Abstract
The hydrogen molecule (H2), which has low redox potential, is produced by colonic fermentation. We examined whether increased H2 concentration in the portal vein in rats fed high amylose maize starch (HAS) helped alleviate oxidative stress, and whether the transplantation of rat colonic microbiota with high H2 production can shift low H2-generating rats (LG) to high H2-generating rats (HG). Rats were fed a 20% HAS diet for 10 days and 13 days in experiments 1 and 2, respectively. After 10 days (experiment 1), rats underwent a hepatic ischemia–reperfusion (IR) operation. Rats were then categorized into quintiles of portal H2 concentration. Plasma alanine aminotransferase activity and hepatic oxidized glutathione concentration were significantly lower as portal H2 concentration increased. In experiment 2, microbiota derived from HG (the transplantation group) or saline (the control group) were orally inoculated into LG on days 3 and 4. On day 13, portal H2 concentration in the transplantation group was significantly higher compared with the control group, and positively correlated with genera Bifidobacterium, Allobaculum, and Parabacteroides, and negatively correlated with genera Bacteroides, Ruminococcus, and Escherichia. In conclusion, the transplantation of microbiota derived from HG leads to stable, high H2 production in LG, with the resultant high production of H2 contributing to the alleviation of oxidative stress.
Keywords: hydrogen, resistant starch, microbiota transplantation, antioxidant effect, rats
1. Introduction
Many bacteria—estimated at around 1013–1014—reside in the large intestine, where they have important effects on human health. Most of the bacteria are helpful, but some are harmful. Due to the low redox potential in the large intestine, many of the bacteria are anaerobes: oxygen is not used as the final electron and hydrogen acceptor. Fermentation products such as succinate, lactate, and short chain fatty acids (SCFA) act as the electron and hydrogen acceptors. A byproduct of the fermentation is hydrogen gas, some of which is excreted in the breath and flatus [1]. H2 is a stable, non-reactive product, which was classically thought to have no effect in the body. However, Osawa et al. [2] demonstrated that H2 gas has a specific antioxidant effect on hydroxyl radicals.
Excess oxidative stress, induced by perturbation of the redox balance, triggers many diseases, such as diabetes, cardiovascular disease, and chronic kidney disease. Therefore, it is important to control redox balance using foods containing antioxidants, such as ascorbic acid, α-tocopherol, and polyphenols. H2 could play an important role as an antioxidant, participating in the inhibition of some diseases, such as diabetes [3] and cardiovascular diseases [4]. We demonstrated that colonic H2 derived from various non-digestible saccharides, such as high amylose maize starch (HAS) [5], pectin [5], fructooligosaccharides [6], and inulin [6], suppressed hepatic oxidative stress and adipose inflammation in rats. Colonic H2, derived from fermentable, non-digestible saccharides, could help supply large amounts of H2 for 24 h, and maintain an appropriate redox balance in the liver compared with H2 water, because it is not easy to deliver large amounts of H2 in the body by the administration of H2 water in a continuous manner. However, the amount of H2 generated by colonic microbes varies, because colonic microbiota in humans [7] and in rats [5] differ among individuals.
Net H2 production is determined by the balance between H2-producing and H2-utilizing microbes in the large intestine [8]. The microbiota of laboratory animals have been reported to differ among breeders and breeding colonies [9], and fermentation patterns may vary, even with the administration of the same fermentation substrate. The differences would be dependent on maternal microbiota in the breeder and colony. During our preliminary experiments, H2 concentration in the portal vein increased only slightly in rats, even after the administration of HAS. These rats were assumed to be low H2-generating (LG). The production of different amounts of H2 among individual rats is suggested to be dependent on colonic microbiota. The formation of high H2-producing microbiota could provide a preventive effect against certain diseases, because microbiota derived from high H2-generating rats (HG) could contribute to the alleviation of oxidative stress. Recently, investigators have attempted to change the inherent colonic microbiota in humans [10] and rodents [11] by fecal microbiota transplantation in order to inhibit certain diseases.
In the present study, we examined whether different H2 production amounts in rats fed HAS affects the alleviation of hepatic oxidative stress in a model of acute hepatic oxidative stress, and whether the transplantation of rat cecal microbiota with high H2 production could shift LG to HG.
2. Materials and Methods
2.1. High Amylose Maize Starch
High amylose maize starch (Hi-maize 1043; total dietary fiber, 64.5% [12]; resistant starch, 45.7% [12], and amylose, 70%) was kindly supplied by Nippon NSC Ltd. (Tokyo, Japan). We previously confirmed, using ileorectostomized rats, that 53% of HAS was digested in the small intestine, and the remainder reached the large intestine [13].
2.2. Animals and Diets
This study was approved by the Nayoro City University Animal Use Committee, and animals were maintained in accordance with the Guidelines for the Care and Use of Laboratory Animals, Nayoro City University (approval number 10-01 and 11-04). Eight-week-old, male Sprague-Dawley rats, weighing 210–230 g, were obtained from Japan SLC (Haruno colony or Ohara colony; Shizuoka, Japan). We used rats from the Haruno colony as HG, and those from the Ohara colony as LG, because in our preliminary experiment, we noticed that many rats from the Haruno colony produced high amounts of colonic H2, while many rats from the Ohara colony showed low H2 production. Rats were housed in individual cages with screen bottoms made of stainless steel, in a room maintained at 23 ± 2 °C, and humidity ranging from 50% to 70%, under lighting conditions of 12 h of light (0700 to 1900) and 12 h of darkness. Rats were acclimated by feeding a lab chow (CE-2, Japan Clea, Tokyo, Japan) for three days in all of the experiments.
2.3. Portal H2 Concentration and Alleviation of Oxidative Stress among Individual Rats (Experiment 1)
To clarify that even rats fed the same diet produce different amounts of colonic H2, and that the difference could affect the alleviation of oxidative stress, we examined whether higher portal H2 concentration alleviated oxidative damage in rats after a hepatic ischemia–reperfusion (IR) operation. After the acclimation period, 66 rats (33 rats from the Haruno colony and 33 from the Ohara colony) were fed a diet supplemented with 200 g of HAS per kg for 10 days. The composition of the HAS diet was as described previously [5]. At the end of the feeding period, all of the rats underwent a hepatic IR operation (ischemia time, 30 min; reperfusion time 45 min) under anesthesia, as described previously [5]. Briefly, under pentobarbital sodium anesthesia (70 mg/kg body weight ip), the hepatic artery and portal vein to the left lateral and median lobe was occluded using a bulldog clamp for 30 min while allowing blood flow through the remaining sections. The clamps were removed after ischemia, and hepatic reperfusion was initiated. Rats were killed 45 min after reperfusion by exsanguination under anesthesia. During surgery, the abdominal incision site was wrapped in plastic wrap to prevent tissues from drying out. Rats were placed over an isothermal pad to maintain their body temperature at 37 °C.
