Abstract
Background and Aims
Amazonia is a major world centre of plant domestication, but little is known about how the crops were dispersed across the region. Manioc (Manihot esculenta) was domesticated in the south-western Amazon basin, and is the most important staple food crop that originated in Amazonia. Current contrasting distributions may reflect distinct histories of dispersal of bitter and sweet manioc landraces. To produce new insights into the evolutionary history of the crop, we investigated the contemporary genetic diversity and structure of bitter and sweet manioc along major Amazonian rivers.
Methods
The patterns of genetic structure and diversity of wild and cultivated sweet and bitter manioc with four chloroplast and 14 nuclear microsatellite markers were evaluated. Results were interpreted in terms of the crop’s dispersal.
Key results
No phylogeographic patterns among rivers were detected, and genetic structure among rivers was confounded by the bitter–sweet divergence. However, differences in the distribution of nuclear diversity and somewhat distinctive patterns of genetic structure across rivers were observed within bitter and sweet manioc.
Conclusions
Various pre-Columbian and post-European conquest events in the history of Amazonian occupation may explain the absence of clearer patterns of genetic structure. However, the wide distribution of the most common chloroplast haplotype agrees with an early dispersal of manioc across Brazilian Amazonia. Furthermore, differences in genetic structure and in the spatial distribution of genetic diversity suggest that bitter and sweet manioc had distinct dispersal histories. Knowledge about how prehistoric and contemporary Amazonian peoples manage their crops is valuable for the maintenance and conservation of the impressive diversity of their native crops.
Keywords: Amazonian crops, chloroplast SSR, genetic structure, Manihot esculenta, nuclear SSR, population genetics
INTRODUCTION
Plant domestication is a long-term co-evolutionary process in which human and natural selection results in plants more useful to humans and better adapted to domesticated landscapes (Clement, 1999a). A number of crops of global importance, such as maize (Zea mays), manioc (Manihot esculenta) and potato (Solanum tuberosum), were domesticated in the Americas (Pickersgill, 2007). Four independent regions of crop domestication in this continent are recognized: eastern North America, Mesoamerica, the Andes and the tropical lowlands of South America (Pickersgill, 2007; Meyer et al., 2012). In this latter region, Amazonia is a major world centre, where at least 83 native species were domesticated to some degree (Clement, 1999a), especially in peripheral parts of the basin (Clement et al., 2010). Among Amazonian food crops, manioc is of great importance in the modern world as the main source of carbohydrates for about 800 million people, and is cultivated across all tropical regions (Lebot, 2009).
Molecular studies have contributed to the elucidation of the botanical and geographic origins of many domesticated plants (Meyer and Purugganan, 2013; Larson et al., 2014). Indeed, the most compelling evidence for the origins of manioc is from genetic studies. The genetic variability found in cultivated manioc is a subset of the genetic variability found in populations of Manihot esculenta ssp. flabellifolia that occur in south-western Amazonia (Olsen and Schaal, 1999, 2001; Olsen, 2004). These studies strongly suggest that cultivated manioc was domesticated only once from M. esculenta ssp. flabellifolia populations in what is now Rondônia, and adjacent areas of Acre and Mato Grosso in Brazil, and possibly northern Bolivia (Schaal et al., 2006). Léotard et al. (2009) confirmed the previous studies by analysing a larger sample of ssp. flabellifolia’s distribution and including other putative hybridizing Manihot species.
Manioc was domesticated as early as 10 000 years before present (BP) (Olsen and Schaal, 1999) and was dispersed quickly after the initial domestication. Archaeobotanical evidence suggests that manioc crossed the Andes by at least 8000 BP and that it was widely dispersed throughout the Neotropics by 6500 BP (Isendahl, 2011). Analyses of proto-languages for which it is possible to reconstruct the word ‘manioc’ suggest a wide distribution of manioc in South America and Mesoamerica before the establishment of sedentary agricultural societies between 4000 and 3000 BP (Brown et al., 2013). It is possible that these dates do not necessarily reflect the palaeodistribution of domesticated manioc or other useful wild Manihot, but rather of cultivated or gathered forms. This is because Manihot is widely distributed across the Neotropics, and some other species might have been cultivated or gathered before the arrival of modern manioc (Brown et al., 2013). Despite this evidence, little is known about the genetics of crop dispersal from the centre of origin to other Amazonian regions.
Manioc domestication resulted in two major groups within the crop that differ in their toxicity: sweet manioc and bitter manioc (McKey et al., 2010). While sweet manioc has low amounts of cyanogenic glycosides (<100 ppm fresh weight) and can be safely consumed with simple processing, bitter manioc demands considerable processing for detoxification due to the large amounts of cyanogenic glycosides (>100 ppm fresh weight; McKey et al., 2010). These groups of cultivated manioc are readily recognized by farmers and supported by molecular studies (Mühlen et al., 2000; Elias et al., 2004). Although manioc was domesticated for vegetative propagation, sexual reproduction still plays an important role in the evolution of the crop under traditional cultivation in Amazonia. Flowering opens the possibility of gene flow among distinct landraces, and even with wild species occurring in the surroundings of swiddens (McKey et al., 2012). The seeds produced become part of the soil seed bank and may sprout amid clonally propagated landraces (Martins, 2001; Pujol et al., 2007; Duputié et al., 2009). Farmers, consciously or not, may let the sexual volunteer seedlings grow and after harvesting they may use cuttings of these plants for clonal propagation (Elias et al., 2000; Rival and McKey, 2008). Then, farmers may either incorporate the seedlings into an existing variety or create a new variety (Martins, 2001; Duputié et al., 2009). Sexual reproduction and the incorporation of volunteer seedlings greatly contribute to the maintenance and amplification of genetic diversity of manioc landraces. Moreover, exchange networks of manioc landraces within and among communities of traditional smallholders promote the diffusion of such diversity at a broad geographic scale (Boster, 1986; Chernela, 1986; Oliveira, 2008). These processes, together with the active selection for diversity and the cultivation of many different landraces in swiddens (Boster, 1985; Elias et al., 2000), are of considerable importance for the conservation of Amazonian manioc’s genetic resources.
Currently, bitter and sweet manioc differ in their patterns of occurrence across Amazonia (McKey and Beckerman, 1993; Emperaire, 2001) (Supplementary Data Fig. S1). Cultivation of bitter manioc is more frequent along the major rivers in the Amazon basin and along the north-eastern coast of South America. On the other hand, sweet manioc is more frequently cultivated in western Amazonia, in the headwaters of major rivers and, on a smaller scale, where bitter manioc is more frequent. Although these groups overlap completely in Amazonia, the somewhat distinct distributions of bitter and sweet manioc may result from independent dispersal processes and limited interchange of landraces among human populations during the history of manioc’s domestication (Emperaire, 2001). The expansion and migrations of Amerindian peoples certainly contributed to manioc’s dispersal, even if the origin and dispersal of some ethnic groups occurred much later than manioc’s domestication (Eriksen, 2011).
The order in which bitter and sweet manioc were selected would have important consequences for the crop’s dispersal and for the current patterns of occurrence across Amazonia. McKey and Beckerman (1993) summarize four hypotheses: (1) sweet wild manioc gave rise to sweet manioc, from which bitter manioc was selected; (2) bitter wild gave rise to bitter manioc, from which sweet manioc was selected; (3) sweet wild gave rise to sweet manioc independently of bitter wild, which gave rise to bitter manioc; (4) wild, possibly of intermediate toxicity, gave rise to bitter and sweet manioc simultaneously. Recently, Arroyo-Kalin (2010) proposed that sweet manioc arose first due to selection by small-scale Amazonian populations of forager-incipient agriculturalists. Bitter manioc would have arisen later to sustain larger sedentary societies during the Formative period, starting around 4000–3000 BP, when agriculture was intensified in Amazonia and technology for detoxification was widespread among Amerindians (Arroyo-Kalin, 2010). Perrut-Lima et al. (2014) showed that current populations of ssp. flabellifolia within the centre of origin of manioc are classified as bitter (tuberous roots containing >100 ppm of HCN on a fresh weight basis). If the populations evaluated in this latter study are typical of the populations of ssp. flabellifolia from which domesticated manioc arose, their results make it possible to discard hypotheses of a wild sweet manioc presented by McKey and Beckerman (1993). A fifth hypothesis may be formulated based on Arroyo-Kalin (2010) and Perrut-Lima et al. (2014): (5) wild, possibly of intermediate to high toxicity, gave rise to sweet manioc, from which bitter manioc was selected. Mühlen et al. (2013), evaluating the genetic diversity of bitter and sweet manioc from 11 ecogeographic regions in Brazil, favoured Arroyo-Kalin’s (2010) hypothesis and also suggested that from manioc’s centre of domestication sweet and bitter manioc were dispersed independently. While sweet manioc would have been dispersed in all directions, bitter manioc would have been dispersed towards the Brazilian coast, following the major Amazonian rivers. However, this study did not include samples of manioc from the region around the centre of domestication, and some ecogeographic regions, including some in Amazonia, were under-represented.
Knowledge of genetic structure and variation of cultivated populations of a crop is essential for the efficient use of its genetic resources, as well as for the better understanding of its evolutionary history (Vencovsky et al., 2007; Clement et al., 2010). This study aimed to evaluate how the genetic variability of bitter and sweet manioc is structured along the major rivers of Brazilian Amazonia in order to present new insights into the evolutionary history of the crop. Genetic variability was assessed with chloroplast (cpSSR) and nuclear (nSSR) microsatellites. Because cpSSRs are haploid, generally maternally inherited and non-recombinant, they provide information about ancient historical relationships among matrilineages, while nSSRs, which are codominant and hypervariable markers, are suitable for the evaluation of contemporary patterns of genetic structure (Freeland et al., 2011). The information presented in this study is discussed in terms of manioc’s dispersal along major Amazonian rivers from its centre of origin in south-western Amazonia.
