Abstract
COPII-coated vesicles carry proteins from the endoplasmic reticulum to the Golgi complex. This vesicular transport can be reconstituted by using three cytosolic components containing five proteins: the small GTPase Sar1p, the Sec23p/24p complex, and the Sec13p/Sec31p complex. We have used a combination of biochemistry and electron microscopy to investigate the molecular organization and structure of Sec23p/24p and Sec13p/31p complexes. The three-dimensional reconstruction of Sec23p/24p reveals that it has a bone-shaped structure, (17 nm in length), composed of two similar globular domains, one corresponding to Sec23p and the other to Sec24p. Sec13p/31p is a heterotetramer composed of two copies of Sec13p and two copies of Sec31p. It has an elongated shape, is 28–30 nm in length, and contains five consecutive globular domains linked by relatively flexible joints. Putting together the architecture of these Sec complexes with the interactions between their subunits and the appearance of the coat in COPII-coated vesicles, we present a model for COPII coat organization.
Transport of proteins in eukaryotic cells from the endoplasmic reticulum (ER) to the Golgi complex proceeds by deformation of specialized portions of the donor membrane to form carrier vesicles (1–3). A group of cytosolic proteins collectively known as COPII carry out a programmed set of sequential interactions, leading to cargo sorting and vesicle budding (4). Vesicular transport can be reconstituted by using three cytosolic components containing five proteins: the small GTPase Sar1p, the Sec23p/24p complex, and the Sec13p/Sec31p complex (5). These proteins will support a cargo-carrying budding reaction from isolated ER membranes. Sar1p, a GTP-binding protein, initiates coat formation (6). The GDP-bound form of Sar1p is normally cytosolic. It is recruited to the ER membrane by interaction with Sec12p, an ER-bound membrane protein that serves as its guanine exchange factor (7). Sar1p-GTP then recruits cytosolic Sec23p/24p complex, most likely through its interaction with Sec23p (8). In addition to recruiting Sec23p/24p, the GTP-bound form of Sar1p stabilizes Sec23p and binds to certain ER/Golgi SNARE proteins involved in the specificity of targeting and in the fusion reaction of vesicles with acceptor membranes (9). The interaction of Sar1p-GTP with Sec23p also facilitates the association of the Sec23p/24p complex with cargo proteins (10); Sec24p is probably the component responsible for cargo recognition (11). ER membranes with Sec23p/24p and Sar1p can then recruit Sec13p/31p, a complex that is likely to act as a scaffold, like clathrin, to effect membrane deformation and vesicle budding. Completing the cycle, Sec23p acts as a GTPase-activating protein for Sar1p (8). It is thought that on GTP hydrolysis, Sar1p-GDP is released, leading to uncoating before fusion of the vesicle to the target membrane and recycling of COPII components.
The formation of COPII vesicles described by this model synthesizes information from a large number of biochemical and genetic observations. We know relatively little, however, about the structure and modes of association of the COPII components, as needed to explain the mechanics of coat formation and membrane budding. In the case of clathrin-coated vesicles, the equivalent information for components of the clathrin coat has led to an understanding of the molecular basis for internal cargo sorting and for coat assembly (1).
We investigated the molecular organization and structure of Sec23p/24p and Sec13p/31p complexes isolated from yeast cells. The results reported here—obtained from a combination of biochemical and biophysical methods—reveal distinct shapes and quaternary structures for each type of complex. These results lead us to propose a model for assembly of a COPII coat.
Materials and Methods
Proteins.
The 6 × His-tagged versions of yeast Sec24p, Sec23p/24p, and Sec13p/31p were expressed in yeast cells and purified as described (12). In the case of Sec13p, a 6 × His tag was fused to the amino-terminal side of the TEV protease cleavage site, which was then fused to the amino terminus of Sec13p. This protein was expressed in Escherichia coli and purified on a Ni2+-NTA-agarose column (Qiagen, Chatsworth, CA), followed by proteolysis with GST-TEV and subsequent collection of the flow-through from two sequential chromatography steps, the first over a Ni2+-NTA-agarose column and the second over a glutathione-agarose column. Aliquots of the samples were flash frozen in liquid nitrogen and stored at −80°C.
Chemical Crosslinking.
