Abstract
Background
Wheat genetic resources have been used for genetic improvement since 1876, when Stephen Wilson (Transactions and Proceedings of the Botanical Society of Edinburgh 12: 286) consciously made the first wide hybrid involving wheat and rye in Scotland. Wide crossing continued with sporadic attempts in the first half of 19th century and became a sophisticated scientific discipline during the last few decades with considerable impact in farmers’ fields. However, a large diversity of untapped genetic resources could contribute in meeting future wheat production challenges.
Perspectives and Conclusion
Recently the complete reference genome of hexaploid (Chinese Spring) and tetraploid (Triticum turgidum ssp. dicoccoides) wheat became publicly available coupled with on-going international efforts on wheat pan-genome sequencing. We anticipate that an objective appraisal is required in the post-genomics era to prioritize genetic resources for use in the improvement of wheat production if the goal of doubling yield by 2050 is to be met. Advances in genomics have resulted in the development of high-throughput genotyping arrays, improved and efficient methods of gene discovery, genomics-assisted selection and gene editing using endonucleases. Likewise, ongoing advances in rapid generation turnover, improved phenotyping, envirotyping and analytical methods will significantly accelerate exploitation of exotic genes and increase the rate of genetic gain in breeding. We argue that the integration of these advances will significantly improve the precision and targeted identification of potentially useful variation in the wild relatives of wheat, providing new opportunities to contribute to yield and quality improvement, tolerance to abiotic stresses, resistance to emerging biotic stresses and resilience to weather extremes.
Keywords: Wheat genetic resources, genomics, single nucleotide polymorphism (SNP), wild species, wide-hybridization
INTRODUCTION
Bread wheat (Triticum aestivum L.) belongs to the tribe Triticeae (syn. Hordeae) and is one of the most important food crops, cultivated on about 220 million ha, providing food to one-third of the global population and providing 20 % of the global caloric requirements. Triticeae has more than 150 different species of various ploidy, among which bread wheat, durum wheat (T. turgidum L.), einkorn wheat (T. monococcum L.), rye (Secale cereale L.) and barley (Hordeum vulgare L.) are cultivated for food and commercial purposes (Ortiz et al., 2008). Together, these species and their close and distant relatives constitute an important reservoir of genetic resources that include 434 358 accessions collected and stored in 23 gene banks and harbouring many beneficial alleles for wheat improvement. The knowledge of and manipulation of these resources has shaped a discipline called ‘wheat wide crosses’ or ‘wide hybridization’, which like other scientific disciplines, evolved significantly in recent decades due to significant advances in cytogenetics, genomics and other allied disciplines (Mujeeb-Kazi et al., 2013; Dolezel et al., 2014). This discipline has potential to bring about ‘super-domestication’ because it could lead to a domesticate with dramatically increased yield that could not be selected in natural environments from naturally occurring variations without resource to new technologies (Vaughan et al., 2007).
Five decades ago we witnessed the demonstrable impact of the ‘green revolution’ (1967–1970) based on dwarfing genes, photoperiod insensitivity and stem rust resistance catalysed through the work in the International Maize and Wheat Improvement Center (CIMMYT) by conventional breeding methods; this was led by late Nobel laureate Dr Norman Borlaug and colleagues with international partnerships involving the National Agriculture Research Systems (NARS) in developing countries. CIMMYT wheat germplasm has impacted almost every wheat improvement programme globally. However, it was sensed very early after the Nobel Prize was awarded to Dr Borlaug in 1970 that there was a need to accelerate more efficiently the use of unique genetic diversity in wheat collections and wheat relatives for future yield improvement. This heralded the wide crossing programme at CIMMYT (Mexico), Kansas State University (USA) and a few other centres of wheat research and development, with emphasis on wheat progenitor species, such as Triticum monococcum, Aegilops tauschii and T. dicoccoides, as well as more distant relatives including Agropyron elongatum, Haynaldia villosa and cultivated cereal rye (S. cereale). Until recently progenies of one derivative of cereal rye (1BL.1RS translocation) were grown on millions of hectares worldwide. Significant outcomes such as this encouraged donors in public and corporate sectors to invest in pre-breeding, and specific international projects like the Synthetic Evaluation Project in Australia (www.caigeproject.org.au), Wheat Initiative Strategic Partnership (WISP; www.wheatisp.org) in the UK, Seeds of Discovery (SeeD) in Mexico and DivSeek (www.divseek.org) were initiated to introduce genes of wild relatives into elite germplasm, making such resources available to wheat breeding programmes. Pay-offs from the wide crossing are quite apparent, but much greater effort must now be made if we are to harness the enormous but untapped potential genetic diversity that can be used not only to improve the yield and nutritional quality of wheat, but also to maintain and protect hard-fought yield increases from the many biotic and abiotic stresses that threaten the crop worldwide.
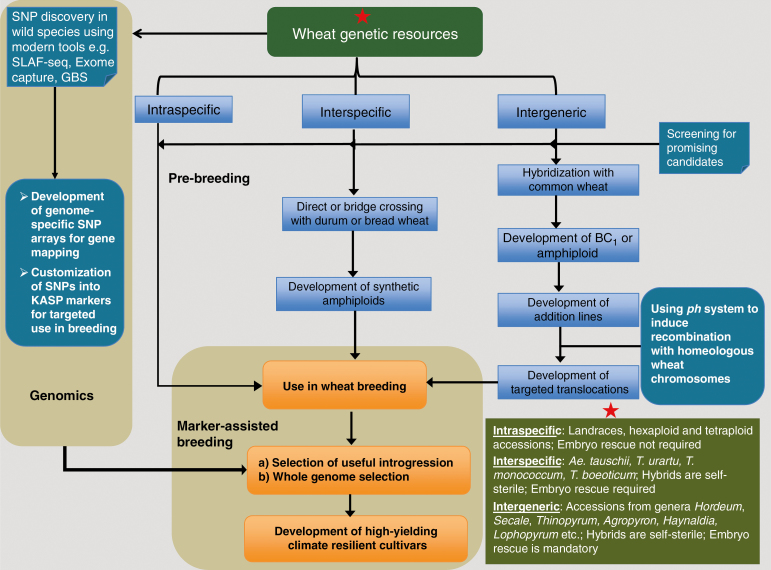
We are now facing the challenge of the two-fold increase in productivity target for a projected human population of 9.2 billion by 2050. This is further compounded by the challenges of climate change, stagnant wheat yields in many countries, outbreaks of new pathogen/pest races, and invasive weed species that delay progress and pose serious threats to world food security. It is therefore prudent to approach the challenges of food security by placing emphasis on outcome-based research approaches. This should embrace strategies that focus on rapid returns by stringent targeting of alien genetic resources to be used ‘anew’ or exploitation of current user-friendly genetic stocks. The scientific community should be well aware of bandwagons to be avoided (Bernardo, 2016), and should adapt a realistic approach in using appropriate genomics and phenotyping technologies and analytical tools to harness actual outputs (Fig. 1).
Fig. 1.
A workflow depicting types of wheat genetic resources, pre-breeding and genomics-assisted breeding strategies to exploit in crop improvement.
We provide here an overview on the use of genomics to exploit wheat genetic resources (WGRs) and discuss the bottlenecks in utilizing those resources.
HISTORICAL ACHIEVEMENTS ARE HOPE FOR THE FUTURE: KEY CONSIDERATIONS
Moore (2015) argued that we are now equipped with new marker tools that provide increased efficiency for breeding, so various issues come to mind in discussing pragmatic approaches that are outcome-focused. The strict requirements for modern plant varieties has meant that breeders are reluctant to go back to wild relatives or even land races for new variation. However, the diversity present in the unadapted gene pool has provided many useful traits now deployed widely in elite germplasm, and expanded use of this germplasm resource is seen as crucial to accelerating the rate of genetic gain in plant breeding (McCouch et al., 2013). This raises an important question of whether pre-breeding should restart with new hybrids of distant wild relatives or should we attempt to exploit what is currently available and set priorities that will result in early practical returns? We posit that the major impact will come from ‘homologous’ gene transfer reliant on genetic recombination involving the primary gene pool of wheat relatives. The potential impact is exemplified by the success of using the D genome (Aegilops tauschii)-derived user friendly genetic stocks (synthetic wheats, SH or SHW) produced at CIMMYT and elsewhere (Ogbonnaya et al., 2013). Advanced derivatives of these materials have positively impacted yield and yield components, enhanced micronutrient content, provided new genes for resistance to major biotic and abiotic stresses, and improved the processing quality of elite varieties (Ogbonnaya et al., 2013). Additional to the D genome donor there are A genome synthetics (2n = 6x = 42, AABBAuAu/AmAm), the AABB tetraploids (T. dicoccum, T. dicoccoides, T. polonicum and T. carthlicum) and less used AABBDD hexaploids (e.g. T. spelta and T. sphaerococcum) representing unharnessed variability of the primary gene pool (Xie and Nevo, 2008; Mujeeb-Kazi et al., 2013). Many leading varieties in Europe (e.g. ‘Robigus’) are derived from unknown introgressions from T. dicoccoides. Similarly, an introgression from Ae. umbellulata saved US wheat production from leaf rust in 1960; and a gene from Ae. ventricrosa conferring resistance to eyespot has been exploited in breeding programmes (Garcia-Olmedo et al., 1977). More recently, early results from whole genome re-sequencing of four elite cultivars suggest that two of the cultivars carry a 20-Mbp translocation on chromosome 2A representing the Vpm-1 introgression from Ae. ventricosa (Pozniak et al., 2017). Beyond these, there is the extensive diversity of the tertiary gene pool (species of distant relatives of wheat that tend to produce anomalous, lethal or completely sterile hybrids when crossed with wheat; embryo rescue may be necessary to obtain viable hybrids) (Fig. 1).
