansT. Transcript levels of pelA (A), pelD (B) and wcaG (C) genes were measured by quantitative polymerase chain reaction (qPCR) and then normalized using DNA 16S and map genes. Values represent the average of four independent experiments ± standard deviation. Significant differences made by a one-way ANOVA test (p < 0.05) are noted (*).
3.3. The PelD Null Mutation Changes At. thiooxidansT Biofilm Structure on Sulfur Surface
To better understand the function of Pel exopolysaccharide in biofilm formation by At. thiooxidans, the construction of ∆pelD null-mutant strain was challenged. A suicide vector harboring a kanamycin (kanR) cassette and 5′ and 3′ ends of At. thiooxidansT pelD gene was constructed and introduced by conjugation in At. thiooxidansT. Two hundred recombinant colonies were analyzed by PCR experiments against pelD, kanR and DNA 16S genes to discriminate single recombinant (pelD+, kanR+, DNA 16S+) and double recombinant (pelD−, kanR+, DNA 16S+) strains. Four clones were selected as double recombinants (Figure S4) and Southern Blot analysis were performed to corroborate the gain of ∆pelD null-mutant strain of At. thiooxidansT (Figure S5).
EPS production from sulfur-grown cells of wild type and ∆pelD strains was examined. As expected for the absence of Pel exopolysaccharide, a six fold decrease of total carbohydrates quantity was observed for ∆pelD S0-attached cells compared to wild type (Table 1). Surprisingly, the measurement of total protein fraction revealed an increase of 33.6% in ∆pelD cells compared to wild-type (WT) strain (Table 1).
Table 1.
Quantification of carbohydrates and proteins levels into Acidithiobacillus thiooxidansT S°-attached cells obtained from wild type ATCC 19377 and ΔpelD strains.
| ATCC 19377 | ΔpelD | |
|---|---|---|
| Carbohydrates (µg/g cells) | 1596.80 ± 67.71 | 272.92 ± 45.88 |
| Proteins (µg/g cells) | 245.58 ± 58.33 | 331.42 ± 52.13 |
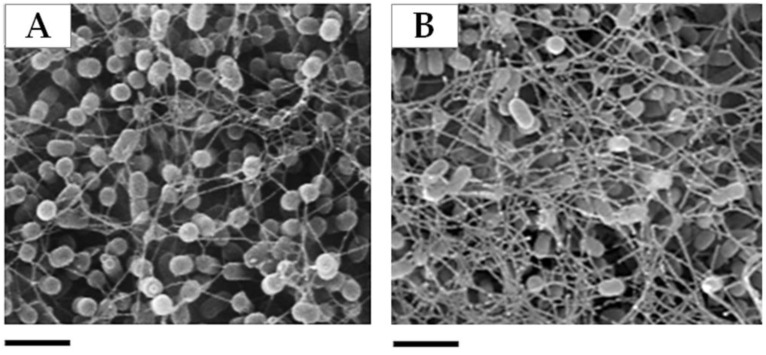
In addition, 5-days old biofilms developed on S°-coupons surface by At. thiooxidans ΔpelD and WT cells were visualized separately by SEM microscopy. As shown in Figure 4, ΔpelD null-mutant strain overexpressed a filamentous structure compared to wild type. Both results strongly indicated that biofilm composition and structure of At. thioooxidans ∆pelD null-mutant strain are modified in comparison to WT strain. Although further studies are still necessary to characterize the nature of this filamentous structure, the EPS quantification result suggests that it could be proteinaceous.
Figure 4.
Biofilm structure of At. thiooxidans is affected by pelD deletion. Compared to wild type strain At. thiooxidansT (A), ∆pelD null-mutant strain of At. thiooxidansT (B) overexpressed a filamentous structure. Bars represent 2 µm.