2.4. Sampling
At the end of the experimental period, 1 mL of blood from the portal vein was successively collected into sealed heparin vials and microtubes, under anesthesia (pentobarbital 50 mg/kg body weight), for H2 analysis and plasma preparation, respectively. After the incubation for 1 min at 37 °C, a 1 mL sample of the gaseous phase was withdrawn using a gas-tight syringe, and H2 concentration was determined using a gas chromatography (GC) (lower detection limit, 0.10 ppm; quantification range, 0.30–50 ppm; Biogas analyzer BAS-1000; Mitleben, Osaka, Japan). The remaining blood sample was separated by centrifugation (1200× g for 20 min at 4 °C), and plasma samples were stored at −80 °C until alanine aminotransferase (ALT) analysis. After blood withdrawal, the liver was perfused immediately with 20 mL cold saline at 4 °C via the portal vein. Immediately after perfusion, the median lobe (ischemic area) was removed, and a portion was rapidly frozen in liquid N2. Samples were stored at −80 °C until the glutathione assays. The cecum was removed and weighed, and the cecal content was collected and stored at −80 °C in air-tight tubes until SCFA analysis and bacterial DNA analysis. The cecal tissue was blotted and weighed after rinsing in an ice-cold saline solution.
2.5. Assessment of Oxidative Stress and Damage in the Liver
Hepatic reduced glutathione (GSH) and oxidized glutathione (GSSG) levels were determined using the method of Anderson [14] and Raman et al. [15]. Briefly, 1 volume of liver tissue was homogenized in 9 volumes of 5% 5-sulfosalicylic acid, and centrifuged (10,000× g for 5 min at 4 °C). The supernatant was used in the 5,5′-dithiobis(2-nitrobenzoic acid)-glutathione reductase recycling assay to determine total glutathione and GSSG concentrations. GSH concentration was calculated from the difference between total glutathione and GSSG. Plasma ALT activity was measured using a commercial kit, Transaminase CII-test (Wako Pure Chemical Industries, Tokyo, Japan), according to the manufacturer’s instructions.
2.6. SCFA Analysis
After homogenization of the cecal contents, cecal organic acids were measured using an HPLC system LC-10A (Shimadzu, Kyoto, Japan) equipped with a Shim-pack SCR-102H column, 8 mm i.d. 30 cm long (Shimadzu, Kyoto, Japan) and an electroconductivity detector CDD-6A (Shimadzu, Kyoto, Japan) [16]. Briefly, 300 mg cecal content was homogenized in 2 mL of 10 mmol/L NaOH containing 0.5 g/L crotonic acid as an internal standard, and then centrifuged at 10,000× g at 4 °C for 15 min. The supernatant was subjected to HPLC analysis after deproteinization with an equal volume of chloroform.
2.7. Preparation of Inoculum from the Cecal Contents of High H2-Generating Rats (Experiment 2)
We prepared cecal microbiota derived from HG fed the HAS diet. After the acclimation period, 12 rats (eight rats from the Haruno colony, and four from the Ohara colony) were given a diet supplemented with 200 g of HAS per kg for 7 days. At the end of the feeding period, portal H2 concentration was immediately measured in rats under anesthesia (pentobarbital 50 mg/kg body weight) using a GC analyzer, Biogas analyzer BAS-1000 (Mitleben, Osaka, Japan). Using the portal H2 concentration data, the three rats with the highest portal H2 concentration (>7.4 μmol/L) were selected as the HG, while the two rats with the lowest portal H2 concentration (<2.0 μmol/L) were selected as the LG. Rats in between were treated as the middle H2-generating rats. Rats were killed by exsanguination under anesthesia. The cecum was then immediately removed and weighed, and the cecal content was collected under a CO2 gas stream. SCFA concentration and the number of total anaerobes were assayed in each rat’s cecal contents. Cecal contents collected from HG were pooled, and then diluted 1 to 10 with pre-reduced sterile dilution buffer (pH 7.2) containing 4.5 g/L KH2PO4, 6.0 g/L K2HPO4, 0.5 g/L l-cysteine hydrochloride monohydrate and 0.5 g/L Tween 80 [17]. The suspension was centrifuged (30× g, 2 min, 4 °C) to remove debris, and the upper layer was collected as rat microbiota. Sterile glycerol was added to the upper layer to produce a 10% final glycerol concentration. The total anaerobe count, measured using the culture method described below, was 1.8 × 109 cfu/mL. The inoculum was dispensed into a vial, immediately frozen in liquid nitrogen, and stored at −80 °C until further use. The total anaerobe count in the inoculum was verified before inoculation into rats to check that the bacterial count had not been adversely affected by the storage procedure. The preparation was performed under a CO2 gas stream, and pre-reduced solutions, which were deoxygenated with CO2, were used. The same method was also used on the cecal contents from LG for comparison. Microbiota compositions were analyzed in the two inoculum, using the method described below.
2.8. Effects of Transplantation of HG Cecal Microbiota into LG on Portal H2 Concentration (Experiment 2)
We determined whether transplanting rat cecal microbiota derived from HG changed H2 production and cecal microbiota in LG. After the acclimation period (day 0), 21 rats (Ohara colony) were divided into two groups based on body weight, and all of the rats were given a diet supplemented with 200 g of HAS per kg for 13 days. All of the rats were administered omeprazole (50 mg/kg/day) through oral gavage for 3 days (days 0, 1 and 2) at the start of the experimental period to inhibit the secretion of gastric acid. After omeprazole administration, the inoculum (1.8 × 109 cfu/rat/day; 1 mL) was administered for 2 days (days 3 and 4) through oral gavage to the transplantation group (n = 11), according to the method of Manichanh et al. [18]. Sterile, pre-reduced saline was given to the control group (n = 10) using the same inoculum volume. H2 excretion (Breath + flatus) was measured using a GC analyzer (Biogas analyzer BAS-1000), as described in our previous study [5,6]. H2 excretion was determined on days 0, 3, 6, 10, and 13.