The most significant measurable output from a tertiary germplasm source was the fortuitous spontaneous wheat-alien chromosome translocation 1BL.1RS that occurred in the 1930s, and greatly contributed to world wheat production after its release in Eastern and Western Europe, Russia, China and CIMMYT. Various varieties were distributed by CIMMYT from the mid-1970s, initially with VEERY ‘S’, and national selections of it such as Pakistan 81 (Pak-81) in Pakistan and elsewhere. Varieties carrying the 1BL.1RS translocation occupied more than 50 % of the wheat area in China and a significant proportion across Europe, Asia, Africa and other wheat growing regions in the 1990s, and continue to cover around 25 % of the present area in China. Another example is Xiaoyan 6, an alleged derivative of a cross between wheat and the 70 chromosome grass species Thinopyrum ponticum, and its derivatives have also occupied millions of hectares in China. Besides these, three genera further exemplify the complexity of using distant relatives despite having the advantage of being diploids and preferred for wide hybridization programmes: Haynaldia villosa, Thinopyrum elongatum and Thinopyrum bessarabicum. However, we assert that the true measure of success in pre-breeding using distant relatives can only be measured by their contribution to improved or maintained wheat productivity in farmer fields; that is, the magnitude of the practical impact on yield improvement measured by the tonnes/hectare of output.
ADVANCES IN CYTOGENETICS TO FACILITATE ALIEN INTROGRESSIONS AND RECOMBINATION
Continuing advances in wheat improvement will rely on effective mining of genetic variation, which is dependent on how efficiently we can make genetic recombination and exploiting the biological mechanisms controlling recombination and meiosis. Wheat has a long history of cytogenetics research, and is also one of the best suited crops for cytogenetics studies due to the huge array of aneuploid stock (Sears, 1954; Law et al., 1987; Endo and Gill, 1996). The outcomes of advances in wheat cytogenetics could be categorized, but not limited to (1) development of precise wheat–alien genetic stocks for gene discovery and breeding (Mujeeb-Kazi et al., 2013; Khlestkina, 2014), (2) advancements in molecular cytology to study polyploidization, genome evolution and structure (Ali et al., 2016; Patokar et al., 2016), (3) initiation, control and enhancing the rate and distribution of homoeologous recombination between wheat and alien chromosomes (Moore, 2014) and (4) advances in chromosome flow-sorting to develop chromosome-specific libraries as a conduit to reducing complexity for further genomics and sequencing applications (Dolezel et al., 2014). Previously, an extensive review was published on the protocols and list of genetic stocks, for example amphpiploids, chromosome addition, and substitution lines and translocations carrying genes of breeding interest (Mujeeb-Kazi et al., 2013). These genetic stocks were developed by key centres such as Kansas State University (http://www.k-state.edu/wgrc/), Kyoto University, Japan (https://shigen.nig.ac.jp/wheat/komugi/), John Innes Centre (https://www.jic.ac.uk/GERMPLASM/Wheat-Precise-Genetic-Stocks.htm), CIMMYT wide crossing programme, University of Adelaide, Australia (http://www.agwine.adelaide.edu.au/research/germplasm/), and Nanjing Agriculture University, China. Therefore, here we briefly discuss the genomics interventions for homoeologous recombination and use of flow cytometry for discovery of single nucleotide polymorphisms (SNPs) from wild species facilitating gene discovery.
Bread and durum wheat are tetra- and hexaploid but behave as diploid because synapsis and crossover only occurs between homologous chromosomes, despite highly similar gene order and content between homoeologous chromosomes. Both Riley and Sears observed that homeologous recombination is possible in the 5B chromosome deletion line (Riley and Chapman, 1958; Sears and Okamoto, 1958), which was later defined as the Ph1 locus present on the long arm of chromosome 5B. Moore and co-workers presented a series of experiments on molecular characterization of the Ph1 gene and localized the Ph1 locus to a 2.5-Mb region in smaller chromosomal deletions (Griffiths et al., 2006) containing a cluster of cdc2-like genes (also known as Hyp3 gene), and a segment of heterochromatin within the cluster (Griffiths et al., 2006; Al-Kaff et al., 2008). Greer et al. (2012) also suggested that the cdc2 cluster may alter chromatin structure. Consistent with these results is that treatment with okadaic acid, an inhibitor of phosphatase activity, increases cdk2-type phosphorylation and phenocopies the ph1b allele by inducing crossovers (Knight et al., 2010). Contrary to these reports, Bhullar et al. (2014) proposed C-Ph1 as a candidate for Ph1, as they used virus-induced gene silencing (VIGS) and RNA interference (RNAi) to suppress expression from all C-Ph1 homoeologous copies in wheat, observing meiotic metaphase I chromosome clumping and chromosome associations, similar to the effects occurring in the absence of Ph1. Recently, Hyp3 was annotated as ZIP4 by EMBL-EBI, which is known as a major controller of crossovers in rice (Shen et al., 2012). Sears (1977) developed a single Ph1-deleted mutant referred to as CS ph1B, which has been extensively used globally to introgress chromosome segments from wild relatives into bread wheat. Several other Ph1 mutants have been described, such as ph1c in tetraploid wheat (Giorgi et al., 1978), and the PhI gene from T. speltoides transferred to ‘Chinese Spring’ to develop genetic stock ‘CS PhI, which has shown remarkable crossover between wheat and H. villosa chromosomes (Chen et al., 1994). Recently, two TaZIP4-B2 (Ph1) mutant lines (Cadenza1691 and Cadenza0348) were selected from a Cadenza-mutant population and these exhibited high levels of homeologous crossovers and are claimed to be more suitable than Ph1 locus deletion lines (CS ph1b) (Rey et al., 2017). As the sequence underpinning variation in the ZIP4 gene within the Ph1 locus is clearly known, this demonstrated the potential use of modern genomics tools to precisely develop desired genetic stocks for wheat pre-breeding.
Besides ph1-mediated homoeologous recombination of wheat with wild species, understanding the rate and distribution of recombination in conventional wheat hybridization remains a major challenge. However, little is known about the variation of the genome-wide recombination rate within plant species. Such information could assist in manipulating those loci to break unfavourable linkage blocks and create desirable recombination. Recently, a nested association mapping (NAM) population comprising 60 bi-parental populations from Watkin’s landraces and ‘Paragon’ was characterized and used to build a consensus map, and 114 quantitative trait loci (QTLs) for crossover counting were identified. About 50 % of QTLs with increasing effects were from non-reference parents, and landrace alleles of QTL regions on chromosomes 2D, 3B, 5A and 7A could be good candidates to increase recombination rates (Wingen et al., 2017). However, no practical example to manipulate these QTLs for increasing the rate of recombinations in breeding programmes has yet been demonstrated.
Since Wang et al. (1992) reported the first successful wheat chromosome-enriched library using flow cytometry, it has witnessed remarkable progress due to its promise in reducing the complexity associated with genomics and sequencing application. The analysis of sequences at the single chromosome level has provided new insights into the structure of the complex and polyploid wheat genome, where comparisons between homoeologous chromosomes were the main bottleneck to sequencing and assembly of the genome. Efforts by the International Wheat Genome Sequencing Consortium were based largely on the construction of ready-to-sequence chromosome arm-specific BAC libraries, which indicated that chromosome genomics can contribute materially to the analysis of genomes lacking a high-quality reference sequence (Dolezel et al., 2014). Sequencing single chromosomes has been highly productive in the context of marker development and validation, especially in wheat–alien hybrids. In some cases, alien chromosome differs in DNA content from those of host species; its peak should be recognizable and can therefore be sorted, or chromosome addition or substitution lines are used to sort alien chromosome. Tiwari et al. (2014) provide a classic example, in which a wheat/Ae. geniculata disomic substitution line [DS5Mg S(5D)] was used to sort the 5MgS chromosome and then subjected to next generation sequencing (NGS) for discovery of SNPs specific to the 5MgS chromosome. These 5MgS-specific SNP markers were then used to screen translocations with smallest introgressions carrying genes resistance to leaf rust (Lr47) and stripe rust (Yr40). Hence, powerful molecular marker tools with least ascertainment bias were developed and then used to track the smallest useful introgressions to avoid linkage drag.
MODERN GENOTYPING PLATFORMS FOR WHEAT GENETIC RESOURCES
In the genomics era, rapid advances in biotechnological techniques should be better targeted to mine genetic diversity as part of pre-breeding and be more closely integrated with conventional breeding programmes to achieve better and faster breeding outcomes. Effective genotyping tools should cover complete genomes and should be high-throughput and cost-effective. The key question remains as to what extent have advances in genomics dealt with the two biggest constraints in exploiting genes from distant relatives, namely ascertainment bias and linkage drag? Solving this is only possible if the genotyping platforms are unbiased to structural variations [large insertions or deletions (indels) and copy number variations] and SNPs in WGRs and abundant enough to target the recombination points in introgression lines.
The NGS of mRNA (RNA-seq) in diverse wheat germplasm produces transcript assemblies permitting development of high-density SNP arrays amenable for genotyping large populations for gene mapping and discovery (Cavanagh et al., 2013). However, SNPs in those arrays were biased to the genetic backgrounds of accessions used for RNA-seq, which were predominantly improved varieties and few landraces. Earlier SNP arrays such as the 9K (Cavanagh et al., 2013) and 90K (Wang et al., 2014) arrays had very few SNPs to detect polymorphism in WGRs (Wang et al., 2013). Hence these genotyping arrays remain ineffective in capturing rare variants among diverse alien genetic resources due to ascertainment bias, and result in hampered identification of introduced chromosomal segments and/or alleles from distantly related genetic resources. Winfield et al. (2016) used a different approach by targeted re-sequencing of wheat exomes for identification of SNPs from 43 bread wheat and wild relatives. They were able to develop a wheat 820K Axiom SNP array consisting of polymorphic SNPs from bread wheat varieties, T. monococcum, Ae. tauschii, Th. bessarabicum, Th. poncticum, Th. intermedium, Th. elongatum, S. cereale, Ae. speltoides, Ae. markgrafii, Ae. mutica, Ae. variabilis, T. timopheevii and T. dicoccoides. King et al. (2016) later developed a 35K wheat relative array from a subset of the 820K Axiom array and demonstrated its functionality by developing high-density linkage maps of introgression lines from wheat/Ambylopyrum muticum derivatives. The marker array and millions of data points available through WISP (www.wheatisp.org) are huge resources (Moore, 2015), of which the Axiom 820K and Axiom 35K wheat relative arrays are capable of tracking introgressions from a range of species in the secondary and tertiary gene pools (Winfield et al., 2016).