Stock solutions of dithiobis succinimidyl propionate (DSP, Pierce) (50 mg/ml in DMSO) and bis(sulfosuccinidyl) suberate (BS3, Pierce) (8 mg/ml in B1 buffer: 0.5 M Sodium acetate/20 mM Hepes, pH 7.4/0.1 mM EGTA) were made immediately before use and mixed with 5–50 μg of the protein samples dissolved in B1 buffer for 40 min at room temperature. The crosslinking reactions were stopped by addition of Laemmli sample buffer (without reducing agent in the case of DSP), immediately boiled, followed by SDS/PAGE fractionation. For reversal of crosslinking, the lanes of interest were excised from the first non-reducing gel, incubated in Laemmli running buffer (with 50 mM DTT) at 37°C for 15 min, and used as a stacking gel on a reducing 10% SDS/PAGE.
Sedimentation Equilibrium by Analytical Centrifugation.
Analytical centrifugation was performed at 4°C in a Beckman XL-I analytical ultracentrifuge equipped with UV absorption optical cells. Equilibrium conditions were achieved by using three different sample concentrations and different centrifugation speeds. The molecular weights were determined by a least-square fitting procedure. Before analysis, the buffer in the samples was exchanged by gel filtration chromatography into B1 and aggregates removed by centrifugation for 20 min at 4°C and 90,000 rpm (Beckman TLX-100 centrifuge).
Gel Filtration Chromatography.
Analytical gel filtration chromatography was performed at room temperature by using 20-μl samples applied to a 3-ml H2/30 Superose 6 column (Smart System, Amersham Pharmacia) calibrated with a mixture of globular molecular weight markers. Before chromatography, aggregates were removed by centrifugation at 90,000 rpm for 30 min at 4°C (Beckman TLX-100 centrifuge).
Quantitative Amino Acid Analysis.
Duplicate samples of Sec23p/24p (6 μg) or Sec13p/31p (13 μg) were fractionated by 10% SDS/PAGE, transferred to a poly(vinylidene difluoride) (PVDF) membrane and stained with Ponceau S. The portions of the membrane containing the protein bands were excised and subjected to acid hydrolysis followed by quantitative amino acid analysis. The transfer efficiency of each subunit from the gel to the PVDF membrane was determined from the transfer of [125I]-labeled Sec 23p/24p and Sec13/31p loaded in adjacent lanes. The molar ratio of the subunits within the Sec23p/24p or Sec13p/31p complexes was calculated from the total mass of a given subunit present in the original gel (13). This technique is insensitive to the physical state of the samples.
Electron Microscopy and Image Processing.
Samples were adsorbed to glow-discharged carbon-coated copper grids, washed with two drops of deionized water, and stained with two drops of freshly prepared 0.75% uranyl formate. Images were taken using low-dose procedures with a Philips Tecnai 12 electron microscope (Philips, Eindhoven, The Netherlands) at 120 kV with a magnification of ×52,000 and a defocus of 1.5 μm. After inspection with a JEOL JFO-3000 laser diffractometer, suitable images were digitized with a Zeiss SCAI scanner by using a pixel size of 0.4 nm at the specimen level and processed with spider (14).
Results
The Sec23p/24p Complex Is a Heterodimer.
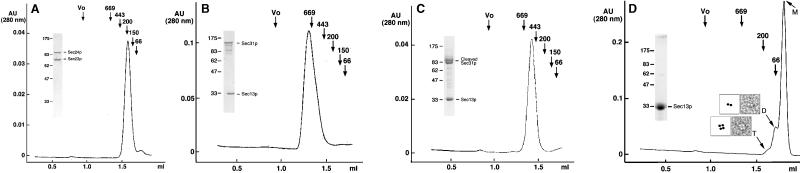
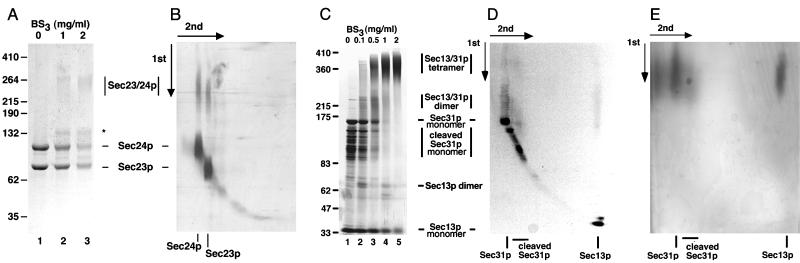
Sec23p/24p eluted on gel filtration chromatography as a single species corresponding to a globular protein of about 200 kDa (Fig. 1A). The masses of Sec23p and Sec24p deduced from their cDNAs are 85 and 104 kDa, and the molar ratio of Sec23p to Sec24p in the complex is 1:1, as assessed by a combination of SDS/PAGE and quantitative amino acid analysis (Table 1). Sec23p/24p must therefore be a heterodimer. To confirm this conclusion, we subjected Sec23p/24p complexes to chemical crosslinking by using increasing amounts of BS3 (Fig. 2A). After crosslinking and SDS/PAGE fractionation, we detected a relatively broad band whose electrophoretic mobility corresponded to a protein of ≈260 kDa; no other bands of larger size could be detected even when using significantly higher ratios of crosslinker to protein (not shown). As expected for the crosslinking product from a heterodimer, the 260-kDa band contained similar amounts of Sec23p and Sec24p (Fig. 2B). This relative composition was established by using the reversible crosslinker DSP and by including the reducing agent DTT during the second dimension of SDS/PAGE fractionation. Final confirmation that Sec23p/24p is a heterodimer was obtained by determining its molecular mass as 195(+/−2) kDa by using sedimentation equilibrium in the analytical centrifuge. This molecular mass is in close agreement with the value of 189 kDa calculated from the corresponding cDNA sequences of Sec23p and Sec24p.