2.9. Total Anaerobe Count in the Inoculum and Cecal Contents (Experiments 2)
The cecum was obtained immediately after the collection of portal blood for analysis of H2. An aliquot of the cecal contents was immediately collected in pre-weighed sterile tubes under a CO2 gas stream, and weighed. Nine volumes of pre-reduced dilution buffer (pH 7.2), containing 6 g/L Na2HPO4, 4.5 g/L KH2PO4, 0.5 g/L l-cysteine hydrochloride monohydrate and 0.5 g/L Tween 80 [17], was added to the cecal contents before homogenization. Serial 10-fold dilutions of the homogenates were prepared from each sample. Aliquots (100 μL) of each diluent were applied to pre-reduced glucose blood liver (BL) agar (Nissui Pharmaceutical, Tokyo, Japan) as a non-selective medium for anaerobes. The plates were incubated anaerobically at 37 °C for 48 h before the bacterial colonies were counted.
2.10. Bacterial DNA Extraction from the Inoculum and the Cecal Content (Experiments 2)
Bacterial DNA from the two inoculum and the cecal contents was isolated from frozen cecal contents using a DNA extraction kit (ISOFECAL for Beads Beating kit, Nippon Gene, Tokyo, Japan), according to the manufacturer’s protocol, except for sample disruption. Frozen cecal contents (200 mg) were disrupted at 2700 rpm for 90 s using a Multi-Beads shocker (MB601U, Yasui Kikai, Osaka, Japan). Extracted DNA samples were dissolved in DNase-free Tris-EDTA buffer (pH 8.0; 10 mmol/L Tris-HCl, 1 mmol/L EDTA), and stored at −30 °C until 16S rDNA sequencing analysis.
2.11. Library Preparation and DNA Sequencing
First, 16S rDNA sequencing using a MiSeqTM system (Illumina, San Diego, CA, USA) was conducted according to a previously described method [19]. PCR amplification of the 16S rRNA V3–V4 region was conducted using primers 341F and 805R. The processing of sequencing data, including quality filtering, chimera check, operational taxonomic unit (OTU) definition, and taxonomy assignment was carried out using the Quantitative Insights Into Microbial Ecology (QIIME) [20], USEARCH (https://www.drive5.com/usearch/) [21], and UCHIME (https://www.drive5.com/usearch/manual/uchime_algo.html) [22], exactly as previously described [19].
2.12. Statistical Analysis
A power analysis was performed with G Power version 3.1 to determine an adequate sample size for studying a significant difference between portal H2 concentration and plasma ALT activity as the primary outcome. The sample size was calculated using the power procedure for Student’s t-test, considering an α probability of 0.05 with a power of 0.80, and an effect size was calculated using the results from our previous study [5]. This power analysis determined that a sample size of 13 to 14 rats (experiment 1) and 10 to 11 rats (experiment 2) per group was required for studies. Values obtained from the experiments were expressed as means, with their standard errors or medians with range. In experiment 1, rats were categorized into quintiles of portal H2 concentration at the end of the experiment. The number of rats from the respective colony in quintiles 1 to 5 was analyzed using Pearson’s Chi-square test. The Jonckheere–Terpstra trend test was used to assess the effect of colonic H2 production on oxidative stress and damage. The medians of quintiles 2 through to 5 were compared with those of quintile 1 using the Steel test. In other experiments, data were subjected to Bartlett’s test for homogeneity of variances, and data with unequal variances were log transformed. For samples with equal variances, Student’s t-test was used for comparisons between individual group means. If sample variances were still unequal after log transformation, we used Welch’s t-test. Then, α-diversity metrics (Chao1 index, Shannon index and Observed OTU index) were computed, β-diversity metrics were measured using unweighted and weighted UniFrac, and principal coordinate analysis was performed based on the UniFrac distances. The difference of sample clustering was analyzed by permutational multivariate analysis of variation (PERMANOVA). The relationship between portal H2 concentration and the population of bacteria was determined by Spearman’s rank correlation coefficient. The statistical analyses, except for the Jonckheere–Terpstra trend test and PERMANOVA, were performed using SAS JMP software (version 9.0.2; Tokyo, Japan). The Jonckheere–Terpstra trend test and PERMANOVA were performed using R software (version 3.2.3, http://www.r-project.org; R Foundation for Statistical Computing, Vienna, Austria) and QIIME (version 1.9.0, http://qiime.org/), respectively. Significance was defined as p < 0.05.
3. Results
3.1. Experiment 1
We examined the relationship between colonic H2 production and hepatic oxidative damage. Rats were categorized into quintiles of portal H2 concentration at the end of the experiment (Table 1). In quintiles 4 and 5, which had the highest portal H2 concentrations, more than 90% of the rats were from the Haruno colony. Portal H2 concentrations in quintiles 4 and 5 were 5.5 and 11.3 times that of quintile 1, respectively. Food intake, HAS intake, and body weight gain did not differ among the quintiles. Plasma ALT (alanine aminotransferase) activity was significantly lower when the portal H2 concentration was higher, and it was significantly lower in quintile 3 compared with quintile 1. Hepatic GSSG (reduced glutathione) concentration was significantly lower when the portal H2 concentration was high. In addition, the GSH (oxidized glutathione)/GSSG ratio tended to be higher (p = 0.0928) when the portal H2 concentration was high. Quintiles with high portal H2 concentration showed higher cecal acetate and butyrate concentrations, and lower cecal propionate and succinate concentrations. These differences in organic acids were statistically significant between quintiles 4 and 5, and quintile 1.
Table 1.
Liver redox status, plasma alanine aminotransferase (ALT) activity, and cecal organic acid concentration, according to the quintiles of rat portal H2 concentration.