MATERIALS AND METHODS
Sampling sites
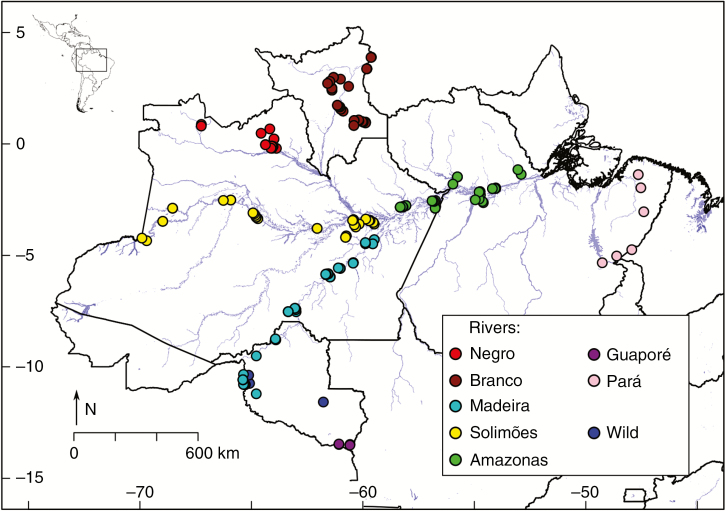
Communities of smallholder farmers in 44 municipalities along major Amazonian rivers (Negro, Branco, Madeira, Solimões and Amazonas), in southern Rondônia (hereafter Guaporé) and in north-eastern Pará (hereafter Pará) states were visited (Fig. 1; Supplementary Data Table S1). A total of 596 individuals were collected: 325 bitter manioc, 226 sweet manioc, 17 non-designated (found outside swiddens and home gardens, and thus without associated traditional knowledge) and 28 Manihot esculenta ssp. flabellifolia (hereafter wild manioc). For cultivated manioc, one individual of each landrace planted in swiddens and/or home gardens was collected. Wild manioc from Rolim de Moura was collected in the surroundings of the experimental station of the Federal University of Rondônia (UNIR), where Perrut-Lima et al. (2014) also collected, and the other three accessions were opportunistically collected when seen near fields of cultivated manioc. The collections were performed by different people of the same research group between July 2009 and September 2014 (Supplementary Data Table S1). Geographic locations of collection sites were recorded with a GPS. Leaf samples were dehydrated with silica gel and, after fieldwork, they were maintained at −20 °C until DNA extraction. Samples were exclusively of leaves and photographs to meet Resolution 21 requirements for basic research and were exempted from authorization by Brazil’s Council for Genetic Patrimony (CGEN in the Brazilian acronym), which was consulted before fieldwork. Considering Resolution 21, it is also important to state that collecting was performed only in communities of riverine smallholder farmers (caboclos) because sampling among other cultural groups (e.g. Amerindians) requires a different authorization.
Fig. 1.
Geographic locations of wild and cultivated manioc (Manihot esculenta) sampled in communities of smallholder farmers along major Amazonian rivers (Negro, Branco, Madeira, Solimões and Amazonas) and adjacent areas (Guaporé and Pará) in Brazil.
DNA extraction and quantification
Total genomic DNA was extracted from 50 mg of manioc leaf tissue using the CTAB protocol (Doyle and Doyle, 1990). DNA quality and quantity were evaluated by electrophoresis in agarose 1 % (w/v) gels stained with SYBR Safe DNA (Invitrogen) by comparison with phage λ molecular size standards (Invitrogen).
Chloroplast microsatellites
Four universal cpSSR primers for dicotyledons were used: ccmp05, 06, 10 (Weising and Gardner, 1999) and ccSSR07 (Chung and Staub, 2003). Polymerase chain reactions (PCRs) contained 10 ng of genomic DNA, 1 × buffer (10 mm Tris–HCl, pH 8.3; 50 mm KCl; 1.5 mm MgCl2), 2.5 ng of bovine serum albumin, 2.5 mm MgCl2, 20 µm of each dNTP, 1.6 pmol of forward primer, 2 pmol of reverse primer, 1.5 pmol of M13(-29) primer (5′-CACGACGTTGTAAAACGAC-3′) and 1 U of Taq DNA polymerase. Amplifications were performed in a final volume of 10 µL in a ProFlex (Applied Biosystems) thermocycler with the following steps: 4 min at 94 °C; 30 cycles of 1 min at 94 °C, 1 min at 58 °C and 1 min at 72 °C. An additional ten cycles of 40 s at 94 °C, 40 s at 53 °C and 40 s at 72 °C, and a final step of 10 min at 72 °C were used to label PCR products with M13(-29) modified with either IRDye700 or IRDye800 infrared dyes (LI-COR). Polymorphisms were detected in 6.5 % acrylamide gel matrix (KB Plus, LI-COR) using the semi-automated LI-COR 4300 DNA analyser. Allele sizes were determined with Saga™ 3.3 (LI-COR) using 50–350 bp IRDye 700 and 800 (LI-COR) sizing standards.
Nuclear microsatellites
A set of 14 nSSRs was used: GA12, 21, 126, 131, 134, 136, 140, GAGG5 (Chavarriaga-Aguirre et al., 1998), SSRY13, 20, 32, 70, 89 and 164 (Mba et al., 2001). Each nSSR forward primer was modified with a specific fluorescent dye (6-FAM, NED or HEX). Amplifications of nSSRs were performed in a final volume of 10 µL in a ProFlex (Applied Biosystems) thermocycler as described by Alves-Pereira et al. (2011). PCR products were multiplexed and nSSR fragments were detected in an ABI 3500xL (Applied Biosystems) automated DNA sequencer. Alleles were scored by size of PCR fragments with GeneMapper v.4.0 (Applied Biosystems) with the aid of the GeneScan 500 ROX Size Standard (Applied Biosystems).
Haplotype diversity analysis
Haplotypes of cpSSRs and estimates of haplotype diversity, described as the total number of haplotypes (NH) and haplotype diversity (HEhap), were obtained with GenAlEx 6.5 (Peakall and Smouse, 2012). The distribution of haplotype variation within and among hierarchical groups of bitter and sweet manioc was evaluated by analysis of molecular variance (AMOVA; Michalakis and Excoffier, 1996) with Arlequin 3.5 (Excoffier and Lischer, 2010). The significance of divergence estimates was tested with 20 000 permutations. Haplotype relationships were examined with a median-joining network (Bandelt et al., 1999) built into Network 5 (http://www.fluxus-engineering.com). Alleles were coded as the length of cpSSR fragments, and a weight of 1 was given for each 1-bp mutation. This haplotype network is a visual representation of both similarities and frequencies of cpSSR haplotypes across sweet, bitter and wild manioc. Estimates of haplotype diversity, AMOVAs and the haplotype network considered only the individuals with no missing data (N = 581).
Genetic structure based on haplotype variation was evaluated using discriminant analysis of principal components (DAPC; Jombart et al., 2010) with adegenet 2.0.0 (Jombart and Ahmed, 2011) for R (R Core Team, 2015). DAPC does not have strong assumptions, such as minimization of linkage disequilibrium and Hardy–Weinberg equilibrium within clusters (Jombart et al., 2010). DAPC searches for linear combinations of alleles that summarize the maximum variation between groups, while minimizing within-group differences (Roullier et al., 2013). DAPC requires a number of a priori groupings of individuals, and adegenet proposes the use of K-means clustering of principal components, based on the Bayesian information criterion (BIC), to assess the best number of clusters. This procedure was performed to assign individuals to genetic clusters and to investigate the genetic relationships and admixture among individuals of bitter, sweet and wild manioc.
Genetic diversity analysis
Genetic diversity revealed by nSSRs was described by estimating the number of multilocus genotypes (MLGs, considering only individuals with no missing data, N = 551), total number of alleles (A), number of private alleles (AP), allelic richness [AR = , where N is the number of occurrences of the ith allele among the N sampled g genes by rarefaction (El Mousadik and Petit, 1996)] and observed (HO) and expected (HE) heterozygosities. Wright’s (1965) inbreeding coefficient (F) was also estimated. While MLGs are an estimate of clonal diversity, A, AP and AR are used to describe the diversity of alleles present at nSSR loci, and HO, HE and F measure the mean intra-individual diversity across manioc groups. Estimations and confidence intervals for AR and F, based upon 1000 bootstrap replicates, were obtained with diveRsity (Keenan et al., 2013) and poppr (Kamvar et al., 2014) for R (R Core Team, 2015).
Spatial representations of diversity based on the numbers of MLGs and allelic richness were evaluated for bitter and sweet manioc with point-to-grid analyses in DIVA-GIS (www.diva-gis.org). Considering the sampling locations of bitter and sweet manioc, grids with 10′ diameter cells (~18 km), and a circular neighbourhood of 0.45045° (~50 km) were constructed. With these options, each cell receives the value of the number of MLGs or alleles found within the cell alone (18 km) plus adjacent areas within a circle of 50 km (Scheldeman and van Zonneveld, 2010). In this way, circular neighbourhoods are expected to produce grids with low probability of losing spatial resolution (Scheldeman and van Zonneveld, 2010). For the distribution of MLGs, grids were constructed based on Shannon indexes (Brown and Weir, 1983) considering the total number of MLGs within cells. For the spatial analysis of allelic richness, DIVA-GIS employs the method described by Petit et al. (1998), which is the same method of estimation of AR from El Mousadik and Petit (1996). Therefore, this statistic will be referred to hereafter as ‘spatial allelic richness’ (spAR) when referring to the spatial analyses performed in DIVA-GIS. Representation of spAR was performed building grids of 0.45045° diameter cells because it cannot be estimated with circular neighbourhoods in DIVA-GIS. Hierarchical distribution of genetic variation within and among groups of manioc was evaluated using locus-by-locus AMOVA with Arlequin 3.5 (Excoffier and Lischer, 2010). The significance of divergence estimates was tested with 20 000 permutations. Pairwise genetic differentiation among bitter, sweet and wild manioc and among manioc from different rivers were estimated with Weir and Cockerham’s (1984)FST. Estimations and tests for significance based upon 1000 bootstrap replicates were performed with diveRsity (Keenan et al., 2013). Heat maps were drawn with R (R Core Team, 2015) for the graphical representation of pairwise FST estimates.
Genetic structure based on nSSR variation was evaluated with DAPCs, in the same way as described for cpSSRs. DAPCs were performed considering all the 596 wild and cultivated maniocs; only the 325 bitter maniocs; and only the 226 sweet maniocs. Spatial patterns of genetic structure of bitter and sweet maniocs obtained with DAPCs were evaluated with point-to-grid analyses in DIVA-GIS (www.diva-gis.org). Grids with 10′ diameter cells and circular neighbourhoods of 0.45045° were generated. Each cell contained the average individuals’ membership coefficients to genetic clusters identified in DAPCs. All the maps were produced with DIVA-GIS (www.diva-gis.org) and drawn with maptools (Bivand and Lewin-Koh, 2015) for R (R Core Team, 2015).
Genetic relationships and divergence among individuals and rivers were investigated with neighbour-joining (Saitou and Nei, 1987) dendrograms built with PHYLIP 3.5 (Felsenstein, 2005), based on the chord distance (DCE) of Cavalli-Sforza and Edwards (1967) obtained with MSA 4.05 (Dieringer and Schlötterer, 2003). DCE is based on geometric distances and performs well for the reconstruction of relationships among closely related individuals (Goldstein and Pollock, 1997; Reif et al., 2005). Confidence of relationships was assessed with 1000 bootstrap replicates. Final trees were formatted in FigTree 1.4.1 (http://tree.bio.ed.ac.uk/software/figtree/).