Figure 1.
Gel filtration chromatography of Sec23p/24p, Sec13p/31p, and Sec13p. The elution profiles correspond to samples of (A) Sec23p/24p, (B) Sec13p/31p, (C) partially cleaved Sec13p/31p, and (D) Sec13p. (Insets) Representative images of negatively stained dimers (D) and tetramers (T) of Sec13p. Molecular masses and position of elution peaks for globular proteins used as standards are indicated. Insets show the Coomassie blue staining of the input samples after fractionation by SDS/PAGE.
Table 1.
Polypeptide chain stoichiometry of Sec complexes
| Polypeptide chain | Blotting efficiency (%) | Total protein in membrane, pmol
|
Total protein in gel, pmol
|
||
|---|---|---|---|---|---|
| Sample A | Sample B | Sample A | Sample B | ||
| Sec23p | 46.81 | 48.88 | 44.91 | 104 | 96 |
| Sec24p | 46.52 | 43.48 | 48.84 | 93 | 105 |
| Molar ratio | 1.11 | .91 | |||
| Sec13p | 48.93 | 64.68 | 132 | ||
| Sec31p (intact) | 32.21 | 28.82 | 44 | ||
| Sec31p (cleaved) | 76.99 | 53.68 | 70 | ||
| Molar ratio | 1.16 | ||||
| Sec13p | 29.99 | 32.63 | 109 | ||
| Sec31p (intact) | 31.07 | 15.95 | 51 | ||
| Sec31p (cleaved) | 45.54 | 23.50 | 51 | ||
| Molar ratio | 1.07 | ||||
Figure 2.
Chemical crosslinking. (A) Chemical crosslinking of Sec23p/24p with the nonreducible crosslinker BS3. Sec23p/24p (4.5 μg) was incubated with increasing amounts of BS3 followed by fractionation on a SDS/(5 to 10% gradient) PAGE and visualization by silver staining. The molecular masses (in kDa) are indicated for the following standards: BSA, crosslinked BSA, clathrin triskelions (containing its heavy and light chains), and crosslinked clathrin triskelions. The electrophoretic mobilities for Sec23p, Sec24p, and the species resulting from chemical crosslinking of Sec23p/24p are indicated. A crosslinking product that elutes ahead of the Sec24p band (star) probably represents the product of an intramolecular crosslinking event, because a similar band appeared when pure Sec24p was subjected to the same conditions of crosslinking (not shown). (B) Chemical crosslinking of Sec23p/24p with the reducible crosslinker DSP. Sec23p/24p (6 μg) was incubated with DSP (2.5 mg/ml) followed by fractionation on a nonreducing SDS/(5 to 10% gradient) PAGE. After excision, the lane containing the products of the first crosslinking reaction (first dimension) was layered on top of a second gel and subjected to fractionation on a reducing SDS/10% PAGE (second dimension) followed by silver staining visualization. (C) Chemical crosslinking of Sec13p/31p with the nonreducible crosslinker BS3. Sec13p/31p (6.5 μg) was incubated with the indicated amounts of BS3 and then fractionated by using a gradient SDS/PAGE as in A. The electrophoretic mobilities of Sec13p, Sec31p, cleaved Sec31p, and the crosslinked species of Sec13/31p are indicated. The crosslinked species of about 65 kDa might represent dimers of Sec13p. (D) Chemical crosslinking of Sec13p/31p with the reducible crosslinker. Sec13p/31p complex (40 μg) was crosslinked with 0.2 mg/ml DSP and subjected to two-dimensional SDS/PAGE as in B. (E) Chemical crosslinking experiment performed with Sec13p/31p as in D, but by using 2 mg/ml of DSP.