| Q1 (Low) | Q2 | Q3 | Q4 | Q5 (High) | χ2 (p) | |
|---|---|---|---|---|---|---|
| Haruno colony | 4 | 1 | 3 | 12 | 13 | <0.0001 |
| Ohara colony | 9 | 12 | 10 | 1 | 1 | <0.0001 |
| Ptrend | ||||||
| Portal H2, μmol/L | 1.54 (0.771–1.89) |
3.39 (2.75–3.92) |
6.08 ** (5.02–6.56) |
8.45 *** (8.02–9.06) |
17.4 *** (13.2–19.9) |
<0.0001 |
| Food intake, g/10 day | 198 (190–204) |
192 (182–208) |
199 (188–206) |
202 (187–209) |
196 (190–200) |
0.4594 |
| HAS intake, g/10 day | 39.6 (38.1–40.9) |
38.3 (36.4–41.7) |
39.7 (37.7–41.3) |
40.3 (37.5–41.9) |
39.2 (37.9–40.0) |
0.4752 |
| Body weight gain, g/10 day | 70 (61–85) |
78 (68–84) |
72 (64–77) |
72 (66–78) |
71 (60–82) |
0.2687 |
| Liver, μmol/g tissue | ||||||
| GSSG | 0.137 (0.111–0.157) |
0.171 * (0.139–0.185) |
0.157 (0.143–0.174) |
0.145 (0.123–0.159) |
0.122 (0.106–0.135) |
0.0314 |
| GSH | 7.20 (6.77–7.88) |
7.13 (6.92–7.93) |
7.22 (6.91–8.40) |
6.83 (6.31–7.30) |
7.01 (6.74–7.25) |
0.1087 |
| GSH/GSSG | 55.4 (46.4–69.5) |
45.9 (40.1–48.6) |
47.9 (40.8–55.2) |
48.1 (42.2–58.2) |
59.6 (50.8–67.2) |
0.0928 |
| Plasma ALT, μkat/L | 7.96 (3.84–8.90 |
6.33 (0.717–9.43) |
3.54 * (0.526–4.92) |
2.03 (0.582–7.88) |
3.99 (0.750–9.88) |
0.0422 |
| Cecal organic acid, μmol/g | ||||||
| Acetate | 42.1 (37.4–59.3) |
41.6 (35.8–43.4) |
47.2 (33.3–60.6) |
71.4 * (60.0–78.5) |
87.7 *** (66.0–97.0) |
<0.0001 |
| Propionate | 8.77 (8.01–10.3) |
11.4 (7.90–15.3) |
8.75 (5.60–10.0) |
4.13 ** (3.32–6.23) |
4.83 ** (3.59–5.55) |
<0.0001 |
| Butyrate | 4.79 (2.97–6.84) |
4.25 (3.56–5.88) |
6.60 (3.85–9.64) |
9.34 * (7.15–12.1) |
12.3 *** (10.3–15.7) |
<0.0001 |
| Succinate | 60.8 (38.9–69.3) |
45.3 (41.6–48.5) |
45.4 (27.5–51.2) |
25.8 ** (18.8–35.0) |
30.7 ** (24.9–37.0) |
<0.0001 |
Data are expressed as the medians (25%–75%), n = 13 or 14. All of the rats were fed high amylose maize starch for 10 days, and then underwent ischemia–reperfusion (IR) treatment (30 min of ischemia + 45 min of reperfusion) at the end of the experiment. Rats were categorized into quintiles of portal H2 concentration at the end of the experiment. Data were analyzed using the Jonckheere–Terpstra trend test (all of the parameters except for the number of rats) and Steel test (all of the parameters except for the number of rats), or the Chi-square test (the number of rats). *, **, *** Median values were significantly different from those of the Q1 (p < 0.05, p < 0.01, p < 0.001) quintile; HAS, high amylose maize starch; GSH, reduced glutathione; GSSG, oxidized glutathione.
3.2. Experiment 2
The portal H2 concentration in rats used to prepare the inoculum for the transplantation experiment was 9.27 μmol/L (Supplementary Materials Table S1). The population of microbiota in the prepared inoculum differed between HG and LG. The phylum-level microbial community structure of the high H2 inoculum was 34.0% Bacteroidetes, 24.8% Firmicutes, 33.0% Actinobacteria, 3.7% Proteobacteria, and 4.5% Verrucomicrobia. The population of the phyla Bacteroidetes and Actinobacteria in the inoculum derived from HG were 0.5 and 1.2 times that of the population from LG, respectively (Supplementary Materials Table S2).
We examined whether the transplantation of colonic microbiota derived from HG could shift LG to HG (Table 2). No significant difference was observed in food intake and body weight gain. Until day 6 (2 days after final inoculation), low H2 excretion (breath + flatus) was maintained in both groups. On and after day 10, the H2 excretion in the transplantation group was more than twice the value of the control group, although no significant difference was observed. Portal H2 concentration was significantly higher in the transplantation group compared with the control group, and was comparable to that in donor rats (Supplementary Materials Table S2). The cecal weight of content and tissue, and concentrations of propionate and butyrate did not differ between the groups. Cecal acetate concentration was significantly higher in the transplantation group compared with the control group. Cecal succinate concentration tended to be lower in the transplantation group compared with the control group (p = 0.0701).
Table 2.
Changes in body weight, food intake, cecal H2, organic acid production, and cecal counts of total anaerobes in HAS-fed rats transplanted with high H2-producing microbiota.
| Control | Transplantation | p Value | |
|---|---|---|---|
| Initial body weight (g on day 0) | 238 ± 2 | 237 ± 2 | 0.9753 |
| Body weight gain (g/13 day) | 87 ± 4 | 78 ± 4 | 0.1348 |
| Food intake (g/13 day) | 287 ± 7 | 261 ± 8 * | 0.0493 |
| Net H2 excretion (μmol/5 min) | |||
| Day 0 | 0.103 (0.064–0.338) | 0.146 (0.101–0.176) | 0.8931 |
| Day 3 | 0.579 (0.447–1.13) | 0.405 (0.196–0.830) | 0.1690 |
| Day 6 | 0.334 (0.212–0.679) | 0.304 (0.151–0.622) | 0.8786 |
| Day 10 | 0.554 (0.199–0.878) | 1.13 (0.252–2.92) | 0.1734 |
| Day 13 | 0.721 (0.347–3.55) | 2.34 (1.65–2.96) | 0.3910 |
| AUCday 6–13 (mmol) | 0.966 (0.665–2.64) | 3.04 (1.41–6.06) | 0.1627 |
| Portal H2 (μmol/L) | 3.07 ± 1.00 | 9.95 ± 1.78 ** | 0.0041 |
| Cecum (g) | |||
| Contents | 9.31 ± 0.58 | 9.59 ± 0.61 | 0.7443 |
| Tissue | 1.87 ± 0.11 | 1.81 ± 0.10 | 0.7253 |
| Cecal organic acids (μmol/g) | |||
| Acetate | 43.5 ± 3.3 | 57.8 ± 2.7 ** | 0.0038 |
| Propionate | 9.80 ± 0.98 | 8.17 ± 1.07 | 0.2777 |
| n-Butyrate | 5.65 ± 1.05 | 7.06 ± 0.80 | 0.2999 |
| Succinate | 61.4 ± 4.2 | 48.5 ± 5.2 | 0.0701 |
| Cecal bacteria | |||
| Total anaerobes (log10cfu/g) | 12.7 ± 0.5 | 12.6 ± 0.3 | 0.7592 |
Data are expressed as the means ± standard error (SE) or medians (25%–75%), control group, n = 10 and transplantation groups, n = 11. Data were analyed using the Student’s t-test (data expect for net H2 excretion and area under the curve (AUC)) or Welch’s test (data for net H2 excretion and AUC). Transplantation of the inoculum was performed on days 3 and 4. Net H2 excretion in breath and flatus was measured. AUC was calculated from changes in net H2 excretion from day 6 to day 13. *, ** Mean values were significantly different from those of the control group (p < 0.05 and p < 0.01, respectively).