We recognize that it may be difficult to regard sets of manioc landraces as true ‘populations’. However, these analyses aimed to describe the genetic diversity found within ‘synthetic groups’ of manioc according to their toxicity and/or river. Although manioc is a clonally propagated crop, sexual reproduction still occurs (McKey et al., 2010), and social networks contribute to the exchange of landraces even over large distances (Delêtre et al., 2011), which may contribute to some degree of connectivity among the synthetic groups discussed below. Because some bias may be introduced by the presence of samples with the same MLG, clustering analyses, AMOVAs and FST estimations were also performed retaining only one individual for each MLG. Nonetheless, results followed the same general patterns (data not shown) and are available upon request from the corresponding author. Passport data and cpSSR and nSSR genotypes used to perform the analyses of this study are in Supplementary Data Table S1.
RESULTS
Haplotype diversity and structuring of manioc based on cpSSR variation
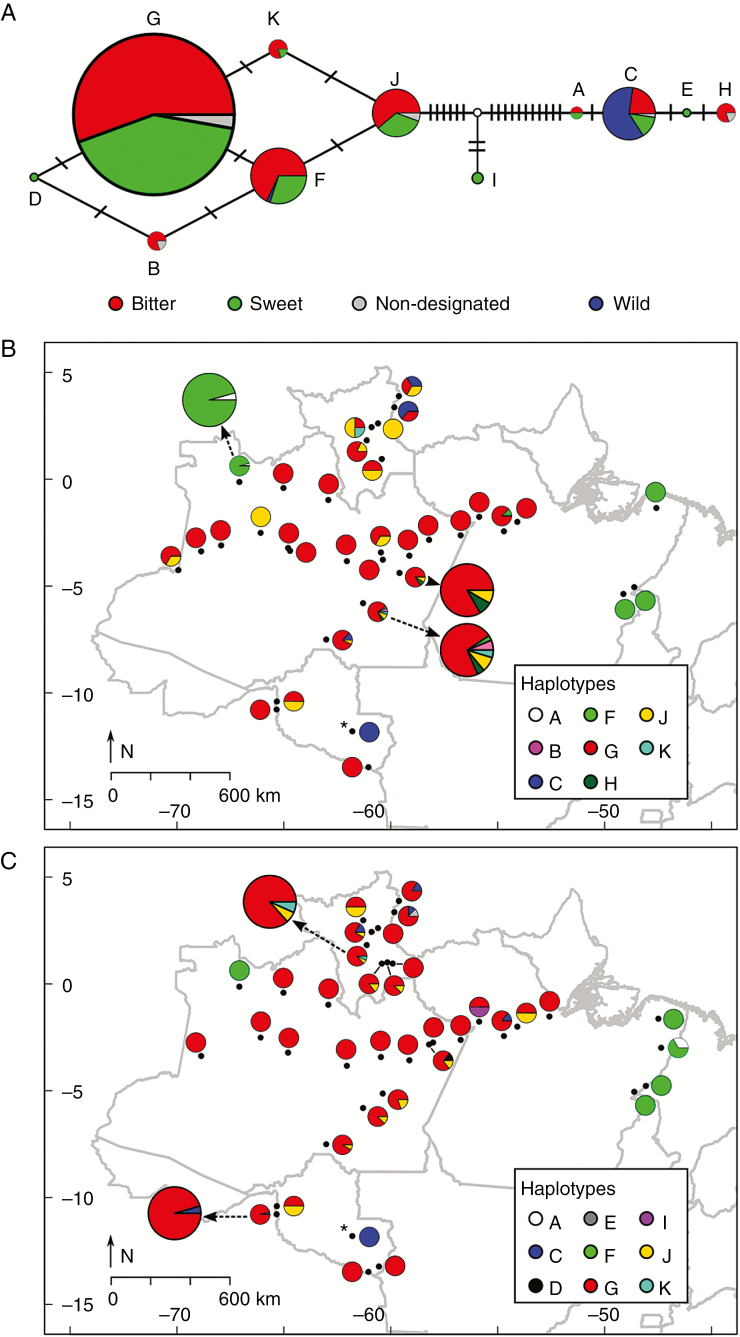
A total of 11 distinct cpSSR haplotypes were found (Fig. 2A). Of these, six were shared among bitter and sweet manioc, including haplotype G, the most frequent one. Haplotype C occurred in all but one individual of wild manioc, which had haplotype F. These two haplotypes were also present in bitter and sweet manioc from different rivers (Fig. 2B, C). Sweet manioc had nine haplotypes while bitter manioc had eight, and haplotype diversity was higher for cultivated manioc than for wild manioc, certainly because of the smaller sample size for the latter (Table 1). Disregarding non-designated samples, three haplotypes occurred only in sweet manioc, while two haplotypes occurred only in bitter manioc. These exclusive haplotypes were all of minor frequency (Fig. 2A, Supplementary Data Table S2).
Fig. 2.
Haplotype network and distribution of chloroplast genetic variation for wild and cultivated manioc (Manihot esculenta) along major Amazonian rivers and adjacent locations in Brazil. (A) Median-joining network where each circle represents a distinct cpSSR haplotype and the size of circles is proportional to their frequencies. Transverse dashes represent 1-bp mutations between haplotypes, and the white circle represents an unobserved haplotype. Frequencies of cpSSR haplotypes across municipalities (dots) are represented for (B) bitter and (C) sweet manioc. In (B) and (C) circle sizes are not proportional to sampling sizes. Some of the pies are enlarged for better visualization and identified with an arrow. The municipality from where most wild manioc was sampled is indicated with an asterisk.
Table 1.
Estimates of chloroplast (cpSSR) and nuclear (nSSR) diversity for wild and cultivated manioc (Manihot esculenta) along major Amazonian rivers and adjacent locations in Brazil
| cpSSR | nSSR | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | NH | H Ehap | N | MLG | A | A P | A R (95 % CI) | H O | H E | F | |
| Manioc | |||||||||||
| Bitter | 318 | 8 | 0.419 | 325 | 227 | 95 | 9 | 4.69 (4.21, 5.21) | 0.573 | 0.613 | 0.065* |
| Sweet | 218 | 9 | 0.319 | 226 | 104 | 93 | 10 | 4.27 (3.71, 4.86) | 0.691 | 0.594 | −0.164* |
| Cultivated | 553 | 11 | 0.383 | 568 | 330 | 113 | 58 | 5.32 (4.86, 5.86) | 0.623 | 0.633 | 0.016 |
| Wild | 28 | 2 | 0.071 | 28 | 21 | 68 | 13 | 4.26 (3.21, 4.79) | 0.465 | 0.511 | 0.088 |
| Rivers and adjacent locations | |||||||||||
| Madeira bitter | 108 | 7 | 0.388 | 111 | 87 | 75 | 2 | 2.40 (1.86, 2.79) | 0.619 | 0.626 | 0.011 |
| Madeira sweet | 49 | 3 | 0.190 | 51 | 33 | 70 | 3 | 2.34 (1.64, 2.79) | 0.701 | 0.583 | −0.202* |
| Negro bitter | 83 | 3 | 0.412 | 83 | 58 | 68 | 1 | 2.16 (1.57, 2.50) | 0.532 | 0.546 | 0.027 |
| Negro sweet | 19 | 2 | 0.281 | 19 | 7 | 49 | – | 2.12 (1.64, 2.50) | 0.671 | 0.505 | −0.331* |
| Branco bitter | 34 | 4 | 0.658 | 36 | 21 | 60 | 1 | 2.20 (1.57, 2.71) | 0.560 | 0.551 | −0.017 |
| Branco sweet | 67 | 5 | 0.296 | 69 | 31 | 72 | 1 | 2.31 (1.64, 2.64) | 0.683 | 0.587 | −0.163* |
| Solimões bitter | 56 | 2 | 0.166 | 58 | 40 | 72 | – | 2.16 (1.57, 2.50) | 0.535 | 0.558 | 0.041 |
| Solimões sweet | 18 | 1 | 0 | 18 | 13 | 51 | – | 2.32 (1.64, 2.64) | 0.754 | 0.567 | −0.331* |
| Amazonas bitter | 26 | 2 | 0.077 | 26 | 20 | 75 | 1 | 2.30 (1.64, 2.64) | 0.594 | 0.597 | 0.005 |
| Amazonas sweet | 39 | 5 | 0.325 | 43 | 22 | 70 | 4 | 2.29 (1.64, 2.64) | 0.667 | 0.565 | −0.180* |
| Guaporé bitter | 2 | 1 | 0 | 2 | 1 | 23 | – | 1.64 (1.64, 1.64) | 0.643 | 0.321 | −1.000* |
| Guaporé sweet | 13 | 1 | 0 | 13 | 8 | 55 | 1 | 2.34 (1.64, 2.79) | 0.753 | 0.595 | −0.266* |
| Pará bitter | 9 | 1 | 0 | 9 | 4 | 46 | – | 2.16 (1.57, 2.71) | 0.611 | 0.538 | −0.136 |
| Pará sweet | 13 | 2 | 0.154 | 13 | 8 | 55 | – | 2.28 (1.57, 2.64) | 0.657 | 0.594 | −0.106 |
| Overall | 581 | 11 | 0.436 | 596 | 351 | 126 | 0.616 | 0.650 | 0.052* | ||
N, number of individuals.
Genetic diversity: NH, number of haplotypes; HEhap, haplotype diversity; MLG, number of multilocus genotypes; A, number of alleles; AP, number of private alleles; AR (95 % CI), allelic richness (95 % confidence interval); observed (HO) and expected (HE) heterozygosities; F, Wright’s inbreeding coefficient.
*Significant based upon 1000 bootstrap replicates.
In general, haplotype G was widely dispersed along the Negro, Madeira, Solimões and Amazonas rivers, leading to a somewhat homogeneous distribution of haplotypes across these different rivers (Fig. 2B, C). Bitter and sweet maniocs from the Branco river were more diverse than those from other rivers, still with the predominance of haplotype G, but with greater frequency of occurrence of haplotypes C and J. Bitter manioc from mid-lower Madeira river also had a high number of haplotypes. Remarkable exceptions to the large predominance of haplotype G are most of the bitter and sweet maniocs from Pará and from the municipality of São Gabriel da Cachoeira, in the upper Negro River, which shared haplotype F (distinct from haplotype G by a 1-bp mutation). Interestingly, haplotype F was also shared by one wild manioc individual (Fig. 2A). Although the primary wild haplotype was present in minor frequencies in other rivers, most cultivated maniocs with haplotype C occurred along the Branco River. The spatial distribution of haplotypes was reflected in genetic clusters found in DAPC, which revealed no genetic structure among rivers (Supplementary Data Fig. S2, Supplementary Data Table S3).
