Abstract
Cancers are caused by mutations that may be inherited, induced by environmental factors, or result from DNA replication errors (R). We studied the relationship between the number of normal stem cell divisions and the risk of 17 cancer types in 69 countries throughout the world. The data revealed a strong correlation (median = 0.80) between cancer incidence and normal stem cell divisions in all countries, regardless of their environment. The major role of R mutations in cancer etiology was supported by an independent approach, based solely on cancer genome sequencing and epidemiological data, which suggested that R mutations are responsible for two-thirds of the mutations in human cancers. All of these results are consistent with epidemiological estimates of the fraction of cancers that can be prevented by changes in the environment. Moreover, they accentuate the importance of early detection and intervention to reduce deaths from the many cancers arising from unavoidable R mutations.
It is now widely accepted that cancer is the result of the gradual accumulation of driver gene mutations that successively increase cell proliferation (1–3). But what causes these mutations? The role of environmental factors (E) in cancer development has long been evident from epidemiological studies, and this has fundamental implications for primary prevention. The role of heredity (H) has been conclusively demonstrated from both twin studies (4) and the identification of the genes responsible for cancer predisposition syndromes (3, 5). We recently hypothesized that a third source—mutations due to the random mistakes made during normal DNA replication (R)—can explain why cancers occur much more commonly in some tissues than others (6). This hypothesis was based on our observation that, in the United States, the lifetime risks of cancer among 25 different tissues were strongly correlated with the total number of divisions of the normal stem cells in those tissues (6, 7). It has been extensively documented that approximately three mutations occur every time a normal human stem cell divides (8, 9). We therefore inferred that the root causes of the correlation between stem cell divisions and cancer incidence were the driver gene mutations that randomly result from these divisions. Recent evidence from mouse models supports the notion that the number of normal cell divisions dictates cancer risk in many organs (10).
This hypothesis has generated much scientific and public debate and confusion, in part because our analysis was confined to explaining the relative risk of cancer among tissues rather than the contribution of each of the three potential sources of mutations (E, H, and R) to any single cancer type or cancer case. Determination of the contributions of E, H, and R to a cancer type or cancer case is challenging. In some patients, the contribution of H or R factors might be high enough to cause all the mutations required for that patient’s cancer, whereas in others, some of the mutations could be due to H, some to R, and the remainder to E. Here we perform a critical evaluation of the hypothesis that R mutations play a major role in cancer. Our evaluation is predicated on the expectation that the number of endogenous mutations (R) resulting from stem cell divisions in a tissue, unlike those caused by environmental exposures, would be similarly distributed at a given age across human populations. Though the number of stem cell divisions may vary with genetic constitution (e.g., taller individuals may have more stem cells), these divisions are programmed into our species’ developmental patterns. In contrast, deleterious environmental and inherited factors, either of which can directly increase the mutation rate or the number of stem cell divisions, vary widely among individuals and across populations.
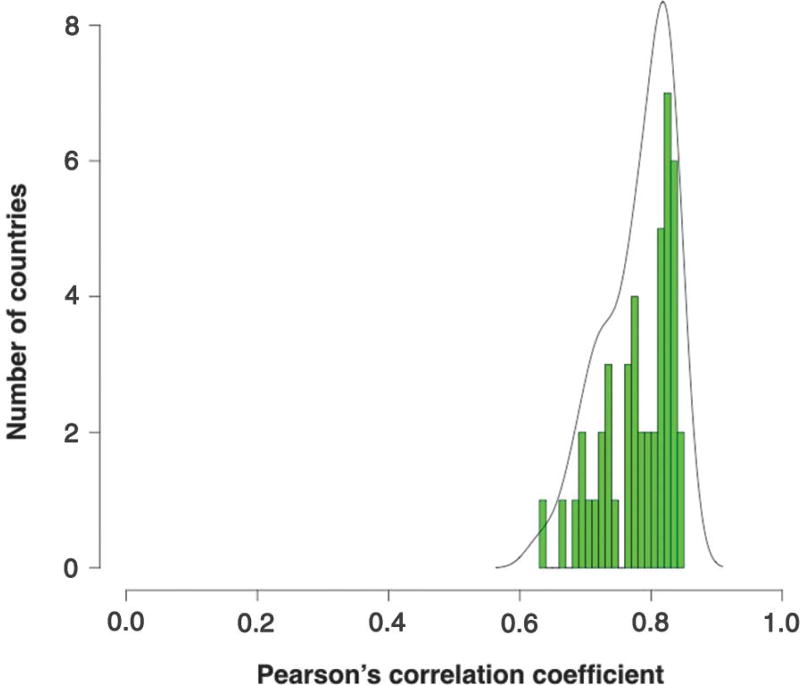