4. Discussion
Insights into the c-di-GMP pathway in Acidithiobacillus genus have recently started to emerge [24,25]. These studies indicate that c-di-GMP signaling is a widespread pathway into Acidithiobacillus genus [25]. As it occurs for At. ferrooxidans [24] and At. caldus [25], higher c-di-GMP levels were observed in attached cells of At. thiooxidansT compared to planktonic cells (Figure 2), supporting that the c-di-GMP pathway regulates biofilm formation by this acidophilic leaching species. With twenty-five genes encoding proteins with GGDEF alone (9), EAL alone (3), both GGDEF/EAL (12) and HD-GYP (1) domains as well as fourteen genes encoding putative effector proteins, At. thiooxidansT possesses the c-di-GMP pathway with the highest complexity currently known among species of this genus [24,25]. Moreover, At. thiooxidans has a HD-GYP domain encoding gene which was transcribed (Figure S1) suggesting that unlike At. ferrooxidans and At. caldus, PDE activity should be generated by both EAL and HD-GYP domains in this bacterial species. To date no c-di-GMP riboswitches [32] have been identified in Acidithiobacillus genomes [60] suggesting that c-di-GMP effector proteins are the predominant way for c-di-GMP signal transduction in this genus. Indeed, this work allowed the identification of five different proteins families for c-di-GMP effectors in At. thiooxidans (Table S3). Altogether, these data suggest that the c-di-GMP pathway signaling has a specific molecular network At. thiooxidans, even different to other sulfur-oxidizing species such as At. caldus.
The MshE ATPase from Vibrio cholerae has been recently characterized as a new c-di-GMP effector protein involved in the biosynthesis and function of Type IVa MshA pili which is a relevant extracellular and adhesive appendage for initial attachment to surfaces by bacterial cells [32]. The binding of c-di-GMP occurs through the MshE N-terminal domain that is the longest nucleotide-binding motif identified yet [61]. Here we identified a mshE orthologue (ATHIO_RS0109755) that encodes a MshE ATPase-like with a canonical MshE N-terminal domain that was transcribed in At. thiooxidans cells grown on elemental sulfur (Figure S1). In addition, the mining of At. thiooxidansT genome sequence revealed the presence of several type IV pilin-like protein encoding genes (Table S3). Thus, it is possible to hypothesize that the initial attachment to solid energetic substrates by At. thiooxidansT may be regulated by c-di-GMP pathway through MsHE/Type IVa pilin system.
FleQ was first characterized in P. aeruginosa as a transcriptional regulator for genes related to flagellar-based motility and mucin adhesion [62]. Then it was identified as a c-di-GMP effector involved in Pel exopolysaccharide biosynthesis [35] and its pivotal role for production of biofilm matrix components such as cellulose as well as the regulation of flagellar motility has been well documented [63,64,65,66]. Most of these c-di-GMP-regulated machineries have been identified in At. thiooxidans. As reported for At. caldus [25], At. thiooxidans also possesses a bcsAB operon involved in cellulose biosynthesis. Moreover, the transcription level of bcsA gene (ATHIO_RS0101475), encoding for the cellulose synthase catalytic subunit (BcsA), involved in the regulation of cellulose biosynthesis through the binding of c-di-GMP to its PilZ domain [28,33,67,68] was increased in At. thiooxidans S0-biofilm cells compared to planktonic ones (Figure 5) suggesting that cellulose participates to the biofilm architecture in this Acidithiobacillus species. In addition, a flagellar encoding operon has been also identified in At. thiooxidans [46]. Thus, the identification of two transcribed fleQ-like genes in At. thiooxidansT (Figure S1) strongly points out that in addition to Pel exopolysaccharide, c-di-GMP levels could regulate the biosynthesis of cellulose and flagella in this Acidithiobacillus species. Interestingly, At. ferrooxidans is a primary colonizer that increases mineral colonization by sulfur-oxidizing species such as At. thioooxidans [69] but it does not have the genetic capacity to produce Pel exopolysaccharide, cellulose and flagellum (Table S3). This suggests that capsular exopolysaccharide whose expression is induced in At. ferrooxidans cells attached to mineral [14] and/or a not yet identified matrix component have to play a pivotal role for biofilm formation by this iron/sulfur-oxidizing species.
Figure 5.