According to AMOVA, cultivated and wild manioc were highly divergent (ΦST = 0.667; Table 2), while bitter and sweet maniocs were not very divergent (ΦST = 0.005), since they shared the most frequent haplotypes. Divergence among different rivers was low (ΦST = 0.084) yet significant, but most of it may be attributed to divergence between bitter and sweet maniocs within rivers (ΦSC = 0.058). When maniocs from the Guaporé and Pará were included in the latter analysis, an increased divergence (ΦST = 0.247) was observed due to the predominance of haplotype F in Pará.
Table 2.
Analysis of molecular variance (AMOVA) based on chloroplast (cpSSR) and nuclear (nSSR) diversity for different hierarchical levels of wild and cultivated manioc (Manihot esculenta) grown along major Amazonian rivers and adjacent locations in Brazil
| Source of variation | cpSSR | nSSR | ||||
|---|---|---|---|---|---|---|
| N | Percentage of variation | Φ statistics | N | Percentage of variation | Φ statistics | |
| Between wild and cultivated | 581 | 66.62 | Φ ST = 0.667* | 596 | 27.89 | Φ ST = 0.279* |
| Within wild and cultivated | 33.38 | 72.11 | ||||
| Between bitter and sweet | 536 | 0.54 | Φ ST = 0.005 | 551 | 8.35 | Φ ST = 0.083* |
| Within bitter and sweet | 99.46 | 91.65 | ||||
| Among rivers§ | 499 | 2.64 | Φ ST = 0.084* | 514 | −4.63 | Φ ST = 0.081* |
| Between bitter and sweet varieties within rivers | 5.73 | Φ SC = 0.058* | 12.82 | Φ SC = 0.122* | ||
| Within rivers | 91.63 | Φ CT = 0.026* | 91.82 | Φ CT = -0.047* | ||
| Among all locationsǂ | 536 | 20.56 | Φ ST = 0.247* | 551 | −4.11 | Φ ST = 0.083* |
| Between bitter and sweet manioc within locations | 4.11 | Φ SC = 0.052* | 12.37 | Φ SC = 0.119* | ||
| Within locations | 75.33 | Φ CT = 0.205* | 91.74 | Φ CT = -0.041* | ||
N, sample size at each hierarchical level.
*Significant at P < 0.05.
§Disregarding Pará and Guaporé, non-designated and wild manioc.
ǂDisregarding non-designated and wild manioc.
Genetic diversity and structuring of manioc based on nSSR variation
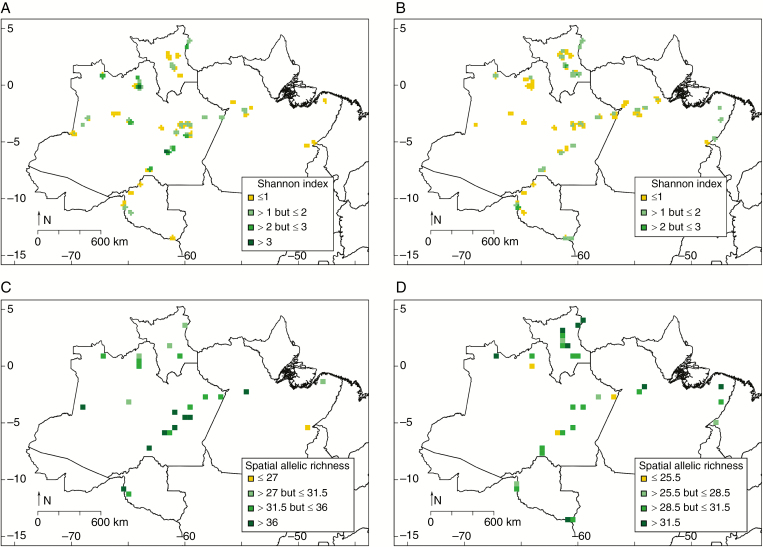
Considering the 551 individuals with no missing data, 351 MLGs were found (Table 1). All the wild manioc individuals (N = 21) had distinct MLGs, and bitter manioc had proportionally more MLGs in relation to sample size (N = 304, MLG = 227) than sweet manioc (N = 209, MLG = 104), again perhaps due to sampling differences. The spatial distribution of MLG diversity was somewhat different between bitter and sweet manioc (Fig. 3A, B). For bitter manioc, the diversity of MLGs was higher along the middle Madeira and mid-upper Negro Rivers than in other locations (Fig. 3A). Sweet manioc from the Branco River showed higher diversity of MLGs than other locations (Fig. 3B). Contrasting spatial patterns were also observed for spatial allelic richness (spAR) of bitter and sweet manioc (Fig. 3C, D). For bitter manioc, high spAR was observed along the Madeira River and in a few locations along the Solimões and Amazonas Rivers (Fig. 3C). Just as for MLG diversity, sweet manioc from the Branco River showed higher spAR than sweet manioc from other locations (Fig. 3D).
Fig. 3.
Spatial distribution of multilocus genotype (MLG) diversity and spatial allelic richness (spAR) for bitter and sweet manioc (Manihot esculenta) across major Amazonian rivers and adjacent locations in Brazil, based on the variation of 14 nSSR. Maps at the top show the diversity measured with the Shannon index, based on the number of MLGs, estimated for 10′ cells (~18 km), applying a 0.45045° (~50 km) circular neighbourhood of (A) bitter and (B) sweet manioc. Maps at the bottom show the diversity measured as allelic richness estimated for 0.45045° cells, based on the total number of alleles of (C) bitter and (D) sweet manioc.
Bitter and sweet manioc had similar numbers of alleles and allelic richness, but sweet manioc had a significant excess of heterozygotes, while bitter manioc had a low, yet significant, heterozygote deficit (F = −0.164 and 0.065, respectively). Cultivated manioc had higher genetic diversity than wild manioc, although the difference decreased when comparing wild with bitter and sweet manioc separately. Wild manioc had a greater number of private alleles than bitter and sweet manioc, but had the greatest heterozygote deficit (Table 1). Sweet manioc from all the rivers, Guaporé and Pará had a remarkable excess of heterozygotes.
Vernacular names of the sampled landraces were neither widely distributed nor associated with specific MLGs. Most of these were identified simply as macaxeira or mandioca: sweet (75 of 209) or bitter (54 of 304) manioc, respectively. For bitter manioc, the most common vernacular names were registered no more than 14 times, and were locally concentrated, while the most common MLG occurred only eight times. For sweet manioc, both the most frequent vernacular names and the most frequent MLGs appear to be spread across the Madeira and Branco Rivers, but still with low counts (Supplementary Data Fig. S3).
Only a few of the genetic distances among bitter and sweet manioc MLGs were small (DCE <0.1), and the great majority of pairwise comparisons had intermediate values (0.4 < DCE < 0.6). Some of the most frequent MLGs of bitter and sweet manioc were very similar (DCE <0.05), but most of the pairwise genetic distances were intermediate to high (Supplementary Data Fig. S3).
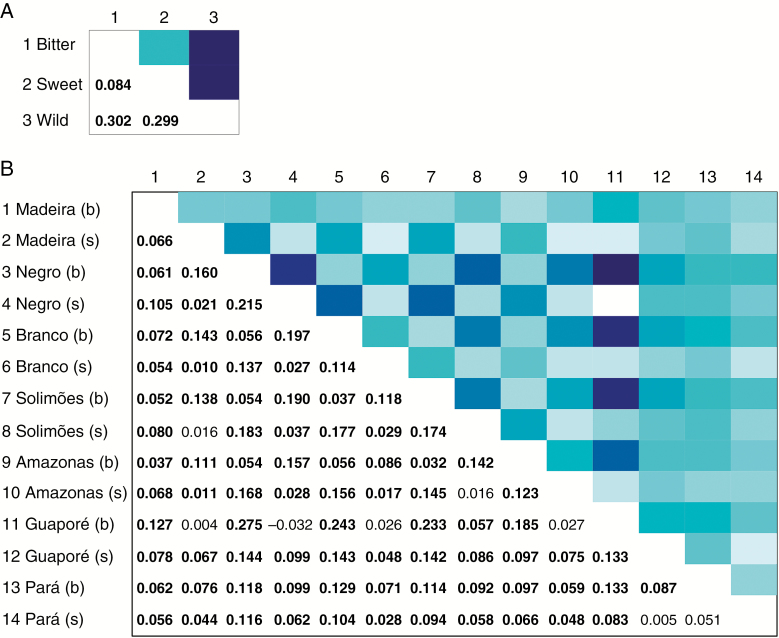
AMOVAs revealed that the highest divergence based on nSSR was between wild and cultivated manioc (ΦST = 0.279), followed by the divergence between bitter and sweet manioc (ΦST = 0.083; Table 2). Similar degrees of divergence were found among rivers and among all locations, but most of these may be attributed to genetic differences between bitter and sweet manioc within locations, given the ΦSC estimates (0.122 for rivers, 0.119 for all locations; Table 2). Pairwise FST estimates also showed high divergence between cultivated and wild manioc (Fig. 4). Most FST estimates were low, yet significant. In general, manioc from Guaporé showed the highest levels of genetic differentiation in relation to the major Amazonian rivers. Additionally, divergence among bitter and sweet manioc from different rivers was generally higher than the comparisons among bitter (or among sweet) manioc from different rivers.
Fig. 4.
Pairwise FST (Weir and Cockerham, 1984) estimates (below diagonal) and heat maps of FST values (above diagonal) among manioc (Manihot esculenta) cultivated along major Amazonian rivers and adjacent locations in Brazil, based on the variation of 14 nSSRs. Genetic divergence is represented among (A) bitter, sweet and wild manioc; and (B) bitter (b) and sweet (s) manioc within rivers and adjacent locations. Boldfaced values are significant based upon 1000 bootstrap replicates. For each heat map, the darker the blue shade the greater is FST.
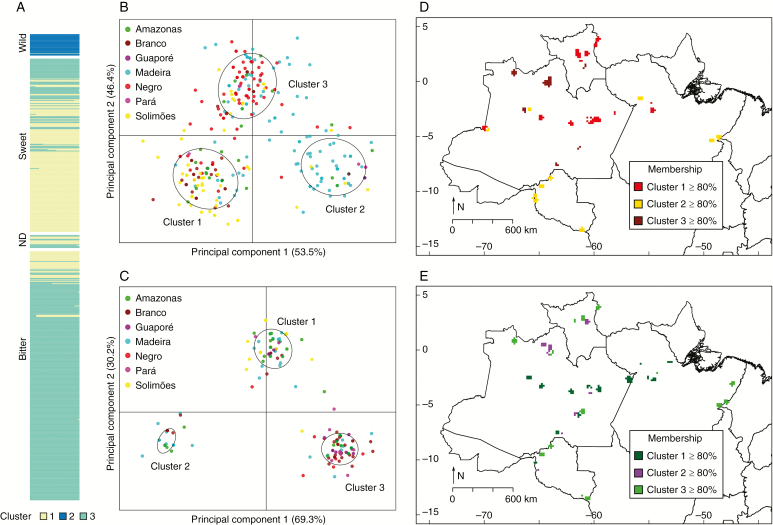
The K-means algorithm suggested the existence of three clusters in the DAPC performed for the whole set of individuals (Fig. 5A), which had good concordance with wild, bitter and sweet manioc. Among sweet maniocs, 72.5 % were allocated in the first cluster, while 89.5 % of the bitter maniocs were in the third cluster. Except for one individual, which was more related to bitter manioc, all wild maniocs formed a very distinct cluster (Fig. 5A, Supplementary Data Fig. S4, Supplementary Data Table S3). DAPC also suggested a considerable degree of admixture among bitter and sweet maniocs (Supplementary Data Fig. S4).