Our previous analyses were confined to the U.S. population, which could be considered to be exposed to relatively uniform environmental conditions (6). In this study, we have evaluated cancer incidence in 69 countries, representing a variety of environments distributed throughout the world and representing 4.8 billion people (two-thirds of the world’s population). Cancer incidences were determined from analysis of 423 cancer registries that were made available by the International Agency for Research on Cancer (IARC) (http://ci5.iarc.fr/CI5-X/Pages/download.aspx). All 17 different cancer types recorded in the IARC database for which stem cell data are available were used for this analysis (see supplementary materials). The Pearson’s correlation coefficients of the lifetime risk of cancer in a given tissue with that tissue’s lifetime number of stem cell divisions are shown in Fig. 1. Strong, statistically significant correlations were observed in all countries examined (median P value = 1.3 × 10−4; full range: 2.2 × 10−5 to 6.7 × 10−3). The median correlation was 0.80 (95% range: 0.67 to 0.84), with 89% of the countries having correlations >0.70 in the 0 to 85+ age interval (Table 1). This correlation of 0.80 is nearly identical to that observed for a somewhat different set of tissues, which did not include those of the breast or prostate, in the U.S. population (6). Details of the incidence data and correlations for each evaluated country and registry are provided in tables S1 to S4.
Fig. 1. Correlations between stem cell divisions and cancer incidence in different countries.
For each country, the correlation between the number of stem cell divisions in 17 different tissues and the lifetime incidence of cancer in those tissues was calculated. This resulted in correlation coefficients, which were grouped and plotted into a histogram. In this histogram, the x axis represents the correlation coefficients and the y axis represents the number of countries with the corresponding correlation coefficient. For example, there were seven countries in which the correlation between the number of stem cell divisions and cancer incidence was between 0.82 and 0.83; these seven countries are represented by the tallest green bar in the histogram. The median correlation coefficient over all countries was 0.8. The black line represents the density for the observed distribution of the correlation coefficient among different countries.
Table 1. Correlations between the lifetime risk of cancers in 17 tissues and the lifetime number of stem cell divisions in those tissues.
The median Pearson’s correlation coefficients and 95% range in various geographic regions are listed. The values in columns CR 0–85+, CR 0–85, CR 0–80, and CR 0–75 represent the correlations when the lifetime risk of each cancer (cumulative risk, CR) could be determined from birth to age 85+, birth to age 85, birth to age 80, and birth to age 75, respectively. No cancer incidence data were available for individuals older than 80 years in African countries (tables S1 to S4). NA, not applicable.
| Geographic regions | CR 0–85+ | CR 0–85 | CR 0–80 | CR 0–75 |
|---|---|---|---|---|
| Overall | 0.80 | 0.78 | 0.76 | 0.75 |
| (0.67–0.84) | (0.67–0.83) | (0.64–0.82) | (0.63–0.81) | |
| North America | 0.81 | 0.79 | 0.78 | 0.76 |
| (0.80–0.81) | (0.79–0.80) | (0.77–0.78) | (0.75–0.76) | |
| Latin America and Caribbean | 0.73 | 0.72 | 0.70 | 0.66 |
| (0.69–0.78) | (0.67–0.77) | (0.64–0.75) | (0.63–0.73) | |
| Europe | 0.82 | 0.81 | 0.80 | 0.78 |
| (0.74–0.84) | (0.74–0.83) | (0.72–0.82) | (0.70–0.81) | |
| Asia | 0.72 | 0.71 | 0.70 | 0.67 |
| (0.64–0.77) | (0.63–0.78) | (0.62–0.77) | (0.60–0.77) | |
| Africa | NA | NA | 0.72 | 0.72 |
| (0.71–0.76) | (0.69–0.74) | |||
| Oceania | 0.83 | 0.82 | 0.81 | 0.79 |
| (0.82–0.83) | (0.81–0.83) | (0.80–0.81) | (0.78–0.79) |
The correlations in Fig. 1 were derived for the largest age interval available (0 to 85+ in Table 1). Data on individuals from the same countries at younger ages indicate that the larger the age range considered, the higher the correlation. Note that cancer incidence increases exponentially with age (11), but stem cell divisions do not increase proportionally with age in tissues with low or no cell turnover, such as bone and brain. An increase in the evaluated age range would therefore be expected to be associated with an increase in the correlation between the lifetime number of stem cell divisions and cancer incidence, as was observed (Table 1).