In addition to fixed array-based genotyping platforms there are de novo genotyping-by-sequencing (GBS)-based platforms that are applicable for various crops regardless of prior knowledge of genomics, genome size, organization or ploidy. In comparison with whole-genome sequencing, reduced representational sequencing has many advantages, such as reducing genome complexity, avoiding inherent ascertainment bias in current fixed SNP arrays, and lower cost. It has been applied in studies on evolutionary genomics, genome-wide association studies (GWAS) and marker-assisted breeding. Parallel efforts have been made to exploit specific locus amplified fragment sequencing (SLAF-seq) in Agropyron (Zhang et al., 2015) and Thinopyrum (Chen et al., 2013) for genome-specific SNP marker discovery. These efforts have demonstrated that successful strategies do exist to identify and exploit rare diversity present in wheat relatives. However, these academic demonstrations should be translated on an applied scale for practical outcomes. We now have high-throughput, cost-effective uniplex genotyping platforms such as Kompetitive Allele-specific PCR (KASP), which can enhance the selection process among wide cross progenies; these capabilities were previously impossible (Rasheed et al., 2016).
These new genotyping platforms have facilitated genetic mapping and gene discovery in wheat (Table 1). Their major application areas are for association of available natural diversity with traits of agronomic importance and improved understanding of the genetics of important traits, but gaps remain in the integration of phenotype–genotype–environment cues, as do the challenges of appropriate statistical design and analysis models to optimize outcomes.
Table 1.
Modern genomics platforms for wheat genetic resources with different breeding and genetics objectives
| Method | Species | Genomics technology/strategy | Reference |
|---|---|---|---|
| Draft or whole-genome sequencing | Aegilops tauschii | Illumina HiSeq2000, Roche 454 | Jia et al. (2013) |
| Triticum turgidum (subsp. dicoccoides) | Whole genome sequencing | Avni et al. (2017) | |
| T. urartu | Illumina HiSeq2000, Roche 454 | Ling et al. (2013) | |
| T. monococcum, T urartu, Ae. sharonensis, Ae. speltoides, Ae. tauschii | Illumina HiSeq2000 | Marcussen et al. (2014) | |
| Transcriptome sequencing | Ae. tauschii | Roche 454 | Iehisa et al. (2014) |
| T. urartu | Roche 454 | Ling et al. (2013) | |
| Ae. sharonensis | Roche 454 | Bouyioukos et al. (2013) | |
| T. urartu | Illumina HiSeq2000 | Krasileva et al. (2013) | |
| T. monococcum | Illumina HiSeq2000 | Fox et al. (2014) | |
| Ae. variabilis | Illumina HiSeq2000 | Xu et al. (2012) | |
| T. turgidum subsp. dicoccoides | Illumina HiSeq2000 | Akpinar et al. (2015) | |
| Genotyping platforms for diversity studies | Ae. tauschii | 10K SNP infinium | Wang et al. (2014) |
| 10 wild species | 820K Affymetrix | Winfield et al. (2016) | |
| Ambylopyrum muticum but applicable to many species | 35K wheat relative array from Affymetrix | King et al. (2016) | |
| Landraces | 35K wheat breeder’s array | Allen et al. (2017) | |
| Thinopyrum elongatum | SLAF-seq | Chen et al. (2013) | |
| Agropyron cristatum | SLAF-seq | Zhang et al. (2015) | |
| Creole landraces | DArTseq | Vikram et al. (2016) | |
| CIMMYT landraces collection | Genotyping-by-sequencing | Sehgal et al. (2015) | |
| Watkins landrace collection | Illumina 90K SNP | Jordan et al. (2015) | |
| Vavilov landrace collection | DArTseq | Riaz et al. (2016) | |
| Watkins landrace collection | Exome capture | Shi et al. (2017) | |
| Wild emmer | Illumina GoldenGate | Ren et al. (2013a, b) | |
| Wheat/Aegilops geniculata | Chromosome-specific SNP discovery | Tiwari et al. (2014) | |
| Gene/allele discovery | Ae. tauschii | Sr33 | Periyannan et al. (2013) |
| T. urartu | Sr22 | ||
| Secale cereale | Sr50 | Mago et al. (2015) | |
| T. dicoccoides | Gpc-B1 | Trick et al. (2012) | |
| Watkins landrace collection | Pin-D1 | Qamar et al. (2014) | |
| Sumai 3 landrace | Fhb1 | Rawat et al. (2016) | |
| Genome-wide association studies | Synthetic hexaploids | Illumina 9K for stripe rust response | Zegeye et al. (2014) |
| Synthetic hexaploids | Illumina 9K for tolerance to boron toxicity | Emebiri and Ogbonnaya (2015) | |
| Watkins landrace collection | Illumina 90K SNP array for rust resistance | Jordan et al. (2015) | |
| Ae. tauschii | 10K Infinium array/agronomic traits | Liu et al. (2015b) | |
| Ae. tauschii | 10K Infinium array/drought tolerance | Qin et al. (2016) | |
| Ae. tauschii | 10K Infinium array/phosphorus deficiency traits | Liu et al. (2015a ) | |
| T. spelta | 15K SNP array/agronomic traits | Würschum et al. (2017) | |
| Ae. sharonensis | Stem rust resistance | Yu et al. (2017) | |
| Wild emmer | Illumina SNP GoldenGate/agronomic traits | Hu et al. (2015) | |
| Genomic predictions | Mexican and Iranian landraces | DArTseq genotyping/agronomic traits | Crossa et al. (2016) |
| Th. intermedium | Genotyping-by-sequencing | Zhang et al. (2016) | |
| Synthetically derived wheats | GBS/heat and drought adaptability | Jafarzadeh et al. (2016) | |
| Landraces | Minerals/GBS | Manickavelu et al. (2017) | |
| Landraces | Rust resistance | Pasam et al. (2017), Daetwyler et al. (2014) |
NEW INSIGHTS INTO GENOME EVOLUTION AND GENETIC DIVERSITY
There is a consensus that bread wheat evolved through two polyploidization events between T. urartu and Ae. speltoides-related species 0.5 Mya forming T. turgidum, and between T. turgidum and Ae. tauschii 10 000 years ago forming modern hexaploid wheat. However, the phylogeny and evolutionary history of progenitor species and Aegilops–Triticum are more complex than our current understanding from nuclear (Marcussen et al., 2014) and chloroplast genome assemblies (Li et al., 2015a). The genome assemblies of bread wheat and five diploid relatives provided novel insight and helped to estimate the evolutionary relatedness and divergence times of ancestral components of bread wheat (Marcussen et al., 2014). Divergence of the A and B genomes from a common ancestor is estimated to have occurred almost 7 Mya, and these genomes gave rise to the D genome through homoploidy 5–6 Mya. These findings laid the foundation for a new framework to understand bread wheat genome as a multi-level phylogenetic mosaic.
This initiated a debate on the origin of hexaploid wheat. Marcussen et al. (2014) proposed a scenario of a homoploid origin of Ae. tauschii through hybridization of ancient A- and B-genome species 5 Mya. Gornicki et al. (2014) later analysed the 25 chloroplast genomes from 13 species, but they did not address the origin of the D genome. Li et al. (2015b) re-evaluated the origin of Ae. tauschii based on data from nuclear and chloroplast genomes and concluded that the homoploid origin of Ae. tauschii was far more complex than envisaged by Marcussen et al. (2014). The nested topology of Ae. tauschii suggested its origin involved multiple rounds of both ancient and recent hybridizations. Sandve et al. (2015) responded to the findings and reported that the disagreement between Marcussen et al. (2014) and Li et al. (2015b) was due to the differences in the nomenclature used to describe the major Triticum/Aegilops clades and that the homoploid hybridization reported by Marcussen et al. (2014) gave rise to an ancestor of all D+S*+M clade species, including Ae. tauschii and Ae. sharonensis, contrary to just Ae. tauschii as reported by Li et al. (2015b). Similarly, the origin of the B genome remains a subject of ongoing debate (Feldman and Levy, 2015), and numerous phylogenetic studies have been carried out since the early work of Zohary and Feldman (1962). Taken together, the findings of these studies suggested two hypotheses. The first was that the progenitor of the B genome is a unique, ancient Aegilops species that remains unknown [i.e. monophyletic origin and ancestor closely related to Ae. speltoides (Sitopsis section)]. The second hypothesis was that the B genome resulted from introgression of several unknown parental Aegilops species (i.e. polyphyletic origin) from the Sitopsis section. Furthermore, El Baidouri et al. (2017) proposed a reconciled evolutionary scenario based on transposable elements which complemented the earlier studies. They concluded that more complete genome sequences from diploid, tetraploid and hexaploid wheat will offer long-term opportunities to improve upon the currently proposed evolutionary scenario to explain how the modern bread wheat genome evolved from its diploid progenitors (El Baidouri et al., 2017).
The past three years have also witnessed an increasing number of reports on using modern genomics technologies to characterize the wheat germplasm collections. SNP arrays and de novo GBS platforms have been used in characterizing landraces (Wingen et al., 2014; Sehgal et al., 2015; Vikram et al., 2016; Riaz et al., 2017), and these offer great potential. If these technologies were integrated with choice of germplasm and appropriate analytical approaches, they could provide solutions to the problems of linkage drag. Vikram et al. (2016) investigated ‘Creole’ wheat accessions held in CIMMYT’s gene bank using the GBS platform, and identified opportunities to harness these materials in developing the next generation of high-yielding wheat varieties. Most of the 8416 ‘Creole’ landraces were genetically similar, but some were adapted to extreme environments, and a reference set of 1133 accessions captured 89 % of the rare alleles present in the whole collection. Parallel studies on characterization of the century-old Vavilov (Riaz et al., 2017) and Watkins (Wingen et al., 2014; Jordan et al., 2015) collections, and of the CIMMYT landrace collection (Sehgal et al., 2015) aimed at mobilizing unused diversity in wheat breeding have also been recently reported.