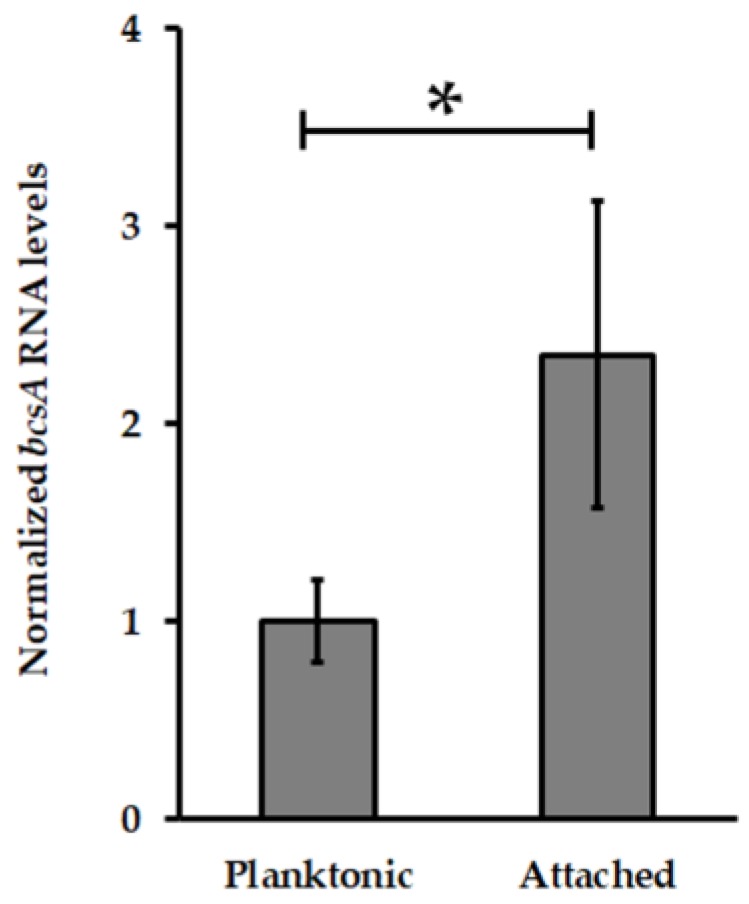
Transcription levels analysis of BcsA (cellulose synthase catalytic subunit) encoding gene from At. thiooxidansT. Transcript levels of bcsA were measured by qPCR and then normalized using DNA 16S and map genes. Values represent the average of four independent experiments ± standard deviation. Significant differences made by a one-way ANOVA test (p < 0.05) are noted (*).
Due to the huge difficulties for genetic manipulation of Acidithiobacillus spp., very few (up to five) knockout mutant strains have been reported [54,70,71,72,73]. Recently, an Acidithiobacillus DGC-defective mutant strain has been developed by our research group [25]. This At. caldus DGC null-mutant strain revealed that c-di-GMP pathway is directly involved in the regulation of motility and adherence to sulfur surfaces in this Acidithiobacillus species. However, the identification of molecular players connecting the decreased amounts of c-di-GMP intracellular levels with phenotypical observations is still an open question. Castro et al. [25] hypothesized that Pel exopolysaccharide should be involved in biofilm formation by Acidithiobacillus species that can only oxidize RISCs, namely At. caldus and At. thiooxidans and this hypothesis was tested. The results obtained from EPS quantification (Table 1) and qPCR assays demonstrating that transcription levels of several genes from pel operon including the c-di-GMP effector protein encoding gene pelD were enhanced in biofilm cells compared to planktonic cells (Figure 3), as well as the mutagenesis and SEM experiments allowing the visualization of a filamentous structure which was overexpressed in ΔpelD null-mutant strain compared to wild type strain (Figure 4) clearly revealed that Pel exopolysaccharide is involved in biofilm architecture developed by At. thiooxidans. Because the proteinaceous fraction was increased in ΔpelD null-mutant strain (Table 1) we infer that these filamentous compounds should be proteinaceous. It has been reported that flagellum and amyloid curli fibers are involved in macrobiofilm architecture developed by E. coli [74,75]. Bioinformatic search on At. thiooxidansT genome sequence revealed that canonical csgD-like and csgBAC-like genes involved in the synthesis of the most characterized amyloid fibers are absent from all Acidithiobacillus genomes. This suggests that Acidithiobacillus species either do not produce amyloid fibers or produce some yet uncharacterized amyloid fibers related to its acidophilic lifestyle. In contrast, predicted genes for flagella formation has been identified in At. thiooxidans, At. caldus and At. ferrivorans but not in At. ferrooxidans [17,46]. On the other way, because we demonstrated that the transcription level of bcsA gene are increased in biofilm cells (Figure 5), this overproduced filamentous structure could be a mesh of flagella used as scaffold for the formation of cellulose filaments as it has been reported in E. coli macrocolony biofilm [75]. However further studies are still necessary to decipher the nature of this filamentous structure and the molecular network involved in its biosynthesis.
Acknowledgments
We thank Mario Vera for critical reviewing the manuscript. M.D. acknowledges CONICYT to support his doctoral studies (scholarship 21120064, 2012). This work was supported by FONDECYT project 1160702. N.G. and S.C. are supported by Universidad de Chile (UCH).