The Sec23p/24p Complex Has a Bone-Like Shape.
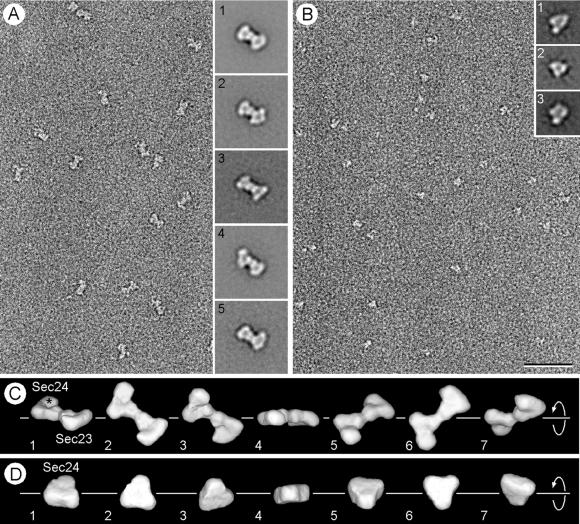
We calculated three-dimensional (3D) maps for Sec23p/24p and Sec24p at a resolution of about 2.5 nm by single-particle averaging techniques by using image pairs recorded at tilts of 0° and 60°. A typical micrograph obtained from an untilted specimen containing negatively stained Sec23p/24p complexes is shown in Fig. 3A. Correspondence analysis performed on 7,826 untilted particles revealed that they form a continuous data cloud with no obvious clustering (data not shown). We then used multireference alignment to define 50 classes of images and found that the averages of the four largest classes appeared indistinguishable from each other (average 1 in Fig. 3A). This average shows that the Sec23p/24p complex has a bone-like outline, the outside dimensions of which are about 11 × 17 nm. Inspection of the remaining 46 class averages revealed that most of the differences were in the appearance of the two end domains (Fig. 3A, averages 2 and 3) and in a flexing of the structure along its long axis (Fig. 3A, averages 4 and 5). These variations could be explained by uneven staining of the particles, by different orientations in which the particles adsorb to the carbon film, or perhaps by different conformational states of the Sec23p/24p complex.
Figure 3.
Electron microscopy and 3D maps of negatively stained Sec23p/24pand Sec24p. (A) The electron micrograph of an untilted specimen reveals the bone-shaped structure of the Sec23p/24p complex. Insets show five representative averages obtained by multireference alignment by using 7,826 particles, revealing variations in the structure of the two end domains and in their orientation relative to each other. (B) Negative staining of pure Sec24p reveals the triangular shape of the molecule, although its small size gives rise to only a weak contrast. Insets represent three typical class averages that resulted from multireference alignment performed on 6,553 particles. (C) Different views of the 3D map of the Sec23p/24p complex obtained from 1,605 particles that contributed to average 1 in A. The asterisk depicts the enlarged domain that led us to assign this half of the complex to Sec24p. The corresponding quicktime movie is published as supporting information on the PNAS web site (www.pnas.org). The close match between the different views of the 3D map and the class averages (Insets in A) suggested that the variations in the projection averages are because of different orientations of adsorption of the complex to the carbon film (compare for example view 3 in C with average 4 in A). (D) The 3D map of Sec24p was calculated from 1,295 particles, and the views correspond to a rotation around the marked axis. The triangular overall shape of Sec24p looks very similar to one half of the Sec23p/24p complex. The three domains are, however, not as well resolved as in the 3D map of the complex, and the enlarged domain is not apparent. This is most likely because of the small size and the triangular shape of Sec24p, which made alignment of the individual particles not sufficiently precise for these features to be resolved in the 3D map. (Bar = 50 nm.) The side length of the Insets in A and B are 40 and 24 nm, respectively.