The universally high correlations between normal stem cell divisions and cancer incidences shown in Table 1 are surprising given the voluminous data indicating large differences in exposures to environmental factors and associated cancer incidences across the world (12–16). To explore the basis for this apparent discrepancy, we sought to determine what fractions of cancer-causing mutations result from E, H, or R. As these fractions have not been estimated for any cancer type, we developed an approach to achieve this goal. A theoretical example that illustrates the underlying conceptual basis of this approach is as follows. Imagine that a population of humans in which all inherited mutations have been corrected move to Planet B, where the environment is perfect. On this planet, E and H are zero, and the only somatic mutations are caused by R. Note that the number of R mutations in all tissues is >0, regardless of the environment, because perfect, error-free replication is incompatible with basic biologic principles of evolution. Suppose that a powerful mutagen, E, was then introduced into the environment of Planet B, and all inhabitants of Planet B were equally exposed to it throughout their lifetimes. Assume that this mutagen substantially increased the somatic mutation rate in normal stem cells, causing a 10-fold increase in cancer risk, i.e., 90% of all cancer cases on this planet were now attributable to E. Therefore, 90% of all cancer cases on Planet B would be preventable by avoiding exposure to E. But even in this environment, it can be shown that 40% of the driver gene mutations are attributable to R (Fig. 2A and supplementary materials). This extreme example demonstrates that even if the vast majority of cancer cases were preventable by reducing exposure to environmental factors, a large fraction of the driver gene mutations required for those cancers can still be due to R—as long as the number of mutations contributed by normal stem cell divisions is not zero. In other words, the preventability of cancers and the etiology of the driver gene mutations that cause those cancers are related but have different metrics (see supplementary materials for their mathematical relationship).
Fig. 2. Mutation etiology and cancer prevention in a hypothetical scenario and in real life.
Patients exposed to environmental factors, such as cigarette smoke, are surrounded by a cloud. The driver gene mutations calculated to be attributable to environmental (E), hereditary (H), and replicative (R) factors are depicted as gray, blue (containing an “H”), and yellow circles, respectively. For simplicity, each cancer patient is shown as having three driver gene mutations, but the calculations are based on percentages. Thus, if there are three driver gene mutations in a cancer and R accounts for 33% of them, then one mutation is assigned to R. (A) A hypothetical scenario consisting of an imaginary place, Planet B, where all inherited mutations have been corrected and where the environment is perfect. A powerful mutagen is then introduced that increases cancer risk 10-fold, so that 90% of cancers on this planet are preventable. In some individuals on this planet, all mutations are due to E, whereas in the two individuals in the bottom right corner, all are due to R. In the other patients, only some of the somatic mutations in their tumors result from E. Even though 90% of the cancers are preventable by eliminating the newly introduced mutagen, 40% of the total driver gene mutations are due to R. (B to D) Real-life examples of mutation etiology and cancer prevention. (B) The approximate proportion of driver gene mutations in lung adenocarcinomas that are due to environmental versus nonenvironmental factors are shown as gray and yellow circles, respectively. Even though 89% of lung adenocarcinomas are preventable (17) by eliminating E factors, we calculate that 35% (95% CI: 30 to 40%) of total driver gene mutations are due to factors unrelated to E or H and presumably are due to R. (C) The approximate proportion of driver gene mutations in pancreatic ductal adenocarcinomas, in which hereditary factors are known to play a role. It has been estimated that ~37% (17) of pancreatic ductal adenocarcinomas are preventable, but at most 18% and 5% of the driver gene mutations in these cancers are estimated to be due to E and H, respectively. The remaining 77% (95% CI: 82 to 94%) of the total driver gene mutations are due to factors other than E or H, presumably R. (D) The approximate proportion of driver gene mutations in prostate cancers, in which environmental factors are thought to play essentially no role (17) and hereditary factors account for 5 to 9% of cases (see supplementary materials). None of these cancers are preventable, and less than 5% of the driver mutations in these cancers are due to E or H. The remaining 95% of the total driver gene mutations are due to factors other than E or H, presumably R. [Image: The Johns Hopkins University]