Fig. 5.
Patterns of genetic structure of cultivated and wild manioc (Manihot esculenta) along major Amazonian rivers and adjacent locations in Brazil, based on the variation of 14 nSSRs. (A) Bar plot of DAPC performed for the whole dataset (N = 596), showing individuals’ assignments to genetic clusters. Different colours represent different clusters. ND, non-designated. Scatterplots show individuals’ dispersion across genetic clusters of DAPCs performed for (B) bitter (N = 325) and (C) sweet (N = 226) manioc. Maps show 10′ cells (~18 km), applying a 0.45045° (~50 km) circular neighbourhood, with the average membership coefficients of samples in genetic clusters identified by DAPC for (D) bitter and (E) sweet manioc. Considering the arbitrary threshold of 80 %, locations represented in the maps contain landraces with higher membership coefficients in one of the genetic clusters. In relation to Fig. 1, locations that are not represented in these maps have individuals with admixed ancestry.
When DAPC was performed separately for bitter and for sweet manioc, each analysis showed three clusters (Fig. 5B, C). There were no clear spatial concordances between sampling locations and genetic clusters within bitter and sweet maniocs (Fig. 5D, E). However, the spatial distribution of membership coefficients revealed some tendencies. In both analyses, bitter and sweet maniocs from the Madeira River were present in all clusters, and sweet maniocs of the Amazonas and Branco Rivers were also present in the three clusters (Fig. 5B, C). Considering the arbitrary threshold of 80 % for the membership coefficients, bitter maniocs from the mid-lower Solimões, upper Amazonas and upper Branco Rivers were preferentially assigned to the first cluster (Fig. 5B, D). Bitter maniocs from the upper Madeira River and the Guaporé and Pará regions were preferentially distributed in the second cluster. Bitter maniocs from the upper Negro River tended to be allocated to the third cluster (Fig. 5B, D). Tendencies observed for the genetic structure of sweet manioc were somewhat less evident than those observed for bitter manioc (Fig. 5C, E). Sweet maniocs from the mid-lower Solimões and Amazonas Rivers had high membership coefficients in the first cluster, which also presented individuals from the middle Madeira River with high memberships. Some sweet maniocs scattered along the Negro, Branco and Madeira Rivers had higher membership coefficients in the second DAPC cluster (Fig. 5C, E). Sweet manioc from the upper Madeira River was more frequent in the third cluster, which also contained maniocs from the Negro, Branco and middle Madeira Rivers and from Guaporé and Pará.
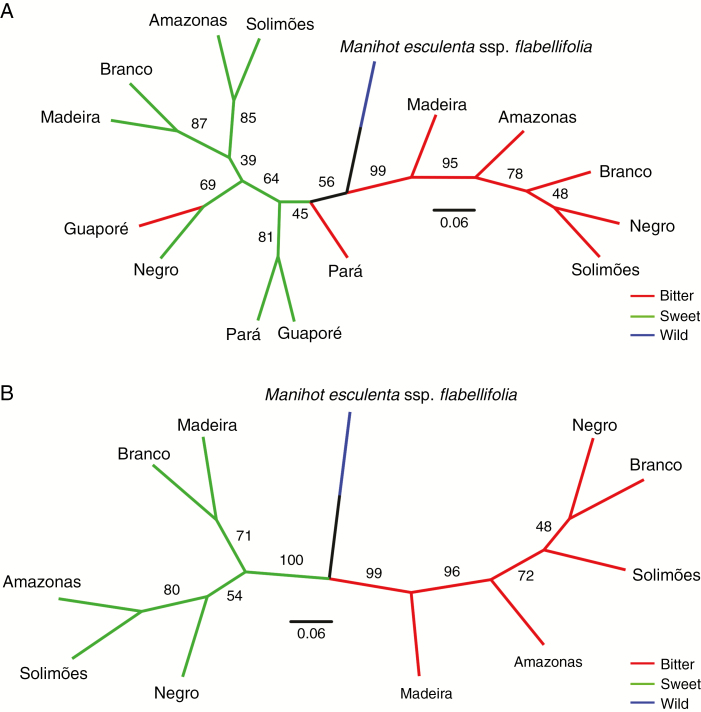
The dendrogram demonstrated that bitter manioc from major Amazonian rivers formed a consistent group that is closely related to wild manioc (Fig. 6A). Sweet manioc from major Amazonian rivers formed a less consistent group, in which bitter manioc from Guaporé is inserted. This surprising group is closely related to the sweet manioc from Guaporé and Pará, followed by bitter manioc from Pará. Because major Amazonian rivers were likely important for dispersals of manioc, and since the numbers of samples from Guaporé and Pará were much smaller than the number of samples from the rivers, another dendrogram was built including only manioc from the major Amazonian rivers. In this latter dendrogram, bitter and sweet manioc form two consistent and well-supported groups, which are equally related to wild manioc (Fig. 6B). Additionally, sweet manioc from the Madeira and Branco Rivers showed a closer relationship to wild manioc in comparison with Fig. 6A. In both dendrograms, bitter manioc from the Madeira River is closer to wild manioc than are the bitter maniocs from the other rivers. However, higher internal support was found in Fig. 6B than in Fig. 6A. A dendrogram depicting relationships among individuals agreed with DAPC results, showing a consistent grouping of wild manioc and admixture among bitter and sweet maniocs (Supplementary Data Fig. S5).
Fig. 6.
Neighbour-joining dendrograms showing (A) groupings among bitter, sweet and wild manioc (Manihot esculenta) grown along the major Amazonian rivers and adjacent locations in Brazil, and (B) only along the major Amazonian rivers. Consensus topologies, based on Cavalli-Sforza and Edwards’ (1967) chord distance estimated from 14 nuclear microsatellites, are shown with internal support tested based upon 1000 bootstrap replicates.
DISCUSSION
Diversification of bitter and sweet manioc in Amazonia
Sweet manioc had greater observed heterozygosity and lower inbreeding coefficient (HO = 0.691, F = −0.164) than bitter manioc (HO = 0.573, F = 0.065). Although little discussed, similar findings were reported in previous studies with Brazilian maniocs (Elias et al., 2004; Peroni et al., 2007), but not for maniocs from different countries (Bradbury et al., 2013). At small time scales (successive cycles of cultivation), many factors may lead to increasing heterozygosity in manioc. Among these, the indirect selection for heterozygotes in manioc swiddens (Pujol et al., 2005; Pujol and McKey, 2006), the retention of sexual reproduction and the possibility of recombination among landraces (McKey et al., 2010) are key factors. At a large time scale (the history of domestication and diversification of manioc), these factors and the occurrence of somatic mutations (McKey et al., 2012) may lead to an even more pronounced increase in the heterozygosity. Arroyo-Kalin (2010) suggested early selection of sweet manioc after the initial domestication, and subsequent selection of bitter manioc, which may imply a temporal gap between dispersals and diversification times of sweet and bitter manioc. Therefore, the greater levels of heterozygosity found for sweet manioc may be a signature of a greater diversification period after its creation than that of bitter manioc. Although far from being a general tendency, the occurrence of the most frequent sweet manioc MLGs in a wider spatial range and at higher frequencies than the bitter manioc MLGs may corroborate this scenario. Considering that bitter manioc became more important than sweet manioc in Brazilian Amazonia, the former may have been subjected to more intensive selection pressures and may have undergone a greater number of genetic bottlenecks than the latter. This may have led to the higher deficit of heterozygotes observed for bitter than for sweet manioc.
On the other hand, neighbour-joining analysis of the whole set of individuals (Fig. 6A, Supplementary Data Fig. S5) suggests that bitter manioc is less divergent from wild manioc than is sweet manioc. These results agree with Perrut-Lima et al. (2014), who classified as bitter the roots of M. esculenta ssp. flabellifolia from Rondônia state in Brazil, including populations from the municipality where most of the wild manioc was sampled in our study. However, this pattern is not present in Fig. 6B, where bitter and sweet maniocs from the major Amazonian rivers were equally related to wild manioc. Although these results failed to support Arroyo-Kalin’s (2010) hypothesis, it cannot be discarded because sweet manioc from other locations around the centre of domestication may be genetically closer to wild manioc than was sweet manioc sampled in this study, as observed by Mühlen et al. (2013).
The spatial distribution of genetic diversity is different for bitter and sweet manioc (Figs 2 and 3). High haplotype diversity of bitter and sweet manioc from the Branco River may be the result of the great environmental heterogeneity of this region, including areas of Amazonian rainforest and savannahs. This may also result from the great socio-diversity that existed in this area during pre-Columbian times (i.e. before 1492), with the occurrence of Arawak, Carib and other unassociated languages (Eriksen, 2011). High diversity of MLGs in bitter manioc from the Negro River agrees with the results of Mühlen et al. (2013), and also may reflect the occurrence of pre-Columbian Arawak speakers (Eriksen, 2011).
It may be worth asking whether manioc landraces sampled from riverine non-indigenous communities of smallholder farmers reflect the indigenous diversity of manioc that was once dispersed and selected by native Amerindian peoples. Some authors have commented on the importance of matrilineal and kin heritage, and exchange networks of local landraces among indigenous and non-indigenous communities for obtaining and managing of manioc (Salick et al., 1997; Heckler and Zent, 2008; Coomes, 2010). It is reasonable to suggest that during the post-conquest occupation of Amazonia (after the 15th century), inter-ethnic marriages between Amerindians and Europeans was accompanied by the inheritance of indigenous manioc landraces. This probably occurred many times in different parts of Amazonia. The indigenous manioc landraces, as well as the management practices, were likely the original sources of the current landraces cultivated by smallholder farmers in Amazonia. This appears to be supported by genetic evidence. Elias et al. (2004) showed considerable overlap and close genetic relationships among manioc landraces from indigenous, mixed-race and riverine populations of Brazilian Amazonia and Guyana. It is therefore reasonable to assume that the current manioc landraces cultivated across Amazonia are largely a legacy of Native Amazonians. Therefore, the current patterns of genetic structure and diversity of manioc landraces cultivated by smallholder farmers may be a good approximation of that expected for Native Amazonian manioc landraces. Nevertheless, studies including indigenous landraces would improve our understanding of the genetic structure and diversity of manioc in Amazonia.