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4425/9/2/113/s1, Table S1: Strains and plasmids used in this work, Table S2: Primers used in this work, Table S3: Type IV pilin-like protein and pili apparatus subunits encoding genes in At. thioooxidansT. Table S4: Putative molecular players for biofilm architecture identified in two iron/sulfur- and two sulfur-oxidizing species of Acidithiobacillus, Figure S1: RT-PCR analysis of c-di-GMP metabolism and effectors encoding genes identified in At. thiooxidansT, Figure S2: Domain organization of At. thiooxidans ATCC 19377 proteins involved in c-di-GMP metabolism, Figure S3: Comparative analysis of pel operon structures, Figure S4: PCR analysis of At. thiooxidans ATCC 19377 and the four double recombinant ΔpelD mutant strains to check double-recombination, Figure S5: Southern blot analysis of At. thiooxidans ATCC 19377 and the four double recombinant ΔpelD mutant strains.
Author Contributions
N.G. conceived the experiments. M.D. designed and performed experiments. M.C. contributed to genome analysis and red congo assays. Chemical analysis of c-di-GMP were done by M.D. in collaboration with S.C. Manuscript was written by N.G. and M.D. M.C. and S.C. reviewed and improved the manuscript quality.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Vera M., Schippers A., Sand W. Progress in bioleaching: Fundamentals and mechanisms of bacterial metal sulfide oxidation-part A. Appl. Microbiol. Biotechnol. 2013;97:7529–7541. doi: 10.1007/s00253-013-4954-2. [DOI] [PubMed] [Google Scholar]
- 2.Johnson D.B. Biomining-biotechnologies for extracting and recovering metals from ores and waste materials. Curr. Opin. Biotechnol. 2014;30:24–31. doi: 10.1016/j.copbio.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Johnson D.B., Hallberg K.B. Acid mine drainage remediation options: A review. Sci. Total Environ. 2005;338:3–14. doi: 10.1016/j.scitotenv.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Rohwerder T., Sand W. Oxidation of Inorganic Sulfur Compounds in Acidophilic Prokaryotes. Eng. Life Sci. 2007;7:301–309. doi: 10.1002/elsc.200720204. [DOI] [Google Scholar]
- 5.Harneit K., Göksel A., Kock D., Klock J.-H., Gehrke T., Sand W. Adhesion to metal sulfide surfaces by cells of Acidithiobacillus ferrooxidans, Acidithiobacillus thiooxidans and Leptospirillum ferrooxidans. Hydrometallurgy. 2006;83:245–254. doi: 10.1016/j.hydromet.2006.03.044. [DOI] [Google Scholar]
- 6.Africa C.J., van Hille R.P., Harrison S.T. Attachment of Acidithiobacillus ferrooxidans and Leptospirillum ferriphilum cultured under varying conditions to pyrite, chalcopyrite, low-grade ore and quartz in a packed column reactor. Appl. Microbiol. Biotechnol. 2013;97:1317–1324. doi: 10.1007/s00253-012-3939-x. [DOI] [PubMed] [Google Scholar]
- 7.Baker B.J., Banfield J.F. Microbial communities in acid mine drainage. FEMS Microbiol. Ecol. 2003;44:139–152. doi: 10.1016/S0168-6496(03)00028-X. [DOI] [PubMed] [Google Scholar]
- 8.Rawlings D.E. Characteristics and adaptability of iron- and sulfur-oxidizing microorganisms used for the recovery of metals from minerals and their concentrates. Microb. Cell Fact. 2005;4:13. doi: 10.1186/1475-2859-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nuñez H., Moya-Beltrán A., Covarrubias P.C., Issotta F., Cárdenas J.P., González M., Atavales J., Acuña L.G., Johnson D.B., Quatrini R. Molecular Systematics of the Genus Acidithiobacillus: Insights into the Phylogenetic Structure and Diversification of the Taxon. Front. Microbiol. 2017;8:30. doi: 10.3389/fmicb.2017.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelly D.P., Wood A.P. Reclassification of some species of Thiobacillus to the newly designated genera Acidithiobacillus gen. nov., Halothiobacillus gen. nov. and Thermithiobacillus gen. nov. Int. J. Syst. Evol. Microbiol. 2000;50:511–516. doi: 10.1099/00207713-50-2-511. [DOI] [PubMed] [Google Scholar]