This theoretical example is not very different from what occurs on Earth with respect to the etiology of the most common form of lung cancer, adenocarcinoma. Epidemiologic studies have estimated that nearly 90% of adenocarcinomas of the lung are preventable and that tobacco smoke is by far the major component of E. Secondhand smoking, occupational exposures, ionizing radiation, air pollution, and diet play important but smaller roles (17, 18). Moreover, no hereditary factors have been implicated in lung adenocarcinomas (19). To determine the fraction of mutations attributable to nonenvironmental and nonhereditary causes in lung adenocarcinomas, we developed an approach based on the integration of genome-wide sequencing and epidemiological data. The key insight is the recognition of a relationship between mutation rates in a cancer type and the etiology of the somatic mutations that are detected in that cancer. Specifically, if an environmental factor causes the normal somatic mutation rate to increase by a factor x, then (x−1)/x of the somatic mutations found in a cancer can be attributed to that environmental factor (see supplementary materials). For example, if patients exposed to a factor E have a mutation rate that is three times higher than that of patients not exposed to it, then two-thirds of the mutations in the exposed patients can be attributed to factor E. This method is completely independent of any data or knowledge about normal stem cell divisions. We applied this approach to representative patients with lung adenocarcinomas as depicted in Fig. 2B. In 8 of the 20 depicted patients, all of the driver gene mutations can be attributed to E. In 10 of the 20 depicted patients, a portion of the driver gene mutations are attributable to E. And in the two patients depicted on the bottom right of Fig. 2B, none of the driver gene mutations are attributable to E. We calculate that 35% [95% confidence interval (CI): 30 to 40%] of the total driver gene mutations are due to factors that, according to current exhaustive epidemiologic studies, are unrelated to H or E and thus are presumably due to R. These data are based on conservative assumptions about the risk contributed by factors other than smoking. For example, we assumed that the increase in mutations resulting from poor diet is identical to the increase resulting from smoking cigarettes. Thus, Cancer Research UK estimates that the great majority (89%) of lung adenocarcinoma cases are preventable (17), but even so, more than a third (35%) of the driver gene mutations in lung cancers can be attributed to R.
This same analytic approach can be applied to cancer types for which epidemiologic studies have indicated a less pronounced role of environmental factors. Figure 2C depicts pancreatic ductal adenocarcinomas. About 37% of these cancers are thought to be preventable (versus 89% for lung adenocarcinomas) (17). Using exome sequencing data and extremely conservative assumptions about the influence of environmental factors, we calculated that 18% of the driver gene mutations were due to environmental factors, at most 5% were due to hereditary factors, and the remaining 77% (95% CI: 67 to 84%) were due to nonenvironmental and nonhereditary factors, presumably R (see supplementary materials). As with lung adenocarcinomas, these results are independent of any assumptions about, or measurements of, stem cell divisions.
A third class of cancers comprise those in which only a very small effect of E or H has been demonstrated (17), such as those of the brain, bone, or prostate. For example, a very high fraction of the driver gene mutations in prostate cancers can be attributed to R (95%; Fig. 2D and supplementary materials). In the past, the causes of cancer types like these were obscure, as there was no evidence that the two most well-recognized causes of cancer—environment and heredity—play a substantial role. The recognition that a third source of mutations, i.e., those due to R, are omnipresent helps explain the pathogenesis of these malignancies. Even if future epidemiological or genetic studies identify previously unknown E or H factors that permit 90% of cancers of the prostate to be prevented, the percentage of mutations due to R will still be very high, as illustrated by the Planet B analogy in Fig. 2A.