Some expected results were observed in this study. Incomplete lineage sorting due to common ancestry with M. esculenta ssp. flabellifolia (Olsen and Schaal, 1999) may explain the occurrence of the primary haplotype of wild manioc in bitter and sweet manioc. Selection for different characteristics due to local preferences of farmers (Miller and Schaal, 2005) and sampling of a small fraction of wild manioc’s distribution (Léotard et al., 2009) may explain the higher haplotype and genetic diversity found in cultivated manioc than in wild manioc. The high levels of inbreeding in wild manioc may be due to the occurrence of auto-compatibility and short-distance dispersal of seeds (Olsen and Schaal, 2001).
Naming of manioc landraces is influenced by many different factors, and different farmers may manage the same genotype under different vernacular names (Boster, 1985; Salick et al., 1997; Elias et al., 2000; Alves-Pereira et al., 2011, 2017). Therefore, it is not surprising that vernacular names are not associated with specific MLGs. Given the predominance of bitter manioc cultivation in Brazilian Amazonia, it is very likely that the collection of many landraces named simply as macaxeira (sweet) or mandioca (bitter) is a reflection of the preference for cultivation of either manioc type.
Evidence of hybridization between bitter and sweet manioc is commonly observed in molecular studies (Bradbury et al., 2013; Mühlen et al., 2013). The great overlap observed among cpSSR matrilineages of bitter and sweet manioc was also observed for nuclear diversity, suggesting that gene flow and incorporation of volunteer seedlings (Martins, 2001; Duputié et al., 2009) occurs frequently along the major Amazonian rivers. Probability of gene flow increases when bitter and sweet manioc are cultivated in the same field, which was observed sometimes during sampling. However, bitter manioc is commonly cultivated in swiddens farther from house units than in home gardens, where sweet manioc is typically cultivated (McKey and Beckerman, 1993; Elias et al., 2000). It is possible that pollination by small insects (Lebot, 2009) and even seed dispersal by ants (Elias and McKey, 2000) are effective in connecting swiddens and home gardens within communities of smallholder farmers. Exchange networks of manioc stem cuttings (Coomes, 2010; Delêtre et al., 2011) extend gene flow and hybridization among unrelated manioc landraces over greater geographic scales. Nonetheless, genetic divergence between bitter and sweet manioc is still considerable (ΦST(nSSR) = 0.083), corroborating their recognition by smallholder farmers (Martins, 2001). The intermediate to high genetic distances observed among bitter and sweet manioc MLGs also shows this differentiation, and suggests that bitter and sweet manioc are actively selected for diversity and distinction (Boster, 1985; Duputié et al., 2009). Differential management and selective pressures, such as cultivation in separated areas (Martins, 2001) and selection for low/high toxicity levels (McKey and Beckerman, 1993), explains why hybridization has not effaced the genetic differentiation between sweet and bitter manioc.
Genetic structure of bitter and sweet manioc: considerations on the crop’s dispersal
The wide distribution of the most common haplotype of cultivated manioc suggests ample dispersal along the major Amazonian rivers long before European arrival in the Americas at the end of the 15th century, just as observed for sweet potato (Roullier et al., 2011). The strong genetic divergence observed between wild (M. esculenta ssp. flabellifolia) and cultivated manioc may also reflect the antiquity of manioc domestication (10 000–9 000 BP; Olsen and Schaal, 1999). These results agree with evidence for an early ample dispersal of manioc across the Neotropics by 6500 BP (Isendahl, 2011).
Contemporary patterns of genetic diversity and structure of cultivated plants reflect prehistoric processes associated with domestication, such as those affecting patterns of dispersal from the geographic origins of crops (Miller and Schaal, 2005; Roullier et al., 2011). The absence of clear patterns of haplotype and genetic structure among rivers observed in this study does not permit direct inferences about how the dispersal of manioc occurred. The ample dispersion of manioc from the Madeira River in the DAPC clusters (Fig. 5B, C), and their closer position in relation to wild manioc in the dendrogram (Fig. 6B) suggest that this river was important for northward dispersal of bitter and sweet manioc to the other major Amazonian rivers. The Madeira River was a route for northward and eastward dispersals of peach palm (Bactris gasipaes) (Cristo-Araújo et al., 2013), another important Amazonian crop. According to these authors, molecular data suggest a single domestication for peach palm somewhere in the upper Madeira River and adjacent areas in Peru, an area that partially overlaps manioc’s centre of domestication. Given their prominence as centres of crop diversity (Clement, 1999b), the major Amazonian rivers were probably important routes for the dispersals of many crops that were domesticated in the periphery of the Amazon basin (Clement et al., 2010).
The somewhat different spatial patterns of distribution of genetic diversity and structure of bitter and sweet manioc may result from distinct dispersal histories, which agrees with Mühlen et al. (2013). For peach palm (B. gasipaes), different molecular analyses identified a deep split between eastern Amazonian landraces and those from western and central Amazonia (Rodrigues et al., 2004; Hernández-Ugalde et al., 2011; Cristo-Araújo et al., 2013). Just as for manioc, the genetic structure and diversity of peach palm across these regions may also reflect distinct patterns of dispersals, although they may also reflect selection for different preferences, as discussed below. The wide dispersal of Brazil nut (Bertholletia excelsa) across Amazonia has been recognized as human-mediated (Shepard and Ramirez, 2011), and genetic studies contributed to this conclusion (Kanashiro et al., 1997; Gribel et al., 2007). However, the sampling of very disjunct populations and the high levels of genetic structure (Sujii et al., 2015) complicate inferences on how the dispersal of Brazil nut occurred.
In this study, no phylogeographic patterns were detected for matrilineages of manioc from different Amazonian rivers, and the genetic divergence among rivers was superimposed on the bitter–sweet distinction. However, somewhat different genetic structure within bitter and sweet manioc was observed (Fig. 5D, E). The distinct DAPC cluster of bitter manioc from the Negro River may reflect adaptations to the low-fertility and highly acidic sandy soils of this region. This might not be a general tendency, though, since sweet maniocs from the Negro River were allocated into two different genetic clusters (Fig. 5E). These patterns of genetic structure within bitter and sweet manioc may also reflect the great diversity of ethnic groups in pre-Columbian Amazonia. Manioc cultivation, distribution and consumption were primary factors for the sustenance of Amazonian complex societies (Heckenberger, 1998). The expansion and diffusion of indigenous populations in Amazonia occurred much later than the domestication of manioc (Eriksen, 2011), but it is possible that different ethnic populations created distinct sets of manioc landraces in response to different cultural preferences (Boster, 1985; Heckler and Zent, 2008; Peña-Venegas et al., 2014). Eriksen (2011) discusses the great ethnolinguistic diversity in pre-Columbian Amazonia and highlights the fact that Arawakan groups were prevalent along the Negro River: these peoples might have been responsible for the distinctiveness of bitter manioc in this region. However, both bitter and sweet manioc from the upper Madeira River tended to form a consistent geographic cluster (Fig. 5D, E), but a mosaic of Arawak, Tupi and Panoan peoples occupied this region at the time of European conquest (Eriksen, 2011). Thus, prehistoric linguistics alone cannot explain the patterns observed and certainly does not explain the lack of patterns.
It is probable that ethnic preferences were important for the dispersal patterns of Amazonian crops (Clement et al., 2010). However, it is also possible that more recent historical processes than the dispersal of manioc by native Amazonian peoples have blurred the signatures of different dispersals. The rivers included in this study had great importance as pre-Columbian centres of Amazonian crop diversity (Clement et al., 2010). Valleys of these rivers presented high population densities and intensification of agriculture, which created centres of crop genetic diversity (Clement, 1999b). Through exchange networks, these centres and regions of crop diversity might have collaborated to homogenize sets of manioc landraces created by different pre-Columbian peoples. The post-European conquest period (1541–1616) and subsequent colonization (1616–1850) also had an enormous impact on Amazonian indigenous populations, which were reduced by 90–95 % during the two centuries following European colonization (Eriksen, 2011). This drastic population decrease was very likely associated with the loss of a great proportion of the diversity of Amazonian crops, and resulted in the movement of survivor populations (Clement, 1999b). The movement of indigenous peoples during the colonial period in Brazil (1616–1850) certainly altered the distribution of their manioc landraces, and may have contributed to the weak genetic structure among rivers. Even more recently, the ‘rubber boom’ (1850–1920) caused massive human migration from north-eastern Brazil to different regions in Amazonia. The great influence of north-eastern Brazilian migrants in the production of manioc flour in the upper Juruá River in south-western Amazonia (Emperaire et al., 2012) exemplifies the role of migrants in the management of Amazonian manioc landraces. Migrants certainly contributed to the diffusion of manioc landraces, which may also have reduced genetic structure among rivers. The occurrence of haplotype F in manioc from the upper Negro River and Pará (Fig. 2B, C) may be an example of the impact of human movements on the introduction of manioc from one region to the other. In this case, homoplasy and hybridization with wild manioc are also possible explanations. Frequent germplasm exchange and the migration of human populations are important factors for the absence of clear regional structure of manioc, as suggested by several other studies (Siqueira et al., 2009; Oliveira et al., 2014; Carmo et al., 2015). Recently, Neves (2013) proposed that the advent of predominantly agriculture-based economies in Amazonia occurred only after European conquest in the 15th century. Extensive manioc-based agriculture, typical of Amazonia, would be associated with demographic and technological changes brought by European peoples (Neves, 2013). In this scenario, it is more likely that the patterns of genetic diversity and structure of Amazonian manioc are explained by post-European conquest events in the history of domestication rather than by any signatures of the crop’s early dispersal. Approaches relying on ancient DNA, such as those employed by Roullier et al. (2013) with sweet potato, may be useful to better understand the role of post-European conquest historical processes in shaping current patterns of genetic structure of manioc and other Amazonian crops.