The final 3D map of Sec23p/24p shown in Fig. 3C was calculated with the 1,605 particles included in average 1 and contoured to include 100% of the Sec23p/24p mass (189 kDa), assuming a specific density for protein of 0.81 Da/10−3 nm3. The images in Fig. 3C present views of the 3D map complex as seen from different orientations, and a corresponding movie is published as supporting information on the PNAS web site (www.pnas.org). The complex consists of two triangularly shaped halves, each containing three approximately globular domains. The shapes of the two halves are similar, and their positions in the complex are related by a pseudo-2-fold axis. The shapes of these halves are in turn similar to the 3D map of Sec24p calculated on the basis of 1,295 particles (Fig. 3 B and D). We therefore propose that one of the halves of the complex corresponds to Sec23p and the other to Sec24p. We believe that the similarity in the shapes of Sec23p and Sec24p implies that they have similar overall 3D folds, a conclusion that is consistent with statistically significant agreement of their protein sequences along the entire length of the two polypeptides (http://www.ch.embnet.org/software/ClustalW.html). This analysis did not include the divergent amino termini, which correspond to the regions where they interact with each other (15). The most salient difference between the two halves is that one of them contains a relatively enlarged domain (marked by an asterisk in Fig. 3C). This half was therefore assigned to the larger Sec24p (molecular mass 104 kDa), whereas the second half was assigned to the smaller Sec23p (molecular mass 85 kDa). We note that the remaining class averages are likely to represent different orientations of adsorption of the complexes to the carbon film rather than uneven staining or different conformational states of the Sec23p/24p complex (compare, for example, view 3 in Fig. 3C with average 4 in Fig. 3A). This conclusion was verified by using all 7,826 particles to calculate a new 3D map that looked identical to the one shown in Fig. 3C (data not shown).
The Sec13p/31p Complex Is a Heterotetramer.
During gel filtration chromatography, Sec13p/31p eluted as a single monodisperse species corresponding to a globular protein of about 700 kDa in mass (Fig. 1B). Because of the marked disparity with the added molecular masses of its Sec13p and Sec31p subunits (33 and 140 kDa, respectively), it has been proposed that the Sec13p/31p complex might be a heterodimer of elongated shape (16), although the possibility that the complex could contain more subunits was not explored. As described below, Sec13p/31p is a relatively asymmetric heterotetramer. We first established that the Sec13p and Sec31p subunits are present within the complex in a molar ratio of 1:1 (Table 1). We then determined its quaternary organization by chemical crosslinking with increasing amounts of BS3 (Fig. 2 C–E). Lanes 3–5 in Fig. 2C show the appearance of new bands, most notably in the regions around 380, 220, and 65 kDa. The size of the 220-kDa species suggested a crosslinking of one Sec13p and one Sec31p subunit. Indeed, the 220-kDa species contained both Sec13p and Sec31p, as shown in Fig. 2D, by using the reversible crosslinker DSP and nonreducing SDS/PAGE for the first dimension of fractionation, followed by crosslinking reversal with DTT during the second dimension of SDS/PAGE fractionation. We propose that the 380-kDa species results from the association of two Sec13p and two Sec31p subunits, because its size is approximately twice that of the 220-kDa species, and because it also contains about the same ratio of Sec13p and Sec31p as intact Sec13p/Sec31p complex (Fig. 2E).
Isolated Sec13p eluted as a globular protein of 30 kDa and an estimated diameter of 3.5 nm, that is, as a monomeric β-propeller (M in Fig. 1D), the structure predicted from the seven WD40 repeats in the Sec13p amino acid sequence. Two minor fractions of Sec13p eluted as globular species of ≈60 and 120 kDa (Fig. 1D), probably corresponding to dimers (D) and tetramers (T) of Sec13p, respectively. As shown in the examples in Fig. 1, Sec13p dimers and tetramers were also visualized by electron microscopy of Sec13p alone. The relatively faint 65-kDa species seen by crosslinking of the Sec13p/31p complex (seen best in Fig. 2C, lane 2) may represent dimers of Sec13p, reflecting a contact between these subunits within the complex.
The Elongated and Flexible Structure of Sec13p/31p.