The A. E. Watkins global landrace collection, consisting of 826 landraces from 32 countries, was established to develop resources for wheat research and breeding (Wingen et al., 2014). The Watkins collection is also a core pillar of the WISP project initiated to capture useful diversity from exotic sources. More insightful analysis of diversity in the Watkins collection using the 35K SNP Breeder’s array identified substantial numbers of novel SNP variants which either have not been captured in current breeding programmes or have been lost through previous selection pressure (Winfield et al., 2017). Genetic diversity studies on this collection led to a core set of 120 accessions capturing most of the inherent diversity. Each of these lines was crossed with variety ‘Paragon’ to generate segregating populations amenable to NAM. These populations, comprising over 9000 unique individuals, were genotyped, to produce more than three million data points (Moore, 2015). These efforts enabled identification of 130 loci for nitrogen-use efficiency and biomass accumulation, and marker-assisted selection (MAS) will allow some of these genes to be deployed in wheat breeding programmes. The Watkins collection was also assessed for diversity at the Pina-D1 and Pinb-D1 loci (Qamar et al., 2014), and for resistance to rusts (Bansal et al., 2013), eyespot (Burt et al., 2014) and root-lesion nematode (Thompson and Seymour, 2011). Similarly, the Vavilov landrace collection of 295 accessions from 25 countries was assessed for diversity using DArT-seq markers and later screened for leaf rust reaction (Riaz et al., 2016). Accessions resistant to leaf rust and phenotyped for other useful characters were hybridized with selected varieties to exploit useful variations through speed-breeding (L. Hickey, University of Queensland, personal communication). Manickavelu et al. (2014) analysed genetic diversity in 446 landraces from Afghanistan using GBS markers and similarly highlighted the importance of exotic alleles present in these resources.
With regard to other species in the Triticeae, Ren et al. (2013a) analysed genetic diversity in wild emmer wheat from Israel and Turkey, and found genetic diversity to be correlated with ecological factors. Similarly, Ren et al. (2013b) evaluated genetic diversity in a worldwide collection of 150 durum accessions and concluded that the richest genetic diversity occurred in South America, North America and Europe. Wang et al. (2013) analysed diversity in global Ae. tauschii collections using the 10K Infinuim SNP array and established two major lineages with little genetic contact. Each of the two lineages has two sub-lineages and it was concluded that lineage 2 in the south-western and southern Caspian region is the main source of the wheat D genome. Winfield et al. (2016) developed a 820K SNP array specific for wheat secondary and tertiary gene pool species, but did not assess diversity in large collections of these species. Thus, the 820K SNP array and its subsequent version, the ‘wheat relative 35K SNP array’, hold promise for studies of genetic diversity to target useful variation in large gene bank collections, and derivatives of wild relatives such as amphiploids and partial amphiploids, chromosome addition lines and translocation lines. These reports on genetic diversity studies are timely and necessitate discussion on prioritization of WGRs, especially landraces, and the use of available innovative genomics technologies to more effectively harness diversity in ancient and close relatives of wheat.
GENOME-WIDE ASSOCIATION STUDIES AND GENOMIC PREDICTIONS IN WGR
Conventional linkage mapping using bi-parental mapping populations is the most common method to detect QTLs for complex traits in plants. However, GWAS is considered a powerful tool for resolving complex trait variations and preliminary identification of loci for novel traits in natural populations. Compared to classical linkage mapping, GWAS provides a more representative gene pool and a higher mapping resolution because all historical meiotic events that have occurred in ancestors of a diverse panel of germplasm can be assessed. Furthermore, GWAS mapping bypasses the expense and time of developing mapping populations, and enables the mapping of many traits in one set of genotypes, making the method more efficient and less expensive than linkage mapping (Huang and Han, 2014). Thus, GWAS mapping is now routinely used in genetic studies.
While linkage mapping and GWAS are the most widely used approaches to understand the genetic architecture of quantitative traits in WGRs, very few examples have led to gene discovery of complexly inherited traits and/or variety development through MAS. For example, the wheat grain gene database lists 1527 QTLs as at the time of this study (https://wheat.pw.usda.go, accessed 30 April 2017). This is consistent with the opinion expressed by Bernardo (2016) that this gap persists despite early optimism that increased knowledge on genetic architecture of quantitative traits through gene mapping will increase our understanding of underlying genes for exploitation in breeding. Successful examples of genes or QTLs from WGR studies being used in breeding include, but are not limited to, the Fhb1 gene from Sumai 3, Gpc-B1 from T. dicoccoides and Pm21 from Hynalida villosa. Others such as Sr2 and Lr34 were selected in conventional breeding by phenotypic markers before the underlying genetics and function were discovered.
The number of studies using GWAS and genomic predictions continue to increase rapidly. SHWs and landraces have been central to such studies (Börner et al., 2015; Jordan et al., 2015), but there have been reports on derivatives of tertiary gene pools such as advanced lines derived from Th. intermedium and Ae. sharonensis. GWAS have been conducted in hexaploid landraces for rust resistances (Jordan et al., 2015; Pasam et al., 2017), agronomic traits and grain mineral contents (Manickavelu et al., 2017). Similarly, tetraploid landraces were studied for agronomic traits (Hu et al., 2015). SHWs were studied for rust resistance (Zegeye et al., 2014), root architecture and multiple disease resistances (Mulki et al., 2013), insect resistances (Joukhadar et al., 2013), boron tolerance (Emebiri and Ogbonnaya, 2015), pre-harvest sprouting response (Imtiaz et al., 2008) and grain morphology (Rasheed et al., 2014) using DArT and SNP markers.
Among primary gene pool species, Ae. tauschii collections have been widely targeted for GWAS of morphological traits (Liu et al., 2015b), phosphorus deficiency (Liu et al., 2015a) and drought tolerance (Qin et al., 2016), and several genomic regions were identified. Spelt wheat (T. spelta), now gaining favour in several parts of the world (Longin and Wurschum, 2016), was also used in GWAS for agronomic and disease resistance traits using a 15K SNP array (Würschum et al., 2017). Among tertiary gene pool species, 125 accessions of the diploid species Ae. sharonensis were used to identify stem rust resistance genes by GWAS, with mapping populations used to validate two Sr genes (Yu et al., 2017). Intermediate wheatgrass (Th. intermedium) has high grain yield, large seed weight and several other desirable agronomic traits. Genomic analysis in this grass species using GBS markers indicated high prediction ability for biomass and grain weight (Zhang et al., 2016). This could be helpful to increase the efficiency of recurrent selection and accelerate domestication and improvement in this species. It has also been argued that associations between SNP alleles and environment of origin in crop landraces reflect adaptation, and these could be used to predict phenotypic variation for adaptive traits. Thus, envirotyping is essential, as evidenced from several recent studies which detected population genetic signatures of adaptation around GWAS loci. Examples can be found in other crops such as African rice, where Meyer et al. (2016) identified 11 significant loci, four of which are within ~300 kb of genomic regions that possess signatures of positive selection for six salt tolerance traits as identified from whole-genome re-sequencing of 93 landraces. Similarly, Lasky et al. (2015, and references therein) reported that genomic studies in sorghum landraces have demonstrated that genome–environment associations (i.e. associations between SNP alleles and environment of origin of accessions) can be used to identify adaptive loci and predict phenotypic variation.
In contrast to GWAS, genomic prediction or genomic selection (GS) studies have gained more importance in breeding, because unlike GWAS and QTL mapping it is not used to identify trait-associated genomic regions, but rather to predict the performance of germplasm based on genomics-estimated breeding values (GEBVs) (Heffnera et al., 2009). GS has emerged as a valuable tool for improving complex traits controlled by QTLs with small effects. Various simulation models for predicting selection accuracy depend largely on marker density, size of training populations (a population subset used to train a best prediction model) and trait heritability. A detailed GS experiment on 97 synthetically derived introgression populations indicated several candidates with higher GEBVs than the respective recurrent bread wheat parents with a clear contribution of synthetic parents in improving grain yield in heat-stressed environments (Jafarzadeh et al., 2016). Similar results were reported by Dunckel et al. (2017) in a GS study on synthetic derivatives from a cross with the bread wheat parent ‘Opata M85’. Optimization of GS in WGRs is now becoming routine in many breeding programmes focused on harnessing new diversity from alien species, and has been explored for domesticating new crops such as Th. intermedium (Zhang et al., 2016), and landraces for rust resistance (Daetwyler et al., 2014; Pasam et al., 2017), mineral contents (Manickavelu et al., 2017), and heat and drought stress adaptation (Crossa et al., 2016). Compared to QTL mapping and GWAS, GS has more promise in harnessing the genetic gains from WGRs for quantitative traits and is seen as a more reliable and useful approach (Bernardo, 2016). However, the key challenges in successful practice of GS depend on: cost-effectiveness and less biased approaches for genotyping; software for handling, quality control and joint analysis of genotypic, phenotypic and environment data; and a streamlined work flow for using GS within the overall breeding pipeline.
GENE DISCOVERY AND ISOLATION USING MODERN GENOMICS TOOLS
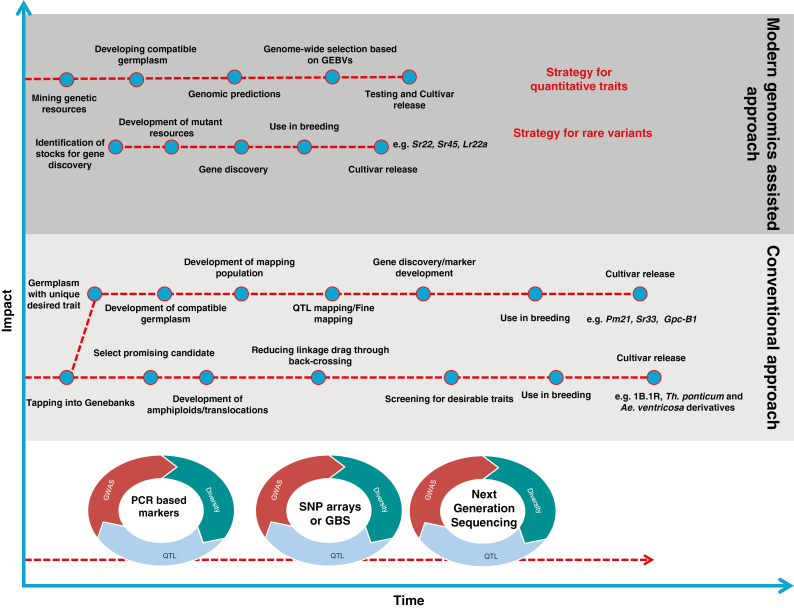
Gene discovery in WGRs is a relatively long process compared to gene discovery in adapted germplasm. This is mainly due to the time required for introgression into adapted wheat backgrounds, followed by QTL mapping, validation, fine mapping and subsequent gene cloning (Fig. 2). Given previous reviews in the use of WGRs (Mujeeb-Kazi et al., 2013; Longin and Reif, 2014), we will focus here on recent advances in genomics that have significantly shortened the gene discovery process in WGRs. Due to affordable, high-throughput sequencing technologies innovative approaches are emerging to discover genes in wild relatives. For example, well-annotated genes with distinctive functions and sequences can be captured using gene family-specific oligonucleotide probes, which are then sequenced and assembled to provide the genetic information of the gene in wild relatives. This strategy has been successfully used to discover and clone genes underlying stem rust resistance in Ae. tauschii (Steuernagel et al., 2016). This recently developed technology is fast and does not rely on recombinant populations or fine mapping. It combines mutagenesis and genome complexity reduction and is referred to as resistance-gene enrichment sequencing (RenSeq or MutRenSeq). The stem rust resistance gene Sr50 derived from S. cereale was also cloned using mutant populations and sequencing of BAC clones (Mago et al., 2015). Thind et al. (2017) reported ‘targeted chromosome-based cloning via long range assembly’, which combines complexity-reduction via chromosome flow sorting and also works in chromosome regions with reduced recombination rates. They cloned the broad-spectrum leaf rust resistance gene Lr22a in wheat with de novo genome assembly, marker information and mutant population. Another strategy that holds promise for gene discovery in WGRs is through RNAseq, which involves SNP discovery in bulks with contrasting phenotypes (referred to as bulked segregant analysis) and has recently been used to fine map the Gpc-B1 (Trick et al., 2012) and Yr15 (Ramirez-Gonzalez et al., 2015) genes from T. turgidum subsp. dicoccoides and T. dicoccoides, respectively.