Understanding how genetic diversity of manioc is distributed and structured across Amazonia also has practical importance, since it may be useful for prospection of manioc landraces aiming to conserve the crop’s genetic resources. Moreover, the results found here are important not only for the better understanding of manioc’s evolutionary history. It is possible that other Amazonian crops that were domesticated near the area of manioc origin and were important for indigenous peoples as well, such as peanuts (Arachis hypogaea) and Capsicum peppers, spread in association with manioc (Pickersgill, 2007). Whether the same patterns exist for these crops remains to be tested, but investigations like this would be valuable for understanding how prehistoric Amazonian peoples managed their crops. At the same time, such approaches serve to illustrate the valuable role of current populations of smallholder farmers in the maintenance of the impressive diversity of their native Amazonian crops.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Figure S1: map of occurrence of bitter and sweet manioc (Manihot esculenta) landraces in Amazonia. Figure S2: discriminant analysis of principal components (DAPC) for 596 manioc (Manihot esculenta) landraces, based on four chloroplast microsatellites (cpSSRs). Figure S3: distribution of the most common vernacular names and the most frequent MLGs, and genetic divergence among MLGs of manioc (Manihot esculenta) landraces along major Amazonian rivers and adjacent locations in Brazil. Figure S4: discriminant analysis of principal components (DAPC) for 596 manioc (Manihot esculenta) landraces, based on 14 nuclear microsatellites (nSSRs). Figure S5: neighbour-joining dendrogram for 596 individuals of wild and cultivated manioc (Manihot esculenta), based on 14 nuclear microsatellites (nSSRs). Table S1: spreadsheet with cpSSR and nSSR genotypes and passport data on 596 manioc (Manihot esculenta) landraces sampled along major Amazonian rivers and adjacent locations in Brazil. Table S2: chloroplast haplotypes of manioc (Manihot esculenta) landraces grown along major Amazonian rivers and adjacent locations in Brazil. Table S3: distribution of manioc landraces among discriminant analysis of principal components (DAPC) clusters.
ACKNOWLEDGEMENTS
The authors thank all smallholder farmers who shared their time and permitted sampling. A.A.-P. (2013/11137-7), G.D. (2013/08884-5) and S.L.F.R. (2014/-109478) thank Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) for scholarships. C.R.C., E.A.V. and M.I.Z. (PQ-CNPq) thank Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for fellowships. The authors thank three anonymous reviewers for criticism and suggestions that improved the manuscript. This study was funded by FAPESP (2013/00003-0 to M.I.Z.) and supported by CNPq (CT-Amazônia 575588/08-0 to C.R.C.) and FAPESP (2012/08307-5 to E.A.V.).
LITERATURE CITED
- Alves-Pereira A, Peroni N, Abreu AG, Gribel R, Clement CR. 2011. Genetic structure of traditional varieties of bitter manioc in three soils in Central Amazonia. Genetica 139: 1259–1271. [DOI] [PubMed] [Google Scholar]
- Alves-Pereira A, Peroni N, Cavallari MM, Lemes MR, Zucchi MI, Clement CR. 2017. High genetic diversity among and within bitter manioc varieties cultivated in different soil types in Central Amazonia. Genetics and Molecular Biology 40: 468–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo-Kalin M. 2010. The Amazonian Formative: crop domestication and anthropogenic soils. Diversity 2: 473–504. [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16: 37–48. [DOI] [PubMed] [Google Scholar]
- Bivand R, Lewin-Koh N. 2015. Maptools: tools for reading and handling spatial objects. R package version 0.8–36 http://CRAN.R-project.org/package=maptools. Accessed 25 December 2015.
- Boster JS. 1985. Selection for perceptual distinctiveness: evidence from Aguaruna cultivars of Manihot esculenta. Economic Botany 39: 310–325. [Google Scholar]
- Boster JS. 1986. Exchange of varieties and information between Aguaruna manioc cultivators. American Anthropologist 88: 428–436. [Google Scholar]
- Bradbury EJ, Duputié A, Delêtre M et al. . 2013. Geographic differences in patterns of genetic differentiation among bitter and sweet cassava (Manihot esculenta: Euphorbiaceae). American Journal of Botany 100: 857–866. [DOI] [PubMed] [Google Scholar]
- Brown AHD, Weir BS. 1983. Measuring genetic variability in plant populations. In: Tanksley SD, Orton TJ eds. Isozymes in plant genetics and breeding. Amsterdam: Elsevier, 219–239. [Google Scholar]
- Brown CH, Clement CR, Epps P, Luedeling E, Wichmann S. 2013. The paleobiolinguistics of domesticated manioc (Manihot esculenta). Ethnobiology Letters 4: 61–70. [Google Scholar]
- Carmo CD, Santos DB, Alves LB, Oliveira GAF, Oliveira EJ. 2015. Development of TRAP (target region amplification polymorphism) as new tools for molecular genetic analysis in cassava. Plant Molecular Biology Reporter 33: 1953–1966. [Google Scholar]
- Cavalli-Sforza LL, Edwards AWF. 1967. Phylogenetic analysis: models and estimation procedures. American Journal of Human Genetics 19: 233–257. [PMC free article] [PubMed] [Google Scholar]
- Chavarriaga-Aguirre P, Maya MM, Bonierbale MV et al. . 1998. Microsatellites in cassava (Manihot esculenta Crantz): discovery, inheritance and variability. Theoretical and Applied Genetics 97: 493–501. [Google Scholar]
- Chernela JM. 1986. Os cultivares de mandioca na área do Uaupés. In: Ribeiro B. ed. Suma Etnológica Brasileira. Vol. 1.Petrópolis: Vozes/FINEP, 151–158. [Google Scholar]
- Chung S-M, Staub JE. 2003. The development and evaluation of consensus chloroplast primer pairs that possess highly variable sequence regions in a diverse array of plant taxa. Theoretical and Applied Genetics 107: 757–767. [DOI] [PubMed] [Google Scholar]
- Clement CR. 1999a. 1492 and the loss of Amazonian crop genetic resources. I. The relation between domestication and human population decline. Economic Botany 53: 188–202. [Google Scholar]
- Clement CR. 1999b. 1492 and the loss of Amazonian crop genetic resources. II. Crop biogeography at contact. Economic Botany 53: 203–216. [Google Scholar]
- Clement CR, Cristo-Araújo M, Coppens D’Eeckenbrugge G, Alves Pereira A, Picanço-Rodrigues D. 2010. Origin and domestication of native Amazonian crops. Diversity 2: 72–106. [Google Scholar]
- Coomes OT. 2010. Of stakes, stems and cuttings: the importance of local seed systems in traditional Amazonian societies. Professional Geographer 62: 323–334. [Google Scholar]
- Cristo-Araújo M, Reis VM, Picanço Rodrigues D, Clement CR. 2013. Domestication of peach palm in southwestern Amazonia. Tipití 2: 74–80. [Google Scholar]
- Delêtre M, McKey DB, Hodkinson TR. 2011. Marriage exchanges, seed exchange, and the dynamics of manioc diversity. Proceedings of the National Academy of Sciences of the USA 108: 18249–18254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieringer D, Schlötterer C. 2003. Microsatellite analyzer (MSA): a platform independent analysis tool for large microsatellite data sets. Molecular Ecology Notes 3: 167–169. [Google Scholar]
- Doyle JJ, Doyle JL. 1990. Isolation of plant DNA from fresh tissue. Focus 1: 13–15. [Google Scholar]
- Duputié A, David P, Debain C, McKey D. 2007. Natural hybridization between a clonally propagated crop, cassava (Manihot esculenta Crantz) and a wild relative in French Guiana. Molecular Ecology 16: 3025–3038. [DOI] [PubMed] [Google Scholar]
- Duputié A, Massol F, David P, Haxaire C, McKey D. 2009. Traditional Amerindian cultivators combine directional and ideotypic selection for sustainable management of cassava genetic diversity. Journal of Evolutionary Biology 22: 1317–1325. [DOI] [PubMed] [Google Scholar]
- Elias M, McKey D. 2000. The unmanaged reproductive ecology of domesticated plants in traditional agroecosystems: an example involving cassava and a call for data. Acta Oecologica 21: 223–230. [Google Scholar]
- Elias M, Rival L, McKey D. 2000. Perception and management of cassava (Manihot esculenta Crantz) diversity among Makushi Amerindians of Guyana (South America). Journal of Ethnobiology 20: 239–265. [Google Scholar]
- Elias M, Mühlen GS, McKey D, Roa AC, Tohme J. 2004. Genetic diversity of traditional South American landraces of cassava (Manihot esculenta Crantz): an analysis using microsatellites. Economic Botany 58: 242–256. [Google Scholar]
- El Mousadik A, Pettit RJ. 1996. High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.) Skeels] endemic to Morocco. Theoretical and Applied Genetics 92: 832–839. [DOI] [PubMed] [Google Scholar]
- Emperaire L. 2001. Elementos de discussão sobre a conservação da agrobiodiversidade: o exemplo da mandioca (Manihot esculenta Crantz) na Amazônia brasileira. In: Capobianco J.-P. ed. Biodiversidade da Amazônia brasileira, avaliação e ações prioritárias para a conservação, uso sustentável e repartição dos benefícios. São Paulo: ISA, 225–234. [Google Scholar]
- Emperaire L, Eloy L, Cunha MC et al. . 2012. D’une production localisée à une indication géographique en Amazonie: les enjeux écologiques de la production de farinha de Cruzeiro do Sul. Cahiers Agricultures 21: 25–33. [Google Scholar]
- Eriksen L. 2011. Nature and culture in prehistoric Amazonia. Using G.I.S. to reconstruct ancient ethnogenetic processes from archaeology, linguistics, geography, and ethnohistory. Doctoral thesis, Lund University, Sweden. [Google Scholar]
- Excoffier L, Lischer HEL. 2010. Arlequin suit ver. 3.5: a new series of programs to perform population genetics analyses under Linux or Windows. Molecular Ecology Resources 10: 564–567. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 2005. PHYLIP (Phylogeny Inference Package), version 3.6 http://evolution.genetics.washington.edu/phylip.html. Accessed 15 January 2013.