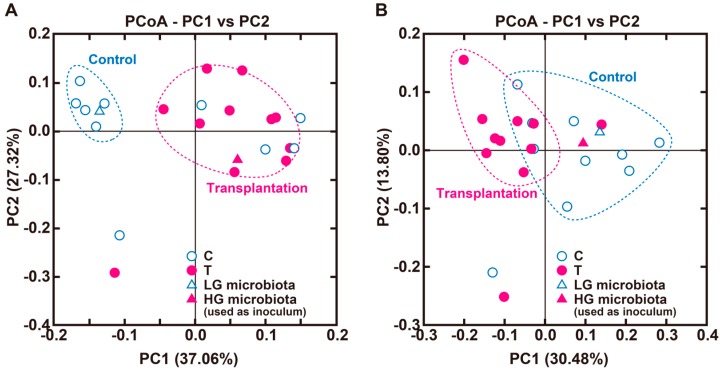
Cecal anaerobe count did not differ between the groups. For microbiota composition analysis, a total of 312,451 sequence reads were obtained after data processing (average 14,202 reads/sample; standard deviation 10,048). For α-diversity, the Shannon index was significantly lower in the transplantation group than in the control group (control group, 2.47 ± 0.13; transplantation group, 1.97 ± 0.06; p = 0.0051), while the Chao1 index and observed OTU index did not differ between the groups (Chao1 index, 72.6 ± 3.1 and 69.5 ± 3.4; p = 0.5236; observed OTU index, 63.0 ± 1.9 and 60.4 ± 3.3; p = 0.4947 in the control and transplantation groups, respectively). Although microbiota communities tented to be different between both groups (p = 0.086; Figure 1) by β-diversity analyses using weighted UniFrac, a specific cluster was formed in the transplantation group that was similar to HG microbiota. β-diversity analyses using unweighted UniFrac revealed that microbiota communities were significantly distant from each other (p = 0.006; Figure 1). The cecal population of the Bacteroidales family S24-7 and the genera Oscillospira and Escherichia were significantly greater in the transplantation group compared with the control group, while the genus Bacteroides was significantly lower (Table 3). There was no significant difference in the population of the Desulfovibrionaceae family, which utilizes H2 to produce hydrogen sulfide, between both groups (data not shown), and the populations were less than 0.01% in both groups. The genus Desulfovibrio, which belongs to the Desulfovibrionaceae family, was not detected. Portal H2 concentration positively correlated with genera Bifidobacterium, Parabacteroides, and Allobaculum, and negatively correlated with genera Bacteroides, Ruminococcus, and Escherichia.
Figure 1.
Principle coordinate analysis plot based on weighted (A) and unweighted (B) UniFrac distances of microbial communities in the cecum of control and transplantation rats. Sample clustering was significant between both groups (p = 0.086 and p = 0.006, respectively, PERMANOVA). control group, n = 10; transplantation group, n = 11. C, control group; HG, high H2 generators; LG, low H2 generators; PCoA, principal coordinate analysis; PERMANOVA, permutational multivariate analysis of variation; T, transplantation group.
Table 3.
Changes in the population of cecal microbiota in HAS-fed rats transplanted with high H2-producing microbiota.
| Correlation ‡ (vs. Portal H2) | ||||||
|---|---|---|---|---|---|---|
| Order | Family | Genus | C (%) | T (%) | r | p |
| Actinobacteria | 12.0 ± 5.5 | 18.3 ± 4.4 | 0.789 | 4.82 × 10−5 | ||
| Bifidobacteriales | Bifidobacteriaceae | Bifidobacterium | 12.0 ± 5.4 | 18.0 ± 4.3 | 0.791 | 4.49 × 10−5 |
| Bacteroidetes | 58.0 ± 7.3 | 39.8 ± 4.1 * | −0.408 | 0.0757 | ||
| Bacteroidales | Bacteroidaceae | Bacteroides | 40.2 ± 9.0 | 8.0 ± 3.1 ** | −0.507 | 0.0240 |
| Bacteroidales | s24-7 | 15.7 ± 2.7 | 30.7 ± 4.7 * | −0.084 | 0.724 | |
| Bacteroidales | Porphyromonadaceae | Parabacteroides | 1.7 ± 0.5 | 1.0 ± 0.4 | 0.316 | 0.0190 |
| Firmicutes | 23.8 ± 3.2 | 34.3 ± 4.6 † | −0.0992 | 0.677 | ||
| Lactobacillales | Lactobacillaceae | Lactobacillus | 9.3 ± 3.1 | 9.1 ± 1.9 | 0.368 | 0.111 |
| Clostridiales | Lachnospiraceae | Blautia | 1.1 ± 0.2 | 3.1 ± 0.9 | −0.107 | 0.653 |
| Clostridiales | Lachnospiraceae | Clostridium | 1.2 ± 0.3 | 2.1 ± 0.5 | 0.383 | 0.0957 |
| Clostridiales | Lachnospiraceae | Other | 5.8 ± 0.9 | 1.9 ± 0.8 | −0.397 | 0.0841 |
| Clostridiales | Ruminococcaceae | Ruminococcus | 1.1 ± 0.5 | 6.8 ± 4.9 | −0.567 | 0.0103 |
| Clostridiales | Ruminococcaceae | Oscillospira | 1.1 ± 0.1 | 2.1 ± 0.3 * | −0.0391 | 0.871 |
| Clostridiales | Ruminococcaceae | Other | 1.2 ± 0.2 | 0.6 ± 0.1 | −0.112 | 0.678 |
| Erysipelotrichales | Erysipelotrichaceae | Allobaculum | 1.2 ± 0.4 | 6.1 ± 2.5 | 0.666 | 1.77 × 10−3 |
| Erysipelotrichales | Erysipelotrichaceae | Eubacterium | 0.1 ± 0.0 | 0.1 ± 0.0 | −0.405 | 0.0780 |
| Proteobacteria | 4.1 ± 1.2 | 4.4 ± 0.7 | −0.370 | 0.109 | ||
| Enterobacteriale | Enterobacteriaceae | Escherichia | 0.3 ± 0.2 | 3.1 ± 0.7 ** | −0.453 | 0.0466 |
| Enterobacteriales | Enterobacteriaceae | Other | 3.0 ± 1.3 | 0.0 ± 0.0 | − | − |
| Burkholderiales | Alcaligenaceae | Sutterella | 0.8 ± 0.3 | 1.3 ± 0.2 | 0.341 | 0.141 |
| Verrucomicrobia | 1.9 ± 1.9 | 3.1 ± 2.7 | −0.383 | 0.0960 | ||
| Verrucomicrobiales | Verrucomicrobiaceae | Akkermansia | 1.9 ± 1.9 | 3.1 ± 2.7 | −0.383 | 0.0960 |
Data are expressed as the means ± SE, n = 10 and transplantation groups, n = 11. Data that were less than 0.1% in both the control and transplantation groups were omitted from the table. Data were analyzed using the Student’s t-test or Welch’s test. Transplantation of the inoculum was performed on days 3 and 4. C, control group; T, transplantation group. *, ** Mean values were significantly different from those of the control group (p < 0.05, p < 0.01). † Mean values tended to be higher than those of the control group (p = 0.0777). ‡ Correlations were determined by Spearman’s rank correlation coefficient.