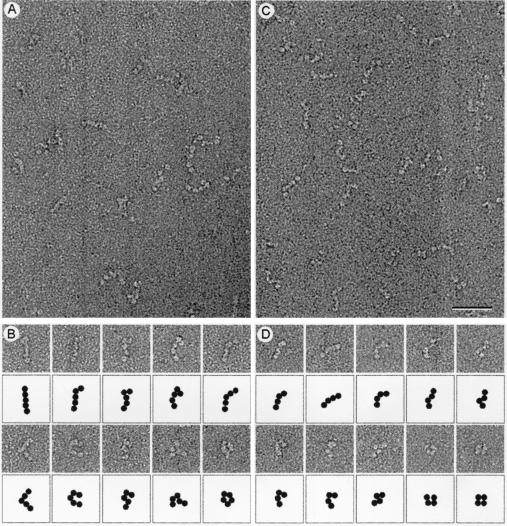
Micrographs of the negatively stained Sec13p/31p complex revealed elongated flexible molecules of variable length (Fig. 4). The longest molecules were about 28–30 nm in length and consisted of five linearly arranged globular domains that may represent intact Sec13p/31p complex. A gallery of such molecules illustrates the variety of shapes that Sec13p/31p can adopt (Fig. 4B). Each globular domain accommodates a mass of about 75 kDa if approximated by a sphere of 2.8 nm in radius with a density for protein of 0.81 Da/10−3 nm3. The mass of the complete Sec13p/31p complex would therefore be 375 kDa, a value that is in good agreement with the molecular mass of 343 kDa calculated for the heterotetramer model. The simplest organization that is consistent with the elongated shape of Sec13p/31p is a side-by-side arrangement of its subunits, such that the globular domain at one end of the complex corresponds to two Sec13p subunits, and the globular domain at the opposite end contains the carboxyl-terminal part of Sec31p. We obtained evidence in support of this assignment by a combination of gel filtration chromatography and visualization of partially cleaved Sec13p/31p. We took advantage of a contaminating protease copurifying with the complex to digest Sec31p into fragments of about 100 kDa; Sec13p remained resistant to the protease. The elution profile (Fig. 1C) shows that the cleaved complex is significantly smaller than intact Sec13p/31p. The Sec31p fragments remained in association with Sec13p during gel filtration chromatography. They must therefore contain the amino terminus of Sec31p, because this is the part of Sec 31p known to interact with Sec13p (15). The smaller size of the partially cleaved Sec13p/31p complex was also evident by electron microscopy (Fig. 4D). The predominant species had four globular domains and assumed a variety of shapes (Fig. 4D). The size of the missing globular domain (estimated from the micrographs to be 75 kDa) corresponds closely to the added size of two 30- to 40-kDa carboxyl terminal Sec31p fragments. This assignment for the cleaved Sec13p/31p is also consistent with its molecular mass of 291(+/−5) kDa, as determined by sedimentation equilibrium in the analytical ultracentrifuge.
Figure 4.
Electron microscopy of negatively stained Sec13p/31p. (A) Electron micrographs of negatively stained Sec13p/31p revealed the complex to be linear and flexible. The largest molecules consisted of five globular domains that are likely to be connected by flexible joints. (B) Gallery of full-length Sec13p/31p molecules and their schematic representation illustrating the many conformations the complex can adopt. (C) Electron micrographs of partially cleaved Sec13p/31p. (D) Gallery of partially cleaved Sec13p/31p and their schematic representation showing the loss of one of the five globular domains. (Bar = 50 nm.)
The N-terminal 400 residues of Sec31p appear to contain WD40 repeats. They may therefore, like Sec13p, fold as a β-propeller. Two of these, associated laterally, would form the second globular domain of the heterotetramer. The amino acid sequence of the rest of Sec31p suggests that the remaining three globular units have other folds. Each unit in the Sec13p/31p complex is thus a side-by-side association of equivalent segments from two polypeptide chains (Fig. 5A).
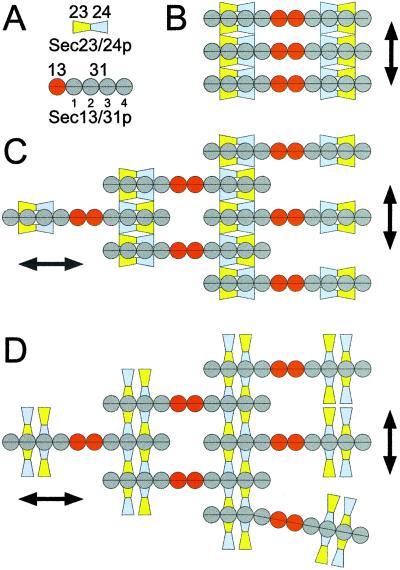
Figure 5.
Possible models for the assembly of Sec23p/24p and Sec13p/31p to form a COPII coat. A shows the domain organization of Sec23p/24p and Sec13p/31p, taking into account the biochemical data and the electron microscopic images obtained in this work. B–D depict schematic representations corresponding to possible arrangements of the complexes as they form lattices on the surface of the ER membrane. The models are viewed from the cytosol toward the membrane.