Fig. 2.
Research and breeding activities surrounding wheat genetic resources plotted against time (x-axis) and impact on wheat improvement (y-axis). QTL mapping, GWAS and genetic diversity studies will continue with discovery of new technologies and have given low impact as very little has been converted into resources that are actually used in breeding and gene discovery. The major impact of genomics technologies will be in reducing the time to characterize and deploy useful variation with reduced linkage drag.
GENOME EDITING TOOLS HOLD PROMISE FOR GENE DISCOVERY IN WGR
The recent developments in genome editing based on site-specific nucleases offer exciting potential to precisely edit targeted genes with greater speed. These methods include zinc finger nuclease (ZFN), transcription activator-like effector nuclease (TALEN) and clustered regulatory interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9), which were originally developed in non-plant systems and are now being adopted to modify genes in crop plants (Lozano-Juste and Cutler, 2014). These technologies make use of sequence-specific designer nucleases that cleave targeted loci to create small indels of novel DNA, and even replacement of individual alleles. ZFN and TALEN suffer from difficulties in design, construction, cost and uncertain success rates. By contrast, CRISPR-Cas9 is robust, affordable and easy to use and has proved useful across a range of plant species (Jiang et al., 2013). Although reports on the use of CRISPR-Cas9 are limited, it is expected that large-scale germplasm-characterization efforts in conjunction with CRISPR-based genome-editing technologies will herald a new era whereby crop plants can be precisely modified without necessarily needing to use physical seed samples that contain important traits. Starting with genetically less complex traits and large-effect alleles, breeders will increasingly be able to ‘overwrite’ undesirable allelic variants in otherwise elite material using beneficial variants discovered through genetic analyses of other plant genetic resources. It is relatively simple to demonstrate correlative associations between sequence variants and phenotypic traits, but it can take years to demonstrate causation using laborious approaches such as fine mapping, expression analysis, biochemical modelling and other functional genomics tools. CRISPR-Cas9 offers a shortcut to establish causation quickly by editing candidate causal variants (Li et al., 2013). As ‘digitalization’ of genetic resources unfolds, the role and significance of gene banks will grow in that they will transform into biological discovery platforms and enhance the role of crop diversity as a source of haplotypes encoding desirable traits. Another area where gene editing could hold promise is in inactivating undesirable dragged genes, a major impediment in the use of wild relatives in breeding because introduced chromosome segments come as variably sized linkage blocks that may carry genes conferring negative impacts on traits of economic interest. A classic example is the 1RS component of the T1BL.1RS translocation carrying a gene(s) for sticky dough.
REGULATIONS ON GERMPLASM ACCESS AND DATA SHARING
Ownership of genetic resources and genes, and better access to genomics database resources in an era of information technology must be resolved to ensure easier access to genetic resources worldwide. A fundamental principle for granting access to genetic resources must be an assurance of fair and equitable sharing of the benefits arising from their use. The huge investments made by private companies in recombinant DNA technologies in the 1980s and release of commercial genetically modified (GM) crops greatly contributed to strengthening intellectual property rights (IPR), a trend that has gained strength since adoption of the Trade-Related Aspects of Intellectual Property Rights (TRIPS) Agreement. In response to concerns that IPR were being detrimental to global progress in plant improvement, new policy frameworks emerged to regulate access to, and use of, genetic resources. The first legally binding intergovernmental agreement was the convention of biological diversity (CBD) in 1993, which established access and benefit-sharing principles by which anyone wishing to access genetic resources from a particular country must obtain prior informed consent (PIC) from relevant authorities and establish mutually agreed terms (MAT) specifying the conditions under which the resources can be used. The CBD was seen as a significant treaty for consolidating efforts to conserve biodiversity, but was also seen as a barrier to sharing genetic material, including domesticated plants and their wild relatives. The Plant Treaty also known as the International Treaty on Plant Genetic Resources for Food and Agriculture (ITPFGR) that came into force in 2004 was designed to operate in harmony with the CBD, and was the first agreement to recognize farmers’ rights. It established a multilateral system of access and benefit sharing regulated by a standard material transfer agreement (SMTA) that defines non-negotiable MAT. The Plant Treaty’s objectives are the conservation and sustainable use of PGRFAs and the fair and equitable sharing of benefits arising out of their use for sustainable agriculture and food security.
With current advancements in the genomics of wheat genetic resources, we can now predict agronomic potential using genome-wide marker scans or select plants with specific genes for biotic and abiotic resistances without exposing them to the relevant stresses. The need for wheat genetic resources will grow as a result of these technologies, which will enable breeders to eliminate 70–80 % of individuals in any generation without having to invest in laborious multi-environmental field testing. Therefore, a more efficient regulatory framework for better access to germplasm for both public and commercial breeding purposes will facilitate the deployment of recent and emerging advances so that global impacts that were previously not possible could be catalysed. Initiatives such as WISP and SeeD are classical examples where germplasm resources are freely available on request along with genotyping data. For example, a toolkit comprising 40 lines derived from landrace/’Paragon’ crosses, and capturing diversity in specific traits, has been provided to breeders, and is freely available to all on request in the WISP project. These resources could be particularly useful for less developed countries that lack sophisticated genotyping facilities. Out-sourcing is also an increasingly common practice that is both time- and cost-effective (Rossetto and Henry, 2014). As the germplasm and genomic data could be easily available, more investment is needed to train scientists from developing countries in data analysis and decision support tools to enable global exploitation of WGRs. Considerable investment in data management and informatics systems will be required to ensure that gene banks can fulfil their roles as custodians of the expanding genetic knowledge linked to their physical resources. This view was echoed by Shaw et al. (2017), who reported that the ability to effectively manage genetic resource collections and integrate unique and diverse data types is crucial in exploring, understanding and exploiting the diversity contained within gene banks. A ‘one shoe fits all’ approach that promotes a common platform may negate the achievement of that goal. Instead we posit that there is some sense of urgency in directing efforts towards ensuring that the disparate gene bank databases are interoperable, ontologies for collection and characterization are harmonized, and ego-geographical, edaphic and biotic information for the accessions be strengthened at points of collection. While recent advances in genotyping technologies will enhance the use of genetic resources, that by itself is not enough, and must be coupled with advances in phenotyping tools integrated with advances in sensing technologies that link environmental cues with phenotypic plasticity and genotypic (DNA, proteomic and metabolic) signatures if the goals of achieving food security and improving current agricultural output are to be met.
TIME FACTOR IN EXPLOITATION OF GENETIC RESOURCES: MAKING LONG JOURNEY SHORTER
A major constraint in using WGRs in breeding programmes is the time factor associated with difficulties in evaluating WGRs with widely different phenologies of growth habit, flowering time and height. These differences make accurate assessments and comparisons difficult, if not impossible. Even if useful characteristics can be identified the difficulties of transferring desirable traits into cultivated species are considerable, often requiring embryo rescue and cytological expertise. The time required to transfer traits from WGRs and to enable their use in breeding often may exceed that in conventional breeding programmes. The most important impact from innovative genomics technologies in utilizing WGRs will be reducing the time to harness beneficial alleles with reduced linkage drag and increased capacity to do so (Fig. 2).
The recent advances in the development of high-throughput, time-saving methods fall into five categories:
Rapid generation advancements: CIMMYT introduced the ‘shuttle breeding’ strategy, which moves germplasm between contrasting environments: Ciudad Obregon in north-western Mexico and Toluca in the highlands of Central Mexico, enabling two generations per year. This has enabled screening for a range of traits such as photo-period sensitivity, heat tolerance and arrangement of important biotic and abiotic stresses (Ortiz et al., 2007). Double haploids (DHs) and single seed descent rapidly bring about homozygosity, but usually without selection. A new generation advancement method called ‘speed breeding’ uses constant light and precisely controlled temperature to accelerate plant growth and development (Hickey et al., 2012, 2017). Screening protocols can be combined with speed breeding (Riaz et al., 2016; L. Hickey, personal communication). Several advantages in using speed breeding include: (a) 7 weeks to complete one cycle, (b) controlled environmental factors for homogenous treatment of populations for screening, and (c) can be practised year-round. Based on these points, speed breeding holds significant promise to accelerate exploitation of WGRs in a time-efficient manner. Similarly, Zheng et al. (2013) reported a procedure which combines embryo culture with management of watering regimes, lighting intensity and duration, temperature and quantity of potting mixture that allows the production of up to eight generations of wheat and nine generations of barley per year. However, a drawback may be its limitation in mimicking diverse environmental factors attainable under field conditions.