4. Discussion
We previously found that colonic H2 suppressed hepatic oxidative stress and damage in IR rats that were fed either the HAS or pectin diets compared with those fed a control diet [5]. In the present study, liver damage due to a hepatic IR operation was improved in HAS-fed rats with a high portal H2 concentration (Table 1). These results suggest that the alleviating effect of colonic H2 on hepatic oxidative stress and damage is dependent on portal H2 concentration. In our previous study, we screened and used only HG: the portal H2 concentrations were 4.5–11 μmol/L, with the high range of portal H2 concentration leading to suppressed oxidative stress and damage [5]. The range in the previous study is comparable with the range in quintiles 3 and 4 in the present study (Table 1). Although the difference in the hepatic GSH/GSSG ratio did not reach statistical significance (p = 0.0928), hepatic oxidative stress (liver GSSG concentration) and damage (plasma ALT activity) after the IR operation were significantly improved when the portal H2 concentration was high: greater than 4.3 μmol/L, which corresponded to quintiles 3–5. Hepatic oxidative damage was significantly improved in quintile 3 compared with quintile 1. Colonization by high H2 producing microbiota, as well as the administration of high fermentable, non-digestible saccharides, would be required for a stable supply of large amounts of H2 in the body, because fermentation substrate and colonic bacteria cause colonic fermentation.
Net H2 production is determined by a balance between H2-producing and H2-utilizing microbes in the large intestine [8]. Colonic H2 is mainly produced by members of the phylum Firmicutes and the phylum Bacteroidetes, and utilized predominantly by reductive acetogens, sulfate-reducing bacteria, and methanogens to produce acetate, hydrogen sulfide, and methane, respectively [8,23]. Since the microbiota of laboratory animals also differ among the breeders and breeding colonies depending on maternal microbiota [9], fermentation patterns would vary, even with the administration of the same fermentation substrate. In the present study, we observed large differences in portal H2 concentration, which is a marker of colonic H2 production, among the colonies of the same breed or individuals in the same colony. Therefore, the different H2 productions in the present study reflect the composition of colonic microbiota. Manichanh et al. showed that the transplantation of cecal microbiota to rats without antibiotic treatment produced a similar composition of microbiota between the transplanted rats and the donor rats [18]. Using the same method, we determined that the transplantation of microbiota derived from HG into LG caused high H2 production in the transplanted rats (Table 2 and Figure 1). Previous investigators reported that bacteria used as probiotics did not colonize the large intestine over the long term unless the bacteria were continuously administered [24]. Although the shift by transplantation may be temporary, this result shows that the presence of high H2-producing microbiota in the large intestine causes an increase in H2 production. In the present study, we used rat cecal microbiota as the inoculum. Li et al. demonstrated successful fecal microbiota transplantation in humans [25]. The results suggest that the microbiota colonization, using the same species of animal, might be relatively easy; however, colonization using a single species of bacteria may not be successful.
Inoculum was prepared from rats showing high H2 production in their large intestine. In comparison with the microbiota from LG, the high H2-producing inoculum showed a higher population of the phylum Actinobacteria (predominantly bifidobacteria) and lower population of the genus Bacteroides. The differences were maintained in the cecal content of rats transplanted with the inoculum (Supplementary Materials Figure S2); populations of the phyla Actinobacteria and Bacteroidetes in the transplantation group were 153% and 69% higher than in the control group, respectively. HAS containing resistant starch selectively promotes Bifidobacterium spp. according to its availability [26,27]. HAS is strongly utilized by a limited type of bacteria, such as Bifidobacterium spp. [27,28,29], but not Bacteroides spp. [28]. The change in these bacteria, due to transplantation, is suggested to be reasonable to promote the utilization of HAS. Bifidobacteria cannot produce H2 gas [23,30], because they do not have hydrogenase, although they can degrade resistant starch to produce acetate and lactate. Lactate is utilized by other bacteria to produce acetate and butyrate [31]. Therefore, even though bifidobacteria cannot produce H2 gas, a 50% increase in the population of the genus Bifidobacterium in the transplantation group could promote the colonic fermentation of HAS and stimulate high H2 production through metabolite cross-feeding by any type of H2-producing bacteria [8,32,33]. It would be difficult to identify the H2-producing bacteria involved in high portal H2 concentration due to transplantation, because 66% of the total bacteria in human feces are estimated to produce H2 [23]. Although not all of the bacteria in the inoculum reached the large intestine of the recipient rats alive and colonized, we suggest that bacteria contributing to the increased H2 production came from the inoculum, and were successful in colonizing the large intestine. In the present study, the higher H2 quintiles and the transplantation group showed higher colonic acetate concentration. These results could reflect the finding that H2-producing bacteria convert pyruvate to acetate, carbon dioxide, and H2 [34]. However, H2 is also produced by other reactions. Moreover, H2 and acetate are utilized by some bacteria to produce other metabolites, such as methane and butyrate [34]. Therefore, further investigation need to be performed to determine relationships between colonic production of H2 and organic acids.