- 11.Hallberg K.B., González-Toril E., Johnson D.B. Acidithiobacillus ferrivorans, sp. nov.; facultatively anaerobic, psychrotolerant iron-, and sulfur-oxidizing acidophiles isolated from metal mine-impacted environments. Extremophiles. 2010;14:9–19. doi: 10.1007/s00792-009-0282-y. [DOI] [PubMed] [Google Scholar]
- 12.Hedrich S., Johnson D.B. Acidithiobacillus ferridurans sp. nov., an acidophilic iron-, sulfur- and hydrogen-metabolizing chemolithotrophic gammaproteobacterium. Int. J. Syst. Evol. Microbiol. 2013;63:4018–4025. doi: 10.1099/ijs.0.049759-0. [DOI] [PubMed] [Google Scholar]
- 13.Falagán C., Johnson D.B. Acidithiobacillus ferriphilus sp. nov., a facultatively anaerobic iron- and sulfur-metabolizing extreme acidophile. Int. J. Syst. Evol. Microbiol. 2016;66:206–211. doi: 10.1099/ijsem.0.000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bellenberg S., Leon-Morales C.F., Sand W., Vera M. Visualization of capsular polysaccharide induction in Acidithiobacillus ferrooxidans. Hydrometallurgy. 2012;129–130:82–89. doi: 10.1016/j.hydromet.2012.09.002. [DOI] [Google Scholar]
- 15.Barahona S., Dorador C., Zhang R., Aguilar P., Sand W., Vera M., Remonsellez F. Isolation and characterization of a novel Acidithiobacillus ferrivorans strain from the Chilean Altiplano: Attachment and biofilm formation on pyrite at low temperature. Res. Microbiol. 2014;165:782–793. doi: 10.1016/j.resmic.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 16.Zhang R., Bellenberg S., Neu T.R., Sand W., Vera M. The Biofilm Lifestyle of Acidophilic Metal/Sulfur-Oxidizing Microorganisms. In: Rampelotto P., editor. Biotechnology of Extremophiles Advances and Challenges. Volume 1. Springer; Cham, Switzerland: 2016. pp. 177–213. [Google Scholar]
- 17.Liljeqvist M., Ossandon F.J., González C., Rajan S., Stell A., Valdes J., Holmes D.S., Dopson M. Metagenomic analysis reveals adaptations to a cold-adapted lifestyle in a low-temperature acid mine drainage stream. FEMS Microbiol. Ecol. 2015;91 doi: 10.1093/femsec/fiv011. [DOI] [PubMed] [Google Scholar]
- 18.Menzel P., Gudbergsdóttir S.R., Rike A.G., Lin L., Zhang Q., Contursi P., Moracci M., Kristjansson J.K., Bolduc B., Gavrilov S., et al. Comparative Metagenomics of Eight Geographically Remote Terrestrial Hot Springs. Microb. Ecol. 2015;70:411–424. doi: 10.1007/s00248-015-0576-9. [DOI] [PubMed] [Google Scholar]
- 19.Chen L.X., Hu M., Huang L.N., Hua Z.S., Kuang J.L., Li S.J., Shu W.S. Comparative metagenomic and metatranscriptomic analyses of microbial communities in acid mine drainage. ISME J. 2015;9:1579–1592. doi: 10.1038/ismej.2014.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farah C., Vera M., Morin D., Haras D., Jerez C.A., Guiliani N. Evidence for a functional quorum-sensing type AI-1 system in the extremophilic bacterium Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 2005;71:7033–7040. doi: 10.1128/AEM.71.11.7033-7040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez A., Bellenberg S., Mamani S., Ruiz L., Echeverria A., Soulere L., Doutheau A., Demergasso C., Sand W., Queneau Y., et al. AHL signaling molecules with a large acyl chain enhance biofilm formation on sulfur and metal sulfides by the bioleaching bacterium Acidithiobacillus ferrooxidans. Appl. Microbiol. Biotechnol. 2013;97:3729–3737. doi: 10.1007/s00253-012-4229-3. [DOI] [PubMed] [Google Scholar]
- 22.Mamani S., Moiner D., Denis Y., Soulere L., Queneau Y., Talla E., Bonnefoy V., Guiliani N. Insights into the Quorum Sensing Regulon of the Acidophilic Acidithiobacillus ferrooxidans Revealed by Transcriptomic in the Presence of an Acyl Homoserine Lactone Superagonist Analog. Front. Microbiol. 2016;7:1365. doi: 10.3389/fmicb.2016.01365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valdes J., Pedroso I., Quatrini R., Holmes D.S. Comparative genome analysis of Acidithiobacillus ferrooxidans, A. thiooxidans and A. caldus: Insights into their metabolism and ecophysiology. Hydrometallurgy. 2008;94:180–184. doi: 10.1016/j.hydromet.2008.05.039. [DOI] [Google Scholar]
- 24.Ruiz L.M., Castro M., Barriga A., Jerez C.A., Guiliani N. The extremophile Acidithiobacillus ferrooxidans possesses a c-di-GMP signalling pathway that could play a significant role during bioleaching of minerals. Lett. Appl. Microbiol. 2012;54:133–139. doi: 10.1111/j.1472-765X.2011.03180.x. [DOI] [PubMed] [Google Scholar]