Rapid gene cloning through mutation and genomics: WGRs are largely seen as sources of rare variants controlled by major genes, not specifically for quantitative traits. The discovery of such genetic factors in WGRs is now a rapid process due to the advancements in genomics, and we have seen discovery of several rust resistance genes from Ae. tauschii and T. monococcum within a few years (Periyannan et al., 2013; Steuernagel et al., 2016). The three-step method (MutRenSeq) that combines mutagenesis with exome capture and sequencing provided a rapid way to clone R genes from wheat wild relatives, a classical example being discovery of Sr22 from T. monococcum and Sr45 from Ae. tauschii. MutRenSeq is fast, cheap and takes <24 months to complete because it is independent of fine mapping, uses the generation of a physical contig across the map interval and is easily scalable. This approach can be applied to most crops or their wild relatives, and will allow the cloning of R genes that could be used in multi-R gene pyramids, a strategy that promises more durable disease resistance in crops. Thind et al. (2017) reported ‘targeted chromosome-based cloning via long read assembly (TACCA)’, which offers great flexibility with respect to gene validation and could be used for traits with partial phenotypes such as partial disease resistance or abiotic stress tolerance. They also compared various rapid gene cloning methods and concluded that TACCA holds significant promise and is equivalent to positional cloning. Unfortunately, gene discovery for quantitative traits is still lagging behind because it depends on heritability of trait, number of genes, extent of quantitative variation, and size and position of recombinants in the genome. Bulk-segregation analysis by making bulks of contrasting phenotypes followed by exome-capture or NGS could be an effective strategy to underpin genes for quantitative traits in WGRs, and it has been successfully used to identify the genetic basis of kernel numbers per row in maize (Yang et al., 2015) and holds promise for underlying gene discovery for quantitative traits in WGRs.
-
High-throughput genotyping and phenotyping platforms: In recent years, there have been rapid advances in high-throughput genotyping arrays or GBS. It is now possible to genotype hundreds of samples for high-density markers within a couple of days (Rasheed et al., 2017). The 820K and 35K wheat SNP chips, GBS and DArTseq platforms are classical examples. However, accurate phenotyping of growth, yield and stress responses in both the field and controlled environments remains a major bottleneck in both pre-breeding and breeding.
Recent developments in high-throughput phenotyping based on imaging (Fahlgren et al., 2015), spectral reflectance and remote-sensing (Araus and Cairns, 2014) will make significant impacts with regard to characters such as variation for leaf surface temperature associated with osmotic components of salinity stress and quantified by high-throughput infrared thermography (Z. Khan et al., Quaid-i-Azam University, Pakistan, unpublished data). Although there is still no demonstration on the application of high-throughput phenomics on WGRs or wild species, they are likely to bridge the phenome–genome gap. Recent and continuing advances in computing, robotics, machine vision and image analysis to the wider field of plant biology will make significant contributions in phenotying (Furbank and Tester, 2011). The ultimate benefit will be to what extent they contribute to achieving improved rates of genetic gain by significantly influencing the key components: selection intensity, selection accuracy, and identification and introgression of desirable genetic variation within a short period of time.
High-throughput marker-assisted selection strategies: Progress in developing high-throughput single marker genotyping was relatively slow compared to current high-density genotyping platforms. Single marker genotyping is very important for wheat pre-breeding programmes because breeders are much more interested in deploying specific alleles from WGRs: for example, screening for the presence of alien translocations, or new rust resistance genes. A high-throughput KASP marker toolkit for wheat breeding that includes markers for the 1B.1R translocation, Lr37, Yr15 and several other alleles for agronomic traits was recently developed (Rasheed et al., 2016). KASP markers have scalable flexibility without compromising data throughput and it is possible to obtain 150K data points in one day. KASP is a commercial technology involving chemical reagents from LGC Genomics, and it is a relatively expensive procedure. A step change in lowering costs of single marker genotyping is semi-thermal asymmetric reverse PCR (STARP) markers, which unlike KASP markers, can be used with many types of commercial chemical reagents (Long et al., 2016).
Rapid gene editing technologies: The establishment of efficient and specific CRISPR/Cas9-mediated gene editing methods with decreased off-target mutations in a short period of time is a major focus for researchers. Liang et al. (2017) recently established a DNA-free gene editing method using CRISPR/Cas9 ribonucleoproteins (RNPs), taking only 7–9 weeks with 4–5 independent mutants produced from 100 immature wheat embryos. The main steps of this method include RNP preparation, RNP functional validation, RNP coating and delivery, plantlet regeneration and mutant identification. The most important advantage of CRISPR/Cas9 RNP-mediated genome editing is elimination of transgene integration and the small DNA insertions that can be generated. This is highly desirable for public acceptance of genome-edited plants and holds significant promise for exploitation of WGRs within reduced time periods.
CONCLUSION AND FUTURE PROSPECTS
The success of a programme, in our view, is global impact using new diversity that can easily be manipulated to generate user-friendly genetic stocks that are unique, readily accessible to both public and private sectors, amenable to immediate use in breeding, and globally distributed, as originally occurred with synthetic wheats produced at CIMMYT and elsewhere. It was those collective features that catalysed their worldwide use and subsequent release of more than 30 high-yielding varieties in Mexico, China, Pakistan, USA and other countries. Synthetically derived germplasm currently constitutes about 30 % of entries in international nurseries distributed annually to over 150 countries by CIMMYT and ICARDA. Despite the proven evidential value of synthetic hexaploid wheats and their derivatives in contributing novel alleles for wheat improvement, such materials have scarcely been characterized using current genomics resources and statistical genetic methods. In addition, there are also few extensive wide crossing programmes at present, and most scientists previously trained in cytogenetics have moved to other areas. In a world that is reluctant to accept transformation technologies in our major food crops, training in cytogenetics could give rapid returns in translating academic research into breeding outcomes. The value of deploying advances in genomics, phenotyping, envirotyping and statistical tools to optimize the benefits in SHW as a valuable genetic diversity for wheat improvement also lies in the fact that they can be directly hybridized with cultivated wheat. Widely shared germplasm derived from wide crosses and open-access databases for WGRs will enhance more expanded collaborations including developing countries. It is timely to undertake a critical appraisal of the prioritization of WGRs that will be appropriate to meet production targets set by policy-makers around current production constraints within reasonable time lines by harnessing and optimizing existing resources. The urgency is now more compelling if the sustainability goals are to be attained.
ACKNOWLEDGEMENTS
We are grateful to Prof. Bob McIntosh (University of Sydney, Australia) and Dr Peter Langridge (University of Adelaide, Australia) for reviews of the manuscript.
LITERATURE CITED
- Akpinar BA, Lucas SJ, Vrana J, Dolezel J, Budak H. 2015. Sequencing chromosome 5D of Aegilops tauschii and comparison with its allopolyploid descendant bread wheat (Triticum aestivum). Plant Biotechnology Journal 13: 740–752. [DOI] [PubMed] [Google Scholar]
- Al-Kaff N, Knight E, Bertin I et al. 2008. Detailed dissection of the chromosomal region containing the Ph1 locus in wheat Triticum aestivum: with deletion mutants and expression profiling. Annals of Botany 101: 863–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen AM, Winfield MO, Burridge AJ et al. 2017. Characterization of a Wheat Breeders’ Array suitable for high-throughput SNP genotyping of global accessions of hexaploid bread wheat (Triticum aestivum). Plant Biotechnology Journal 15: 390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali N, Heslop-Harrison JP, Ahmad H et al. 2016. Introgression of chromosome segments from multiple alien species in wheat breeding lines with wheat streak mosaic virus resistance. Heredity 117: 114–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araus JL, Cairns JE. 2014. Field high-throughput phenotyping: the new crop breeding frontier. Trends in Plant Science 19: 52–61. [DOI] [PubMed] [Google Scholar]
- Avni R, Nave M, Barad O et al. 2017. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357: 93–97. [DOI] [PubMed] [Google Scholar]
- Bansal UK, Arief VN, DeLacy IH, Bariana HS. 2013. Exploring wheat landraces for rust resistance using a single marker scan. Euphytica 194: 219–233. [Google Scholar]
- Bernardo R. 2016. Bandwagons I, too, have known. Theoretical and Applied Genetics 129: 2323–2332. [DOI] [PubMed] [Google Scholar]
- Bhullar R, Nagarajan R, Bennypaul H et al. 2014. Silencing of a metaphase I-specific gene results in a phenotype similar to that of the Pairing homeologous 1 (Ph1) gene mutations. Proceedings of the National Academy of Sciences of the United States of America 111: 14187–14192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner A, Ogbonnaya FC, Röder MS, Rasheed A, Periyannan S, Lagudah ES. 2015. Aegilops tauschii introgressions in wheat. In Molnár-Láng M, Ceoloni C, Doležel J, eds. Alien introgression in wheat. Switzerland: Springer International. [Google Scholar]
- Bouyioukos C, Moscou MJ, Champouret N, Hernandez-Pinzon I, Ward ER, Wulff BB. 2013. Characterisation and analysis of the Aegilops sharonensis transcriptome, a wild relative of wheat in the Sitopsis section. PLOS One 8: e72782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt C, Griffe LL, Ridolfini AP, Orford S, Griffiths S, Nicholson P. 2014. Mining the Watkins collection of wheat landraces for novel sources of eyespot resistance. Plant Pathology 63: 1241–1250. [Google Scholar]
- Cavanagh CR, Chao SM, Wang SC et al. 2013. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proceedings of the National Academy of Sciences of the United States of America 110: 8057–8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PD, Tsujimoto H, Gill BS. 1994. Transfer of PhI genes promoting homoeologous pairing from Triticum speltoides to common wheat. Theoretical and Applied Genetics 88: 97–101. [DOI] [PubMed] [Google Scholar]
- Chen S, Huang Z, Dai Y et al. 2013. The development of 7E chromosome-specific molecular markers for Thinopyrum elongatum based on SLAF-seq technology. PLOS One 8: e65122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossa J, Jarquin D, Franco J et al. 2016. Genomic prediction of gene bank wheat landraces. G3 (Bethesda) 6: 1819–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler HD, Bansal UK, Bariana HS, Hayden MJ, Hayes BJ. 2014. Genomic prediction for rust resistance in diverse wheat landraces. Theoretical and Applied Genetics 127: 1795–1803. [DOI] [PubMed] [Google Scholar]
- Dolezel J, Vrana J, Capal P, Kubalakova M, Buresova V, Simkova H. 2014. Advances in plant chromosome genomics. Biotechnology Advances 32: 122–136. [DOI] [PubMed] [Google Scholar]
- Dunckel S, Crossa J, Wu S, Bonnett D, Poland J. 2017. Genomic selection for increased yield in synthetic-derived wheat. Crop Science 57: 713–725. [Google Scholar]
- El Baidouri M, Murat F, Veyssiere M et al. 2017. Reconciling the evolutionary origin of bread wheat (Triticum aestivum). New Phytologist 213: 1477–1486. [DOI] [PubMed] [Google Scholar]
- Emebiri LC, Ogbonnaya FC. 2015. Exploring the synthetic hexaploid wheat for novel sources of tolerance to excess boron. Molecular Breeding 35: 68. [Google Scholar]
- Endo TR, Gill BS. 1996. The deletion stocks of common wheat. Heredity (Edinb) 87: 295–307. [Google Scholar]
- Fahlgren N, Gehan MA, Baxter I. 2015. Lights, camera, action: high-throughput plant phenotyping is ready for a close-up. Current Opinion in Plant Biology 24: 93–99. [DOI] [PubMed] [Google Scholar]
- Feldman M, Levy AA. 2015. Origin and evolution of wheat and related Triticeae species. In. Alien introgression in Wheat. Springer International. [Google Scholar]
- Fox SE, Geniza M, Hanumappa M et al. 2014. De novo transcriptome assembly and analyses of gene expression during photomorphogenesis in diploid wheat Triticum monococcum. PLOS One 9: e96855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furbank RT, Tester M. 2011. Phenomics – technologies to relieve the phenotyping bottleneck. Trends in Plant Science 16: 635–644. [DOI] [PubMed] [Google Scholar]
- Garcia-Olmedo F, Delibes A, Sanchez-Monge R. 1977. Transfer of resistance to eyespot disease from Aegilops ventricosa to wheat Proceedings of the 8th Congress of Eucarpia.