In the present study, sulfate-reducing bacteria, such as the Desulfovibrionaceae family, which consumed H2 to produce hydrogen sulfate, were detected at very low levels. A negative correlation between portal H2 concentration and the genus Ruminococcus, part of which includes reductive acetogens, were observed after the transplantation of inoculum. These results suggest that some H2-utilizing bacteria may be related to the supply amount of H2 in the body. However, methanogens, which is H2-utilizing archaea, could not be detected, because PCR amplicon of the 16S rRNA V3–V4 region, which was conducted using primers 341F and 805R, was sequenced. Low H2 partial pressure is essential to maintain anaerobic fermentation, because high H2 partial pressure inhibits reduced nicotinamide adenine dinucleotide dehydrogenase and ATP production [35,36]. Therefore, excess accumulation of H2 might be inadequate in regards to the continuing fermentation and proliferation of bacteria. The extent of colonic H2 production that can alleviate oxidative stress without inhibiting fermentation remains unclear. Moreover, changes in H2 concentration may also result in changes in microbiota, because high H2 partial pressure inhibits proliferation of some anaerobes. Further investigation is required to overall verify the physiological significance of high H2 production and mechanisms by which transplantation of high H2 inoculum could lead to a high amount of net H2 production in rats that have been fed HAS. In the present study, transplanted microbiota colonized for 10 days after the final inoculation, and changes in the abundance of the phyla and genera due to transplantation, caused high H2 production. However, it remains unclear whether transplanted microbiota would continue to colonize the large intestine in recipients over longer periods, and continue to produce large amounts of H2. Further study is required to study the long-term effect of transplantation on portal H2 concentration.
In conclusion, portal H2 concentration differs widely among individual rats, even when the same non-digestible saccharides were fed. H2 production was determined to be dependent on the composition of cecal microbiota, which varies between individual rats. Furthermore, we found that LG converted to HG after the transplantation of microbiota derived from HG. High portal H2 concentration correlated with greater suppression of oxidative stress. The formation of an appropriate composition of colonic microbiota, and the supply of suitable fermentation substrate to the large intestine, would lead to high colonic H2 production, which may contribute to the alleviation of acute oxidative stress induced by hepatic IR operation.
Acknowledgments
This research was supported in part by a Grant-in-Aid for Scientific Research (B) (16H03036) from the Japan Society for the Promotion of Science, Japan. We thank NAI, Inc. (Yokohama, Japan) for their assistance in manuscript editing.
Supplementary Materials
The following are available online at http://www.mdpi.com/2072-6643/10/2/144/s1, Figure S1: Phyla level distribution of rat cecal contents in rats transplanted with high H2-producing microbiota, Figure S2: Genus level distribution in rat cecal contents after transplantation with high H2-producing microbiota, Table S1: H2 production and cecal organic acid concentration in rats with different colonic H2 producing ability, Table S2: Population of cecal microbiota in the inoculum.
Author Contributions
N.N. conducted the study, analyzed the dataset and wrote the manuscript. H.T., E.K. and T.Y. conducted the biological analysis. Y.S. performed the IR operation. R.I. performed the 16S rDNA sequencing analysis. All authors were involved in designing the study, reviewing and interpreting the results, and drafting the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors have no conflict of interest associated with the present study.
References
- 1.Macfarlane G.T., Cummings J.H. The colonic flora, fermentation and large bowel digestive function. In: Phillips S.F., Pemberton J.H., Shorter R.G., editors. The Large Intestine: Physiology, Pathophysiology and Disease. Raven Press; New York, NY, USA: 1991. pp. 51–92. [Google Scholar]
- 2.Ohsawa I., Ishikawa M., Takahashi K., Watanabe M., Nishimaki K., Yamagata K., Katsura K., Katayama Y., Asoh S., Ohta S. Hydrogen acts as a therapeutic antioxidant by selectively reducing cytotoxic oxygen radicals. Nat. Med. 2007;13:688–694. doi: 10.1038/nm1577. [DOI] [PubMed] [Google Scholar]
- 3.Wu F., Qiu Y., Ye G., Luo H., Jiang J., Yu F., Zhou W., Zhang S., Feng J. Treatment with hydrogen molecule attenuates cardiac dysfunction in streptozotocin-induced diabetic mice. Cardiovasc. Pathol. 2015;24:294–303. doi: 10.1016/j.carpath.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Hayashida K., Sano M., Ohsawa I., Shinmura K., Tamaki K., Kimura K., Endo J., Katayama T., Kawamura A., Kohsaka S., et al. Inhalation of hydrogen gas reduces infarct size in the rat model of myocardial ischemia-reperfusion injury. Biochem. Biophys. Res. Commun. 2008;373:30–35. doi: 10.1016/j.bbrc.2008.05.165. [DOI] [PubMed] [Google Scholar]
- 5.Nishimura N., Tanabe H., Sasaki Y., Makita Y., Ohata M., Yokoyama S., Asano M., Yamamoto T., Kiriyama S. Pectin and high-amylose maize starch increase caecal hydrogen production and relieve hepatic ischaemia-reperfusion injury in rats. Br. J. Nutr. 2012;107:485–492. doi: 10.1017/S0007114511003229. [DOI] [PubMed] [Google Scholar]
- 6.Nishimura N., Tanabe H., Adachi M., Yamamoto T., Fukushima M. Colonic hydrogen generated from fructan diffuses into the abdominal cavity and reduces adipose mrna abundance of cytokines in rats. J. Nutr. 2013;143:1943–1949. doi: 10.3945/jn.113.183004. [DOI] [PubMed] [Google Scholar]
- 7.Matsumoto M., Fujita A., Yamashita A., Kameoka S., Shimomura Y., Kitada Y., Tamada H., Nakamura S., Tsubota K. Effects of functional milk containing galactooligosaccharide, maltitol, and glucomannan on the production of hydrogen gas in the human intestine. J. Funct. Foods. 2017;35:13–23. doi: 10.1016/j.jff.2017.05.013. [DOI] [Google Scholar]
- 8.Carbonero F., Benefiel A.C., Gaskins H.R. Contributions of the microbial hydrogen economy to colonic homeostasis. Nat. Rev. Gastroenterol. Hepatol. 2012;9:504–518. doi: 10.1038/nrgastro.2012.85. [DOI] [PubMed] [Google Scholar]
- 9.Ohashi Y., Hiraguchi M., Ushida K. The composition of intestinal bacteria affects the level of luminal iga. Biosci. Biotechnol. Biochem. 2006;70:3031–3035. doi: 10.1271/bbb.60164. [DOI] [PubMed] [Google Scholar]