- 25.Castro M., Deane S.M., Ruiz L., Rawlings D.E., Guiliani N. Diguanylate cyclase null mutant reveals that c-di-GMP pathway regulates the motility and adherence of the extremophile bacterium Acidithiobacillus caldus. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0116399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Römling U., Galperin M.Y., Gomelsky M. Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jenal U., Reinders A., Lori C. Cyclic di-GMP: Second messenger extraordinaire. Nat. Rev. Microbiol. 2017;15:271–284. doi: 10.1038/nrmicro.2016.190. [DOI] [PubMed] [Google Scholar]
- 28.Ross P., Weinhouse H., Aloni Y., Michaeli D., Weinberger-Ohana P., Mayer R., Braun S., de Vroom E., van der Marel G.A., van Boom J.H., et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 29.Lee V.T., Matewish J.M., Kessler J.L., Hyodo M., Hayakawa Y., Lory S. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 2007;65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guttenplan S.B., Kearns D.B. Regulation of flagellar motility during biofilm formation. FEMS Microbiol. Rev. 2013;37:849–871. doi: 10.1111/1574-6976.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hengge R. Trigger phosphodiesterases as a novel class of c-di-GMP effector proteins. Philos. Trans. R Soc. Lond. B Biol. Sci. 2016;371 doi: 10.1098/rstb.2015.0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sudarsan N., Lee E.R., Weinberg Z., Moy R.H., Kim J.N., Link K.H., Breaker R.R. Riboswitches in Eubacteria Sense the Second Messenger Cyclic Di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amikam D., Galperin M.Y. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 34.Newell P.D., Monds R.D., O’Toole G.A. LapD is a bis-(3′, 5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc. Natl. Acad. Sci. USA. 2009;106:3461–3466. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hickman J.W., Harwood C.S. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 2008;69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tao F., He Y.W., Wu D.H., Swarup S., Zhang L.H. The cyclic nucleotide monophosphate domain of Xanthomonas campestris global regulator Clp defines a new class of cyclic di-GMP effectors. J. Bacteriol. 2010;192:1020–1029. doi: 10.1128/JB.01253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones C.J., Utada A., Davis K.R., Thongsomboon W., Zamorano Sanchez D., Banakar V., Cegelski L., Wong G.C., Yildiz F.H. C-di-GMP Regulates Motile to Sessile Transition by Modulating MshA Pili Biogenesis and Near-Surface Motility Behavior in Vibrio cholerae. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lori C., Ozaki S., Steiner S., Böhm R., Abel S., Dubey B.N., Schirmer T., Hiller S., Jenal U. Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature. 2015;523:236–239. doi: 10.1038/nature14473. [DOI] [PubMed] [Google Scholar]
- 39.Franklin M.J., Nivens D.E., Weadge J.T., Howell P.L. Biosynthesis of the Pseudomonas aeruginosa Extracellular Polysaccharides, Alginate, Pel, and Psl. Front. Microbiol. 2011;2:167. doi: 10.3389/fmicb.2011.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vasseur P., Vallet-Gely I., Soscia C., Genin S., Filloux A. The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology. 2005;151:985–997. doi: 10.1099/mic.0.27410-0. [DOI] [PubMed] [Google Scholar]