- Freeland JR, Kirk H, Petersen S. 2011. Molecular ecology. Chichester: Wiley-Blackwell. [Google Scholar]
- Goldstein DB, Pollock DD. 1997. Launching microsatellites: a review of mutation processes and methods of phylogenetic inference. Journal of Heredity 88: 335–342. [DOI] [PubMed] [Google Scholar]
- Gribel R, Lemes MR, Bernardes LG, Pinto AE, Shepard GH Jr. 2007. Phylogeography of Brazil-nut tree (Bertholletia excelsa, Lecythidaceae): evidence of human influence on the species distribution. In: Meeting, Association for Tropical Biology and Conservation, Morelia, 281. [Google Scholar]
- Heckenberger MJ. 1998. Manioc agriculture and sedentism in Amazonia: the Upper Xingu example. Antiquity 72: 633–648. [Google Scholar]
- Heckler S, Zent S. 2008. Piaroa manioc varietals: hyperdiversity or social currency?Human Ecology 36: 679–697. [Google Scholar]
- Hernández-Ugalde JA, Mora-Urpí J, Rocha OJ. 2011. Genetic relationships among wild and cultivated populations of peach palm (Bactris gasipaes Kunth, Palmae): evidence for multiple independent domestication events. Genetic Resources and Crop Evolution 58: 571–583. [Google Scholar]
- Isendahl C. 2011. The domestication and early spread of manioc (Manihot esculenta Crantz): a brief synthesis. Latin American Antiquity 22: 452–468. [Google Scholar]
- Jombart T, Ahmed I. 2011. Adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27: 3070–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Balloux F. 2010. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genetics 11: 94. doi:10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamvar ZN, Tabima JF, Grünwald NJ. 2014. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2: e281. doi: 10.7717/peerj.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanashiro M, Harris SA, Simons A. 1997. RAPD diversity in Brazil nut (Bertholletia excelsa Humb. & Bonpl.: Lecythidaceae). Silvae Genetica 46: 219–223. [Google Scholar]
- Keenan K, McGinnity P, Cross TF, Crozier WW, Prodöhl PA. 2013. diveRsity: an R package for the estimation of population genetics parameters and their associated errors. Methods in Ecology and Evolution 4: 782–788. [Google Scholar]
- Larson G, Piperno DR, Allaby RG et al. . 2014. Current perspectives and the future of domestication studies. Proceedings of the National Academy of Sciences of the USA 111: 6139–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebot V. 2009. Tropical root and tuber crops: cassava, sweet potato, yams and aroids. Wallingford: CABI. [Google Scholar]
- Léotard G, Duputié A, Kjellberg F et al. . 2009. Phylogeography and the origin of cassava: new insights from the northern rim of the Amazonian basin. Molecular Phylogenetics and Evolution 53: 329–334. [DOI] [PubMed] [Google Scholar]
- Martins PS. 2001. Dinâmica evolutiva em roças de caboclos amazônicos. In: Vieira ICG, Silva JMC, Oren DC, D’Incao MA eds. Diversidade biológica e cultural da Amazônia. Belém: Museu Paraense Emílio Goeldi, 369–384. [Google Scholar]
- Mba REC, Stephenson P, Edwards K et al. . 1998. Simple sequence repeat (SSR) markers survey of the cassava (Manihot esculenta Crantz) genome: towards an SSR-based molecular genetic map of cassava. Theoretical and Applied Genetics 102: 21–31. [Google Scholar]
- McKey D, Beckerman S. 1993. Chemical ecology, plant evolution and traditional manioc cultivation systems. In: Hladik CM, Linares OF, Pagezy H, Semple A, Hadley M eds. Tropical forests, people and food: biocultural interactions and applications to development. Paris: Parthenon Carnforth and UNESCO, 83–112. [Google Scholar]
- McKey D, Cavagnaro TR, Cliff J, Gleadow R. 2010. Chemical ecology in coupled human and natural systems: people, manioc, multitrophic interactions and global change. Chemoecology 20: 109–133. [Google Scholar]
- McKey D, Elias M, Pujol B, Duputié A. 2012. Ecological approaches to crop domestication. In: Gepts P, Famula TR, Bettinger R. et al. eds. Biodiversity in agriculture: domestication, evolution and sustainability. Cambridge: Cambridge University Press, 377–406. [Google Scholar]
- Meyer RS, DuVal AE, Jensen HR. 2012. Patterns and processes in crop domestication: an historical review and quantitative analysis of 203 global food crops. New Phytologist 196: 29–48. [DOI] [PubMed] [Google Scholar]
- Meyer RS, Purugganan MD. 2013. Evolution of crop species: genetics of domestication and diversification. Nature Reviews Genetics 14: 840–852. [DOI] [PubMed] [Google Scholar]
- Michalakis Y, Excoffier LGL. 1996. A generic estimation of population subdivision using distances between alleles with special reference for microsatellite loci. Genetics 142: 1061–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A, Schaal B. 2005. Domestication of a Mesoamerican cultivated fruit tree, Spondias purpurea. Proceedings of the National Academy of Sciences of the USA 102: 12801–12806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlen GS, Martins PS, Ando A. 2000. Variabilidade genética de etnovariedades de mandioca, avaliada por marcadores de DNA. Scientia Agricola 57: 319–328. [Google Scholar]
- Mühlen GS, Alves-Pereira A, Clement CR, Valle TL. 2013. Genetic diversity and differentiation of Brazilian bitter and sweet manioc varieties (Manihot esculenta Crantz, Euphorbiaceae) based on SSR molecular markers. Tipití 11: 66–73. [Google Scholar]
- Neves EG. 2013. Was agriculture a key productive activity in pre-colonial Amazonia? The stable productive basis for social equality in the Central Amazon. In: Brondízio ES, Moran EF eds. Human-environment interactions: current and future directions. Dordrecht: Springer, 371–388. [Google Scholar]
- Olsen KM. 2004. SNPs, SSRs and inferences on cassava’s origin. Plant Molecular Biology 56: 517–526. [DOI] [PubMed] [Google Scholar]
- Olsen KM, Schaal BA. 1999. Evidence on the origin of cassava: phylogeography of Manihot esculenta. Proceedings of the National Academy of Sciences of the USA 96: 5586–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen KM, Schaal BA. 2001. Microsatellite variation in cassava (Manihot esculenta, Euphorbiaceae) and its wild relatives: further evidence for a Southern Amazonian origin of domestication. American Journal of Botany 88: 131–142. [PubMed] [Google Scholar]
- Oliveira EJ, Ferreira CF, Santos VS, Jesus ON, Oliveira GAF, Silva MS. 2014. Potential of SNP markers for the characterization of Brazilian cassava germplasm. Theoretical and Applied Genetics 127: 1423–1440. [DOI] [PubMed] [Google Scholar]
- Oliveira JC. 2008. Social networks and cultivated plants. Tipití 6: 101–110. [Google Scholar]
- Peakall R, Smouse PE. 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinformatics 28: 2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Venegas CP, Stomph TJ, Verschoor G, Lopez-Lavalle LAB, Struik PC. 2014. Differences in manioc diversity among five ethnic groups of the Colombian Amazon. Diversity 6: 792–826. [Google Scholar]
- Peroni N, Kageyama P, Begossi A. 2007. Molecular differentiation, diversity, and folk classification of “sweet” and “bitter” cassava (Manihot esculenta) in Caiçara and Caboclo management systems (Brazil). Genetic Resources and Crop Evolution 54: 1333–1349. [Google Scholar]
- Perrut-Lima P, Mühlen GS, Carvalho CRL. 2014. Cyanogenic glycoside content of Manihot esculenta subsp. flabellifolia in south-central Rondônia, Brazil, in the center of domestication of M. esculenta subsp. esculenta. Genetic Resources and Crop Evolution 61: 1035–1038. [Google Scholar]
- Petit RJ, El Mousadik A, Pons O. 1998. Identifying populations for conservation on the basis of genetic markers. Conservation Biology 12: 844–855. [Google Scholar]
- Pickersgill B. 2007. Domestication of plants in the Americas: insights from Mendelian and molecular genetics. Annals of Botany 100: 925–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol B, McKey D. 2006. Size asymmetry in intraspecific competition and the density dependence of inbreeding depression in a natural plant population: a case study in cassava (Manihot esculenta Crantz, Euphorbiaceae). Journal of Evolutionary Biology 19: 85–96. [DOI] [PubMed] [Google Scholar]
- Pujol B, David P, McKey D. 2005. Microevolution in agricultural environments: how a traditional Amerindian farming practice favours heterozygosity in cassava (Manihot esculenta Crantz, Euphorbiaceae). Ecology Letters 8: 138–147. [Google Scholar]
- Pujol B, Renoux F, Elias M, Rival L, Mckey D. 2007. The unappreciated ecology of landrace populations: conservation consequences of soil seed banks in cassava. Biological Conservation 136: 541–551. [Google Scholar]
- R Core Team 2015. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; http://www.R-project.org. Accessed 15 December 2015. [Google Scholar]
- Reif JC, Melchinger AE, Frisch M. 2005. Genetical and mathematical properties of similarity and dissimilarity coefficients applied in plant breeding and seed bank management. Crop Science 45: 1–7. [Google Scholar]
- Rival L, McKey D. 2008. Domestication and diversity in manioc (Manihot esculenta Crantz ssp. esculenta, Euphorbiaceae). Current Anthropology 49: 1119–1128. [Google Scholar]
- Rodrigues DP, Astolfi Filho S, Clement CR. 2004. Molecular marker-mediated validation of morphologically defined landraces of pejibaye (Bactris gasipaes) and their phylogenetic relationships. Genetic Resources and Crop Evolution 51: 871–882. [Google Scholar]
- Roullier C, Rossel D, Tay D, McKey DB, Lebot V. 2011. Combining chloroplast and nuclear microsatellites to investigate origin and dispersal of New World sweet potato landraces. Molecular Ecology 20: 3963–3977. [DOI] [PubMed] [Google Scholar]
- Roullier C, Benoit L, McKey DB, Lebot V. 2013. Historical collections reveal patterns of diffusion of sweet potato in Oceania obscured by modern plant movements and recombination. Proceedings of the National Academy of Sciences of the USA 110: 2205–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Salick J, Cellinese N, Knapp S. 1997. Indigenous diversity of cassava: generation, maintenance, use and loss among the Amuesha, Peruvian Upper Amazon. Economic Botany 51: 6–19. [Google Scholar]
- Scheldeman X, van Zonneveld M. 2010. Training manual on spatial analysis of plant diversity and distribution. Rome: Bioversity International. [Google Scholar]
- Schaal BA, Olsen KM, Carvalho LJCB. 2006. Evolution, domestication, and agrobiodiversity in the tropical crop cassava. In: Motley TJ, Zerega N, Hugh H eds. Darwin’s harvest – new approaches to the origins, evolution, and conservation of crops. New York: Columbia University Press, 269–284. [Google Scholar]
- Shepard GH Jr, Ramirez H. 2011. ‘Made in Brazil’: human dispersal of the Brazil nut (Bertholletia excelsa, Lecythidaceae) in ancient Amazonia. Economic Botany 65: 44–65. [Google Scholar]
- Siqueira MVBM, Queiroz-Silva JR, Bressan E et al. . 2009. Genetic characterization of cassava (Manihot esculenta) landraces in Brazil assessed with simple sequence repeats. Genetics and Molecular Biology 32: 104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujii PS, Martins K, de Oliveira Wadt LH, Azevedo VCR, Solferini VN. 2015. Genetic structure of Bertholletia excelsa. Conservation Genetics 16: 955–964. [Google Scholar]
- Vencovsky R, Nass LL, Cordeiro CMT, Ferreira MAJF. 2007. Amostragem em recursos genéticos vegetais. In: Nass LL. ed. Recursos genéticos vegetais. Brasília: Embrapa Recursos Genéticos e Biotecnologia, 231–280. [Google Scholar]
- Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- Weising K, Gardner RC. 1999. A set of conserved PCR primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledonous angiosperms. Genome 42: 9–19. [PubMed] [Google Scholar]
- Wright S. 1965. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 19: 395–420. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.