- Giorgi B. 1978. A homoeologous pairing mutant isolated in Triticum durum cv. Cappelli. Mutation Breeding Newsletter 11: 4–5. [Google Scholar]
- Gornicki P, Zhu H, Wang J et al. 2014. The chloroplast view of the evolution of polyploid wheat. New Phytologist 204: 704–714. [DOI] [PubMed] [Google Scholar]
- Greer E, Martin AC, Pendle A et al. 2012. The Ph1 locus suppresses Cdk2-type activity during premeiosis and meiosis in wheat. Plant Cell 24: 152–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths S, Sharp R, Foote TN et al. 2006. Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439: 749–752. [DOI] [PubMed] [Google Scholar]
- Heffnera EL, Sorrellsa ME, Jannink J. 2009. Genomic selection for crop improvement. Crop Science 49: 1–12. [Google Scholar]
- Hickey LT, Wilkinson PM, Knight CR et al. 2012. Rapid phenotyping for adult-plant resistance to stripe rust in wheat. Plant Breeding 131: 54–61. [Google Scholar]
- Hickey L, Germán SE, Pereyra SA et al. 2017. Speed breeding for multiple disease resistance in barley. Euphytica 213: 64. [Google Scholar]
- Hu X, Ren J, Ren X et al. 2015. Association of agronomic traits with SNP markers in durum wheat (Triticum turgidum L. durum (Desf.)). PLOS One 10: e0130854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Han B. 2014. Natural variations and genome-wide association studies in crop plants. Annual Review of Plant Biology 65: 531–551. [DOI] [PubMed] [Google Scholar]
- Iehisa JCM, Shimizu A, Sato K et al. 2014. Genome-wide marker development for the wheat D genome based on single nucleotide polymorphisms identified from transcripts in the wild wheat progenitor Aegilops tauschii. Theoretical and Applied Genetics 127: 261–271. [DOI] [PubMed] [Google Scholar]
- Imtiaz M, Ogbonnaya FC, Oman J, van G inkel M. 2008. Characterization of quantitative trait loci controlling genetic variation for preharvest sprouting in synthetic backcross-derived wheat lines. Genetics 178: 1725–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafarzadeh J, Bonnett D, Jannink JL, Akdemir D, Dreisigacker S, Sorrells ME. 2016. Breeding value of primary synthetic wheat genotypes for grain yield. PLOS One 11: e0162860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia JZ, Zhao SC, Kong XY et al. 2013. Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496: 91–95. [DOI] [PubMed] [Google Scholar]
- Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP. 2013. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Research 41: e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan KW, Wang S, Lun Y et al. 2015. A haplotype map of allohexaploid wheat reveals distinct patterns of selection on homoeologous genomes. Genome Biology 16: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joukhadar R, El-Bouhssini M, Jighly A, Ogbonnaya FC. 2013. Genome-wide association mapping for five major pest resistances in wheat. Molecular Breeding 32: 943–960. [Google Scholar]
- Khlestkina EK. 2014. Current applications of wheat and wheat–alien precise genetic stocks. Molecular Breeding 34: 273–281. [Google Scholar]
- King J, Grewal S, Yang CY et al. 2016. A step change in the transfer of interspecific variation into wheat from Amblyopyrum muticum. Plant Biotechnology Journal 15: 217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight E, Greer E, Draeger T et al. 2010. Inducing chromosome pairing through premature condensation: analysis of wheat interspecific hybrids. Functional and Integrative Genomics 10: 603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva KV, Buffalo V, Bailey P et al. 2013. Separating homeologs by phasing in the tetraploid wheat transcriptome. Genome Biology 14: R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasky JR, Upadhyaya HD, Ramu P et al. 2015. Genome-environment associations in sorghum landraces predict adaptive traits. Science Advances 1: e1400218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CN, Snape JW, Worland AJ. 1987. Aneuploidy in wheat and its uses in genetic analysis. In Lupton FGH, ed. Wheat breeding. Its scientific basis. London: Chapman & Hall. [Google Scholar]
- Li JF, Norville JE, Aach J et al. 2013. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nature Biotechnology 31: 688–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LF, Liu B, Olsen KM, Wendel JF. 2015a. Multiple rounds of ancient and recent hybridizations have occurred within the Aegilops-Triticum complex. New Phytologist 208: 11–12. [DOI] [PubMed] [Google Scholar]
- Li LF, Liu B, Olsen KM, Wendel JF. 2015b. A re-evaluation of the homoploid hybrid origin of Aegilops tauschii, the donor of the wheat D-subgenome. New Phytologist 208: 4–8. [DOI] [PubMed] [Google Scholar]
- Liang Z, Chen K, Li T et al. 2017. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nature Communications 8: 14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Zhao SC, Liu DC et al. 2013. Draft genome of the wheat A-genome progenitor Triticum urartu. Nature 496: 87–90. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang L, Deng M et al. 2015a. Genome-wide association study of phosphorus-deficiency-tolerance traits in Aegilops tauschii. Theoretical and Applied Genetics 128: 2203–2212. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang L, Mao S et al. 2015b. Genome-wide association study of 29 morphological traits in Aegilops tauschii. Scientific Reports 5: 15562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long YM, Chao WS, Ma GJ, Xu SS, Qi LL. 2016. An innovative SNP genotyping method adapting to multiple platforms and throughputs. Theoretical and Applied Genetics 130: 597–607. [DOI] [PubMed] [Google Scholar]
- Longin CF, Reif JC. 2014. Redesigning the exploitation of wheat genetic resources. Trends in Plant Science 19: 631–636. [DOI] [PubMed] [Google Scholar]
- Longin CF, Wurschum T. 2016. Back to the future – Tapping into ancient grains for food diversity. Trends in Plant Science 21: 731–737. [DOI] [PubMed] [Google Scholar]
- Lozano-Juste J, Cutler SR. 2014. Plant genome engineering in full bloom. Trends in Plant Science 19: 284–287. [DOI] [PubMed] [Google Scholar]
- Mago R, Zhang P, Vautrin S et al. 2015. The wheat Sr50 gene reveals rich diversity at a cereal disease resistance locus. Nature Plants 1: 15186. [DOI] [PubMed] [Google Scholar]
- Manickavelu A, Jighly A, Ban T. 2014. Molecular evaluation of orphan Afghan common wheat (Triticum aestivum L.) landraces collected by Dr. Kihara using single nucleotide polymorphic markers. BMC Plant Biology 14: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manickavelu A, Hattori T, Yamaoka S et al. 2017. Genetic nature of elemental contents in wheat grains and its genomic prediction: toward the effective use of wheat landraces from Afghanistan. PLOS One 12: e0169416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcussen T, Sandve SR, Heier L et al. 2014 Ancient hybridizations among the ancestral genomes of bread wheat. Science 345: 1250092. [DOI] [PubMed] [Google Scholar]
- McCouch S, Baute GJ, Bradeen J et al. 2013. Agriculture: feeding the future. Nature 4: 23–24. [DOI] [PubMed] [Google Scholar]
- Meyer RS, Choi JY, Sanches M et al. 2016. Domestication history and geographical adaptation inferred from a SNP map of African rice. Nature Genetics 48: 1083–1088. [DOI] [PubMed] [Google Scholar]
- Moore G. 2014. The control of recombination in wheat by Ph1 and its use in breeding. Methods in Molecular Biology 1145: 143–153. [DOI] [PubMed] [Google Scholar]
- Moore G. 2015. Strategic pre-breeding for wheat improvement. Nature Plants 1: 15018. [DOI] [PubMed] [Google Scholar]
- Mujeeb-Kazi A, Kazi AG, Dundas I et al. 2013. Genetic diversity for wheat improvement as a conduit to food security. Advances in Agronomy 122: 179–257. [Google Scholar]
- Mulki MA, Jighly A, Ye GY et al. 2013. Association mapping for soilborne pathogen resistance in synthetic hexaploid wheat. Molecular Breeding 31: 299–311. [Google Scholar]
- Ogbonnaya FC, Abdalla O, Mujeeb-Kazi A et al. 2013. Synthetic hexaploids: harnessing species of the primary gene pool for wheat improvement. Plant Breeding Reviews 37: 35–122. [Google Scholar]
- Ortiz R, Braun HJ, Crossa J et al. 2008. Wheat genetic resources enhancement by the International Maize and Wheat Improvement Center (CIMMYT). Genetic Resources and Crop Evolution 55: 1095–1140. [Google Scholar]
- Ortiz R, Trethowan R, Ferrara GO et al. 2007. High yield potential, shuttle breeding, genetic diversity, and a new international wheat improvement strategy. Euphytica 157: 365–384. [Google Scholar]
- Pasam RK, Bansal U, Daetwyler HD et al. 2017. Detection and validation of genomic regions associated with resistance to rust diseases in a worldwide hexaploid wheat landrace collection using BayesR and mixed linear model approaches. Theoretical and Applied Genetics 130: 777–793. [DOI] [PubMed] [Google Scholar]
- Patokar C, Sepsi A, Schwarzacher T, Kishii M, Heslop-Harrison JS. 2016. Molecular cytogenetic characterization of novel wheat-Thinopyrum bessarabicum recombinant lines carrying intercalary translocations. Chromosoma 125: 163–172. [DOI] [PubMed] [Google Scholar]
- Periyannan S, Moore J, Ayliffe M et al. 2013. The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science 341: 786–788. [DOI] [PubMed] [Google Scholar]
- Pozniak C, Hucl PJ, Stein N et al. 2017. Genome assemblies of elite cultivars provides insights into the wheat pan-genome. In: Proceedings of 13th International Wheat Genetics Symposium, April 23–28, Tulln, Austria, 46. [Google Scholar]