- 41.Jennings L.K., Storek K.M., Ledvina H.E., Coulon C., Marmont L.S., Sadovskaya I., Secor P.R., Tseng B.S., Scian M., Filloux A., et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. USA. 2015;112:11353–11358. doi: 10.1073/pnas.1503058112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marmont L.S., Rich J.D., Whitney J.C., Whitfield G.B., Almblad H., Robinson H., Parsek M.R., Harrison J.J., Howell P.L. Oligomeric lipoprotein PelC guides Pel polysaccharide export across the outer membrane of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA. 2017;114:2892–2897. doi: 10.1073/pnas.1613606114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marmont L.S., Whitfield G.B., Rich J.D., Yip P., Giesbrecht L.B., Stremick C.A., Whitney J.C., Parsek M.R., Harrison J.J., Howell P.L. PelA and PelB proteins form a modification and secretion complex essential for Pel polysaccharide-dependent biofilm formation in Pseudomonas aeruginosa. J. Biol. Chem. 2017;292:19411–19422. doi: 10.1074/jbc.M117.812842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colvin K.M., Gordon V.D., Murakami K., Borlee B.R., Wozniak D.J., Wong G.C.L., Parsek M.R. The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 2011;7:e1001264. doi: 10.1371/journal.ppat.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mackintosh M.E. Nitrogen Fixation by Thiobacillus ferrooxidans. J. Gen. Microbiol. 1978;105:215–218. doi: 10.1099/00221287-105-2-215. [DOI] [Google Scholar]
- 46.Valdés J., Ossandon F., Quatrini R., Dopson M., Holmes D.S. Draft genome sequence of the extremely acidophilic biomining bacterium Acidithiobacillus thiooxidans ATCC 19377 provides insights into the evolution of the Acidithiobacillus genus. J. Bacteriol. 2011;193:7003–7004. doi: 10.1128/JB.06281-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M.A., Barrell B. Artemis: Sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 48.Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A., Salazar G.A., Tate J., Bateman A. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016;44:279–285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sigrist C.J.A., de Castro E., Cerutti L., Cuche B.A., Hulo N., Bridge A., Bougueleret L., Xenarios I. New and continuing developments at PROSITE. Nucleic Acids Res. 2012;40:344–347. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 51.McWilliam H., Li W., Uludag M., Squizzato S., Park Y.M., Buso N., Cowley A.P., Lopez R. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 2013;41:597–600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nieto P.A., Covarrubias P.C., Jedlicki E., Holmes D.S., Quatrini R. Selection and evaluation of reference genes for improved interrogation of microbial transcriptomes: Case study with the extremophile Acidithiobacillus ferrooxidans. BMC Mol. Biol. 2009;10:63. doi: 10.1186/1471-2199-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Antoniani D., Bocci P., Maciąg A., Raffaelli N., Landini P. Monitoring of diguanylate cyclase activity and of cyclic-di-GMP biosynthesis by whole-cell assays suitable for high-throughput screening of biofilm inhibitors. Appl. Microbiol. Biotechnol. 2010;85:1095–1104. doi: 10.1007/s00253-009-2199-x. [DOI] [PubMed] [Google Scholar]
- 54.Van Zyl L.J., van Muster J.M., Rawlings D.E. Construction of arsB and tetH mutants of the sulfur-oxidizing bacterium Acidithiobacillus caldus by marker exchange. Appl. Environ. Microbiol. 2008;74:5686–5694. doi: 10.1128/AEM.01235-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dubois M., Gilles K.A., Hamilton J.K., Rebers P.A., Smith F. Colorimetric Method for Determination of Sugars and Related Substances. Anal. Chem. 1956;28:350–356. doi: 10.1021/ac60111a017. [DOI] [Google Scholar]
- 56.Smith P.K., Krohn R.I., Hermanson G.T., Mallia A.K., Gartner F.H., Provenzano M.D., Fujimoto E.K., Goeke N.M., Olson B.J., Klenk D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 57.Zogaj X., Nimtz M., Rohde M., Bokranz W., RömLing U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 2001;39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
- 58.Diaz M., Guiliani N. Molecular regulatory network involved in biofilm structure development by Acidithiobacillus thiooxidans includes Pel exopolysaccharide machinery. Solid State Phenom. 2017;262:330–333. doi: 10.4028/www.scientific.net/SSP.262.330. [DOI] [Google Scholar]
- 59.Castro M., Ruiz L.M., Barriga A., Jerez C.A., Holmes D.S., Guiliani N. C-di-GMP Pathway in Biomining Bacteria. Adv. Mater. Res. 2009;71–73:223–226. doi: 10.4028/www.scientific.net/AMR.71-73.223. [DOI] [Google Scholar]
- 60.Weinberg Z. (Yale University, New Haven, CT, USA). Personal communication. 2015.