A Model for the COPII Coat.
The outer dimensions and structural features of Sec23p/24p and Sec13p/31p reported here, together with the known details of the interactions between Sec23p/24p and Sec13p/31p and the observation that equivalent amounts of these complexes constitute the coat in COPII-coated vesicles, lead to significant constraints in their possible dispositions within a coat. The slightly curved Sec23p/24p is likely to have its concave side facing the membrane, with both subunits making contact with the cytosolic portion of membrane proteins selected for transport by COPII vesicles. Sec23p must also contact the membrane-bound Sar1p-GTP (8). Yeast two-hybrid reactions and pull-down experiments demonstrate that Sec23p and Sec24p interact with different regions of Sec31p, located near its carboxyl terminus (15). We can therefore imagine three modes of association for the complexes. The first two models involve simultaneous contacts between both subunits of a single Sec23p/24p complex and one Sec31p molecule (Fig. 5 B and C). The third model involves simultaneous interactions between the opposite ends of two Sec23p/24p complexes and two antiparallel Sec31p molecules, themselves contributed by two different Sec13p/31p complexes (Fig. 5D). Is it possible to differentiate between these models? We think so. One important characteristic in the growth of any lattice is that it requires the sequential addition of components that involve repetitive interactions. Inspection of the first model of association (Fig. 5B) indicates that it is a dead end for lateral lattice growth, because Sec23p/24p can bind only to one copy of Sec31p. In contrast, the second and third modes of association (Fig. 5 C and D) allow for lattice growth because of the repetitive association of two copies of Sec23p/24p with two antiparallel Sec13p/31p complexes. The model in Fig. 5C also requires dimerization of the Sec23p/24p dimers; we have not observed Sec23p/24p dimerization in any of our experiments. Another important feature in the formation of any coat is the ability of some (if not all) of its elements to self-associate, to allow for two-dimensional growth of the lattice. In the case of the clathrin coat, for example, self association of the clathrin triskelion provides the structural scaffolding required for coated vesicle formation. In the COPII coat, it would seem that Sec13p/31p has such a scaffolding role through Sec13p contacts. Order-of-addition experiments during in vitro budding reactions have shown that Sec23p/24p is first recruited to the donor membrane, followed by the addition of Sec13p/31p (6). Interaction of the elongated flexible dimers of Sec13p/31p would thus appear to drive membrane budding. In addition to the tight longitudinal interactions within a dimer, Sec13p/31p can heterooligomerize (laterally) with Sec23p/24p and homooligomerize (longitudinally) through Sec13p contacts.
We favor our third proposal for contacts between the two complexes, because the first model (Fig. 5B) would lead essentially to dead-end structures rather than propagating lattices (Fig. 5 C and D), and because the second proposal requires dimerization of Sec23p/24p for which we have no evidence. The relatively fixed concave shape of Sec23p/24p can impart local curvature to the lattice. Lateral growth is provided by the molecular crosslinks between Sec23p/24p and the adjacent Sec13p/31p complexes, whereas the head-to-head associations of two Sec13p/31p complexes provide longitudinal growth mediated through Sec13p contacts. The resulting molecular arrangement needs to satisfy the structural requirements of a completely enclosed coat that can eventually lead to vesicle budding. Baskets can be made with linear elements, but local dislocations are required to ensure curvature. We propose that the observed flexibility of Sec13p/31p allows for dislocations and accommodates to variable curvature. In those Sec13p/31p complexes close to the dislocations, not all of the available sites in Sec31p would be bound by Sec23p/24p, leading to a slight excess of Sec13p/31p relative to Sec23p/24p. The arrangements of the coat elements proposed here predict the appearance of striations at the surface of the coat and are consistent with the lack of discernable features of COPII-coated vesicles visualized by negative staining (not shown). They also allow the coat to change its size in response to the overall dimensions of the enclosed cargo. The proposed arrangement also predicts an upper limit of ≈15 nm for the coat thickness, a value that is consistent with the thickness in sectioned samples of COPII-coated vesicles (11). The relatively unconstrained organization of the coat would permit the budding of vesicles surrounding a few copies of very small cargo molecules or many copies of very large molecules, such as lipoproteins and collagen, whose dimensions can exceed 300 nm (17, 18). This requirement for size plasticity contrasts with what appears to be a more constrained geometry for clathrin-coated vesicles, resulting in coats with relatively modest variations in overall size (1). In the model in Fig. 5, Sec13p mediates essential head-to-head interactions of Sec13p/31p complexes. Indeed, null Sec13 mutations are lethal. They can be suppressed, however, by mutations in any one of three other genes (BST 1,2,3), which encode ER membrane proteins (19). The structures and properties of COPII vesicles from such cells will be useful tests of aspects of our model.