- 10.Van Nood E., Vrieze A., Nieuwdorp M., Fuentes S., Zoetendal E.G., de Vos W.M., Visser C.E., Kuijper E.J., Bartelsman J.F., Tijssen J.G., et al. Duodenal infusion of donor feces for recurrent clostridium difficile. N. Engl. J. Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 11.Di Luccia B., Crescenzo R., Mazzoli A., Cigliano L., Venditti P., Walser J.C., Widmer A., Baccigalupi L., Ricca E., Iossa S. Rescue of fructose-induced metabolic syndrome by antibiotics or faecal transplantation in a rat model of obesity. PLoS ONE. 2015;10:e0134893. doi: 10.1371/journal.pone.0134893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCleary B.V., Monaghan D.A. Measurement of resistant starch. J. AOAC Int. 2002;85:665–675. [PubMed] [Google Scholar]
- 13.Nishimura N., Tanabe H., Yamamoto T. Sufficient intake of high amylose cornstarch maintains high colonic hydrogen production for 24 h in rats. Biosci. Biotechnol. Biochem. 2017;81:173–180. doi: 10.1080/09168451.2016.1234929. [DOI] [PubMed] [Google Scholar]
- 14.Anderson M.E. Determination of glutathione and glutathione disulfide in biological samples. Methods Enzymol. 1985;113:548–555. doi: 10.1016/s0076-6879(85)13073-9. [DOI] [PubMed] [Google Scholar]
- 15.Rahman I., Kode A., Biswas S.K. Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat. Protoc. 2006;1:3159–3165. doi: 10.1038/nprot.2006.378. [DOI] [PubMed] [Google Scholar]
- 16.Hoshi S., Sakata T., Mikuni K., Hashimoto H., Kimura S. Galactosylsucrose and xylosylfructoside alter digestive tract size and concentrations of cecal organic acids in rats fed diets containing cholesterol and cholic acid. J. Nutr. 1994;124:52–60. doi: 10.1093/jn/124.1.52. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka Y., Benno Y. Application of a single-colony coculture technique to the isolation of hitherto unculturable gut bacteria. Microbiol. Immunol. 2015;59:63–70. doi: 10.1111/1348-0421.12220. [DOI] [PubMed] [Google Scholar]
- 18.Manichanh C., Reeder J., Gibert P., Varela E., Llopis M., Antolin M., Guigo R., Knight R., Guarner F. Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res. 2010;20:1411–1419. doi: 10.1101/gr.107987.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoue R., Ohue-Kitano R., Tsukahara T., Tanaka M., Masuda S., Inoue T., Yamakage H., Kusakabe T., Hasegawa K., Shimatsu A., et al. Prediction of functional profiles of gut microbiota from 16s rrna metagenomic data provides a more robust evaluation of gut dysbiosis occurring in japanese type 2 diabetic patients. J. Clin. Biochem. Nutr. 2017;61:217–221. doi: 10.3164/jcbn.17-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I., et al. Qiime allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgar R.C. Search and clustering orders of magnitude faster than blast. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 22.Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. Uchime improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolf P.G., Biswas A., Morales S.E., Greening C., Gaskins H.R. H2 metabolism is widespread and diverse among human colonic microbes. Gut Microbes. 2016;7:235–245. doi: 10.1080/19490976.2016.1182288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saxelin M., Pessi T., Salminen S. Fecal recovery following oral administration of lactobacillus strain gg (atcc 53103) in gelatine capsules to healthy volunteers. Int. J. Food Microbiol. 1995;25:199–203. doi: 10.1016/0168-1605(94)00091-J. [DOI] [PubMed] [Google Scholar]
- 25.Li S.S., Zhu A., Benes V., Costea P.I., Hercog R., Hildebrand F., Huerta-Cepas J., Nieuwdorp M., Salojarvi J., Voigt A.Y., et al. Durable coexistence of donor and recipient strains after fecal microbiota transplantation. Science. 2016;352:586–589. doi: 10.1126/science.aad8852. [DOI] [PubMed] [Google Scholar]
- 26.Nishimura N., Tanabe H., Yamamoto T. Isomaltodextrin, a highly branched alpha-glucan, increases rat colonic H2 production as well as indigestible dextrin. Biosci. Biotechnol. Biochem. 2016;80:554–563. doi: 10.1080/09168451.2015.1104237. [DOI] [PubMed] [Google Scholar]
- 27.Regmi P.R., Metzler-Zebeli B.U., Ganzle M.G., van Kempen T.A., Zijlstra R.T. Starch with high amylose content and low in vitro digestibility increases intestinal nutrient flow and microbial fermentation and selectively promotes bifidobacteria in pigs. J. Nutr. 2011;141:1273–1280. doi: 10.3945/jn.111.140509. [DOI] [PubMed] [Google Scholar]
- 28.Wang X., Conway P.L., Brown I.L., Evans A.J. In vitro utilization of amylopectin and high-amylose maize (amylomaize) starch granules by human colonic bacteria. Appl. Environ. Microbiol. 1999;65:4848–4854. doi: 10.1128/aem.65.11.4848-4854.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ze X., Duncan S.H., Louis P., Flint H.J. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 2012;6:1535–1543. doi: 10.1038/ismej.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falony G., Lazidou K., Verschaeren A., Weckx S., Maes D., De Vuyst L. In vitro kinetic analysis of fermentation of prebiotic inulin-type fructans by bifidobacterium species reveals four different phenotypes. Appl. Environ. Microbiol. 2009;75:454–461. doi: 10.1128/AEM.01488-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duncan S.H., Louis P., Flint H.J. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 2004;70:5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamlage B., Gruhl B., Blaut M. Isolation and characterization of two new homoacetogenic hydrogen-utilizing bacteria from the human intestinal tract that are closely related to clostridium coccoides. Appl. Environ. Microbiol. 1997;63:1732–1738. doi: 10.1128/aem.63.5.1732-1738.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chinda D., Nakaji S., Fukuda S., Sakamoto J., Shimoyama T., Nakamura T., Fujisawa T., Terada A., Sugawara K. The fermentation of different dietary fibers is associated with fecal clostridia levels in men. J. Nutr. 2004;134:1881–1886. doi: 10.1093/jn/134.8.1881. [DOI] [PubMed] [Google Scholar]
- 34.Agler M.T., Wrenn B.A., Zinder S.H., Angenent L.T. Waste to bioproduct conversion with undefined mixed cultures: The carboxylate platform. Trends Biotechnol. 2011;29:70–78. doi: 10.1016/j.tibtech.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 35.Mathur R., Kim G., Morales W., Sung J., Rooks E., Pokkunuri V., Weitsman S., Barlow G.M., Chang C., Pimentel M. Intestinal methanobrevibacter smithii but not total bacteria is related to diet-induced weight gain in rats. Obesity. 2013;21:748–754. doi: 10.1002/oby.20277. [DOI] [PubMed] [Google Scholar]
- 36.Samuel B.S., Gordon J.I. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc. Natl. Acad. Sci. USA. 2006;103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.