- 61.Wang Y.C., Chin K.H., Tu Z.L., He J., Jones C.J., Sanchez D.Z., Yildiz F.H., Galperin M.Y., Chou S.H. Nucleotide binding by the widespread high-affinity cyclic di-GMP receptor MshEN domain. Nat. Commun. 2016;7:12481. doi: 10.1038/ncomms12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arora S.K., Ritchings B.W., Almira E.C., Lory S., Ramphal R. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J. Bacteriol. 1997;179:5574–5581. doi: 10.1128/jb.179.17.5574-5581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baraquet C., Harwood C.S. Cyclic diguanosine monophosphate represses bacterial flagella synthesis by interacting with the Walker A motif of the enhancer-binding protein FleQ. Proc. Natl. Acad. Sci. USA. 2013;110:18478–18483. doi: 10.1073/pnas.1318972110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Srivastava D., Hsieh M.L., Khataokar A., Neiditch M.B., Waters C.M. Cyclic di-GMP inhibits Vibrio cholerae motility by repressing induction of transcription and inducing extracellular polysaccharide production. Mol. Microbiol. 2013;90:1262–1276. doi: 10.1111/mmi.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiao Y., Nie H., Liu H., Luo X., Chen W., Huang Q. C-di-GMP regulates the expression of lapA and bcs operons via FleQ in Pseudomonas putida KT2440. Environ. Microbiol. Rep. 2016 doi: 10.1111/1758-2229.12419. [DOI] [PubMed] [Google Scholar]
- 66.Jiménez-Fernández A., López-Sánchez A., Jiménez-Díaz L., Navarrete B., Calero P., Platero A.I., Govantes F. Complex Interplay between FleQ, Cyclic Diguanylate and Multiple σ Factors Coordinately Regulates Flagellar Motility and Biofilm Development in Pseudomonas putida. PLoS ONE. 2016;11:e0163142. doi: 10.1371/journal.pone.0163142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chang A.L., Tuckerman J.R., Gonzalez G., Mayer R., Weinhouse H., Volman G., Amikam D., Benziman M., Gilles-Gonzalez M.A. Phosphodiesterase A1, a Regulator of Cellulose Synthesis in Acetobacter. xylinum, is a Heme-Based Sensor. Biochemistry. 2001;40:3420–3426. doi: 10.1021/bi0100236. [DOI] [PubMed] [Google Scholar]
- 68.García B., Latasa C., Solano C., García del Portillo F., Gamazo C., Lasa I. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol. Microbiol. 2004;54:264–277. doi: 10.1111/j.1365-2958.2004.04269.x. [DOI] [PubMed] [Google Scholar]
- 69.Bellenberg S., Díaz M., Noël N., Sand W., Poetsch A., Guiliani N., Vera M. Biofilm formation, communication and interactions of leaching bacteria during colonization of pyrite and sulfur surfaces. Res. Microbiol. 2014;165:773–781. doi: 10.1016/j.resmic.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 70.Liu Z., Guiliani N., Appia-Ayme C., Borne F., Ratouchniak J., Bonnefoy V. Construction and characterization of a recA mutant of Thiobacillus. ferrooxidans by marker exchange mutagenesis. J. Bacteriol. 2000;182:2269–2276. doi: 10.1128/JB.182.8.2269-2276.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang H., Liu X., Liu S., Yu Y., Lin J., Lin J., Pang X., Zhao J. Development of a markerless gene replacement system for Acidithiobacillus. ferrooxidans and construction of a pfkB mutant. Appl. Environ. Microbiol. 2012;78:1826–1835. doi: 10.1128/AEM.07230-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu Y., Liu X., Wang H., Li X., Lin J. Construction and characterization of tetH overexpression and knockout strains of Acidithiobacillus ferrooxidans. J. Bacteriol. 2014;196:2255–2264. doi: 10.1128/JB.01472-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wen Q., Liu X., Wang H., Lin J. A versatile and efficient markerless gene disruption system for Acidithiobacillus thiooxidans: Application for characterizing a copper tolerance related multicopper oxidase gene. Environ. Microbiol. 2014;16:3499–3514. doi: 10.1111/1462-2920.12494. [DOI] [PubMed] [Google Scholar]
- 74.Serra D.O., Richter A.M., Klauck G., Mika F., Hengge R. Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. mBio. 2013;4:e00103–e00113. doi: 10.1128/mBio.00103-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Serra D.O., Richter A.M., Hengge R. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J. Bacteriol. 2013;195:5540–5554. doi: 10.1128/JB.00946-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.