The work reported here used COPII components from yeast. We imagine that similar dimensions and geometric constraints will be found in the proteins of COPII vesicles from multicellular organisms, because of the extensive similarities of amino acid sequences and biochemical properties.
Supplementary Material
Acknowledgments
The molecular electron microscopy facility at Harvard Medical School was established by generous funds from the Giovanni Armenise Harvard Center for Structural Biology and is maintained by funds from National Institutes of Health Grant GM62580–01. G.L. acknowledges the support for a sabbatical visit by a Dorot Fellowship. This work was supported by National Institutes of Health Grant GM36548 (T.K.).
Abbreviations
- ER
endoplasmic reticulum
- 3D
three dimensional
- DSP
dithiobis succinimidyl propionate
- B3
bis(sulfosuccinidyl) suberate
References
- 1.Kirchhausen T. Nat Rev Mol Cell Biol. 2000;1:187–198. doi: 10.1038/35043117. [DOI] [PubMed] [Google Scholar]
- 2.Schekman R, Orci L. Science. 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- 3.Springer S, Spang A, Schekman R. Cell. 1999;97:145–148. doi: 10.1016/s0092-8674(00)80722-9. [DOI] [PubMed] [Google Scholar]
- 4.Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach M F, Ravazzola M, Amherdt M, Schekman R. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 5.Salama N R, Yeung T, Schekman R W. EMBO J. 1993;12:4073–4082. doi: 10.1002/j.1460-2075.1993.tb06091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuoka K, Orci L, Amherdt M, Bednarek S Y, Hamamoto S, Schekman R, Yeung T. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- 7.Barlowe C, d'Enfert C, Schekman R. J Biol Chem. 1993;268:873–879. [PubMed] [Google Scholar]
- 8.Yoshihisa T, Barlowe C, Schekman R. Science. 1993;259:1466–1468. doi: 10.1126/science.8451644. [DOI] [PubMed] [Google Scholar]
- 9.Springer S, Schekman R. Science. 1998;281:698–700. doi: 10.1126/science.281.5377.698. [DOI] [PubMed] [Google Scholar]
- 10.Kuehn M J, Herrmann J M, Schekman R. Nature (London) 1998;391:187–190. doi: 10.1038/34438. [DOI] [PubMed] [Google Scholar]
- 11.Shimoni Y, Kurihara T, Ravazzola M, Amherdt M, Orci L, Schekman R. J Cell Biol. 2000;151:1–12. doi: 10.1083/jcb.151.5.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimoni Y, Schekman R. In: Guide to Yeast Genetics and Molecular Biology. Guthrie C, Fink G R, editors. San Diego: Academic; 2001. , in press. [Google Scholar]
- 13.Matsui W, Kirchhausen T. Biochemistry. 1990;29:10791–10798. doi: 10.1021/bi00500a011. [DOI] [PubMed] [Google Scholar]
- 14.Frank J, Radermacher M, Penczek P, Zhu J, Li Y H, Ladjadj M, Leith A. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 15.Shaywitz D A, Espenshade P J, Gimeno R E, Kaiser C A. J Biol Chem. 1997;272:25413–25416. doi: 10.1074/jbc.272.41.25413. [DOI] [PubMed] [Google Scholar]
- 16.Salama N R, Chuang J S, Schekman R W. Mol Biol Cell. 1997;8:205–217. doi: 10.1091/mbc.8.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beck K, Boswell B A, Ridgway C C, Bachinger H P. J Biol Chem. 1996;271:21566–21573. doi: 10.1074/jbc.271.35.21566. [DOI] [PubMed] [Google Scholar]
- 18.Davis R A. Biochim Biophys Acta. 1999;1440:1–31. doi: 10.1016/s1388-1981(99)00083-9. [DOI] [PubMed] [Google Scholar]
- 19.Elrod-Erickson M J, Kaiser C A. Mol Biol Cell. 1996;7:1043–1058. doi: 10.1091/mbc.7.7.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.