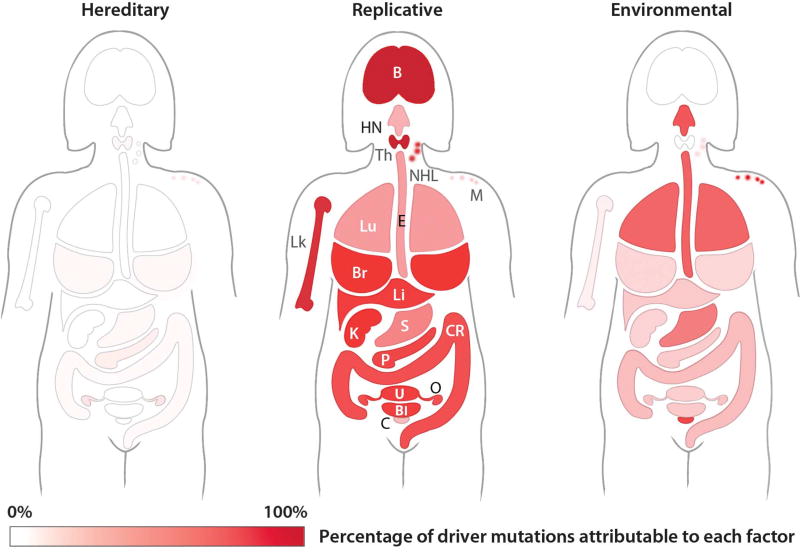
We next calculated the proportion of driver gene mutations caused by E or H in 32 cancer types (see supplementary materials and tables S5 and S6). We considered those mutations not attributable to either E or H to be due to R. These cancers have been studied in depth through sophisticated epidemiological investigations and are reported in the Cancer Research UK database [see (17, 20–22) and references therein]. For the U.K. female population, mutations attributable to E (right), R (center), and H (left) are depicted in Fig. 3 (see fig. S2 for equivalent representations of males and table S6 for the numerical values for both sexes). The median proportion of driver gene mutations attributable to E was 23% among all cancer types. The estimate varied considerably: It was greater than 60% in cancers such as those of the lung, esophagus, and skin and 15% or less in cancers such as those of the prostate, brain, and breast. When these data are normalized for the incidence of each of these 32 cancer types in the population, we calculate that 29% of the mutations in cancers occurring in the United Kingdom were attributable to E, 5% of the mutations were attributable to H, and 66% were attributable to R. Cancer Research UK estimates that 42% of these cancer cases are preventable. Given the mathematical relationship between cancer etiology and cancer preventability (see supplementary materials), the proportion of mutations caused by environmental factors is always less than the proportion of cancers preventable by avoidance of these factors. Thus, our estimate that a maximum of 29% of the mutations in these cancers are due to E is compatible with the estimate that 42% of these cancers are preventable by avoiding known risk factors.
Fig. 3. Etiology of driver gene mutations in women with cancer.
For each of 18 representative cancer types, the schematic depicts the proportion of mutations that are inherited, due to environmental factors, or due to errors in DNA replication (i.e., not attributable to either heredity or environment). The sum of these three proportions is 100%. The color codes for hereditary, replicative, and environmental factors are identical and span white (0%) to brightest red (100%). The numerical values used to construct this figure, as well as the values for 14 other cancer types not shown in the figure, are provided in table S6. B, brain; Bl, bladder; Br, breast; C, cervical; CR, colorectal; E, esophagus; HN, head and neck; K, kidney; Li, liver; Lk, leukemia; Lu, lung; M, melanoma; NHL, non-Hodgkin lymphoma; O, ovarian; P, pancreas; S, stomach; Th, thyroid; U, uterus. [Image: The Johns Hopkins University]
The results described above have important ramifications for understanding the root causes of cancer as well as for minimizing deaths from this disease. Uniformly high correlations between the number of stem cell divisions and cancer risk among tissues were observed in countries with widely different environments. This strongly supports the idea that R mutations make major contributions to cancer (see also fig. S1). However, the actual contribution of R mutations to any particular cancer type cannot be reliably estimated from such correlations. The approaches described here—a combination of cancer sequencing data and conservative analyses of environmental and hereditary risk factors—provide such estimates. They indicated that even in lung adenocarcinomas, R contributes a third of the total mutations, with tobacco smoke (including secondhand smoke), diet, radiation, and occupational exposures contributing the remainder. In cancers that are less strongly associated with environmental factors, such as those of the pancreas, brain, bone, or prostate, the majority of the mutations are attributable to R.
These data and analyses should help clarify prior confusion about the relationship between replicative mutations and cancer (23–28). First, the data demonstrate that the correlation between cancer incidence and the number of stem cell divisions in various tissues cannot be explained by peculiarities of the U.S. population or its environment. This correlation is observed worldwide, as would be expected for a fundamental biological process such as stem cell divisions. Second, these results explicitly and quantitatively address the difference between cancer etiology and cancer preventability. As illustrated in Figs. 2 and 3, these concepts are not equivalent. A cancer in which 50% of the mutations are due to R can still be preventable. The reason for this is that it generally requires more than one mutation to develop the disease. A cancer that required two mutations is still preventable if one of the mutations was due to R and the other due to an avoidable environmental factor.