- Qamar Z, Bansal UK, Dong CM, Alfred RL, Bhave M, Bariana HS. 2014. Detection of puroindoline (Pina-D1 and Pinb-D1) allelic variation in wheat landraces. Journal of Cereal Science 60: 610–616. [Google Scholar]
- Qin P, Lin Y, Hu Y et al. 2016. Genome-wide association study of drought-related resistance traits in Aegilops tauschii. Genetics and Molecular Biology 39: 398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Gonzalez RH, Segovia V, Bird N et al. 2015. RNA-Seq bulked segregant analysis enables the identification of high-resolution genetic markers for breeding in hexaploid wheat. Plant Biotechnology Journal 13: 613–624. [DOI] [PubMed] [Google Scholar]
- Rasheed A, Xia X, Ogbonnaya F et al. 2014. Genome-wide association for grain morphology in synthetic hexaploid wheats using digital imaging analysis. BMC Plant Biology 14: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed A, Wen W, Gao FM et al. 2016. Development and validation of KASP assays for functional genes underpinning key economic traits in wheat. Theoretical and Applied Genetics 129: 1843–1860. [DOI] [PubMed] [Google Scholar]
- Rasheed A, Hao Y, Xia XC et al. 2017. Crop breeding chips and genotyping platforms: progress, challenges and perspectives. Molecular Plant 10: 1047–1064. [DOI] [PubMed] [Google Scholar]
- Rawat N, Pumphrey MO, Liu S et al. 2016. Wheat Fhb1 encodes a chimeric lectin with agglutinin domains and a pore-forming toxin-like domain conferring resistance to Fusarium head blight. Nature Genetics 48: 1576–1580. [DOI] [PubMed] [Google Scholar]
- Ren J, Chen L, Sun D et al. 2013a. SNP-revealed genetic diversity in wild emmer wheat correlates with ecological factors. BMC Evolutionary Biology 13: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Sun D, Chen L et al. 2013b. Genetic diversity revealed by single nucleotide polymorphism markers in a worldwide germplasm collection of durum wheat. International Journal of Molecular Sciences 14: 7061–7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey MD, Martin AC, Higgins J et al. 2017. Exploiting the ZIP4 homologue within the wheat Ph1 locus has identified two lines exhibiting homoeologous crossover in wheat-wild relative hybrids. Molecular Breeding 37: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaz A, Athiyannan N, Periyannan S et al. 2016. Mining Vavilov’s treasure chest of wheat diversity for adult plant resistance to Puccinia triticina. Plant Disease 101: 317–323. [DOI] [PubMed] [Google Scholar]
- Riaz A, Hathorn A, Dinglasan E et al. 2017. Into the vault of the Vavilov wheats: old diversity for new alleles. Genetic Resources and Crop Evolution 64: 531–544. [Google Scholar]
- Riley R, Chapman V. 1958. Genetic control of the cytologically diploid behaviour of hexaploid wheat. Nature 182: 713–715. [Google Scholar]
- Rossetto M, Henry RJ. 2014. Escape from the laboratory: new horizons for plant genetics. Trends in Plant Science 19: 554–555. [DOI] [PubMed] [Google Scholar]
- Sandve SR, Marcussen T, Mayer K et al. 2015. Chloroplast phylogeny of Triticum/Aegilops species is not incongruent with an ancient homoploid hybrid origin of the ancestor of the bread wheat D-genome. New Phytologist 208: 9–10. [DOI] [PubMed] [Google Scholar]
- Sears ER. 1954. The aneuploids of common wheat. University of Missouri Agriculture Experiment Station, Bulletin 572: 1–58. [Google Scholar]
- Sears ER. 1977. Induced mutant with homoeologous pairing in common wheat. Canadian Journal of Genetics and Cytology 19: 585–593. [Google Scholar]
- Sears ER, Okamoto M. 1958. Intergenomic chromosome relationships in hexaploid wheat Proceedings of the Xth Interface Congress on Genetics, Montreal 2: 258–259. [Google Scholar]
- Sehgal D, Vikram P, Sansaloni CP et al. 2015. Exploring and mobilizing the gene bank biodiversity for wheat improvement. PLOS One 10: e0132112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PD, Raubach S, Hearne SJ et al. 2017. Germinate 3: Development of a common platform to support the distribution of experimental data on crop wild relatives. Crop Science 57: 1–15. [Google Scholar]
- Shen Y, Tang D, Wang K et al. 2012. ZIP4 in homologous chromosome synapsis and crossover formation in rice meiosis. Journal of Cell Science 125: 2581–2591. [DOI] [PubMed] [Google Scholar]
- Shi F, Tibbits J, Pasam RK et al. 2017. Exome sequence genotype imputation in globally diverse hexaploid wheat accessions. Theoretical and Applied Genetics 130: 1393–1404. [DOI] [PubMed] [Google Scholar]
- Steuernagel B, Periyannan SK, Hernandez-Pinzon I et al. 2016. Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nature Biotechnology 34: 652–655. [DOI] [PubMed] [Google Scholar]
- Thind AK, Wicker T, Simkova H et al. 2017. Rapid cloning of genes in hexaploid wheat using cultivar-specific long-range chromosome assembly. Nature Biotechnology DOI: 10.1038/nbt.3877]. [DOI] [PubMed] [Google Scholar]
- Thompson JP, Seymour NP. 2011. Inheritance of resistance to root-lesion nematode (Pratylenchus thornei) in wheat landraces and cultivars from the West Asia and North Africa (WANA) region. Crop and Pasture Science 62: 82–93. [Google Scholar]
- Tiwari VK, Wang S, Sehgal S et al. 2014. SNP discovery for mapping alien introgressions in wheat. BMC Genomics 15: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trick M, Adamski NM, Mugford SG, Jiang CC, Febrer M, Uauy C. 2012. Combining SNP discovery from next-generation sequencing data with bulked segregant analysis (BSA) to fine-map genes in polyploid wheat. BMC Plant Biology 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan DA, Balazs E, Heslop-Harrison JS. 2007. From crop domestication to super-domestication. Annals of Botany 100: 893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikram P, Franco J, Burgueño-Ferreira J et al. 2016. Unlocking the genetic diversity of Creole wheats. Scientific Reports 6: 23092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JR, Luo MC, Chen ZX et al. 2013. Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D-genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytologist 198: 925–937. [DOI] [PubMed] [Google Scholar]
- Wang ML, Leitch AR, Schwarzacher T, Heslop-Harrison JS, Moore G. 1992. Construction of a chromosome-enriched HpaII library from flow-sorted wheat chromosomes. Nucleic Acids Research 20: 1897–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SC, Wong DB, Forrest K et al. 2014. Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnology Journal 12: 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfield MO, Allen AM, Burridge AJ et al. 2016. High-density SNP genotyping array for hexaploid wheat and its secondary and tertiary gene pool. Plant Biotechnology Journal 14: 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfield MO, Allen AM, Wilkinson PA et al. 2017. High density genotyping of the AE Watkins Collection of hexaploid landraces identifies a large molecular diversity compared to elite bread wheat. Plant Biotechnology Journal DOI: 10.1111/pbi.12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingen LU, Orford S, Goram R et al. 2014. Establishing the A. E. Watkins landrace cultivar collection as a resource for systematic gene discovery in bread wheat. Theoretical and Applied Genetics 127: 1831–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingen LU, West C, Leverington-Waite M et al. 2017. Wheat landraces genome diversity. Genetics 205: 1657–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würschum T, Leiser WL, Longin C. 2017. Molecular genetic characterization and association mapping in spelt wheat. Plant Breeding 136: 214–223. [Google Scholar]
- Xie WL, Nevo E. 2008. Wild emmer: genetic resources, gene mapping and potential for wheat improvement. Euphytica 164: 603–614. [Google Scholar]
- Xu DL, Long H, Liang JJ et al. 2012. De novo assembly and characterization of the root transcriptome of Aegilops variabilis during an interaction with the cereal cyst nematode. BMC Genomics 13: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Jiang H, Yeh CT et al. 2015. Extreme-phenotype genome-wide association study (XP-GWAS): a method for identifying trait-associated variants by sequencing pools of individuals selected from a diversity panel. Plant Journal 84: 587–596. [DOI] [PubMed] [Google Scholar]
- Yu G, Champouret N, Steuernagel B et al. 2017. Discovery and characterization of two new stem rust resistance genes in Aegilops sharonensis. Theoretical and Applied Genetics 130: 1207–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegeye H, Rasheed A, Makdis F, Badebo A, Ogbonnaya FC. 2014. Genome-wide association mapping for seedling and adult plant resistance to stripe rust in synthetic hexaploid wheat. PLOS One 9: e105593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Sallam A, Gao L et al. 2016. Establishment and optimization of genomic selection to accelerate the domestication and improvement of intermediate wheatgrass. Plant Genome 9 DOI: 10.3835/plantgenome2015.07.0059. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Huang L et al. 2015. A high-density genetic map for P genome of Agropyron Gaertn. based on specific-locus amplified fragment sequencing (SLAF-seq). Planta 242: 1335–1347. [DOI] [PubMed] [Google Scholar]
- Zheng Z, Wang HB, Chen GD, Yan GJ, Liu CJ. 2013. A procedure allowing up to eight generations of wheat and nine generations of barley per annum. Euphytica 191: 311–316. [Google Scholar]
- Zohary D, Feldman M. 1962. Hybridization between amphidiploids and the evolution of polyploids in the wheat (Aegilops-Triticum) group. Evolution 16: 44–61. [Google Scholar]