Our results are fully consistent with epidemiological evidence on the fraction of cancers in developed countries that are potentially preventable through improvements in environment and lifestyle. Cancer Research UK estimates that 42% of cancer cases are preventable (17); the U.S. Centers for Disease Control and Prevention estimates that 21% of annual cancer deaths in individuals <80 years old could be prevented (29).
Of equal importance, these studies provide a well-defined, molecular explanation for the large and apparently unpreventable component of cancer risk that has long puzzled epidemiologists. It is, of course, possible that virtually all mutations in all cancers are due to environmental factors, most of which have simply not yet been discovered. However, such a possibility seems inconsistent with the exhaustively documented fact that about three mutations occur every time a normal cell divides and that normal stem cells often divide throughout life.
Our studies complement, rather than oppose, those of classic epidemiology. For example, the recognition of a third, major factor (R) underlying cancer risk can inform epidemiologic studies by pointing to cancers that cannot yet be explained by R (i.e., those with too few stem cell divisions to account for cancer incidence). Such cancer types seem particularly well suited for further epidemiologic investigation. Additionally, R mutations appear unavoidable now, but it is conceivable that they will become avoidable in the future. There are at least four sources of R mutations in normal cells: quantum effects on base pairing (30), mistakes made by polymerases (31), hydrolytic deamination of bases (32), and damage by endogenously produced reactive oxygen species or other metabolites (33). The last of these could theoretically be reduced by the administration of antioxidant drugs (34). The effects of all four could, in principle, be reduced by introducing more efficient repair genes into the nuclei of somatic cells or through other creative means.
As a result of the aging of the human population, cancer is today the most common cause of death in the world (12). Primary prevention is the best way to reduce cancer deaths. Recognition of a third contributor to cancer—R mutations—does not diminish the importance of primary prevention but emphasizes that not all cancers can be prevented by avoiding environmental risk factors (Figs. 2 and 3). Fortunately, primary prevention is not the only type of prevention that exists or can be improved in the future. Secondary prevention, i.e., early detection and intervention, can also be lifesaving. For cancers in which all mutations are the result of R, secondary prevention is the only option.
Supplementary Material
Acknowledgments
We thank W. F. Anderson [Biostatistics branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute (NCI)], N. Chatterjee [Johns Hopkins University (JHU)], K. W. Kinzler (JHU), B. Mensh [Howard Hughes Medical Institute (HHMI)], and J. T. Vogelstein (JHU) for their comments. We thank A. Blackford (JHU) and R. H. Hruban (JHU) for providing the pancreatic cancer data set. This work was made possible through the support of grants from the John Templeton Foundation, the Virginia and D. K. Ludwig Fund for Cancer Research, the Lustgarten Foundation for Pancreatic Cancer Research, The Sol Goldman Center for Pancreatic Cancer Research, and NIH grants P30-CA006973, R37-CA43460, and P50-CA62924. The opinions expressed in this publication are those of the authors and do not necessarily reflect the views of the John Templeton Foundation. C.T. conceived the ideas for determining the proportions of drivers and developed the mathematical methods and their application. C.T. and B.V. designed and performed the research. C.T. performed the world data analysis. L.L. obtained the estimates in tables S5 and S6. B.V. is on the scientific advisory boards of Morphotek, Exelixis GP, and Sysmex Inostics, and is a founder of PapGene and Personal Genome Diagnostics. Morphotek, Sysmex Inostics, PapGene, and Personal Genome Diagnostics, as well as other companies, have licensed technologies from JHU on which B.V. is an inventor. These licenses and relationships are associated with equity or royalty payments to B.V.
Footnotes
C.T. and B.V. wrote the paper. C.T. and L.L. have nothing to disclose.
The terms of these arrangements are being managed by JHU in accordance with its conflict of interest policies.
REFERENCES AND NOTES
- 1.Garraway LA, Lander ES. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Stratton MR, Campbell PJ, Futreal PA. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogelstein B, et al. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mucci LA, et al. JAMA. 2016;315:68–76. doi: 10.1001/jama.2015.17703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stadler ZK, et al. J. Clin. Oncol. 2010;28:4255–4267. doi: 10.1200/JCO.2009.25.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomasetti C, Vogelstein B. Science. 2015;347:78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomasetti C, Vogelstein B. 2015 https://arxiv.org/abs/1501.05035.
- 8.Lynch M. Proc. Natl. Acad. Sci. U.S.A. 2010;107:961–968. doi: 10.1073/pnas.0912629107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomasetti C, Vogelstein B, Parmigiani G. Proc. Natl. Acad. Sci. U.S.A. 2013;110:1999–2004. doi: 10.1073/pnas.1221068110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu L, et al. Cell. 2016;166:1132–1146.e7. doi: 10.1016/j.cell.2016.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nordling CO. Br. J. Cancer. 1953;7:68–72. doi: 10.1038/bjc.1953.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart BW, Wild CP, editors. World Cancer Report 2014. IARC; Lyon, France: 2014. [Google Scholar]
- 13.Edgren G, Adami HO, Weiderpass E, Nyrén O. Gut. 2013;62:1406–1414. doi: 10.1136/gutjnl-2012-302412. [DOI] [PubMed] [Google Scholar]
- 14.Hoel DG, Wakabayashi T, Pike MC. Am. J. Epidemiol. 1983;118:78–89. doi: 10.1093/oxfordjournals.aje.a113619. [DOI] [PubMed] [Google Scholar]
- 15.Edwards BK, et al. Cancer. 2010;116:544–573. doi: 10.1002/cncr.24760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jemal A, et al. CA Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 17.Cancer Research UK. Statistics on preventable cancers. www.cancerresearchuk.org/health-professional/cancer-statistics/risk/preventable-cancers.
- 18.U.S. Office of the Surgeon General and U.S. Office on Smoking and Health. The Health Consequences of Smoking: A Report of the U.S. Surgeon General. U.S. Department of Health and Human Services; 2004. [Google Scholar]
- 19.Cancer Research UK. Lung cancer risks and causes. www.cancerresearchuk.org/about-cancer/type/lung-cancer/about/lung-cancer-risks-and-causes.
- 20.Cancer Research UK. Inherited cancer genes and increased cancer risk. www.cancerresearchuk.org/about-cancer/causes-of-cancer/inherited-cancer-genes-and-increased-cancer-risk/inherited-genes-and-cancer-types-inherited_genes4.
- 21.Cancer Research UK. Statistics by cancer type. www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type.
- 22.Parkin DM, Boyd L, Walker LC. Br. J. Cancer. 2011;105(suppl. 2):S77–S81. doi: 10.1038/bjc.2011.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashford NA, et al. Science. 2015;347:727. doi: 10.1126/science.aaa6246. [DOI] [PubMed] [Google Scholar]
- 24.Gotay C, Dummer T, Spinelli J. Science. 2015;347:728. doi: 10.1126/science.aaa6462. [DOI] [PubMed] [Google Scholar]
- 25.Potter JD, Prentice RL. Science. 2015;347:727. doi: 10.1126/science.aaa6507. [DOI] [PubMed] [Google Scholar]
- 26.Song M, Giovannucci EL. Science. 2015;347:728–729. doi: 10.1126/science.aaa6094. [DOI] [PubMed] [Google Scholar]
- 27.Tomasetti C, Vogelstein B. Science. 2015;347:729–731. doi: 10.1126/science.aaa6592. [DOI] [PubMed] [Google Scholar]
- 28.Wild C, et al. Science. 2015;347:728. doi: 10.1126/science.aaa6799. [DOI] [PubMed] [Google Scholar]
- 29.U.S. Centers for Disease Control and Prevention. Up to 40 percent of annual deaths from each of five leading US causes are preventable. www.cdc.gov/media/releases/2014/p0501-preventable-deaths.html.
- 30.Kimsey IJ, Petzold K, Sathyamoorthy B, Stein ZW, Al-Hashimi HM. Nature. 2015;519:315–320. doi: 10.1038/nature14227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunkel TA. Cold Spring Harb. Symp. Quant. Biol. 2009;74:91–101. doi: 10.1101/sqb.2009.74.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fromme JC, Verdine GL. Adv. Protein Chem. 2004;69:1–41. doi: 10.1016/S0065-3233(04)69001-2. [DOI] [PubMed] [Google Scholar]
- 33.Collins AR. Eur. J. Cancer. 2005;41:1923–1930. doi: 10.1016/j.ejca.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 34.Ferguson LR, et al. Semin. Cancer Biol. 2015;35(suppl):S5–S24. doi: 10.1016/j.semcancer.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.